Abstract
The outcomes of evolution are determined by which mutations occur and fix. In rapidly adapting microbial populations, this process is particularly hard to predict because lineages with different beneficial mutations often spread simultaneously and interfere with one another’s fixation. Hence to predict the fate of any individual variant, we must know the rate at which new mutations create competing lineages of higher fitness. Here, we directly measured the effect of this interference on the fates of specific adaptive variants in laboratory Saccharomyces cerevisiae populations and used these measurements to infer the distribution of fitness effects of new beneficial mutations. To do so, we seeded marked lineages with different fitness advantages into replicate populations and tracked their subsequent frequencies for hundreds of generations. Our results illustrate the transition between strongly advantageous lineages that decisively sweep to fixation and more moderately advantageous lineages that are often outcompeted by new mutations arising during the course of the experiment. We developed an approximate likelihood framework to compare our data to simulations and found that the effects of these competing beneficial mutations were best approximated by an exponential distribution, rather than one with a single effect size. We then used this inferred distribution of fitness effects to predict the rate of adaptation in a set of independent control populations. Finally, we discuss how our experimental design can serve as a screen for rare, large-effect beneficial mutations.
Keywords: distribution of fitness effects, clonal interference, beneficial mutations, experimental evolution, Saccharomyces cerevisiae
EVOLUTIONARY adaptation is driven by the accumulation of beneficial mutations. One can ask two basic questions about this process. First, what set of mutations is available to the population? That is, what is the overall mutation rate, U, and the distribution of fitness effects, ρ(s), of new mutations? Second, what is the fate of those mutations that occur? In other words, how does the frequency of each mutation change over time until it eventually fixes or goes extinct?
When beneficial mutations are rare, these two questions are independent. Mutations of a given fitness effect, s, occur at rate Uρ(s). The fate of each mutant is then decided entirely on its own merits: it increases in frequency (or is lost due to random drift) at a rate commensurate with its selective effect. Experiments, however, have shown that even for modestly sized laboratory populations of viruses and microbes, multiple beneficial mutations often spread simultaneously and interfere with one another, an effect known as clonal interference (de Visser et al. 1999; Miralles et al. 1999; Joseph and Hall 2004; Desai et al. 2007; Perfeito et al. 2007; Kao and Sherlock 2008; Lee and Marx 2013; see Sniegowski and Gerrish 2010 for a recent review). This means that the fate of each beneficial mutation depends not only on its own effect, but also on its interactions with the rest of the variation in the population (Lang et al. 2011, 2013). In this regime, the mutation rate and the distribution of fitness effects of beneficial mutations [the DFE, ρ(s)] controls the availability of competing mutations, which then play an important role in determining the fate of each new beneficial mutation (Gerrish and Lenski 1998; Good et al. 2012).
These factors highlight the importance of the DFE as a central parameter in adaptation, determining which new mutations occur and influencing their subsequent fate. Some theoretical work has argued that the DFE will typically be exponential (Gillespie 1983; Orr 2003). However, this is fundamentally an empirical question, and in principle the details of the DFE could be highly system specific. Thus extensive experimental effort has been devoted to measuring the DFE of beneficial mutations in a variety of laboratory populations (Imhof and Schlötterer 2001; Rozen et al. 2002; Sanjuán et al. 2004; Barrett et al. 2006; Kassen and Bataillon 2006; Burch et al. 2007; Perfeito et al. 2007; Rokyta et al. 2008; MacLean and Buckling 2009; Bataillon et al. 2011; McDonald et al. 2011; a separate literature has used population genetic methods to infer the DFE in natural populations, reviewed by Keightley and Eyre-Walker 2010).
Experimental efforts to measure the DFE of beneficial mutations in laboratory populations have largely taken one of two complementary approaches. The first approach is to isolate mutants and directly assay their fitness. The difficulty with this method is that beneficial mutations are rare, so many clones must be screened to isolate comparatively few beneficial mutations (Sanjuán et al. 2004; Kassen and Bataillon 2006). To avoid this difficulty, some studies have imposed a harsh selection and studied the survivors, which by definition must have a beneficial mutation (MacLean and Buckling 2009; McDonald et al. 2011). However, this approach is limited to harsh and typically narrow stresses (e.g., treatment with antibiotic), which may not be representative of adaptation to other conditions.
The second common experimental approach is to track the frequencies of genetic markers over time and use the resulting dynamics to infer the underlying DFE. Such “marker divergence” experiments typically use two or more strains that differ by a single neutral genetic marker, which can be easily tracked through time (e.g., antibiotic resistance or a fluorescent reporter). These strains are mixed, usually in equal proportions, and allowed to evolve in competition. The changes in frequencies of the neutral markers then reflect subsequent beneficial mutations that occur in one or the other genetic background (Novick and Szilard 1950; Atwood et al. 1951; Helling et al. 1981; Paquin and Adams 1983; Adams and Oeller 1986; Imhof and Schlötterer 2001; Barrett et al. 2006; de Visser and Rozen 2006; Hegreness et al. 2006; Perfeito et al. 2007; Kao and Sherlock 2008; Barrick et al. 2010; Lang et al. 2011). Inferring the DFE from such data typically requires estimating the fitness effects of many mutations from the dynamics of relatively few markers, which is naturally quite difficult (Hegreness et al. 2006; Pinkel 2007; Illingworth and Mustonen 2012; Zhang et al. 2012; de Sousa et al. 2013). In principle, this difficulty could be removed by reducing the population size to such a degree that only one or zero beneficial mutations usually arise in each population (Perfeito et al. 2007). However, this requires careful tuning of the population size, to make it small enough to minimize multiple mutations but also large enough to ensure that many replicates acquire a beneficial mutation.
Here, we introduce a twist on the traditional design of marker divergence experiments that produce dynamics more directly revealing of the underlying DFE. Rather than using neutral markers, we tracked the frequencies of marked lineages with a fitness advantage relative to a reference strain. We seeded these marked lineages at low frequency into populations of the reference, so that their subsequent dynamics are reflective of the fates of beneficial mutations with a particular selective advantage. Since the DFE controls the availability of competing mutations and hence the likelihood of clonal interference, we can exploit the observed fates of seeded lineages to infer the DFE. Using lineages with different fitness advantages enabled us to probe different corresponding portions of the DFE. This approach is particularly suited to inferring those aspects of the DFE that are most important in determining the fates of new beneficial “driver” mutations, e.g., the high-fitness tail, which is otherwise hard to measure directly. In the process, we also directly measured how clonal interference alters a key quantity in adaptation: the fixation probability of a beneficial mutation as a function of its fitness effect.
Materials and Methods
Strains
All strains used in this study were derived from the base strain DBY15084, a haploid Saccharomyces cerevisiae strain derived from the W303 background with genotype MATa, ade2–1, CAN1, his3–11 leu2–3, 112, trp1–1, URA3, bar1Δ::ADE2, hmlαΔ::LEU2. Each experimental population included a resident and a seeded lineage. The resident lineage was DBY15108, a derivative of DBY15084 in which the fluorescent protein ymCherry was integrated at the URA3 locus (Lang et al. 2011). The seeded lineages were descendants of strain DBY15104 isolated from time points of an earlier long-term evolution experiment (Lang et al. 2011). To allow us to track their frequency using flow cytometry, we amplified a pACT1–ymCitrine pTEF–HISMX6 cassette from plasmid pJHK043 (provided by John H. Koschwanez) and integrated it at the HIS locus using oligos oGW137 (5′-TTGGTGAGCGCTAGGAGTC-3′) and oGW138 (5′-TATGAAATGCTTTTCTTGTTGTTCTTACG-3′) provided by Gregg Wildenberg. From this pool of transformants, we selected strains EFY11–17 based on fitness assays described below.
Experimental procedures
To obtain seeded lineage strains with a range of fitnesses, we isolated a large number of evolved clones and assayed their fitnesses as described in Lang et al. (2011). Briefly, this protocol is to mix each strain in roughly equal proportion with a reference strain that bears a different fluorescent reporter, propagate these mixed populations for 30 generations, and measure the ratio of the strains at generations 10 and 30 using flow cytometry. Relative fitness was calculated as s = (1/20) × log(final ratio/initial ratio). From among these clones, we chose EFY11–17 to use as seeded lineages and remeasured their fitnesses in 10 replicates. These additional assays showed that strains EFY12–14 and EFY15–16 had indistinguishable fitnesses, and so for the purposes of analysis, strains EFY11, EFY12–14, EFY15–16, and EFY17 were respectively grouped into the fitness classes indicated in Figure 1.
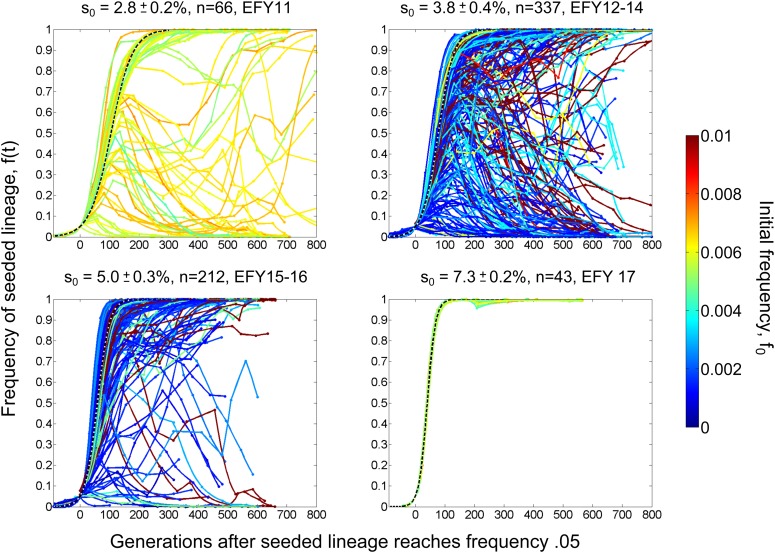
Figure 1.
Trajectories of seeded lineages. Each line represents the frequency over time of a marked lineage with fitness advantage s0 seeded into a replicate resident population. Colors correspond to the initial frequency f0 of the seeded lineage according to the legend at right. Time is measured in generations, with t = 0 defined as the time at which each trajectory reached frequency 0.05. The dashed curves show the expected trajectories in the absence of new beneficial mutations (i.e., without clonal interference). (Note that the seeded lineages for s0 ≈ 4 and 5% consisted of multiple strains; see Materials and Methods.)
To begin the evolution experiment, we grew an individual resident clone to saturation in 3 ml of standard growth media (YPD supplemented with 100 μg/ml ampicillin and 25 μg/ml tetracyclin). We transferred 128μl of this culture into each well of a 96 well-plate, diluted these cultures 210-fold into 12 96-well plates containing fresh media, allowed these cultures to grow for 10 generations, and froze them at −80° in 15% glycerol. Later, these plates were thawed and propagated for 30 generations (as described below) to reacclimate them to this environment. In parallel, we prepared the seeded clones in the same fashion. We then mixed seeded and resident populations to found a total of 1044 populations in 12 96-well plates (see Supporting Information, Table S1). These populations were propagated at 30° in 128μl YPD per well and diluted every 24 hr by a factor of 210 into new plates containing fresh media. This corresponds to an effective population size Ne ≈ 105 (Wahl et al. 2002; Lang et al. 2011). Each plate contained a set of nine empty wells as cross-contamination controls. All control wells remained sterile throughout the experiment except for two accidents involving plate mixing. This contamination was resolved by restarting from glycerol stocks of an earlier time point. Transfers were carried out using a Biomek FX pipetting robot.
At ∼50-generation intervals, seeded lineage frequencies were measured using flow cytometry. In particular, BD Biosciences Fortessa and LSR-II flow cytometers with high-throughput plate samplers counted ∼100,000 cells per population for the initial time point and ∼30,000 cells per population for time points thereafter. Repeated measurement of populations and blanks indicated that ∼100 cell counts per sample were carried over from previous samples. Therefore the uncertainty in frequency at the first time point was ∼0.1% and ∼0.5% thereafter. These raw data were processed in FlowJo v. 9.2. All processed data are provided in Table S1.
We also assayed the fitness of 16 additional control populations founded with only the resident strain. To do so, these populations were thawed from frozen-archive plates, each was duplicated into four replicates, these were propagated for 30 generations to acclimate them, and then their fitness was assayed as described above.
Note that 386 of the populations were later excluded from analysis, leaving a total of 658 replicate populations, apportioned among the seven seeded lineages and controls as described in Table S1. In 232 of these 386 excluded populations, frequency-dependent selection emerged. We identified these by first investigating 15 populations in which lineages coexisted at constant proportion for hundreds of generations. We found that this coexistence was maintained by frequency-dependent selection exclusively in populations having a characteristic pellet morphology, so we excluded from analysis all populations that also had this morphology. In the other 154 cases, the initial frequency of the seeded lineage was so low that it could not be precisely determined or extinction due to drift was common. To exclude these without biasing the statistics of trajectories, we chose a cutoff for the initial frequency of each seeded strain such that in all replicates in which the initial frequency was above the cutoff, the seeded lineage rose to at least 5%. All replicates below the cutoff were excluded.
Results
Tracking the fates of seeded lineages
Any beneficial mutation creates a new lineage that is more fit than the genetic background in which it arose. To systematically study the fates of such lineages, we prepared a set of fluorescently labeled haploid budding yeast strains (the seeded lineages) with measured fitness advantages, s0, of ∼3, 4, 5, and 7% relative to a closely related but separately labeled reference strain. We founded 658 replicate populations of the reference (the resident), and introduced one of the seeded lineages at low frequency into each replicate population. We propagated these populations asexually in batch culture for hundreds of generations at an effective population size of Ne ≈ 105, measuring the frequency of the seeded lineage in each population approximately every 50 generations (see Materials and Methods). This allowed us to track the fate of the seeded lineages over time, as illustrated in Figure 1.
Each seeded lineage was introduced at an initial frequency f0 large enough that genetic drift is expected to be weak relative to natural selection (i.e., ). In the absence of additional mutations, this implies that the frequency f(t) of each seeded lineage should increase deterministically according to the logistic equation, . This expectation is indicated by the dashed curves in Figure 1. As is apparent from the figure, most seeded lineages initially conformed to this expectation (the exceptions are lineages whose initial frequencies were only severalfold greater than , which is low enough that genetic drift could partially reduce their initial rate of increase). Subsequently, many lineages diverged into a variety of qualitatively distinct fates. Since both genetic drift and measurement errors are expected to be small relative to this divergence (see Materials and Methods), the variation in the fates of seeded lineages indicates that their relative fitnesses were modified by new beneficial mutations arising during the experiment.
Fates of seeded lineages reflect supply of competing beneficial mutations
The trajectory of each seeded lineage provides information about the beneficial mutations that did (or did not) arise within the competing resident population. Consider, for example, the case in which a seeded lineage of fitness s0 peaks and then declines in frequency. This reflects a clonal interference event, where one or more new beneficial mutations in the resident population create a competing lineage with fitness >s0 (see Figure 2). By considering the range of outcomes in replicate populations, we can estimate the probability of these events (Figure 3). A higher probability of clonal interference implies a larger supply of beneficial mutations that can generate successful competing lineages.
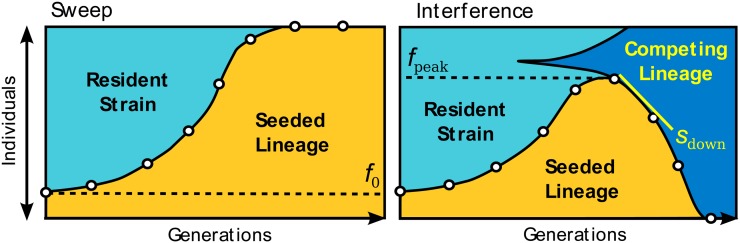
Figure 2.
The fates of seeded lineages. We classified the trajectory of each seeded lineage according to whether it increased monotonically to fixation (a selective sweep, shown left) or peaked and subsequently declined in frequency (clonal interference, shown right). Each clonal interference event implies that the resident adapted fast enough to overtake the seeded lineage in fitness. These cases were further classified by the seeded lineage’s peak frequency, fpeak, and relative fitness after this peak, sdown, as indicated in the right scheme.
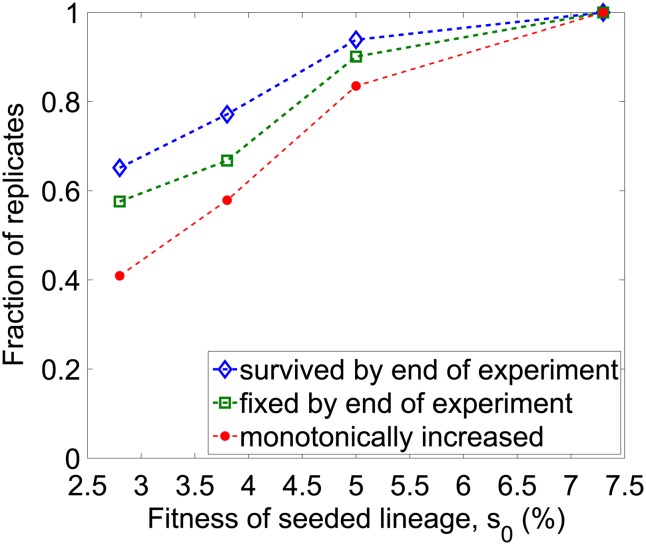
Figure 3.
The fates of seeded lineages as a function of their fitness. We show the fraction of replicate populations in which the seeded lineage had the indicated fate.
Comparing the fates of seeded lineages of different fitnesses provides additional insight into the mutations responsible for clonal interference. For example, the seeded lineage with fitness advantage s0 = 7% always swept to fixation without any detectable deviation from the expectation in the absence of interference. In contrast, the lineage with s0 = 5% swept in 84% of replicates. Together, these two results suggest that clonal interference in the s0 = 5% case was primarily due to beneficial mutations in the resident that created competing lineages with fitness advantages between 5 and 7%. Extending this logic, comparing the fates of seeded lineages with s0 = 5, 4, and 3% provides information about the probabilities that beneficial mutations create competing lineages of fitness between 4 and 5% and between 3 and 4%.
While this intuition is straightforward, quantitative inference of the DFE requires us to connect the rates of individual mutations with the fitnesses of competing lineages. This is complicated because competing lineages may often contain multiple beneficial “driver” mutations. In addition, beneficial mutations may also arise in seeded lineages, despite their initially much smaller population sizes. To fully account for these effects, we now introduce a computational method for inferring the DFE.
DFE inferred from seeded lineage dynamics
We implemented an approximate likelihood method which uses information from the shapes of the trajectories of seeded lineages to infer the DFE of beneficial mutations. Any particular trajectory carries only information about the beneficial mutations that rose to significant frequency in that population (i.e., the “contending” mutations; Rozen et al. 2002), but by modeling the trajectories of many populations together, we can learn about the overall distribution of possible beneficial mutations for the strains in our experiment. To make this inference tractable, we limited ourselves to single-parameter DFE shapes characterized by an average fitness effect and beneficial mutation rate Ub. For concreteness, we considered three canonical distributions commonly used in the literature: an exponential DFE, , a uniform DFE , and a δ-function DFE, where all beneficial mutations have the same fitness effect, . We explain the significance of these choices in the Discussion.
To compute the likelihood of particular DFE parameters, we ran forward-time simulations of the experiment and estimated the likelihood as the fraction of replicate simulations that matched the data (see Appendix). In principle, we could use the complete trajectory of each seeded lineage for this comparison, identifying a match between simulations and data whenever the two were identical. However, in practice this was not computationally tractable. Instead, we focused on two features of the dynamics: the first peak frequency, fpeak, of each seeded lineage (binned into quartiles, including fixed lineages) and the rate at which the seeded lineage declined in frequency following this peak, sdown (binned into 2% intervals). These are illustrated in Figure 2. We chose to focus on these two quantities because we expect them to be particularly sensitive to the DFE: fpeak indicates how quickly a competing lineage arose in the resident population, while sdown measures how much the relative fitness of the resident population increased in this time. In addition, this focus on early time dynamics ensures that most relevant mutations occur in the resident (due to its initially much larger population size), minimizing the effects of potential differences in the DFEs of the seeded genotypes.
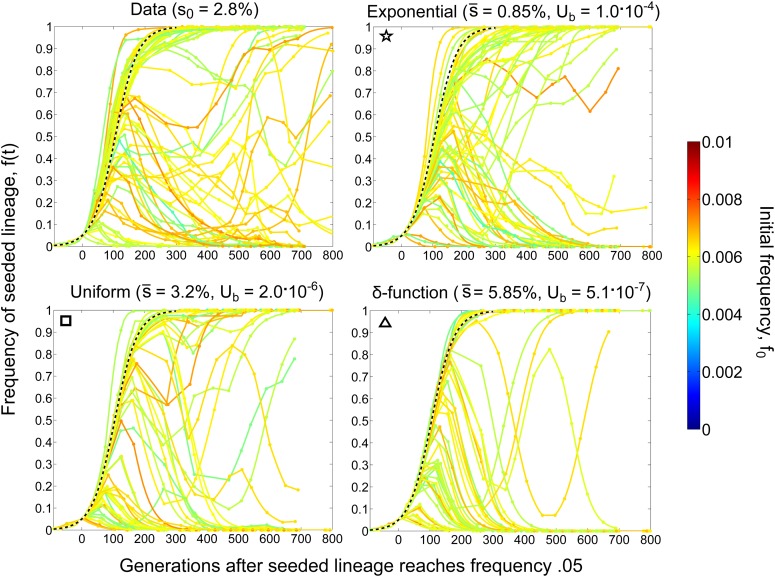
For the three considered DFE shapes, we identified the most-likely parameters Ub and by scanning a grid of candidate values. These parameters are shown in Figure 4, along with confidence bounds estimated by bootstrapping (see Appendix). For each of these most-likely parameters, we show simulations of the s0 = 3% seeded lineage trajectories in Figure 5 and for the s0 = 4, 5, and 7% lineages in Figure S1, Figure S2, and Figure S3. Using a likelihood ratio test, we found that the exponential DFE provided a significantly better fit to the data than either the δ-function (P < 10−4) or uniform distribution (P < 10−4) and that the uniform provided a better fit than the δ (P < 10−4).
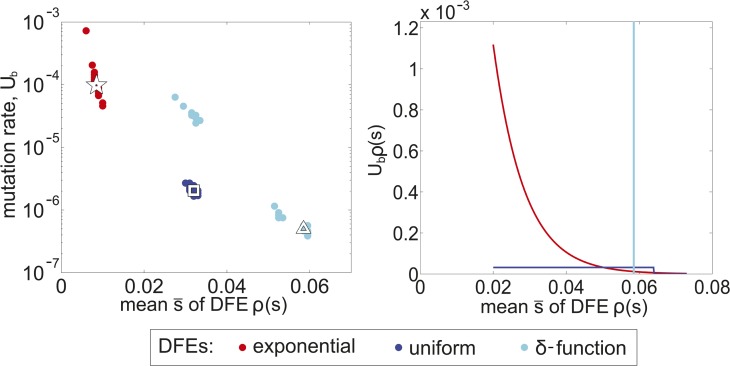
Figure 4.
Inferred DFE parameters. Left: The most-likely parameters Ub and for DFEs ρ(s) given by exponential (star), uniform (square), and δ-function (triangle) distributions. Shaded circles indicate 1% confidence ranges of these parameters as estimated by bootstrapping (see Appendix). Right: The shapes of these distributions shown for s ≥ 2% given their most-likely parameters.
Figure 5.
Lineage dynamics data and simulations for s0 = 2.8%. The trajectories of seeded lineages with initial fitness s0 = 2.8% are shown as observed in the experiment (top left) and as reproduced by simulations assuming the DFE parameters indicated above each part.
Since the seeded lineage with s0 = 7.3% always swept to fixation, indicating that larger-effect mutations must be rare, we checked whether truncating the high fitness end of the exponential DFE would improve its fit to the data. To do so, we considered an exponential DFE truncated at 7.3% and performed the same inference and statistical tests as above. We found that this truncated exponential provided a better fit to the data, but not significantly so (P > 0.08, likelihood-ratio test). We also checked whether truncating the low-fitness end of the exponential would affect its fit to the data. We varied this truncation and found that, for the inferred exponential DFE parameters, discounting mutations with fitness effects <2.1% improved these parameters’ fit to the data, but only marginally so. This indicates that the seeded lineages were not strongly affected by mutations with fitness effects <∼2%.
Measurements of adaptation rate corroborate DFE inference
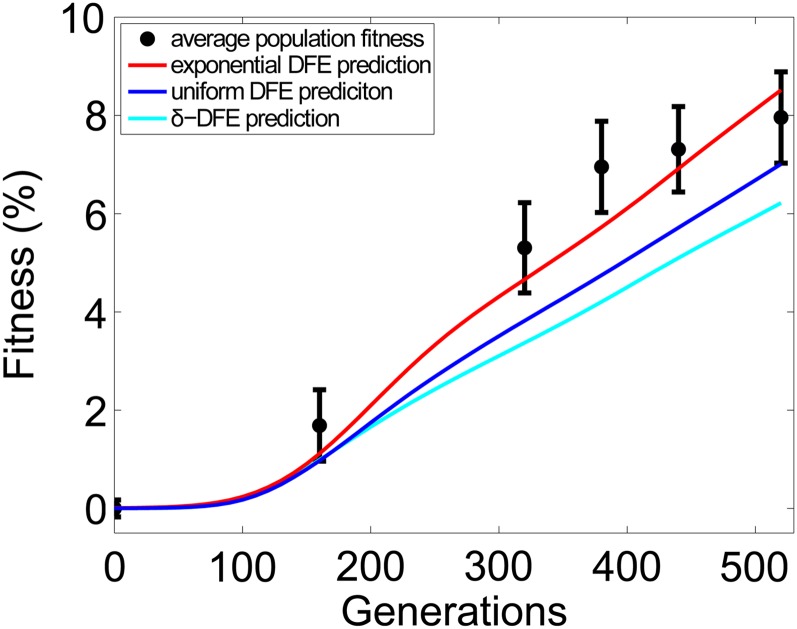
In addition to determining the dynamics of seeded lineages, the DFE determines the rate of adaptation. Thus to test our inferences, we measured the changes in fitness over time of 16 control populations that consisted of the resident strain alone. We compared the average fitness of the control populations with the predictions of the most-likely exponential, uniform, and δ-function DFEs. As seen in Figure 6, the inferred exponential is fairly accurate in predicting these data, whereas the uniform and δ-function are less so.
Figure 6.
The rate of adaptation. The average fitness over time of 16 experimental control populations (± 1 SEM) is shown in black. Solid curves are the predictions of these data given the most-likely exponential, uniform, and δ-function DFEs. The fitnesses of individual populations are shown in Figure S4.
Throughout our analysis, we have implicitly assumed that the DFE remained the same across all genotypes in the experiment, which implies that the fitnesses of populations should increase linearly on average after some initial transient. In contrast, the rate of adaptation slowed after generation 380 (P < 3 × 10−3; see Appendix), which is reminiscent of declines in adaptation rate commonly observed in other evolution experiments (Elena and Lenski 2003). Fortunately, we based our DFE inference on the early features of seeded lineage dynamics, most of which transpired prior to this time. Thus the change in adaptation rate is not inconsistent with our method.
Discussion
Interest in the DFE stems from a desire to know what beneficial mutations are available and which of these drive adaptation. In asexual populations, the DFE also determines the distribution of competing mutations and the frequency of clonal interference. Here, we have described a simple experiment that exploits this connection to infer the DFE in experimental populations of S. cerevisiae. By introducing lineages with different fitnesses and tracking their subsequent dynamics, we inferred the DFE from the statistics of observed interference events. In the process, we directly observed how initial fitness advantages and clonal interference jointly influence the fixation or loss of adaptive lineages.
Previous experimental work has analyzed several other cases where an introduced lineage is outcompeted by a less-fit resident population (Waite and Shou 2012; Gifford and MacLean 2013). Unlike our experiment, these earlier studies focus on the fates of a few key mutations (e.g., antibiotic resistance or microbial “cheaters”) without attempting to infer the underlying DFE. Nevertheless, our results complement this earlier work by showing the transition between fitness effects that are susceptible to clonal interference and those that decisively sweep to fixation, which has previously been studied theoretically (Neher and Shraiman 2011; Schiffels et al. 2011; Good et al. 2012). In our system, this transition occurs when the fitness of the seeded lineage is ∼5%, which represents a critical effect size required for a mutation to drive adaptation. Of course, in natural populations some adaptive variants may arise in populations with substantial standing fitness variation, rather than the homogeneous resident populations employed here. In this case, the transition between mutations that sweep and those that experience interference is determined both by the DFE and by the distribution of fitnesses in the resident population. Further work is needed to address this situation.
Our computational inference method allowed us to distinguish between three representative DFE shapes: exponential, uniform, and δ-function (in which all mutations have the same effect). These represent idealized approximations to the actual DFE, and it is likely that a larger number of replicates or more sophisticated computational techniques could produce other DFE shapes with a significantly better fit. Yet one cannot continue this process indefinitely without reaching a point where further determination of the fine-scale DFE becomes irrelevant for any particular application. In the end, certain features of the DFE matter for predicting certain aspects of the evolutionary process, and the required level of resolution is ultimately determined by the aspect of adaptation one wishes to study. This experiment, which focuses on the fates of advantageous mutants, provides a concrete illustration of this principle. Previous work has suggested that the dynamics of adaptation can be summarized by a single characteristic fitness effect, with a magnitude that depends on the actual DFE and the level of clonal interference within the population (Hegreness et al. 2006; Desai and Fisher 2007; Good et al. 2012). By rejecting the δ-function and uniform DFEs in favor of the exponential, we have shown that this assumption breaks down when one considers more detailed features of the lineage trajectories.
Given these caveats, the DFE that we inferred is worth pondering. We estimated an exponential distribution with mean and total beneficial mutation rate Ub = 1.0 × 10−4. Our modeling indicated that of these mutations, only those with effects >2% affected the fates of seeded lineages and that these mutations are predicted to arise at a rate of order 10−5 per individual per generation. If one assumes a per-genome point mutation rate of roughly 4 × 10−3 (Lynch et al. 2008), this would imply that of order 1 in 1000 mutations confer a fitness advantage of two percent or more. This is consistent with past work in a related system (Desai et al. 2007) and is also similar to DFEs reported for bacteria adapting to rich laboratory media (Kassen and Bataillon 2006; Perfeito et al. 2007; Wiser et al. 2013). In such permissive environments, other studies in yeast that have identified specific adaptive mutations report a mix of loss-of-function vs. other kinds of beneficial mutations (Jansen et al. 2005; Kao and Sherlock 2008; Wenger et al. 2011; Kvitek and Sherlock 2013; Lang et al. 2013). If a large fraction of beneficial mutations in our system are loss of function, and if ∼10% of spontaneous mutations in a gene cause loss of function (Lang and Murray 2008), our results would suggest that ∼1 in 100 genes are beneficial to disrupt. This is at least qualitatively consistent with direct measurements using the yeast deletion collection (Sliwa and Korona 2005; Bell 2010). Together, these results illustrate how inferences from lineage dynamics can combine with other lines of evidence to help build a more complete picture of adaptation.
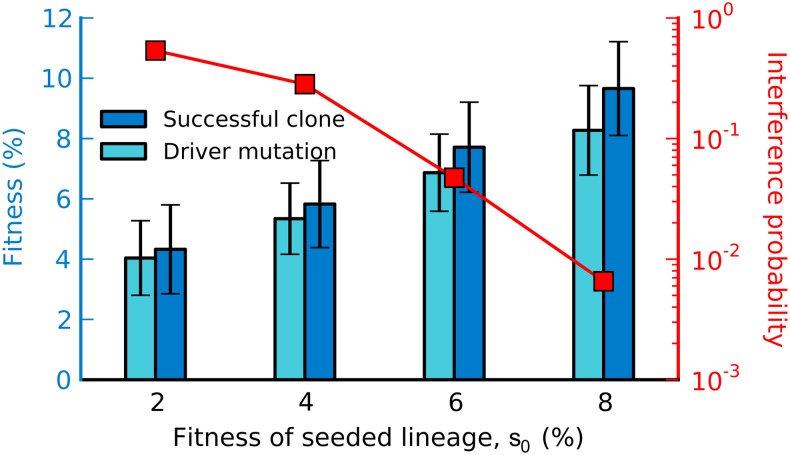
Finally, we note that our experimental design has a potential practical application as a screen for beneficial mutations. Whenever a seeded lineage with fitness advantage s0 experiences clonal interference, the resident must contain a mutant lineage at appreciable frequency with fitness greater than s0. Thus, by picking clones from the resident immediately after a clonal interference event, we should in principle be able to isolate rare large-effect beneficial mutations. This is similar in spirit to earlier studies that used the dynamics of neutral markers to screen for adaptive clones (e.g., Rozen et al. 2002). However, because our seeded lineages are more fit than the resident, we can screen for beneficial mutations with particularly large effects. Further, since the resident must quickly generate a competing lineage, our approach is more likely to find clones with fewer mutations of larger effect rather than many of smaller effect, as well as limit the number of nonbeneficial hitchhiking mutations. To illustrate this idea, we simulated seeded lineage trajectories and then simulated picking a clone from the resident population after observed clonal interference events. In Figure 7, we show the average fitness of each of these simulated clones and of the largest-effect mutation in each clone. As is apparent from the figure, it should be feasible to use this approach with a seeded lineage of the appropriate fitness to isolate large-effect beneficial mutations with specific fitness effects.
Figure 7.
A screen for beneficial mutations. We simulated the evolutionary dynamics for a range of seeded lineages and then simulated picking a single clone at random from the resident population immediately after a clonal interference event. The bars indicate the average fitness of this clone and its largest effect mutation (±1 SD, scale at left). We also show the fraction of replicate populations in our simulations in which clonal interference occurs (scale at right). The simulations assumed the most-likely exponential DFE inferred in the study.
Supplementary Material
Acknowledgments
We thank Sergey Kryazhimskiy, Christopher S. Wylie, Andrew Murray, and Katya Kosheleva for useful discussions and comments on the manuscript; Melanie Muller, Gabriel Perron, John Koschwanez, and Gregg Wildenberg for help with strain construction; and Patricia Rogers for generous technical support of flow-cytometry. Simulations in this article were performed on the Odyssey cluster of the Research Computing Group at Harvard University. This work was supported by training grant GM831324 from the National Institutes of Health (NIH) and grant 1219334 from the NSF Physics of Living Systems graduate student network (E.M.F.), a National Science Foundation Graduate Research Fellowship (B.H.G.), and the James S. McDonnell Foundation, the Alfred P. Sloan Foundation, the Harvard Milton Fund, grant PHY 1313638 from the National Science Foundation, and grant GM104239 from the NIH (M.M.D.).
Footnotes
Communicating editor: J. J. Bull
Literature Cited
- Adams J., Oeller P., 1986. Structure of evolving populations of Saccharomyces cerevisiae: adaptive changes are frequently associated with sequence alterations involving mobile elements belonging to the Ty family. Proc. Natl. Acad. Sci. USA 83: 7124–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood K., Schneider L., Ryan F., 1951. Periodic selection in Escherichia coli. Proc. Natl. Acad. Sci. USA 37: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett R. D., MacLean R. C., Bell G., 2006. Mutations of intermediate effect are responsible for adaptation in evolving Pseudomonas fluorescens populations. Biol. Lett. 2: 236–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J., Kauth M., Strelioff C., Lenski R., 2010. Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol. Biol. Evol. 27: 1338–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataillon T., Zhang T., Kassen R., 2011. Cost of adaptation and fitness effects of beneficial mutations in Pseudomonas fluorescens. Genetics 189: 939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G., 2010. Experimental genomics of fitness in yeast. Proc. Biol. Sci. 277: 1459–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L. D., Cai T. T., DasGupta A., 2001. Interval estimation for a binomial proportion. Stat. Sci. 16: 101–117. [Google Scholar]
- Burch C. L., Guyader S., Samarov D., Shen H., 2007. Experimental estimate of the abundance and effects of nearly neutral mutations in the RNA virus φ6. Genetics 176: 467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa J. A. M., Campos P. R., Gordo I., 2013. An ABC method for estimating the rate and distribution of effects of beneficial mutations. Genome Biol. Evol. 5: 794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser J., Rozen D., 2006. Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli. Genetics 172: 2093–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser J. A., Zeyl C. W., Gerrish P. J., Blanchard J. L., Lenski R. E., 1999. Diminishing returns from mutation supply rate in asexual populations. Science 283: 404–406. [DOI] [PubMed] [Google Scholar]
- Desai M., Fisher D., 2007. Beneficial mutation–selection balance and the effect of linkage on positive selection. Genetics 176: 1759–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M., Fisher D., Murray A., 2007. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17: 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena S. F., Lenski R. E., 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4: 457–469. [DOI] [PubMed] [Google Scholar]
- Gerrish P., Lenski R., 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102: 127–144. [PubMed] [Google Scholar]
- Gifford D. R., MacLean R. C., 2013. Evolutionary reversals of antibiotic resistance in experimental populations of Pseudomonas aeruginosa. Evolution 67: 2973–2981. [DOI] [PubMed] [Google Scholar]
- Gillespie J., 1983. A simple stochastic gene substitution model. Theor. Popul. Biol. 23: 202–215. [DOI] [PubMed] [Google Scholar]
- Good B., Rouzine I., Balick D., Hallatschek O., Desai M., 2012. Distribution of fixed beneficial mutations and the rate of adaptation in asexual populations. Proc. Natl. Acad. Sci. USA 109: 4950–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegreness M., Shoresh N., Hartl D., Kishony R., 2006. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311: 1615–1617. [DOI] [PubMed] [Google Scholar]
- Helling R., Kinney T., Adams J., 1981. The maintenance of plasmid-containing organisms in populations of Escherichia coli. J. Gen. Microbiol. 123: 129–141. [DOI] [PubMed] [Google Scholar]
- Illingworth C., Mustonen V., 2012. A method to infer positive selection from marker dynamics in an asexual population. Bioinformatics 28: 831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof M., Schlötterer C., 2001. Fitness effects of advantageous mutations in evolving Escherichia coli populations. Proc. Natl. Acad. Sci. USA 98: 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M. L., Diderich J. A., Mashego M., Hassane A., de Winde J. H., et al. , 2005. Prolonged selection in aerobic, glucose-limited chemostat cultures of Saccharomyces cerevisiae causes a partial loss of glycolytic capacity. Microbiology 151: 1657–1669. [DOI] [PubMed] [Google Scholar]
- Joseph S. B., Hall D. W., 2004. Spontaneous mutations in diploid Saccharomyces cerevisiae more beneficial than expected. Genetics 168: 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao K., Sherlock G., 2008. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet. 40: 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen R., Bataillon T., 2006. Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nat. Genet. 38: 484–488. [DOI] [PubMed] [Google Scholar]
- Keightley P. D., Eyre-Walker A., 2010. What can we learn about the distribution of fitness effects of new mutations from DNA sequence data? Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek D. J., Sherlock G., 2013. Whole genome, whole population sequencing reveals that loss of signaling networks is the major adaptive strategy in a constant environment. PLoS Genet. 9: e1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Murray A. W., 2008. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178: 67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G., Botstein D., Desai M., 2011. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics 188: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Rice D. P., Hickman M. J., Sodergren E., Weinstock G. M., et al. , 2013. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500: 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-C., Marx C. J., 2013. Synchronous waves of failed soft sweeps in the laboratory: remarkably rampant clonal interference of alleles at a single locus. Genetics 193: 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R., et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean R., Buckling A., 2009. The distribution of fitness effects of beneficial mutations in Pseudomonas aeruginosa. PLoS Genet. 5: e1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M., Cooper T., Beaumont H., Rainey P., 2011. The distribution of fitness effects of new beneficial mutations in Pseudomonas fluorescens. Biol. Lett. 7: 98–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles R., Gerrish P. J., Moya A., Elena S. F., 1999. Clonal interference and the evolution of RNA viruses. Science 285: 1745–1747. [DOI] [PubMed] [Google Scholar]
- Neher R. A., Shraiman B. I., 2011. Genetic draft and quasi-neutrality in large facultatively sexual populations. Genetics 188: 975–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick A., Szilard L., 1950. Experiments with the chemostat on spontaneous mutations of bacteria. Proc. Natl. Acad. Sci. USA 36: 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H., 2003. The distribution of fitness effects among beneficial mutations. Genetics 163: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin C., Adams J., 1983. Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature 302: 495–500. [DOI] [PubMed] [Google Scholar]
- Perfeito L., Fernandes L., Mota C., Gordo I., 2007. Adaptive mutations in bacteria: high rate and small effects. Science 317: 813–815. [DOI] [PubMed] [Google Scholar]
- Pinkel D., 2007. Analytical description of mutational effects in competing asexual populations. Genetics 177: 2135–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D., Beisel C., Joyce P., Ferris M., Burch C., et al. , 2008. Beneficial fitness effects are not exponential for two viruses. J. Mol. Evol. 67: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D. E., de Visser J., Gerrish P. J., 2002. Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 12: 1040–1045. [DOI] [PubMed] [Google Scholar]
- Sanjuán R., Moya A., Elena S., 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an rna virus. Proc. Natl. Acad. Sci. USA 101: 8396–8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S., Szöllősi G. J., Mustonen V., Lässig M., 2011. Emergent neutrality in adaptive asexual evolution. Genetics 189: 1361–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwa P., Korona R., 2005. Loss of dispensable genes is not adaptive in yeast. Proc. Natl. Acad. Sci. USA 102: 17670–17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski P., Gerrish P., 2010. Beneficial mutations and the dynamics of adaptation in asexual populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl L. M., Gerrish P. J., Saika-Voivod I., 2002. Evaluating the impact of population bottlenecks in experimental evolution. Genetics 162: 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite A. J., Shou W., 2012. Adaptation to a new environment allows cooperators to purge cheaters stochastically. Proc. Natl. Acad. Sci. USA 109: 19079–19086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger J. W., Piotrowski J., Nagarajan S., Chiotti K., Sherlock G., et al. , 2011. Hunger artists: yeast adapted to carbon limitation show trade-offs under carbon sufficiency. PLoS Genet. 7: e1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiser M. J., Ribeck N., Lenski R. E., 2013. Long-term dynamics of adaptation in asexual populations. Science 342: 1364–1367. [DOI] [PubMed] [Google Scholar]
- Zhang W., Sehgal V., Dinh D. M., Azevedo R. B., Cooper T. F., et al. , 2012. Estimation of the rate and effect of new beneficial mutations in asexual populations. Theor. Popul. Biol. 81: 168–178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.