Abstract
Stomata, the micro-pores on leaf surface, are formed by a pair of guard cells. In addition to control water loss and gas exchange between the plant and the environment, these cells act as immunity gates to prevent pathogen invasion of the plant apoplast. Here, we report a brief procedure to obtain highly pure guard cell preparations using conditions that preserve the guard cell transcriptome as much as possible for a robust high-throughput RNA sequence analysis. The advantages of this procedure included: 1) substantial shortening of the time required for obtaining high yield of >97% pure guard cell protoplasts (GCP), 2) extraction of enough amount of high quality RNA for direct sequencing, and 3) limited RNA decay during sample manipulation. Gene expression analysis by RT-qPCR revealed that wound-related genes were not induced during release of guard cells from leaves. To validate our approach, we performed a high throughput deep-sequencing of guard cell transcriptome (RNA-seq). A total of 18,994 nuclear-encoded transcripts was detected, which expanded the transcriptome by 70%. The optimized GCP isolation and RNA extraction protocols are simple, reproducible, and fast allowing the discovery of genes and regulatory networks inherent to the guard cells under various stresses.
Guard cells are highly specialized type of cells that surround natural pores on the leaf epidermis forming structures called stomata. The primary function of stomata is to control gas exchange (CO2 and O2) between the leaf interior and the environment and, at the same time, control leaf water loss through transpiration. Thus, the guard cell controls stomatal movement (opening and closure) in response to external (e.g. light, temperature, relative humidity) and internal (e.g. endogenous hormones) stimuli. More recently, another important function of the guard cell was discovered; it can sense and respond to epiphytic microbes and protect the leaf against microbial invasion by closing the stomatal pore (Melotto et al. 2006; Gudesblat et al. 2009; Schellenberg et al. 2010). This phenomenon has been defined as stomatal immunity as it requires well known molecular components of the plant innate immune system (reviewed by Zeng et al. 2010).
Some of the downstream molecular processes in the guard cell after microbe recognition are somewhat overlapping with the ones associated with abiotic stress. For instance, synthesis and signaling of the plant hormone abscisic acid (ABA) are required for stomatal closure in response to drought stress (Schroeder et al. 2001) and are also linked to stomatal immunity (Melotto et al. 2006). Because the guard cells respond to several external factors that can simultaneously stimulate them, it is important to dissect the molecular mechanism(s) underlying these responses.
The guard cell is autonomous making it a useful model to understand cell type responses to stresses. Procedures to isolate guard cell protoplasts (GCPs) for western blotting, reverse transcription polymerase chain reaction (RT-PCR), microarray analysis, and electrophysiological studies have been previously reported (Pandey et al. 2002; Leonhardt et al. 2004). With the advent of novel high throughput methods such as direct RNA sequencing (RNA-seq), the quantity, quality, and differential decay of RNA molecules, as well as preservation of whole cell transcriptomes during protoplasting are critical to the success of functional studies. Three important modifications of the traditional GCP preparation (Pandey et al. 2002) have been devised for microarray analysis. First, transcription inhibitors were added during complete digestion of the cell wall to avoid induction of stress-related genes (Leonhardt et al. 2004). However, the long procedure (>5 h) to release guard cell protoplasts may lead to RNA decay. Second, a partial cell wall digestion with 1h of incubation was performed, in which intact guard cells were still attached to the epidermal tissue (Pandey et al. 2010). Although this short procedure may alleviate extensive RNA decay, stress-related genes such as wounding, can still be induced in a very short period of time (Chung et al. 2008). Wounding response can occur when leaves are blended to release the epidermis. Third, leaf strips were cut, frozen, freeze-dried, and guard-cell pairs were manually dissected to avoid alteration of gene expression due to the action of enzymes and osmotic stress during protoplasting (Bates et al. 2012). This third procedure had not yielded enough RNA for downstream applications and extra steps to amplify RNA were required.
Because stomatal immunity is a fast response to biotic stimuli (within 2 h in intact leaves; Melotto et al. 2006), it likely involves tight regulation of gene expression and signal transduction pathways. Therefore, we sought to develop a protocol that minimizes the manipulation of samples to obtain RNA useful for high throughput sequencing. The newly devised method had the following advantages: (1) it shortened the overall GCP preparation procedure from 6 to 2 hours, while maintaining the purity and yield of isolated guard cell; (2) it increased the amount of RNA extracted by two to three fold; and (3) it recovered short-lived transcripts (<3 h) that might be associated with early stages of biotic stress. Furthermore, during the process of procedure validation through RNA-seq, we extended the current number of genes known to be expressed in guard cell.
RESULTS AND DISCUSSION
Length of incubation for cell wall digestions does not affect GCP purity and yield
High throughput sequencing for transcriptomic analysis requires that RNA samples are of excellent quality for assessing the level of gene expression accurately. GCP preparation, in particular, relies on extensive manipulation of the samples before RNA isolation, and maintaining the integrity of the transcriptome during the procedure may be a challenge. Considering that the half-lives of some transcripts can be as short as 1 to 3 h (Narsai et al. 2007) and commonly used protocols take approximately 6 h to be completed (Pandey et al. 2002; Leonhardt et al. 2004), it is likely that RNA decay will occur during GCP preparation. Thus, we determined the shortest incubation times to completely digest the plant cell wall and still yield pure and healthy GCPs.
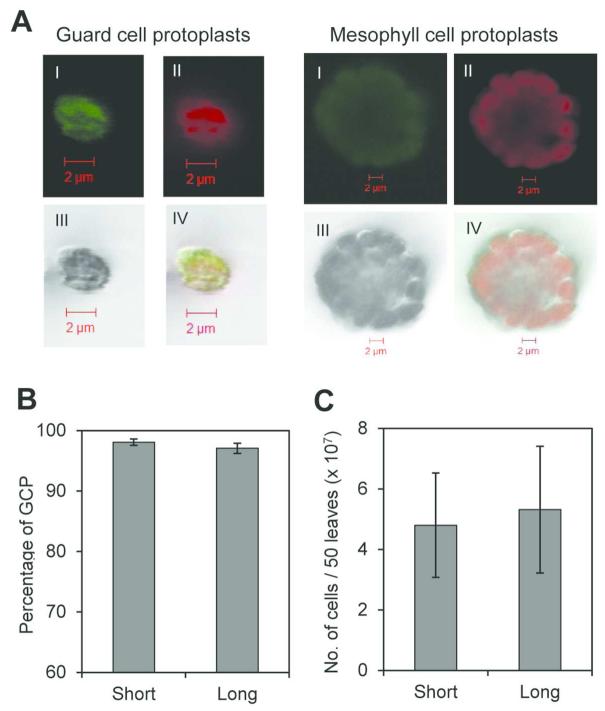
GCPs are approximately ten times smaller than mesophyll cell protoplasts (MCPs) (Fig. 1A) and sample purity can be easily evaluated by observing cell preparation under light microscope and calculating the percentage of GCPs present in the suspension. As intact chloroplasts of these cells auto-fluoresce, cell viability can also be determined using a fluorescence microscope (Fig 1A). Decrease in the incubation times in steps 4 and 7 of the protocol from 3 to 0.5 h and from 2 to 1 h, respectively (Supplementary Fig. S1) does not affect the purity and yield of GCP preparations. Both procedures yielded similar GCP purities with 98% and 97% for the short (2 h) and long protocol (>6 h), respectively (Fig. 1B). This purity is equivalent to other described procedures (Pandey et al. 2002; Leonhardt et al. 2004). Likewise, very similar numbers of GCPs were recovered using either short or long protocol, an average of 4.8 × 107 and 5.3 × 107 cells per 50 leaves, respectively (Fig. 1C), which is approximately ten-fold higher than the yield reported by Pandey et al. (2002). This difference in GCP numbers is not statistically significant.
Fig. 1.
Assessing the yield and purity of GCP preparations. A, Laser scanning confocal micrographs of guard cell and mesophyll cell protoplasts (I=green channel, II=red channel, III=DIC, IV=merged channels). Note the size difference. B, Purity of GCPs extracted using long and short incubation protocols calculated as percentages of total protoplast extracted (MCP and GCP). C, Number of GCPs isolated in long and short methods. Results are shown as means (n=3) ± standard error.
Amount of RNA extracted from GCPs is affected by digestion time, but not by the presence of transcription inhibitors
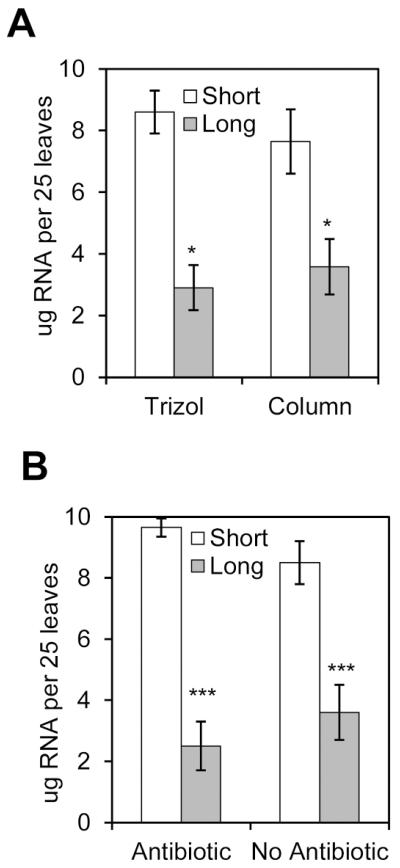
To determine whether the length of the GCP preparation procedure could interfere with the amount of RNA extracted, we isolated GCPs from 50 leaves and divided the GCP suspension in two halves for RNA extraction using two different methods, Trizol® reagent or Qiagen column. Increasing incubation times to digest the plant cell wall negatively affected (P < 0.05) the RNA yield (μg) as determined by NanoDrop® spectroscopy, independent of the RNA extraction method of choice. Two- to three-fold more RNA could be extracted after short cell wall digestion (7-9 μg) as compared to long digestion (3-3.5 μg) (Fig. 2A).
Fig. 2.
Amount of RNA extracted from long and short protocols. A, GCPs were isolated from 50 leaves and GCP suspension was equally divided for total RNA extraction using either the Qiagen column or Trizol reagent, thus yield is expressed in μg per 25 leaves. Transcription inhibitors were not added during guard cell protoplasting. B, Total RNA extracted from GCPs using Qiagen column in presence or absence of the transcription inhibitor antibiotics cordycepin (0.01%) and actinomycin D (0.0033%). Results are shown as means (n=3) ± standard error. Statistical significance between the means (short versus long) was detected with two-tailed Student’s t-test (*** refers to P<0.001, * refers to P <0.05).
Next, we assessed the effect of the transcription inhibitors actinomycin D and cordycepin on the amount of RNA extracted with Qiagen columns. In this experiment, RNA yields were also significantly decreased (P < 0.001) when GCPs were subjected to long digestion periods (Fig. 2B). However, similar RNA yields were obtained with or without the addition of transcription inhibitors during either long or short GCP preparation procedure (Fig. 2B). Taken together, these results suggest that lower RNA yield after longer GCP preparation may be due to RNA decay.
Quality of RNA is affected by extraction protocol, but not by GCP preparation time
To further determine the RNA quality for downstream application, total RNA extracted from GCPs was quantified using BioAnalyzer. We have not observed differences in the RNA amount extracted with either Trizol® reagent or Qiagen column (Fig. 2A) and the A260:280 ratios of all RNA samples ranged from 2.0 to 2.2 based on NanoDrop® readouts. However, BioAnalyzer profiles indicated a significantly low overall quality of the RNA samples extracted with Trizol® reagent. The average RNA integrity number (RIN) for these samples was 4, ranging from 2.7 to 5.9 in four independent trials and the RIN number could not be determined in additional two biological replicates. These results highlight the importance of checking the RNA quantity and integrity using sensitive techniques such as BioAnalyzer profile. Therefore, we have not used Trizol®-extracted RNA for downstream application.
When RNA was extracted from GCPs with the Qiagen column, the RNA integrity based on RIN values averaged around 6 and were not significantly different between the GCP preparation protocols (short and long) or antibiotics addition (Supplementary Fig. S2). Furthermore, the electropherogram profiles (data not shown) and electronic gels for these RNA samples were very similar (Supplementary Fig. S2).
Actinomycin D and cordycepin prevent induction of wound-responsive genes during protoplasting
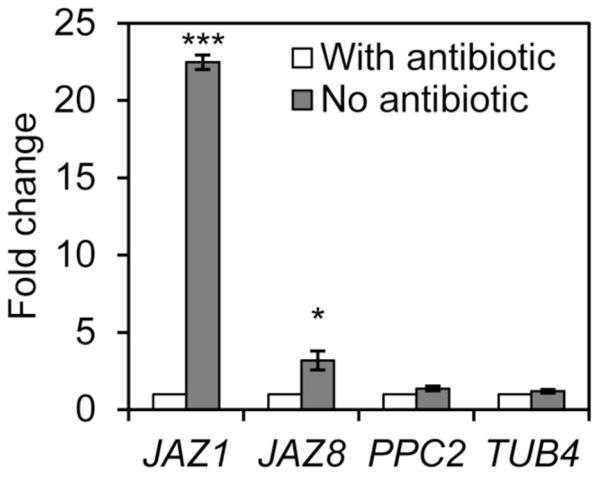
Considering that protoplasting induces the expression of stress-associated genes, (Leonhardt et al. 2004; Wang et al. 2011), we tested whether the transcription inhibitors used during protoplast isolation were efficient in preserving the expression levels of early wound-response genes. First, the quality of the cDNA synthesized with reverse transcriptase was assessed through agarose gel electrophoresis to ensure that only high quality cDNA was used for the gene expression analysis. cDNA smears ranging from 400 to >1000 base pairs were considered of good quality and used for qPCR analysis (Supplementary Fig S3). Second, we evaluated the PCR efficiency according to Schmittgen and Livak (2008) and only reactions with efficiency within 15% of that observed for the reference gene were selected for assessing transcript abundance (Supplementary Fig. S4). Next, we selected two genes that are strongly induced by wounding as fast as 30 min, JAZ1 and JAZ8 (Chung et al. 2008) and determined their transcript abundances in RNA samples extracted from GCPs isolated with short incubation times and in the presence or absence of transcription inhibitors. JAZ1 and JAZ8 transcripts were 23 and 3 times more abundant in samples without antibiotics as compared to samples with antibiotics, respectively (Fig. 3). Furthermore, besides ACT2 that was used as internal control for qPCR, we assessed the expression of two other genes that have predicted half-lives higher than 6 h and are not known to be induced by stresses, PPC2 and TUB4. No differences in transcript abundance were observed for these genes (Fig. 3). These results suggest that the addition of transcription inhibitors during protoplast in fact avoided the induction of genes in the guard cells, which is essential to evaluate global transcriptional changes in response to bacterial treatments.
Fig. 3.
Affect of transcription inhibitor antibiotics (actinomycin and cordycepin) in wound-responsive gene transcription during GCP preparation (2-h procedure). Transcript abundance of the indicated genes relative to the procedure with antibiotics was determined by RT-qPCR analysis. Results are shown as mean (n=6) ± standard error. Statistical significance of the difference between means (with antibiotics versus without antibiotics) was detected with two-tailed Student’s t-test (*** = P<0.001, * = P<0.05).
mRNAs decay in guard cells
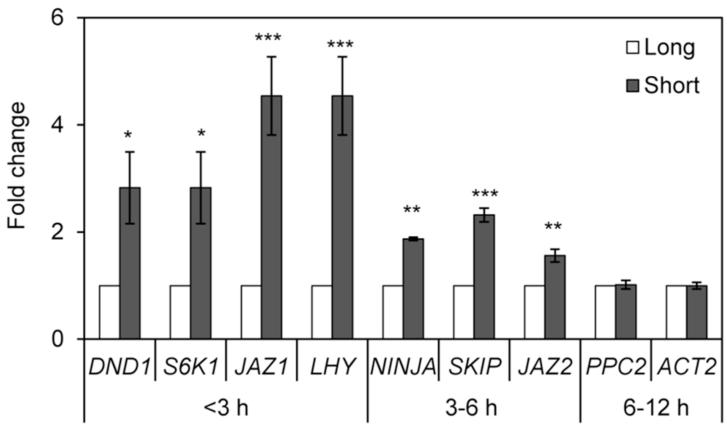
To address the concern of RNA decay (Narsai et al. 2007) due to lengthy procedures for protoplasting, we assessed transcript abundance of ten genes, two of which are commonly used as internal control for qPCR (ACT2 and TUB4), after short and long incubation procedures. These genes were selected based on their half-lives in Arabidopsis cell suspensions (Narsai et al. 2007) and were previously known to be expressed in guard cells (Leonhardt et al. 2004; Wang et al. 2011). We subjected all genes to the same qPCR controls described above (Supplementary Fig. S3 and S4). Consistently, all four transcripts with predicted half-lives shorter than 3 h were three to five fold more abundant in GCP preparations using shorter incubations as compared to long incubation times (Fig. 4). Likewise, three gene transcripts with predicted half-lives between 3-6 h were all significantly more abundant in GCPs released with short incubations; however the fold changes were between 1.5 to 2.3 (Fig. 4). No changes were observed in the abundance of transcripts with half-life longer than 6 h, PPC2, ACT2, and the internal control TUB4 used to create Fig. 4. Genes with shorter half-lives are mostly involved in regulatory functions (Narsai et al. 2007); therefore the time required for isolation of guard cells becomes crucial. Our results indicate that the optimized GCP isolation protocol may yield RNA samples enriched with short-lived transcripts increasing the success to discover genes and regulatory networks of guard cells under biotic and abiotic stresses.
Fig. 4.
Effect of GCP preparation time on transcript abundance. Long procedure takes >6 h whereas the short procedure can be finished in about 2 h. Transcript abundance of the indicated genes relative to the >6 h procedure was determined by RT-qPCR analysis. Results are shown as mean (n=6) ± standard error. Statistical significance of the difference between means (short versus long procedure) was detected with two-tailed Student’s t-test (*** = P<0.001, ** = P<0.01, * = P<0.05).
The guard cell transcriptome
Previously, we have determined that guard cells in intact leaves respond very quickly to the presence of bacteria by closing most of the stomatal pores within 2 h of exposure (Chitrakar and Melotto 2010). Therefore, we devised a procedure for guard cell protoplasting to avoid induction of biotic stress-associated genes and extensive RNA decay, and to obtain high quality and quantity of RNA useful for studying the effects of biotic stress on the guard cell transcription network through direct RNA sequencing. These parameters were optimized by decreasing cell wall digestion time to release isolated GCP, adding transcription inhibitors, and using Qiagen columns to extract RNA directly from frozen GCP suspensions. Furthermore, we were able to perform a high throughput deep-sequencing of the guard cell transcriptome (RNA-seq) to serve as baseline for studying gene regulation of stomatal immunity. Two biological replicates were used for RNA-seq that yielded 36,385,598 and 40,586,179 high quality reads with a 79.6% and 86.1% mapping efficiency to the Arabidopsis reference genome, respectively (Table 1). Once mapped to the reference genomes, we identified the gene transcripts that were present in both biological replicates (Supplementary Table S1).
Table 1.
RNA-seq library concentration and fragment size, number of raw and quality control (QC) reads obtained and percentage of reads mapped to the Arabidopsis gene model (TAIR10) for each biological replicate (BR).
| Library |
|||||||
|---|---|---|---|---|---|---|---|
| BR code |
Index | ng/μl | Fragment Size (bp) |
Yield (Mb) |
# Raw Reads |
# QC reads |
% mapped QCreads |
| CtrlBR4 | CGTACG | 0.281 | 256 | 4,198 | 46,455,276 | 36,385,595 | 79.6 |
| CtrlBR5 | GAGTGG | 0.248 | 255 | 4,239 | 46,986,050 | 40,586,179 | 86.1 |
The most updated list of genes expressed in Arabidopsis guard cells was recently published by Wang et al. (2011). Using microarray analysis, the authors observed that a total of 11,169 unique nuclear-encoded genes were expressed, out of which 1,162 are ABA-responsive and 10,007 ABA-non-responsive. Our optimized GCP preparation, RNA isolation, and RNA-seq allowed the reliable detection of 18,994 (Supplementary Table S1) nuclear genes expressed in the Arabidopsis guard cells that include 10,947 genes listed by Wang et al. (2011) and an additional 8,047 representing a 70% increase on the list previously reported.
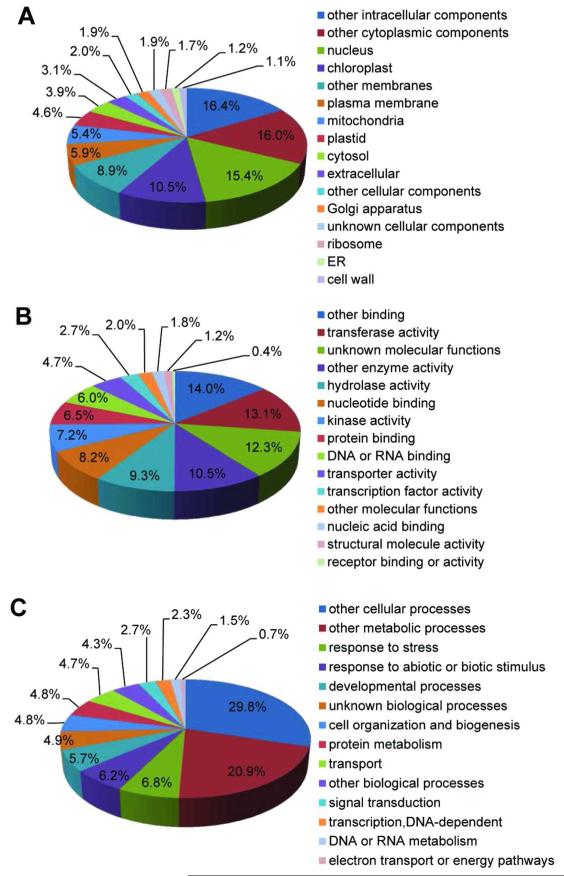
Functional categorization of the 18,994 gene transcripts using the GO Slim classification for plants (TAIR10) revealed the GO terms present in our data set that belong to the three broad GOs, Biological Process, Cellular Component, or Molecular Function (Fig. 5). One fourth (25.9%) of the transcripts encode for proteins targeted to the nucleus and chloroplast (Fig. 5A). The most abundant molecular functions include: other binding (14%; excludes nucleic acid and protein binding), transferase activity (13.1%), and hydrolase activity (9.3%) (Fig. 5B). Response to stress and response to biotic and abiotic stimulus accounted for 13% of the biological process annotations (Fig. 5C).
Fig. 5.
Functional categorization of guard cell expressed genes (18,994) according to the three broad Gene Ontology categories cellular component (A), molecular function (B), and biological process (C) using the GO slim tool available at TAIR.
To further understand the transcriptome of the guard cell, we performed Single Enrichment Analysis (SEA) to identify GOs that are overrepresented in the guard cell transcriptome as compared to the pre-calculated GO frequency in the Arabidopsis reference gene model (TAIR10) using the AgriGO analysis tool (Du et al. 2010). A total of 3,372 GO terms associated with five or more transcripts were identified, out of which 2151, 854, and 367 belong to the broad GOs Biological Process, Molecular Function, and Cellular Component, respectively (Supplementary Table S2 Part A). Abundance of all guard cell transcripts and Arabidopsis gene models within each GO was compared statistically using the Fisher exact test with Benjamini-Hochberg-False Discovery Rate (FDR) correction (Supplementary Table S2 Part B). This analysis revealed that 1,478 GOs are significantly (FDR < 0.01) more abundant in the guard cell as compared to the reference gene model (Supplementary Table S2 Part C). Our results suggest that unique transcriptional patterns occur in the guard cell. Validating our approach to identify metabolic processes in the guard cell, we observed that gene products localized to the chloroplast (GO:0009507) and involved in photosynthesis (GO:0015979) are overrepresented in our dataset. Because we conducted a detailed GO analysis, it was possible to identify specific photosynthetic processes such as light reactions (GO:0019684), photosystem II (PSII) assembly (GO:0010207), photosynthetic electron transport in photosystem I (GO:0009773), PSII associated light-harvesting complex II catabolic process (GO:0010304). Furthermore, GO associated with circadian rhythm (GO:0007623), stomatal development and movement are also overrepresented such as stomatal complex development (GO:0010374), stomatal complex morphogenesis (GO:0010103), stomatal movement (GO:0010118), regulation of stomatal movement (GO:0010119), and stomatal lineage progression (GO:0010440). Other highly represented biological processes were also identified (Supplementary Table S2 Part C) and their biological relevance will become evident as we advance our current understanding of the guard cell physiology.
Finally, the top 30 overrepresented GOs under the Biological Process (FDR ≤ 2.1 × 10−113) includes response to abiotic stimulus (GO:0009628), response to stress (GO:0006950), response to biotic stimulus (GO:0009607), and innate immune response (GO:0045087) (Supplementary Table S2 Part C), which may be due to the fact that guard cells are continuously exposed, and are able to quickly respond, to environmental signals at the leaf surface.
Conclusion
In this study, we demonstrate the feasibility of a robust, straight-forward, and fast procedure to obtain highly pure GCPs and enough high quality RNA to assess the transcriptome of guard cells using direct RNA sequencing. The number of detectable genes expressed in the guard cell was considerably extended providing a unique opportunity to infer the metabolic activities carried out by this special type of cells. The new procedure and protocol adjustments described here will provide new sequence data and increase the likelihood to detect short-lived RNA transcripts involved in the tight regulation of the signal transduction of guard cells under stress conditions, ultimately facilitating the mechanistic understanding of plant-pathogen interactions at the leaf surface.
MATERIAL AND METHODS
Plant material and growth conditions
Arabidopsis thaliana (L. Heyhn.) ecotype Columbia (Col-0, ABRC stock CS60000) seeds were sown in a 1:1:1 (v:v:v) mixture of growing medium (Redi-earth plug and seedling mix, Sun Gro), fine vermiculite, and perlite (Hummert International, Earth City, MO) and grown in controlled environmental chambers at 22°C, 65±5% relative humidity (RH), and a 12-h photoperiod under light intensity of 100 μmol.m−2.s−1. Four- to five-week old plants were used for all experiments.
Guard cell protoplast isolation
Guard cell protoplasts (GCP) were isolated from the second and third layers of rosette leaves using the solutions used by Leonhardt et al. (2004) in the presence or absence of the transcription inhibitors actinomycin D (Sigma, St. Louis, MO) and cordycepin (Sigma). The complete protocol, chemical concentrations, and variations in the incubation times are depicted in the Supplementary Fig. S1. Purity and yield of GCPs were determined by observing and counting cells under Nikon Eclipse 80i fluorescent microscope (Nikon Corporations, Tokyo, Japan) equipped with a digital camera. Cells counts were obtained by using a Petroff Hausser counting chamber (Hausser Scientific, Horsham, PA) using the equation: Total cell number = number of cells counts × dilution factor × 50,000, where 50,000 corresponds to cell depth × cell volume. A minimum of 500 cells were counted for each sample. GCP suspensions were centrifuged at 1000 ×g for 5 min at room temperature and flash frozen in liquid nitrogen for subsequent RNA extraction. A minimum of three biological replicates were performed for each variation of the method and all GCP isolations were performed at 2-3 hours after the lights were turned on in the morning.
Confocal microscopy imaging
Green and red auto-fluorescence and differential interference contrast (DIC) images of the protoplasts were recorded using a confocal laser scanning microscope (LSM 510 Meta, Carl Zeiss Inc., Thornwood, NY) with Argon laser at excitation of 488 nm and emission at 505-550 BP (green) and 560 LP (red). All channels were imaged simultaneously.
RNA extraction
Frozen GCP preparations (~107 cells) were thawed using the lysis buffer supplied with each RNA extraction kit; RNeasy Plant Mini kit including the in-column DNA digestion option (Qiagen, Valencia, CA) or Trizol® reagent (Invitrogen, Carlsbad, CA) following manufacturer’s instructions. The volume of the lysis buffer used was 0.45 ml or 1 ml for the column-based or Trizol-based method, respectively. RNA yield and quality were determined using the NanoDrop-1000 version 3.2 spectrophotometer (Thermo Scientific, Wilmington, DE) and the Agilent 2100 BioAnalyzer RNA 6000 Pico chip (Agilent Technologies, Inc. Wilmington, DE).
Gene expression analysis
Total RNA was synthesized into cDNA in a 20 μl reaction containing 5 μg RNA template, 250 nM oligo dT, and reagents provided with the Takara RNA PCR kit (AMV) (Clontech, Montain View, CA) according to manufacturer’s recommendations. Reverse transcription (RT) reaction was carried out at 50°C for 30 min, 95°C for 5 min, and 4°C for 5 min. Quantitative PCR (qPCR) was performed in 20 μl reaction with iTaq Fast SYBR Green Supermix (BioRad, Hercules, CA) using 0.5 μl of the RT reaction described above and 200 nM of reverse and forward gene-specific primers. Reactions were carried out with the Applied Biosystems 7300 thermocycler (Applied Biosystems, Foster City, CA) using cycling conditions as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 10 sec and 58°C for 30 sec. A dissociation curve was determined for every reaction to confirm the presence of a single amplicon indicating the lack of primer dimers and non-specific products, and that RNA samples were free of DNA contamination. Relative abundance of transcripts was calculated using the ΔΔCT method (Livak and Schmittgen 2001) using the housekeeping genes ACT2 and TUB4 as internal controls. ACT2 and TUB4 have a half-life of 6-12 and 12-24 h, respectively (Narsai et al. 2007) and their transcript levels show no difference among GCP samples as described in the results. All gene-specific primers are described in the Supplementary Table S3. A minimum of two biological replicates and three technical replicates were performed.
PCR efficiency
Gene-specific primer sets that span an intron region were designed using the primer quest software from IDT-SciTools (http://www.idtdna.com/Primerquest/Home/Index) for qPCR analysis. To assess reaction efficiencies, standard curves were created using a five-fold serial dilution of the cDNA pool. A linear regression between the amount of cDNA template and the cycle threshold (CT) value was calculated to obtain a correlation coefficient (R2 T) >0.97. The PCR efficiency was determined according to Schmittgen and Livak (2008).
RNA-seq analysis
Leaves from 35 Arabidopsis plants (4-5 week-old) were used for GCP preparations using the short incubation protocol in the presence of transcription inhibitors. Two biological replicates of >99% pure GCP preparations were performed for RNA extraction using Qiagen columns according to the manufacturer’s recommendation. The in-column DNase treatment with the RNase-free DNase set kit (Qiagen) was carried out for all samples. RNA quality was assessed with the Experion Automated Electrophoresis System (BioRad, Hercules, CA) and 2 ug of total RNA was used for RNA-seq library preparation with the TruSeq RNA v2 kit (Illumina, Inc., San Diego, CA) according to manufacturer’s instructions. Library concentration was measured with the Qubit fluorometer (Invitrogen, Carlsbad, CA) and fragment size was determined with the High Sensitivity DNA kit on a BioAnalyzer 2100 (Table 1). RNA sequences (1×100 bases) were obtained with a HiSeq 2000 system (Illumina Inc., San Diego, CA) at the DNA Core Facility, University of Missouri, Columbia. Sequence reads were subjected to a multi-phase quality control regime as follows: raw reads were trimmed with fastx_trimmer using a minimum quality threshold of 13 and minimum length of 32 bases. Subsequently, reads were filtered with fastq_quality_filter with a quality cut-off of 13 and minimum percentage of 90. Reads were further filtered out by match to mitochondrial and plastid genomes, repeat elements using the bowtie-based TopHat suite (Trapnell et al. 2009). Reads that passed quality control were mapped to the Arabidopsis genome (TAIR10; http://arabidopsis.org) using the default parameter of the TopHat program (http://tophat.cbcb.umd.edu/). Mapped reads assembly and quantitated expression of transcripts were performed with Cufflinks using default parameters (http://cufflinks.cbcb.umd.edu/) (Trapnell et al. 2010). Normalized expression levels of the genes were expressed as Fragments Per Kilobase of exon per Million fragments mapped (FPKM).
Functional annotation of guard cell expressed genes according to plant GO slim categories was retrieved from The Arabidopsis Information Resource database (TAIR10; Arabidopsis.org). Additionally, the AGI (Arabidopsis Genome Initiative) number was used as input for assessing GO enrichment using the Singular Enrichment Analysis (SEA) through AgriGO (Du et al. 2010; http://bioinfo.cau.edu.cn/agriGO/). TAIR10 was used as a background reference for SEA and statistical significance was detected with Fisher exact test with Benjamini-Hochberg false discovery rate (FDR) correction to calculate the P and FDR values. The Illumina RNA-seq data related to this study is available at the NCBI Sequence Read Archive (SRA, Wheeler et al. 2008, http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession number SRP021005 (mRNA-seq).
Supplementary Material
ACKNOWLEDGMENTS
We thank Nathan Bivens and Scott Givan for their support on RNA sequencing and bioinformatic analysis. This study was supported by a grant from the US National Institute of Allergy and Infectious Disease (5R01AI068718).
Footnotes
Author contributions
NO and MM designed the research. NO performed research. NO, SP, and MM analyzed data. NO and MM wrote the manuscript. All authors have read and approved the final manuscript.
Competing interest
The authors declare no competing interests.
LITERATURE CITED
- Bates GW, Rosenthal DM, Sun J, Chattopadhyay M, Peffer E, Yang J, Ort DR, Jones AM. A comparative study of the Arabidopsis thaliana guard cell transcriptome and its modulation by sucrose. PLoS ONE. 2012;7(11):e49641. doi: 10.1371/journal.pone.0049641. doi:10.1371/journal.pone.0049641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitrakar R, Melotto M. Assessing stomatal response to live bacterial cells using whole leaf imaging. J. Vis. Exp. 2010:44. doi: 10.3791/2185. http://www.jove.com/index/details.stp?id=2185, doi: 10.3791/2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, Jones AD, Howe GA. Regulation and function of Arabidopsis JASMONATE ZIM/Domain genes in response to wounding and herbivory. Plant Physiol. 2008;146:952–964. doi: 10.1104/pp.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. AgriGO: a GO analysis tool kit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat GE, Torres PS, Vojnov AA. Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell/to/cell signal/regulated virulence factor. Plant Physiol. 2009;149:1017–1027. doi: 10.1104/pp.108.126870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell. 2004;16:596–615. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Millar AH, O’Toole N, Small I, Whelan J. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell. 2007;19:3418–3436. doi: 10.1105/tpc.107.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Wang X/Q, Coursol SA, Assmann SM. Preparation and applications of Arabidopsis thaliana guard cell protoplasts. New Phytol. 2002;153:517–526. doi: 10.1046/j.0028-646X.2001.00329.x. [DOI] [PubMed] [Google Scholar]
- Pandey S, Wang RS, Wilson L, Li S, Zhao Z, Gookin TE, Assmann SM, Albert R. Boolean modeling of transcriptome data reveals novel modes of heterotrimeric G-protein action. Mol. Syst. Biol. 2010;6:372. doi: 10.1038/msb.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg B, Ramel C, Dudler R. Pseudomonas syringae virulence factor syringolin A counteracts stomatal immunity by proteasome inhibition. Mol. Plant/Microbe Interact. 2010;23:1287–1293. doi: 10.1094/MPMI-04-10-0094. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real/time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ. Guard cell abscisic acid signaling and engineering drought hardiness in plants. Nature. 2001;410:327–410. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA/seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA/Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R/S, Pandey S, Li S, Gookin TE, Zhao Z, Albert R, Assmann SM. Common and unique elements of the ABA/regulated transcriptome of Arabidopsis guard cells. BMC Genomics. 2011;12:216. doi: 10.1186/1471-2164-12-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Ostell J, Pruitt KD, Schuler GD, Shumway M, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Melotto M, He SY. Plant stomata: a checkpoint of host immunity and pathogen virulence. Curr. Op. Biotechnol. 2010;21:599–603. doi: 10.1016/j.copbio.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.