Abstract
Alu elements are ~300 bp sequences that have amplified via an RNA intermediate leading to the accumulation of over 1 million copies in the human genome. Although few of the copies are active, Alu germline activity is the highest of all human retrotransposons and does significantly contribute to genetic disease and population diversity. There are two basic mechanisms by which Alu elements contribute to disease: through insertional mutagenesis and as a large source of repetitive sequences that contribute to non-allelic homologous recombination that cause genetic deletions and duplications.
Introduction
Retrotransposons share various characteristics with retroviruses, as they are DNA sequences that have the ability to generate copies of themselves through the reverse transcription of an RNA intermediate in a process termed retrotransposition [1;2]. However, in contrast to retroviruses, retrotransposons lack an infectious form, thus their method of spread is by vertical transmission through the germ line. Retrotransposons are subdivided into two groups: LTR retrotransposons and non-LTR retrotransposons (also known as Long INterspersed Elements or LINEs). Interestingly, retroviruses are thought to have evolved from LTR retrotransposons that acquired an envelope [3]. Non-LTR retrotransposons are further subdivided into autonomous and non-autonomous elements based on the ability of the element to encode the enzymes and proteins required for retrotransposition. There are three types of non-autonomous elements currently active in the human genome: 1) Alu, which is classified as a Short INterspersed Element (SINE); 2) SVA (SINE/VNTR/Alu) [4;5] ; and 3) the retropseudogenes or processed pseudogenes which are a collective of inserts derived from the retrotransposition of cellular mRNAs [6]. The non-autonomous elements need to “parasitize” the factors they require for retrotransposition from external sources [7-10]. In the human genome, this source derives from the LINE elements, with LINE-1 (L1) being the currently active element. L1 encodes two essential proteins ORF1p and ORF2p [11]. ORF1p is an RNA-binding protein [12] reported to have nucleic acid chaperone activity [13]. ORF2 encodes a protein with multifunctional domains that provide the endonuclease [14] and reverse transcriptase activities [15] critical for retrotransposition [11].
Alu elements
Alu elements are one of the most successful SINEs found in any organism, contributing over a million copies to the human genome [16]. Although both L1 proteins are essential for L1 retrotransposition [11], Alu only strictly requires ORF2p for new insertion events [8;17].
Alu elements insert in the genome using a process termed target primed reverse transcription (TPRT) [18] (see Fig. 1, Inset 3). Inserts generated from RNAs that have undergone TPRT have a 3′ A-tail and are flanked by short, variable-length target site duplications (TSDs). These sequence features are considered the hallmarks of retrotransposition. Because Alu elements insert through an RNA intermediate, each new insertion effectively increases the copy number of Alus, allowing them to currently occupy 11% of the human genome. Alus are approximately 300 base pairs in length, and they make the retrotransposition competent RNA using a bipartite internal RNA polymerase III promoter. However, the vast majority of the Alu copies are neither transcriptionally, nor retrotranspositionally active [19]. There are multiple factors contributing to silencing of most Alu elements. Most Alus in the genome are transcriptionally repressed by methylation or chromatin context [20-22]. Secondly the post-insertion accumulation of mutations contributes to their inactivation by altering its internal pol III promoter or the stability of the RNA structure [23], as well as limiting the RNA’s ability to bind important proteins needed for retrotransposition [24;25]. In addition, the A-tail length is very dynamic and shortens quickly, limiting individual element activity [8;26].
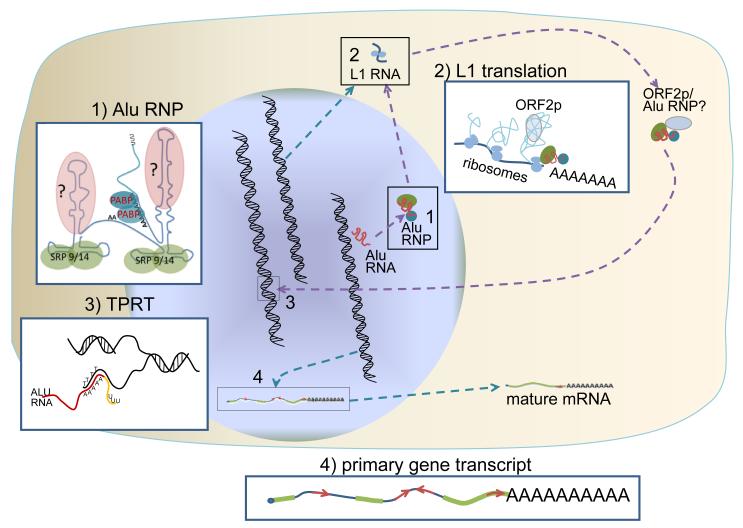
Figure 1. Alu retrotransposition cycle.
Alu RNA is transcribed by RNA polymerase III in the nucleus and associates with several proteins to form an RNP (see Inset 1). Inset 1: The Alu RNA binds two of the signal recognition proteins, SRP9 and SRP14 (green circles), and the polyA binding protein (PABP, blue circles). Other proteins are thought to bind the complex, but are yet to be identified (pink circle marked with a “?”). Because the Alu RNA does not code for proteins, to undergo retrotransposition Alu highjacks L1 ORF2p. The L1 ORF2p is obtained from the active L1 elements in the genome. These L1 elements will generate an mRNA that will go to the cytoplasm for translation (see Inset 2). Inset 2: During translation of the L1 mRNA, it is thought that Alu RNPs associated with ribosomes will recruit the needed L1 protein(s), such as ORF2p. The role of SRP9 and SRP14 has been previously suggested to be the targeting of Alu to ribosomes. The proximity of the Alu RNP to the translating L1 RNA likely favors the recruitment of the L1 ORF2p needed for Alu retrotransposition. However, it is unknown if there is a direct association between the L1 ORF2p and the Alu RNP (indicated by the “?”) or how these components reach the nucleus for insertion. Once in the nucleus, the Alu RNA undergoes the insertion process (see Inset 3). Inset 3: The Alu retrotransposition insertion mechanism is based on a model developed for the R2Bm element. The model proposes a target primed reverse transcription (TPRT) where the 3′ A-rich region of the Alu RNA base pairs with a T-rich region of the cleaved genomic DNA to provide the priming site for reverse transcription by the L1 ORF2p. This eliminates the unique 3′ tail of the Alu RNA (yellow line portion) from the reverse transcription and only inserts the Alu portion. Alu insertions are usually stably maintained in the genome, as there is no known specific mechanism for their removal. Due to the random nature of Alu insertion many inserts are found within genes and both mature and primary mRNA can contain multiple Alu sequences (see Inset 4). Inset 4: Many primary mRNA of genes contain Alu sequences (shown as red arrows) and a significant portion of mRNAs contain one or more Alu elements (located in either orientation) in their 3′ UTR. These Alus located within genes have been implicated in a number of regulatory mechanisms for gene expression.
Alu element insertions, genetic variation and disease
Alu elements are ancestrally derived from 7SL RNA [2], which is part of the signal recognition particle [27]. An Alu element is composed of two non-identical 7SL derived monomers (left half and right half) joined by an A-rich region. Unlike its ancestor, 7SL, Alu elements are unique to primates [19]. Alu amplification initiated around 65 million years ago (mya) peaking about 40 mya. The majority (80%) of the Alu elements present in the human genome inserted before or during this peak of amplification. Most processed pseudogene inserts also occurred about 40 mya [28] suggesting that non-autonomous retrotransposition activity may have been favorable at that point in evolution. Although lower than the peak evolutionary period, Alu amplification still significantly contributes to population diversity [29;30] and genetic disease [31] with studies estimating that approximately one Alu insertion occurs in every 20 births [32;33]. Alu elements can be categorized into subfamilies based on the accumulation of mutations through time. The most ancestral subfamilies, the J and S-families were followed by the young Y-subfamily that emerged after the evolutionary divergence from the great apes. Evidence indicates that only the young Alu elements are currently active. Due to the relatively random nature of insertion, retrotransposition of sequences is estimated to contribute to about 0.3 % of a diverse variety of human genetic diseases [34]. Up to now, there are over 95 examples of insertions of retrotransposons that are either causative or associated with human disease (reviewed in [35;36]), of which Alu contributes to the majority with 60 reported cases. Thus, non-autonomous Alu insertions appear to be occurring at a much higher rate than its autonomous driver element, L1 [31;35].
The Alu ribonucleoprotein
Each Alu locus produces a unique Alu RNA molecule. Transcription from the internal promoter begins at the first base of Alu and transcribes through the entire length of the Alu element and its A-tail region. The transcript continues through the genomic flanking sequences until it reaches an RNA polymerase III terminator, generally thought to be 4 T residues. Thus, each Alu varies within the Alu body because of mutations relative to the consensus [19], has a variable polymorphic length A-tail [37], and also has a unique 3′ region contributed by its specific locus [38]. These sequence variations between Alu elements have been shown to significantly impact the retrotransposition ability of any individual Alu element where specific sequence components need to be maintained for Alu to remain active [25].
Alu elements have no protein coding capacity and therefore carry out their entire life cycle (Fig. 1) utilizing proteins cannibalized from L1 and other cellular processes. Due to their sequence similarity and conservation of key RNA secondary structure relative to 7SL RNA, Alu RNAs have been shown to bind the signal recognition proteins 9 and 14 (SRP 9 and 14) [39], (Fig.1 Inset 1). SRP9 and SRP14 are required for efficient retrotransposition of Alu, although it is unlikely that these proteins interact with the SINEs in other organisms as most are derived from tRNA instead of 7SL. Because these two proteins are involved in recruiting 7SL RNA to a ribosome [40], it has been proposed that they are important in targeting the Alu RNP to the ribosomes. It seems likely that this binding occurs in the nucleolus [41], and thus the Alu may actually be exported with the ribosomes. Any Alu-bound ribosome that translates an L1 RNA would have a proximity advantage that may aid in stealing ORF2 from L1 as it is being translated (Fig.1 Inset 2). Another protein with possible roles in localizing Alu to a ribosome is polyA binding protein (PABP). This protein binds to the A-tail of SINEs [42;43], and based on a previous study [44] it is thought it may also play a role in recruiting Alu to the ribosome. It is likely that other proteins are involved in the retrotransposition, but further studies will be needed to further elucidate these intricate relationships and processes.
Alu RNA is transcribed by RNA polymerase III, probably at very low levels in most cells and tissues [45-48]. However, Alu elements are also present in the introns of RNA polymerase II-transcribed genes and in the 3′ non-coding regions of mature mRNAs (see Fig. 1) at a relatively high abundance [49]. The Alu sequences in gene transcripts have a broad range of impacts on expression of those genes [31;50-52], but do not contribute to retrotransposition. The high level of Alu sequences in gene transcripts makes it very difficult to detect the RNA polymerase III Alu transcripts that are involved in retrotransposition. Relatively few studies have focused on key features to distinguish RNA polymerase III-generated transcripts from the genic transcripts [47;53-55]. These include experiments using primer extension and C-tail RACE [45;47] and the mining of genome-wide ChIP analysis with parallel sequencing (ChIP-seq) to detect the Alus bound by RNA polymerase III factors [48;56-58]. Careful analysis of the pol-III ChIP-seq data show that only a small subset of the pol III-bound Alus have the features required for efficient retrotransposition [48]. However, most have been done in tissue culture cell lines leaving a significant gap in the knowledge of Alu expression in different tissues.
Somatic activity of Alu elements
Much of what we know about Alu insertion activity comes from population and evolutionary analyses [59;60] and there are almost no data on whether Alu elements show significant levels of somatic insertions. If they do insert somatically, they could be important contributors to insertional mutagenesis in contributing to cancer, and perhaps other age-related diseases.
Somatic retrotransposition has the potential to contribute to cancer if the insert mutagenizes a gene necessary for cellular homeostasis such as a tumor suppressor. This event is expected to occur in somatic cells. Although there is clear evidence of Alu retrotransposition in the germline, there are limited data on Alu activity in somatic tissue. There are several reports of de novo Alu inserts linked to cancers, some likely involving germ line insertions (examples are listed in Table 1). Ex vivo studies indicate that Alu can retrotranspose in a variety of cells [8;17] suggesting that somatic cells could support Alu activity. Whole genome sequencing has revealed evidence that supports the activity of L1 in many types of cancer [61-63] but Alu insertions were at much lower levels in a limited set of studies [62]. Due to the limited data available on Alu at this stage, it is unclear if somatic cells are devoid or less supportive of Alu retrotransposition and if Alu insertion activity has a role in cancer.
Table 1.
De novo Alu inserts associated with cancer or cancer related genes
Retrotransposition cycle- regulators
Based on the potential deleterious effects of mobile elements, the cell has developed diverse ways to regulate retrotransposition events. For Alu, most of the regulation methods that have been characterized relate to RNA expression. Methylation, as stated previously, is a way to shut down [global] transcription and gene expression. Specifically, DNA methyltransferase 3-like gene (Dnmt3L) [64], MILI, and MIWI2 [65;66] have been shown to be important for de novo methylation of mobile elements in the mouse germ line. In addition to epigenetic regulation, a number of stress factors have been found to stimulate expression of Alu. These factors include some viral infections [39], heat shock [55] and related stresses [53], and some chemotherapeutic treatments [67].
Culture based assays indicate that Alu activity also appears to be negatively regulated by the microprocessor machinery (Drosha-DGCR8) [68]. However other studies show that the microRNA targets within Alu avoid this type of regulation [69]. In addition, the RNA helicase, MOV10, a component of the RNA-induced silencing complex (RISC) also restricts Alu activity in culture [70]. Although there are many cellular and genetic factors that regulate L1 activity [31], it is striking that most of them do not modulate Alu retrotransposition significantly, despite the dependence of Alu on L1. APOBEC3G has been found to modulate Alu retrotransposition [71], but not L1, whereas different APOBEC3 subtypes regulate L1 without affecting Alu activity. These findings demonstrate that although Alu depends on some aspects of L1, there are significant mechanistic differences that alter the ability of many cellular processes to co-regulate these two different mobile element families. It is quite possible that the higher levels of Alu insertions vs. L1 insertions in the germ line reflect the inability of many of the regulatory processes that repress L1 to effectively inhibit Alu.
Post-insertional impacts of Alu insertions
Even an Alu element that has no immediate genomic impact may later have major consequences for the genome. One of the most devastating is their ability to contribute to non-allelic homologous recombination (NAHR). This has led to many deletions and duplications due to unequal recombination between Alu elements on an evolutionary time frame [60;72], as well as probably being a much bigger contribution to germ line disease than the insertion events [31;35]. It seems very likely the NAHR occurs relatively frequently in somatic tissues and may contribute to Copy Number Variations (CNVs) that lead to cancer progression [72;73]. NAHR events between Alus are a well recognized source of DNA damage (reviewed in [74]) with numerous examples linked to cancers (reviewed in [75;76]). Certain cancer associated genes such as the von Hippel-Lindau tumor suppressor (VHL) and the mixed-lineage leukemia (MLL) seem significantly prone to Alu/Alu NAHR events.
Alu elements also have the ability to influence genes in many ways. Because the Alu sequence is CpG rich insertions can introduce new sites for methylation. For example, one report associates the epigenetic influence of an Alu element variant in the POMC gene with childhood obesity [77]. Alu elements are enriched in introns and are also found very commonly in 3′ UTRs of genes [49;78;79] (Fig. 1, Inset 4). As Alu elements accumulate mutations during evolutionary, a number of cryptic signals within the Alu can be activated. This has included expansion of their A-rich region into triplet repeats leading to triplet-repeat disease [80]. It also involves changes that potentially influence promoters, or defective splicing [31;52]. Alu elements within transcription units have also been shown to provide cryptic polyadenylation sites [81], as well as cryptic splice sites [82], leading to alternative processing of transcripts. In addition, having inverted Alu elements in a mRNA has been shown to be subject to extensive RNA editing by adenosine deaminase reductase (ADAR) that can influence nuclear retention of RNAs, as well as possibly other mRNA regulatory processes [50].
Summary
Alu elements are primate specific elements that are highly abundant in the human genome. Their continued insertion activity contributes to a variety of genetic diseases and genomic variations between different individuals. Alu depends on L1 factors for its mobilization, but is much more active in the germ line than L1 elements. There is still very limited data on Alu expression or activity in either somatic tissues or cancers. The differences between L1 and Alu illustrate that they have different cellular requirements and regulating mechanisms. Alu appears to avoid many of the mechanisms that regulate L1 potentially explaining why Alu insertions contribute more to disease than L1. We need to understand more about Alu expression and the mechanistic differences between Alu and L1 to better predict the conditions under which Alu may have the biggest impact on genome instability and function.
Highlights.
New Alu element insertions continue to cause about 0.1% of new human genetic diseases.
Alu is ancestrally derived from the 7SL RNA and formed a primate-specific SINE family.
Alu RNA interacts with proteins that are needed for efficient retrotransposition.
Alu elements are a major contribute to non-allelic homologous recombination.
Alu elements are involved in a diverse set of RNA processing and regulatory events.
Acknowledgements
This research was supported by National Institutes of Health (NIH) P20 P20GM103518/ P20RR020152 (PLD and AMR-E), R01GM079709A and the National Science Foundation/ EPSCOR PFUND (AMR-E), R01GM45668 and EPSCOR/BORSF grant (PLD). This paper’s contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
References
- 1.Rogers JH, Willison KR. A major rearrangement in the H-2 complex of mouse t haplotypes. Nature. 1983;304:549–552. doi: 10.1038/304549a0. [DOI] [PubMed] [Google Scholar]
- 2.Weiner A, Deininger P, Efstradiatis A. The Reverse Flow of Genetic Information: pseudogenes and transposable elements derived from nonviral cellular RNA. Annual Reviews of Biochemistry. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 3.Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol.Biol.Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 4.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am.J.Hum.Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: a hominid-specific retroposon family. J Mol.Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 6.Goncalves I, Duret L, Mouchiroud D. Nature and structure of human genes that generate retropseudogenes. Genome Res. 2000;10:672–678. doi: 10.1101/gr.10.5.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat.Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 8.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat.Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 9.Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, Lower J, Stratling WH, Lower R, Schumann GG. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum.Mol Genet. 2011;20:3386–3400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 12.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. Embo J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 13.Martin SL. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA.Biol. 2010;7:67–72. [Google Scholar]
- 14.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 15.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 16.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la BM, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 17.Wallace N, Wagstaff BJ, Deininger PL, Roy-Engel AM. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419:1–6. doi: 10.1016/j.gene.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 19.Shen MR, Batzer MA, Deininger PL. Evolution of the master Alu gene(s) J Mol.Evol. 1991;33:311–320. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- 20.Liu WM, Schmid CW. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic.Acids.Res. 1993;21:1351–1359. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Englander EW, Wolffe AP, Howard BH. Nucleosome interactions with a human Alu element. Transcriptional repression and effects of template methylation. J.Biol.Chem. 1993;268:19565–19573. [PubMed] [Google Scholar]
- 22.Slagel V, Deininger P. In vivo transcription of a cloned prosimian primate SINE sequence. Nucleic Acids Res. 1989;17:8669–8682. doi: 10.1093/nar/17.21.8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labuda D, Zietkiewicz E. Evolution of secondary structure in the family of 7SL-like RNAs. J.Mol.Evol. 1994;39:506–518. doi: 10.1007/BF00173420. [DOI] [PubMed] [Google Scholar]
- 24.Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, Devine SE. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL. Diverse cis factors controlling Alu retrotransposition: What causes Alu elements to die? Genome Res. 2009;19:545–555. doi: 10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy-Engel AM, Salem AH, Oyeniran OO, Deininger L, Hedges DJ, Kilroy GE, Batzer MA, Deininger PL. Active alu element "A-Tails": size does matter. Genome Res. 2002;12:1333–1344. doi: 10.1101/gr.384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter P, Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- 28.Ohshima K, Hattori M, Yada T, Gojobori T, Sakaki Y, Okada N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart C, Kural D, Stromberg MP, Walker JA, Konkel MK, Stutz AM, Urban AE, Grubert F, Lam HY, Lee WP, Busby M, Indap AR, Garrison E, Huff C, Xing J, Snyder MP, Jorde LB, Batzer MA, Korbel JO, Marth GT. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011;7:e1002236. doi: 10.1371/journal.pgen.1002236. * This manuscripts performs an in depth evaluation of the Alu polymorphisms from the 1000 Genomes Project. It demonstrates Alu as the major contributor of human diversity..
- 30.Witherspoon DJ, Zhang Y, Xing J, Watkins WS, Ha H, Batzer MA, Jorde LB. Mobile element scanning (ME-Scan) identifies thousands of novel Alu insertions in diverse human populations. Genome Res. 2013;23:1170–1181. doi: 10.1101/gr.148973.112. ** This manuscript presents a next generation sequencing method to identify polymorphic Alu elements. It provides a quantitative evaluation of Alu polymorphisms in human populations highlighting the significant impact and high level of continued activity of Alu elements in germline.
- 31.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 32.Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006 doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 33.Witherspoon DJ, Marchani EE, Watkins WS, Ostler CT, Wooding SP, Anders BA, Fowlkes JD, Boissinot S, Furano AV, Ray DA, Rogers AR, Batzer MA, Jorde LB. Human population genetic structure and diversity inferred from polymorphic L1(LINE-1) and Alu insertions. Hum.Hered. 2006;62:30–46. doi: 10.1159/000095851. [DOI] [PubMed] [Google Scholar]
- 34.Callinan PA, Wang J, Herke SW, Garber RK, Liang P, Batzer MA. Alu retrotransposition-mediated deletion. J Mol.Biol. 2005;348:791–800. doi: 10.1016/j.jmb.2005.02.043. [DOI] [PubMed] [Google Scholar]
- 35.Hancks DC, Kazazian HH., Jr. Active human retrotransposons: variation and disease. Curr.Opin.Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaer K, Speek M. Retroelements in human disease. Gene. 2013;518:231–241. doi: 10.1016/j.gene.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Economou EP, Bergen AW, Warren AC, Antonarakis SE. The polydeoxyadenylate tract of Alu repetitive elements is polymorphic in the human genome. Proc.Natl.Acad.Sci., USA. 1990;87:2951–2954. doi: 10.1073/pnas.87.8.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deininger PL, Jolly D, Rubin C, Friedmann T, Schmid CW. Base Sequence Studies of 300 Nucleotide Renatured Repeated Human DNA Clones. J.Mol.Biol. 1981;151:17–33. doi: 10.1016/0022-2836(81)90219-9. [DOI] [PubMed] [Google Scholar]
- 39.Chang DY, Hsu K, Maraia RJ. Monomeric scAlu and nascent dimeric Alu RNAs induced by adenovirus are assembled into SRP9/14-containing RNPs in HeLa cells. Nucleic Acids Res. 1996;24:4165–4170. doi: 10.1093/nar/24.21.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terzi L, Pool MR, Dobberstein B, Strub K. Signal recognition particle Alu domain occupies a defined site at the ribosomal subunit interface upon signal sequence recognition. Biochemistry. 2004;43:107–117. doi: 10.1021/bi0353777. [DOI] [PubMed] [Google Scholar]
- 41.Jacobson MR, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc.Natl.Acad.Sci.U.S.A. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West N, Roy-Engel A, Imataka H, Sonenberg N, Deininger P. Shared Protein Components of SINE RNPs. J.Mol.Biol. 2002;321:423–432. doi: 10.1016/s0022-2836(02)00542-9. [DOI] [PubMed] [Google Scholar]
- 43.Muddashetty R, Khanam T, Kondrashov A, Bundman M, Iacoangeli A, Kremerskothen J, Duning K, Barnekow A, Huttenhofer A, Tiedge H, Brosius J. Poly(A)-binding Protein is Associated with Neuronal BC1 and BC200 Ribonucleoprotein Particles. J.Mol.Biol. 2002;321:433–445. doi: 10.1016/s0022-2836(02)00655-1. [DOI] [PubMed] [Google Scholar]
- 44.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu.Rev.Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 45.Paulson KE, Schmid CW. Transcriptional inactivity of Alu repeats in HeLa cells. Nucleic.Acids.Res. 1986;14:6145–6158. doi: 10.1093/nar/14.15.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matera AG, Hellmann U, Schmid CW. A transpositionally and transcriptionally competent Alu subfamily. Mol.Cell.Biol. 1990;10:5424–5432. doi: 10.1128/mcb.10.10.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaikh TH, Roy AM, Kim J, Batzer MA, Deininger PL. cDNAs derived from primary and small cytoplasmic Alu (scAlu) transcripts. J Mol.Biol. 1997;271:222–234. doi: 10.1006/jmbi.1997.1161. [DOI] [PubMed] [Google Scholar]
- 48.Oler AJ, Traina-Dorge S, Derbes RS, Canella D, Cairns BR, Roy-Engel AM. Alu expression in human cell lines and their retrotranspositional potential. Mob.DNA. 2012;3:11. doi: 10.1186/1759-8753-3-11. * This manuscript highlights the importance of distinguishing RNA polymerase-III derived Alu transcripts from mRNA containing Alu sequences when evaluating Alu retrotransposition potential. In addition, it makes the first attempt to combining predictions of transcriptionally active loci and sequence feature required for Alu retrotransposition.
- 49.Yulug IG, Yulug A, Fisher EM. The frequency and position of Alu repeats in cDNAs, as determined by database searching. Genomics. 1995;27:544–548. doi: 10.1006/geno.1995.1090. [DOI] [PubMed] [Google Scholar]
- 50.Chen LL, Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 51.Chen LL, Carmichael GG. Altered Nuclear Retention of mRNAs Containing Inverted Repeats in Human Embryonic Stem Cells: Functional Role of a Nuclear Noncoding RNA. Molecular Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaer K, Speek M. Intronic retroelements: Not just "speed bumps" for RNA polymerase II. Mob.Genet Elements. 2012;2:154–157. doi: 10.4161/mge.20774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim C, Rubin CM, Schmid CW. Genome-wide chromatin remodeling modulates the Alu heat shock response. Gene. 2001;276:127–133. doi: 10.1016/s0378-1119(01)00639-4. [DOI] [PubMed] [Google Scholar]
- 54.Li T, Spearow J, Rubin CM, Schmid CW. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239:367–372. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- 55.Li TH, Schmid CW. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene. 2001;276:135–141. doi: 10.1016/s0378-1119(01)00637-0. [DOI] [PubMed] [Google Scholar]
- 56.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, Cairns BR. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Canella D, Praz V, Reina JH, Cousin P, Hernandez N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010;20:710–721. doi: 10.1101/gr.101337.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat.Rev.Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 60.Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB. Mobile DNA elements in primate and human evolution. Am.J Phys.Anthropol. 2007;45(Suppl):2–19. doi: 10.1002/ajpa.20722. [DOI] [PubMed] [Google Scholar]
- 61.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, III, Lohr JG, Harris CC, Ding L, Wilson RK, Wheeler DA, Gibbs RA, Kucherlapati R, Lee C, Kharchenko PV, Park PJ. Landscape of Somatic Retrotransposition in Human Cancers. Science. 2012 doi: 10.1126/science.1222077. ** The authors provide a wide analyses of a variety of tumor samples and demonstrated that mobile element activity varies between different type of cancers. In addition, many identified de novo inserts were within genes normally mutated in cancer suggesting a potential role of mobile element activity and carcinogenesis. Interestingly, the rate of Alu amplification in tumors was much lower than L1.
- 63.Solyom S, Ewing AD, Rahrmann EP, Doucet TT, Nelson HH, Burns MB, Harris RS, Sigmon DF, Casella A, Erlanger B, Wheelan S, Upton KR, Shukla R, Faulkner GJ, Largaespada DA. Kazazian HH: Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012 doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 65.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA Pathway Primed by Individual Transposons Is Linked to De Novo DNA Methylation in Mice. Molecular Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hagan CR, Sheffield RF, Rudin CM. Human Alu element retrotransposition induced by genotoxic stress. Nat.Genet. 2003;35:219–220. doi: 10.1038/ng1259. [DOI] [PubMed] [Google Scholar]
- 68.Heras SR, Macias S, Plass M, Fernandez N, Cano D, Eyras E, Garcia-Perez JL, Caceres JF. The Microprocessor controls the activity of mammalian retrotransposons. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoffman Y, Dahary D, Bublik DR, Oren M, Pilpel Y. The majority of endogenous microRNA targets within Alu elements avoid the microRNA machinery. Bioinformatics. 2013;29:894–902. doi: 10.1093/bioinformatics/btt044. [DOI] [PubMed] [Google Scholar]
- 70.Goodier JL, Cheung LE, Kazazian HH., Jr MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PloS Genetics. 2012 doi: 10.1371/journal.pgen.1002941. * This manuscript shows how cellular factors previously known to regulate retroviruses have also the ability of regulating L1 and Alu element activity, reinforcing the hypothesis that retroviruses and these elements are ancestrally related.
- 71.Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sen SK, Han K, Wang J, Lee J, Wang H, Callinan PA, Dyer M, Cordaux R, Liang P, Batzer MA. Human Genomic Deletions Mediated by Recombination between Alu Elements. Am.J.Hum.Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gebow D, Miselis N, Liber HL. Homologous and nonhomologous recombination resulting in deletion: effects of p53 status, microhomology, and repetitive DNA length and orientation. Mol Cell Biol. 2000;20:4028–4035. doi: 10.1128/mcb.20.11.4028-4035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hedges DJ, Deininger PL. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat.Res. 2006 doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Belancio VP, Roy-Engel AM, Deininger PL. All y’all need to know ’bout retroelements in cancer. Semin.Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Konkel MK, Batzer MA. A mobile threat to genome stability: The impact of non-LTR retrotransposons upon the human genome. Semin.Cancer Biol. 2010;20:211–221. doi: 10.1016/j.semcancer.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuehnen P, Mischke M, Wiegand S, Sers C, Horsthemke B, Lau S, Keil T, Lee YA, Grueters A, Krude H. An Alu element-associated hypermethylation variant of the POMC gene is associated with childhood obesity. PLoS Genet. 2012;8:e1002543. doi: 10.1371/journal.pgen.1002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grover D, Mukerji M, Bhatnagar P, Kannan K, Brahmachari SK. Alu repeat analysis in the complete human genome: trends and variations with respect to genomic composition. Bioinformatics. 2004;20:813–817. doi: 10.1093/bioinformatics/bth005. [DOI] [PubMed] [Google Scholar]
- 79.Tsirigos A, Rigoutsos I. Alu and b1 repeats have been selectively retained in the upstream and intronic regions of genes of specific functional classes. PLoS Comput.Biol. 2009;5:e1000610. doi: 10.1371/journal.pcbi.1000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina G, Monticelli A, Turano M, Filla A, De MG, Cocozza S. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum.Mol.Genet. 1997;6:1261–1266. doi: 10.1093/hmg/6.8.1261. [DOI] [PubMed] [Google Scholar]
- 81.Chen C, Ara T, Gautheret D. Using Alu elements as polyadenylation sites: A case of retroposon exaptation. Mol Biol Evol. 2009;26:327–334. doi: 10.1093/molbev/msn249. [DOI] [PubMed] [Google Scholar]
- 82.Ram O, Schwartz S, Ast G. Multifactorial interplay controls the splicing profile of Alu-derived exons. Mol Cell Biol. 2008;28:3513–3525. doi: 10.1128/MCB.02279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen J, Rattner A, Nathans J. Effects of L1 retrotransposon insertion on transcript processing, localization, and accumulation: lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum.Mol.Genet. 2006 doi: 10.1093/hmg/ddl138. [DOI] [PubMed] [Google Scholar]
- 84.Economou-Pachnis A, Tsichlis PN. Insertion of an Alu SINE in the human homologue of the Mlvi-2 locus. Nucleic Acids Research. 1985;13:8379–8387. doi: 10.1093/nar/13.23.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Halling KC, Lazzaro CR, Honchel R, Bufill JA, Powell SM, Arndt CA, Lindor NM. Hereditary desmoid disease in a family with a germline Alu I repeat mutation of the APC gene. Hum.Hered. 1999;49:97–102. doi: 10.1159/000022852. [DOI] [PubMed] [Google Scholar]
- 86.Teugels E, De Brakeleer S, Goelen G, Lissens W, Sermijn E, De Greve J. De novo Alu element insertions targeted to a sequence common to the BRCA1 and BRCA2 genes. Hum.Mutat. 2005;26:284. doi: 10.1002/humu.9366. [DOI] [PubMed] [Google Scholar]
- 87.Machado PM, Brandao RD, Cavaco BM, Eugenio J, Bento S, Nave M, Rodrigues P, Fernandes A, Vaz F. Screening for a BRCA2 rearrangement in high-risk breast/ovarian cancer families: evidence for a founder effect and analysis of the associated phenotypes. J Clin.Oncol. 2007;25:2027–2034. doi: 10.1200/JCO.2006.06.9443. [DOI] [PubMed] [Google Scholar]
- 88.Miki Y, Katagiri T, Kasumi F, Yoshimoto T, Nakamura Y. Mutation analysis in the BRCA2 gene in primary breast cancers. Nat.Genet. 1996;13:245–247. doi: 10.1038/ng0696-245. [DOI] [PubMed] [Google Scholar]
- 89.Wallace MR, Andersen LB, Saulino AM, Gregory PE, Glover TW, Collins FS. A de novo Alu insertion results in neurofibromatosis type 1. Nature. 1991;353:864–866. doi: 10.1038/353864a0. [DOI] [PubMed] [Google Scholar]