Abstract
The genetic regulators of regressive craniofacial morphologies are poorly understood. To shed light on this problem, we examined the freshwater fish Astyanax mexicanus, a species with surface-dwelling and multiple independent eyeless cave-dwelling forms. Changes affecting the skull in cavefish include morphological alterations to the intramembranous circumorbital bones encircling the eye. Many of these modifications, however, have evolved separately from eye loss, such as fragmentation of the third suborbital bone. To understand the genetic architecture of these eye-independent craniofacial alterations, we developed and scored 33 phenotypes in the context of an F2 hybrid mapping pedigree bred from Pachón cavefish and surface fish. We discovered several individuals exhibiting dramatic left–right differences in bone formation, such as extensive fragmentation on the right side only. This observation, along with well-known eye size asymmetry in natural cave-dwelling animals, led us to further evaluate left–right genetic differences for the craniofacial complex. We discovered three phenotypes, inclusive of bone fragmentation and fusion, which demonstrated a directional heritable basis only on one side. Interestingly, the overall areas of affected bones were genetically symmetric. Phenotypic effect plots of these novel craniofacial QTL revealed that cave alleles are associated with abnormal conditions such as bony fusion and fragmentation. Moreover, many linked loci overlapped with other cave-associated traits, suggesting regressive craniofacial changes may evolve through linkage or as antagonistic pleiotropic consequences of cave-associated adaptations. These novel findings illuminate significant craniofacial changes accompanying evolution in complete darkness and reveal complex changes to the skull differentially influenced by genetic changes affecting the left and right sides.
Keywords: quantitative trait locus analysis, regressive phenotypic evolution, circumorbital bone series, Astyanax, troglomorphy
NATURAL model systems, such as Darwin’s finches (Abzhanov et al. 2004, 2006; Mallarino et al. 2011) and African cichlid fishes (Albertson et al. 2003; Albertson and Kocher 2006; Streelman and Albertson 2006), provide living proof that the skull is remarkably labile over evolutionary time. Dramatic morphological changes accompany, or perhaps enable, expansion and exploitation of niches through rapid evolution of adaptive feeding modes (Anyonge and Baker 2006; Cooper et al. 2010). While a great deal is known regarding the developmental origin of cranial bones mediating these constructive changes (Smith 2006; Streelman et al. 2007; Jheon and Schneider 2009), little is known of the genetic basis for craniofacial changes that evolve in the absence of obvious selective pressures.
To explore this phenomenon, we examined alterations to the craniofacial skeleton of the freshwater fish Astyanax mexicanus. This species consists of an extant surface-dwelling form and multiple independently derived cave-dwelling forms (Strecker et al. 2003). As a consequence of invading the subterranean environment millions of years ago (Bradic et al. 2012), cave-dwelling morphs have evolved a series of regressive (e.g., eye loss) and constructive (e.g., increased lateral line sensitivity) phenotypes (Wilkens 1971; Montgomery et al. 2001; Jeffery 2009; Yoshizawa et al. 2010, 2012). Phenotypic loss is believed to arise through genetic drift (Wilkens 1988), direct selection (Klaus et al. 2013), or indirect selection via linkage or pleiotropy (Yamamoto et al. 2009); however, the evolutionary mechanism that drives regressive loss remains unclear (Gross 2012). Our natural model system enables us to investigate the extent to which evolutionary modifications of the craniofacial complex evolve as an indirect consequence of pleiotropy or close physical linkage between causative gene(s) mediating craniofacial traits and other regressive traits, such as eye loss.
Among the most significant changes affecting the skull in cavefish are alterations to circumorbital bone morphology (Alvaréz 1946, 1947). These modifications demonstrate a spectrum of severity, comprising both bone fragmentation and fusion, much of which is endemic to each independent cavefish population (Mitchell et al. 1977). In surface-dwelling forms, each member of the circumorbital series is present as a single, intact bone (Mitchell et al. 1977). In contrast, fragmentation of the suborbital bones [particularly the first suborbital (SO1) and third suborbital (SO3) bones] has been reported for eight wild cavefish populations, including the Pachón cave (Mitchell et al. 1977). Classic studies, which were essentially descriptive accounts, assumed craniofacial changes in cavefish evolved as a secondary consequence of the loss of the eye.
Yamamoto et al. (2003) tested this hypothesis, using lentectomy and intermorphotype grafting to determine the extent to which craniofacial phenotypes were influenced by experimental removal of the eye. Certain traits were affected, including the distance between the nasal and antorbital bones, the inner sectors of the SO3 and supraorbital bones, and position of the SO3 bone relative to the orbit of the eye (Yamamoto et al. 2003). However, other craniofacial traits were not affected by eye loss, such as number of SO3 bony elements, positioning of SO4–6 bones relative to the opercular bone, and opercular bone shape (Yamamoto et al. 2003). Protas et al. (2008) first investigated the genetic basis for craniofacial defects in Astyanax by evaluating variation in SO3 width on the right side of the face only.
In this study, we searched for additional craniofacial traits demonstrating a genetic basis. We scored 33 phenotypes in a hybrid mapping pedigree derived from blind Pachón cave-dwelling and eyed surface-dwelling forms of A. mexicanus (Supporting Information, Table S1). Prior studies using the same pedigree revealed the genetic basis for a number of cave-associated phenotypes, including eye size reduction, pigmentation loss, and chemical sensitivity (Protas et al. 2008). Evaluation of additional phenotypes in the context of this previous work enabled us to determine whether pleiotropic or tightly linked loci are shared between craniofacial and other cave-associated traits. Certain individuals within our pedigree exhibited dramatic left–right differences in bone formation, such as extensive fragmentation on one side only (Figure 1, A–F). This observation, along with the well-documented asymmetry in eye size in natural cave-dwelling populations (Wilkens 2001, 2010; Pouilly and Miranda 2003), motivated us to extend our studies to evaluate left–right genetic symmetry for craniofacial traits. Indeed, we discovered numerous phenotypes with a genetic basis that was detectable on only one side of the cranium. Significant craniofacial changes have accompanied adaptation to the subterranean habitat, and the work herein reveals that complex evolutionary changes to the skull can be influenced by genetic changes affecting the left and right sides differently.
Figure 1.
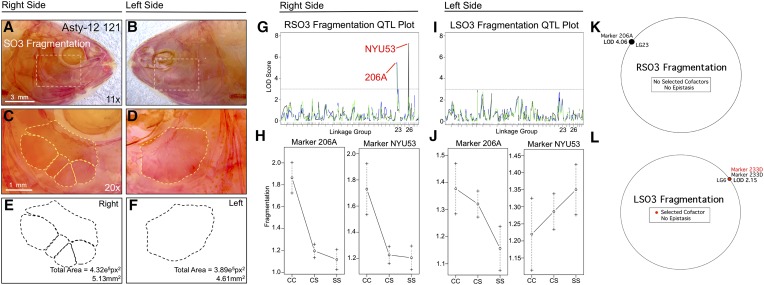
Fragmentation of the third suborbital bone (SO3) demonstrates a right-sided asymmetric genetic basis. Fragmentation of the SO3 bone (dashed box in A and B) was scored separately on the right (A, C, and E) and left sides (B, D, and F), as demonstrated using specimen 121 from our Asty12 pedigree. Bones were present as either unfragmented (the wild-type surface phenotype) (D) or fragmented elements (C). The total area of this bone (E and F) demonstrated a genetic basis in our pedigree (see Figure 5). Two markers, 206A and NYU53, demonstrated a LOD value >3.0 on the right (G), but not the left (I), side of the head. Effect plots reveal a surface-dominant effect for both markers (H), on the right side only (compare with J). MQM analyses reveal one significant QTL associated with fragmentation on the right side with no selected cofactors (K). On the left side, one insignificant QTL (LOD = 2.15) and cofactor were identified (L). Bars: 3 mm in A and B (11×) and 1 mm in C and D (20×). In G and I, black line shows marker regression, green line shows HK, and blue line shows EM mapping methods, respectively.
Materials and Methods
Mapping pedigree
We reanalyzed an F2 pedigree of Pachón cavefish × surface fish hybrid individuals (n = 539) with previously collected genotypic data for 164 microsatellite markers (Protas et al. 2007) (Table S2). These individuals represent a full-sibling F2 pedigree from a sibling cross of F1 individuals derived from a mating of a surface-dwelling male and a Pachón cave-dwelling female. We based our linkage map calculations on supplementary information published in Protas et al. (2007), to which we added genotypic information for 11 markers (see Table S2). Each specimen of this pedigree was cleared and stained using Alizarin red to visualize ossified bone (Protas et al. 2008) and stored in sterile glycerol at 4°.
Craniofacial imaging, phenotypic analyses, and scoring
The left and right sides of each individual were imaged using high-resolution microscopy under identical lighting conditions. For each specimen, a high-resolution “montage” image was consolidated from photographs taken at numerous focal positions, which were aligned and flattened into a single, uniformly focused image, as described in Gross and Wilkens (2013). All micrographs were collected using a Leica (Wetzlar, Germany) M205FA stereomicroscope equipped with a DFC310FX camera. Images were collected utilizing the MultiFocus module within the Leica Application Suite (LAS) v3.8 software package. All subsequent measurements were collected from these montage tiff-formatted images.
We collected lateral images at several magnifications (7.81×, 11.0×, 12.5×, 18.0×, 20.0×, and 30.0×) to evaluate 33 craniofacial phenotypes for both the left and right sides of the cranium, including total lateral head area, supraorbital bone area, suborbital bone area (SO2–6), suborbital bone fusion (SO1+2, SO4+5, and SO5+6), suborbital bone fragment number (for bones SO3, SO4, and SO5), maxillary area, the number of maxillary teeth, opercular area, area of the nares, and antorbital bone area (see Table S1).
Bony area measurements were compiled using the freehand tool in ImageJ (National Institutes of Health, Bethesda, MD). Raw values were obtained as the total number of pixels and converted to square millimeters based on the resolution of each raw image. For bones demonstrating fragmentation (SO3–5), we recorded total number of fragments, as well as individual area measurements for each fragment. In cases of fragmentation, we summed the individual fragment areas to recover the total area of the bone. Finally, we scored certain traits as binary, e.g., the presence (1) or absence (0) of fusion for bone pairs vulnerable to fusion, SO1+2, SO4+5, and SO5+6 (Table S1). We also performed additional analyses of bone area, accounting for size of the fish, in which bony area measurements were regressed against standard length of each specimen. QTL analyses were then performed based on the residual values for each bony area measurement (Table 3).
Table 3. QTL analysis of craniofacial bone areas using residual values against standard length.
| Trait | Regression against standard length (R2 value) | Linkage group(s) | MR | EM | HK | Significance threshold LOD (P < 0.05) | Genotype | N value | Scanone (IM) result: associated marker(s) (LOD value) | MQM result: associated marker (LOD value) | Cofactor(s) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| R SO2 area residuals | 0.626862 | 6, 15 | 106C (4.0), 227A (5.56) | — | 227A (5.38) | LOD 3.62 | CC, larger area; SS, smaller area | 221 | 227A (LOD 3.07) | 227A (LOD 3.16) | 110B, 227A, NYU25, 237E, 223C, 122B |
| L SO2 area residuals | 0.520282 | 15 | 227A (5.69) | — | 227A (5.35) | LOD 3.57 | CC, larger area; SS, smaller area | 218 | 110B (LOD 2.93) | 110B (LOD 1.03) | 110B, 122B, 223C, 214C, 227A |
| R SO3 area residuals | 0.745564 | 5, 14 | 26A (4.90), 136B (4.46) | — | — | LOD 3.71 | 26A, CC, smaller area; SS, larger area; 136B, CC, larger area, SS, smaller area | 222 | 26A (LOD 3.71), 136B (LOD 2.50) | 26A (LOD 3.84), 136B (LOD 2.50) | 26A, 239C, 122B, 223C, NYU33 |
| L SO3 area residuals | 0.722414 | 5, 8, 13, 14 | 26A (4.04), 203F (3.84), 6A (4.23), 136B (4.12) | 203F (3.73) | — | LOD 3.62 | 26A, CC, smaller area; SS, larger area; 203F, 6A, and 136B, CC, larger area; SS, smaller area | 218 | 239C (LOD 3.42), 26A (LOD 2.75) | 239C (LOD 3.07), 26A (LOD 2.88) | 26A, 203F, 217B, 131C, NYU53, 122B, 239A |
| L SO5 area residuals | 0.373122 | 26 | 2B (3.61) | — | — | LOD 3.48 | SS, larger area; CC, smaller area | 200 | 218B (LOD 2.41) | 218B (LOD 2.7) | 237E, NYU53, 122B |
Linkage map construction
We recalculated linkage groups with the R/qtl package, using genotypic information for 175 markers (Protas et al. 2007) (Table S2). We identified 29 linkage groups for a total map length of 2545 cM and an average intermarker distance of 17.4 cM. The karyotypic number of A. mexicanus is 25 (Kavalco and Almeida-Toledo 2007). We attribute the higher number of linkage groups we identified to the reduced number of meiotic events analyzed, as well as potential genotyping errors. Recombination frequencies were collected for 539 individuals; however, craniofacial phenotypes were investigated in 237 cleared-and-stained individuals.
QTL and effect plot analyses
All QTL analyses were carried out using the software program R/qtl (Broman et al. 2003). Each trait was analyzed using marker regression (MR) (Kearsey and Hyne 1994), expectation maximization (EM) (Xu 2010), and Haley–Knott (HK) (Haley and Knott 1992) mapping methods. Most traits were evaluated as parametric or binary phenotypes (above). The number of meioses and genetic markers evaluated in our study was limited and may have reduced our power to detect QTL (Beavis 1998). Therefore, all phenotypes were analyzed using each of the three methods above. To identify prospective QTL from this analysis, we first used a low-stringency significance threshold (LOD = 3) to detect QTL that may otherwise be missed due to the number of genomic markers used and/or the number of hybrid individuals evaluated in our study.
We balanced this approach by discarding certain pseudomarkers, calculated using interval mapping, which harbored inflated LOD values and incongruous genomic positions compared to other mapping methods. These putative “ghost QTL” (Broman and Speed 1999) are artifacts that can arise using interval mapping procedures (Haley and Knott 1992; Martínez and Curnow 1992) and were discarded from further analysis. Effect plots were created for the closest linked markers, using phenotypic information for the left and right sides of each member of the pedigree. Genome-wide LOD significance thresholds (P < 0.05) were calculated in R/qtl for each phenotypic trait (Table 1), using a permutation test (of 1000 permutations) to identify statistically significant associations.
Table 1. Summary of craniofacial and cave-associated QTL evaluated in this study.
| Trait | Linkage group(s) | MR LOD score | EM LOD score | HK LOD score | Significance threshold LOD (P < 0.05) | Genotype | Overlapping traits identified in Protas et al. (2007) |
|---|---|---|---|---|---|---|---|
| Albinism | 5 | OCA2 (33.3) | OCA2 (33.7) | OCA2 (33.2) | 3.9 | CC, albino; SS, wild type | No trait |
| Right eye | 7, 20 | 234B (6.62); NYU14 (6.81) | NYU14 (7.07) | NYU14 (7.12) | 3.9 | CC, reduced eye; SS, wild type | Eye; eye and lens L |
| Left eye | 7, 20 | 234B (6.13); NYU14 (10.7) | 234B (7.44); NYU14 (10.34) | 234B (7.37); NYU14 (10.13) | 3.7 | CC, reduced eye; SS, wild type | Eye; eye and lens L |
| R SO1+2 fusion | 1, 6, 27 | 119C (3.28); 229B (3.48) | 229B (3.47) | 215D (3.33); 229B (4.41) | 3.8 | CC, fusion; SS, wild type | Mel D; eye and MaxTth |
| R SO2 area | 6, 7, 27 | 119C (4.43); 209A (3.42); 229B (3.72) | 110B (3.46); 229B (3.84) | 229B (3.94) | 3.7 | CC, larger area; SS, wild type | Mel D; no trait; eye and MaxTth |
| L SO2 area | 6, 7, 27 | 209A (3.74); 229B (4.15) | NYU27 (3.88); 229B (4.28) | — | 3.7 | CC, larger area; SS, wild type | Lens E; no trait; eye and MaxTth |
| R SO3 area | 18, 27 | 55B (2.56); 229B (3.41) | 55B (3.24); 229B (3.66) | 55B (3.24); 229B (3.66) | 3.7 | CC, larger area; SS, wild type | Tbuds; eye and MaxTth |
| L SO3 area | 18, 27 | 55B (3.11); 229B (3.15) | 55B (3.93); 229B (3.28) | 55B (4.02); 229B (3.02) | 3.8 | CC, larger area; SS, wild type | Tbuds; eye and MaxTth |
| R SO3 no. | 23, 26 | 206A (5.43); NYU53 (7.31) | — | 206A (5.34) | 3.8 | CC, fragmentation; SS, wild type | Mel A; no trait |
| R SO4 area | 26 | 214F (3.08) | — | — | 3.7 | CC, larger area; SS, wild type | No trait; eye and MaxTth |
| L SO4 area | 15, 26, 27 | 222E (4.03); 214F (3.96); 229B (3.75) | 229B (3.80) | — | 3.9 | CC, larger area; SS, wild type | No trait; eye and MaxTth; no trait |
| L SO4+5 fusion | 11 | 112A (3.03) | — | — | 3.8 | CC, fusion; SS, wild type | Maxil |
| R SO5 area | 26 | 216C (3.28) | — | NYU53 (3.11) | 3.6 | CC, larger area; SS, wild type | Dent |
| L SO5 area | 26 | 2B (3.08) | — | — | 3.8 | CC, larger area; SS, wild type | Dent |
| Standard length | 15 | — | 222E (4.53) | — | 3.6 | CS, longest | NA |
| Sex | 25 | 214D (18.3) | — | — | 3.7 | NA | Anal Fin Ray |
L, left; R, right.
Following one-scan analyses, we performed multiple-QTL mapping (MQM), using R/qtl software (as described in Arends et al. 2010). MQM mapping is a powerful analytical tool for further understanding the genetic architecture of complex traits, such as craniofacial phenotypes (see Parnell et al. 2012). The automated MQM mapping procedure we utilized involves first “augmenting” missing genotypic data and then automatically selecting cofactors, using multiple-regression and iterative backward elimination procedures (Arends et al. 2010). Then, QTL were interval mapped using maximum likelihood (Table 2) and represented in circular genome interaction plots for each craniofacial phenotype (see examples in Figure 1, K and L) to identify prospective epistatic interactions between cofactors.
Table 2. Summary of results of multiple-QTL mapping (MQM) analyses of craniofacial phenotypes.
| Trait | Scanone (IM) result: associated marker(s) (LOD value) | MQM result: associated marker(s) (LOD value) | Cofactor(s) | Epistatic interaction |
|---|---|---|---|---|
| R SO1+2 fusion | NYU27 (LOD 3.55) | NYU27 (LOD 3.69), 229B (LOD 4.61) | 223C, 233D | 223C and 233D (+) |
| R SO2 area | NYU27 (LOD 4.16), 229B (LOD 3.7) | NYU27 (LOD 4.35), 229B (LOD 4.2), 209A (LOD 3.42) | 110B, 111A, 227A, 222E, 131C, NYU25, 223C, 116B | 110B and NYU25 (+) |
| L SO2 area | 229B (LOD 4.04), NYU27 (LOD 3.38) | 229B (LOD 4.31), 209A (LOD 3.30) | 110B, 111A, 23C, 232C, 214F, NYU25 | 232C and 110B (−) |
| R SO3 area | 55B (LOD 3.18), 229B (LOD 2.72) | 55B (LOD 3.38), 229B (LOD 2.9) | 119C, 111A, 131C, 213B, 23C, 229B, 223C, 26A | — |
| L SO3 area | 55B (LOD 3.84), 229B (LOD 2.77) | 55B (LOD 4.16), 229B (LOD 3.02) | 218D, 206D, 55B, 229B, 223C, 131C | — |
| R SO3 no. | 206A (LOD 3.83) | 206A (LOD 4.06) | — | — |
| R SO4 area | 223C (LOD 3.07) | 229B (LOD 3.12) | 218D, 223C | — |
| L SO4 area | 229B (LOD 3.57) | 229B (LOD 3.81) | 131B, NYU53, 202E, 223C | — |
| L SO4+5 fusion | 112A (LOD 2.6) | 112A (LOD 2.79) | 232D, 113B, 224E, NYU31 | — |
| R SO5 area | 214F (LOD 2.2) | 214F (LOD 2.29) | 214F, NYU53, 233D | — |
| L SO5 area | 229B (LOD 2.52) | 229B (LOD 2.88) | 8B, 111A, 133B, 16C, 131C, 218B, 229B | — |
| Standard length | 111A (LOD 2.15) | 111A (LOD 2.69) | 55B, 122B | — |
| Sex | 214D (LOD 14.83) | 214D (LOD 14.83) | 214C, 6C, 122B | — |
We also examined the potential effects of sex on our craniofacial phenotypes following procedures established by Broman and Sen (2009). We first performed an analysis of variance for 19 craniofacial trait values, using R/qtl. Accordingly, group means of males and females for each trait were compared (Table S3), followed by a QTL analysis with and without sex as a covariate. This technique utilizes standard interval mapping (Broman and Sen 2009), and therefore traits for which a QTL was detected using only marker regression could not be evaluated. The interval mapping method used in each analysis (EM or HK; Table S3) was selected based on the significant QTL reported for each trait (Table 1 and Table 3). All additional statistical analyses were carried out using Microsoft Excel (v.12.3.4 for Mac) or JMP (v.10).
Syntenic analyses and anchoring to the Danio rerio genome
We identified or estimated positions of linked microsatellite markers for the seven phenotypic traits that yielded significant QTL in this study. Marker position was identified in Danio rerio based on a previously generated “integrated” Astyanax linkage map (Gross et al. 2008). Eleven markers from the present study were not included in the prior analysis, so their homologous positions were estimated, as previously described (Gross et al. 2008). For each marker, we identified or estimated marker positions in the latest draft of the D. rerio genome (Zv9; release 72). Relative marker positions and synteny between the A. mexicanus linkage map and the D. rerio genome were developed as a circular representation, using the Circos software program (Krzywinski et al. 2009).
Results
Seven novel craniofacial phenotypes demonstrate a genetic basis in Astyanax
We scored 33 craniofacial phenotypes on both the left and right sides of the head to determine the extent to which complex craniofacial alterations in a subterranean-dwelling fish are controlled by heritable factors. Seven of these traits yielded a genetic basis, four of which were present on both the left and right sides of the head, including (1) fusion of the first and second suborbital bones (SO1+2), (2) total bony area of the second suborbital bone (SO2), (3) total bony area of the third suborbital bone (SO3), (4) number of SO3 bony fragments, (5) total bony area of the fourth suborbital bone (SO4), (6) fusion of the fourth and fifth suborbital bones (SO4+5), and (7) total bony area of the fifth suborbital bone (SO5). We discovered a previously unappreciated asymmetry in the genetic architecture of three traits: SO1+2 fusion, SO3 fragmentation, and SO4+5 fusion, when comparing these traits on the right and left sides of the cranium (below).
Sex-specific body shape differences are apparent in A. mexicanus (Hinaux et al. 2011); however, dimorphic differences in cranial morphology have not been described. To evaluate the potential influence of sex on these craniofacial phenotypes, we performed a two-part analysis (see Materials and Methods) (Broman and Sen 2009). First, the results of an analysis of variance revealed only three traits differed significantly (P < 0.05) with respect to group means for phenotypic scores between males and females (Table S3). Interestingly, these traits included the residual value QTL identified for the SO2 bone (both right and left sides) and the SO2 area for the left side only (Table S3).
We then performed multiple covariate analyses, in which sex was the assigned covariate, in R/qtl for each trait (1000 permutations; see Broman and Sen 2009). We observed a difference in the LOD value for the QTL detected at genomic marker 227A for both right (R)SO2 area residuals and left (L)SO2 area residuals (Table S3), suggesting that this trait differs between males and females in our hybrid pedigree. However, this analysis did not reveal a difference in the strength of the LOD for the two markers (NYU27 and 229B) associated with LSO2 area. Thus, there appears to be a gender-specific difference in SO2 bone area, after correction for body length. For the majority of craniofacial traits analyzed, however, prior QTL associations did not change (P > 0.05), suggesting most of these traits do not differ between sexes (Table S3). We did detect a change in the LOD values for two other traits, LSO3 area and RSO3 number (Table S3); however, it is unclear whether these are biologically relevant since the ANOVA results for both traits were insignificant.
The majority of the craniofacial traits that we analyzed did not yield significant QTL. These included the total area of the opercular bone, the lateral cranial area of the head, the area of the nasal opening (nares), the area of the antorbital bone, and the area of the supraorbital bone. It was surprising that some of these traits did not yield a genetic effect. For instance, opercular bone shape varies considerably between different Astyanax cave populations (Jeffery 2009) and has undergone marked morphological changes within ray-finned fish (Kimmel et al. 2005). However, unlike that observed for bone area for four of the suborbital bones, we did not observe a genetic effect associated with opercular bone area.
In stickleback fish, this bone demonstrates significant variation between freshwater and oceanic forms (Kimmel et al. 2005, 2012a,b; Arif et al. 2009). Recent morphological analyses of the opercle bone revealed a major-effect locus associated with opercle bone shape in an F2 mapping pedigree of anadromous and lake stickleback fish (Kimmel et al. 2005). Perhaps future morphometric analyses will similarly reveal a genetic effect associated with shape differences in cavefish populations compared to surface morphs. Nonetheless, our collective results indicate that, while some regions of the skull are controlled by heritable genetic changes that evolved following colonization of the subterranean environment, other aspects of the skull remained unchanged.
Fragmentation and fusion phenotypes affecting the suborbital bone series demonstrate genetic asymmetry
Three phenotypes, including the number of SO3 bony fragments and fusion of the SO1+2 and SO4+5 bones, demonstrated a directional genetic basis in our hybrid pedigree. We discovered this pattern by observing that certain individuals with severe fragmentation phenotypes (e.g., five SO3 fragments) on one side of their cranium had less severe fragmentation on the contralateral side (Figure 1). Therefore, we scored both left and right sides of the cranium in our analyses.
The number of SO3 elements in our pedigree ranged from one to five on the right side and from one to three on the left side (Figure S1D). The most common phenotype was presence of a single bony element for both sides of the head. A QTL analysis identified two significantly associated markers, NYU53 (LODMR = 7.31) and 206A (LODMR = 5.43), for the right side SO3 number (Figure 1G). Effect plots revealed a significant effect of genotype for both loci on the right side only (Figure 1H). The heterozygous genotype was highly similar to the surface fish genotype, implying a dominant effect at this locus (Figure 1H). An insignificant phenotypic effect was noted for the same two loci on the left side (Figure 1J). Interestingly, bone size varied minimally between individuals, suggesting that the total area of the bone is constrained on right and left sides, despite uneven fragmentation patterns. We tested whether this result was determined by a few individuals harboring extreme fragmentation (e.g., four and five fragments) by reanalyzing SO3 fragmentation as a binary trait. This analysis identified the same loci, NYU53 and 206A, with lower but significant LODMR values (4.10 and 3.88, respectively). MQM also identified marker 206A (LODMQM = 4.06) as significantly associated with right SO3 fragmentation; however, no cofactors were observed (Figure 1K). For left-sided SO3 fragmentation, MQM analysis identified one marker and cofactor (233D) with an insignificant LOD score (2.15; Figure 1L).
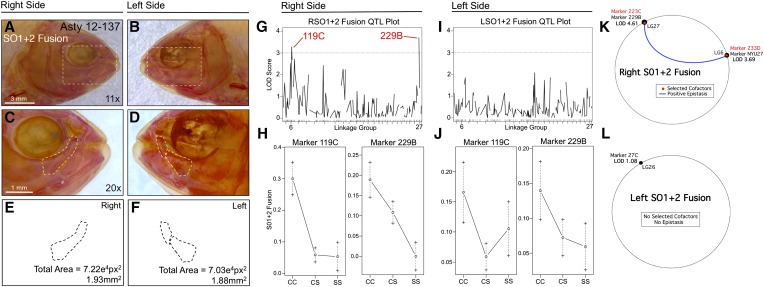
An analysis of SO1+2 bony fusion similarly revealed a right-sided directional genetic effect (Figure 2). Fusion was scored as a binary trait and displayed a similar frequency on both sides of the head (Figure S1A). However, analysis of this trait identified two different loci that exceeded a LOD value of 3.0, 119C (LODMR = 3.28) and 229B (LODHK = 4.41), on the right side only (Figure 2, G and I). Effect plots revealed a dominant effect of the surface allele at marker 119C and an intermediate phenotypic effect at 229B (Figure 2H). MQM analyses supported the results of our one-scan mapping methods, identifying either the neighboring (NYU27; LG6) or the same markers (229B; LG27) for right-sided SO1+2 fusion (Figure 2K). Interestingly, two cofactors were identified on the same linkage groups (223C on LG27; 233D on LG6), which demonstrate a positive epistatic interaction for this trait (blue line, Figure 2K). Consistent with one-scan analyses, no significant markers or cofactors were detected using MQM for left-sided SO1+2 fusion (Figure 2L). As with the SO3 bone, the total area occupied by these bones did not differ between left and right sides.
Figure 2.
Fusion of the first and second suborbital bones (SO1+2) demonstrates a right-sided asymmetric genetic basis. The first and second suborbital bones (dashed box in A and B) were scored separately on the right (A, C, and E) and left sides (B, D, and F), as demonstrated using specimen 137 from our Asty12 pedigree. Bones were present either as separate elements (the wild-type surface phenotype) (D) or fused together (C). The total area of the bones (E and F) did not differ across hybrids. Two markers, 119C and 229B, yielded LOD values >3.0 on the right (G), but not the left (I), side of the head. Effect plots reveal a surface dominant effect for 119C and an intermediate dominant effect for 229B (H), on the right side only (compare with J). MQM analyses reveal the two significant QTL associated with fusion on the right side (NYU27, 229B) with two nearby cofactors (233D, 223C) that demonstrate a positive epistatic interaction (K). On the left side, one insignificant QTL (LOD = 1.08) and no cofactors were identified (L). Bars: 3 mm in A and B (11×) and 1 mm in C and D (20×). Black trace in G and I represents results of MR mapping method.
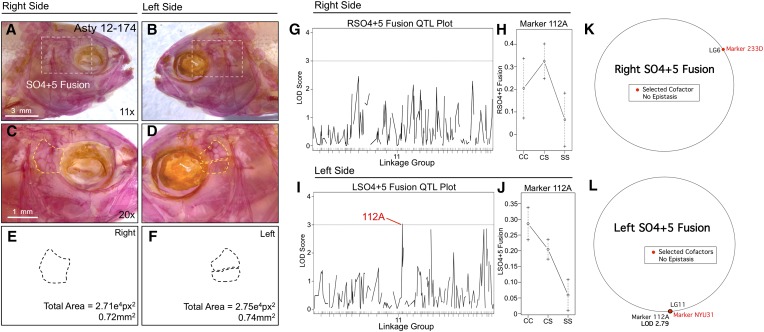
SO4+5 fusion was also scored as a binary trait and revealed an association with marker 112A (LODMR = 3.03) only on the left side of the head (Figure 3). The effect plot for 112A indicates fusion is associated with the homozygous cave condition and that the heterozygous genotype produces an intermediate phenotypic effect (Figure 3, I and J). MQM analyses revealed the same genetic marker (112A; LODMQM = 2.79) as one-scan mapping methods and identified a cofactor (NYU31) that also resides on LG11 (Figure 3L). Consistent with one-scan results, no significant genetic markers were identified for right-sided SO4+5 fusion (Figure 3K). Despite the asymmetric association with fusion of these two elements, the overall area collectively occupied by the SO4 and SO5 bones did not differ across individuals.
Figure 3.
Fusion of the fourth and fifth suborbital bones (SO4+5) demonstrates a left-sided asymmetric genetic basis. The fourth and fifth suborbital bones (dashed box in A and B) were scored separately on the right (A, C, and E) and left sides (B, D, and F), as demonstrated using specimen 174 from our Asty12 pedigree. Bones were present either as separate elements (the wild-type surface phenotype) (D) or fused together (C). The total area of the bones (E and F) demonstrated a genetic basis in our pedigree (see Figure 6 and Figure 7). One marker, 112A, demonstrated a LOD value >3.0 on the left (I), but not the right (G), side of the head. Effect plots reveal an intermediate dominant effect for 112A (J), on the left side only (compare with H). MQM analyses identified one selected cofactor on the right side (233D) (K). On the left side, the same genetic locus (112A) identified through one-scan mapping was identified using MQM, along with one cofactor (NYU31) (L). Bars: 3 mm in A and B (11×) and 1 mm in C and D (20×). Black trace in G and I represents results of MR mapping method.
Genetic symmetry is preserved in several cave-associated and craniofacial phenotypes
In a prior analysis using the pedigree evaluated here, eye size reduction was scored only on the right side of individual fish. We reanalyzed this phenotype and confirmed the identity of loci associated with quantitative changes in eye size for both left and right eye measurements (Table 1). We further validated the same QTL position for albinism, a Mendelian trait (Şadoğlu 1957a,b; Protas et al. 2006), using the previously published data set (Table 1).
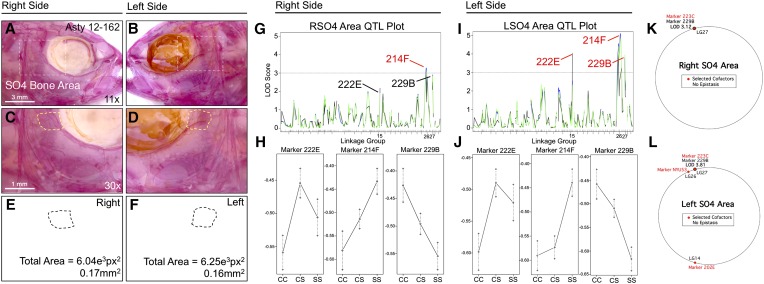
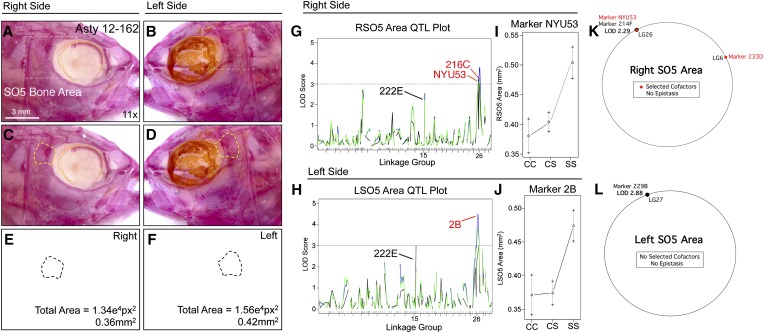
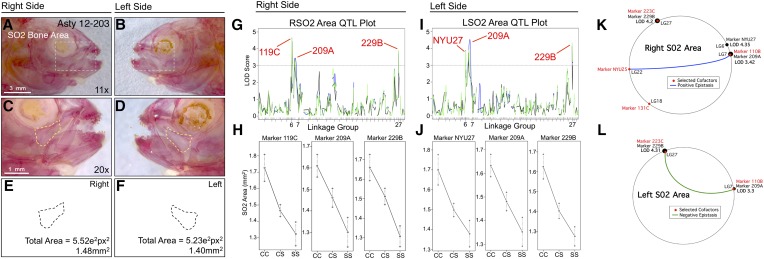
Of the craniofacial traits analyzed, only area measurements for the SO2 and SO3 bones produced QTL that were identical when scored on the left and right sides (i.e., genetically symmetric). Analysis of SO4 and SO5 area revealed partially symmetric results. For the SO4 bone, one QTL was shared for both the right and left sides, while two additional markers were present on the left side only (Figure 6). For the SO5 bone, two nearby markers were detected on the right side (216C and NYU53) and one QTL was detected on the left side (2B); however, all three markers reside on the same linkage group (26; Figure 7). Area measurements for the SO2, SO3, and SO5 bones were distributed normally (Figure S1, B, C, and G); however, SO4 area measurements were positively skewed. We transformed SO4 area measurements to log10 values to create a normal distribution for QTL association studies (Figure S1E).
Figure 6.
The area of the fourth suborbital bone (SO4) demonstrates a partially symmetric genetic basis. The fourth suborbital bone (dashed box in A and B) was scored on the right (A, C, and E) and left sides (B, D, and F), as demonstrated using specimen 162 from our Asty12 pedigree. The total area of this bone (dashed outline, C–F) harbored a genetic basis. One marker, 214F, demonstrated a LOD value >3.0 on both the right (G) and left (I) sides of the head. Two other markers, 222E and 229B, demonstrated LOD values >3.0 on the left (I), but not the right (G), side of the head. Interestingly, effect plots revealed an intermediate dominant effect for 214F on the right side (H), but a cave dominant effect for this marker on the left side of the head (J). Marker 222E demonstrated a surface dominant effect, while marker 229B demonstrated an intermediate dominant effect on the left side (J). MQM analyses revealed the same QTL associated with SO4 area on the right and left sides (229B); however, only one cofactor was shared between the right and left sides (223C) (K and L). On the left side, two additional cofactors (202E, NYU53) were identified (L). Bars: 3 mm in A and B (11×) and 1 mm in C and D (30×). In G and I, black line shows marker regression, green line shows HK, and blue line shows EM mapping methods, respectively.
Figure 7.
The area of fifth suborbital bone (SO5) demonstrates a partially symmetric genetic basis. The fifth suborbital bone (dashed box in A and B) was scored on the right (A, C, and E) and left sides (B, D, and F), as demonstrated using specimen 162 from our Asty12 pedigree. The total area of this bone (dashed outline, C–F) harbored a genetic basis. Markers 216C and NYU53 (which are ∼2 cM apart from one another) both demonstrated a LOD value >3.0 on the right (G) side of the head. A different marker on the same linkage group (2B) demonstrated a significant LOD score for the left side of the head (I). Interestingly, effect plots were nearly identical for the significant markers on the right side (NYU53) (I) and left side (2B) (J). An additional marker, 222E (black, G and H) had a LOD score just below our 3.0 threshold on the left side, and well below 3.0 on the right side. Unexpectedly, MQM analyses revealed two different QTL (on different groups) associated with SO5 area on the right (214F) and left (229B) sides (K and L). On the right, two cofactors were identified (233D and NYU53), while no cofactors were selected on the left (K). Bar, 3 mm (11×). In G and I, black line shows marker regression, green line shows HK, and blue line shows EM mapping methods, respectively.
SO2 area is associated with three loci on both the left and right sides: 119C/NYU27 on linkage group 6, 209A on linkage group 7, and 229B on linkage group 27 (Figure 4). Despite alternative positions for 119C (right side; LODMR = 4.43) and NYU27 (left side; LODEM = 3.88), these loci are merely 3.2 cM apart and effect plots for both markers yield nearly identical patterns (Figure 4, G–J). The markers 209A (left side LODMR = 3.74; right side LODMR = 3.42) and 229B (left side LODEM = 4.28; right side LODHK = 3.94) were associated with increased bony area in cavefish (Figure 4, H and J). MQM analyses largely supported the results of one-scan methods, identifying three markers on the right side (NYU27, 209A, and 229B; Figure 4K), but only two markers on the left side (209A and 229B; Figure 4L). MQM analyses revealed four cofactors (Figure 4K) for right-sided SO2 area, but only two for the left side (Figure 4L). Interestingly, a positive epistatic interaction was observed between two cofactors (NYU25 and 110B) on the right side, and a negative epistatic interaction was found between two markers (223C and 110B) on the left side (Figure 4, K and L). This result demonstrates a complex interaction between the same marker (110B) and two different cofactors (NYU25 and 223C) on different linkage groups, which exert contrasting epistatic effects on the right and left sides, respectively.
Figure 4.
The area of second suborbital bone (SO2) demonstrates a symmetric genetic basis. The second suborbital bone (dashed box in A and B) was scored on the right (A, C, and E) and left sides (B, D, and F), as demonstrated using specimen 203 from our Asty12 pedigree. The total area of this bone (dashed outline in C–F) harbored a genetic basis. Three markers, 119C/NYU27, 209A, and 229B, demonstrated LOD values >3.0 on both the right (G) and left (I) sides of the head. Effect plots revealed an intermediate dominant effect for all three markers, irrespective of whether they were evaluated on the right or left side of the head (H and J). MQM analyses identified two of the same significant QTL for both left and right sides (209A and 229B) (K and L). The marker NYU27 was detected on the right side only (LOD = 4.35) (K). Four cofactors were identified for this trait, two of which were shared (110B, 223C) between right and left sides. Interestingly, a positive epistatic effect was detected between marker NYU25 and 110B on the right (blue line, K), while a negative interaction was found between 223C and 110B on the left (green line, L). Bars: 3 mm in A and B (11×) and 1 mm in C and D (20×). In G and I, black line shows marker regression, green line shows HK, and blue line shows EM mapping methods, respectively.
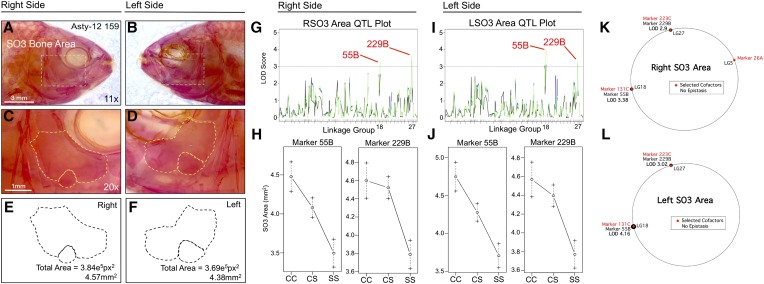
SO3 area is associated with two loci on both right and left sides: 55B on linkage group 18 (left side LODHK = 4.02; right side LODEM+HK = 3.24) and 229B (left side LODEM = 3.28; right side LODEM+HK = 3.66) on linkage group 27 (Figure 5). Interestingly, both SO2 and SO3 areas shared a genetic association with the same genetic marker (229B). Effect plots for both bone area phenotypes reveal the same polarity, wherein two copies of the cave allele are always associated with larger SO2 (Figure 4, H and J) and SO3 bones (Figure 5, H and J). The heterozygous genotype for marker 55B produces an intermediate phenotype between the cave- and surface-dwelling phenotypes (Figure 5, H and J); however, the heterozygous genotype for marker 229B is similar to the cave-dwelling phenotype, possibly indicating dominance of the cave allele at the linked locus (Figure 5, H and J). MQM analyses identified the same QTL as one-scan methods for the both the right (55B, LODMQM = 3.38; 229B, LODMQM = 2.9) and left (55B, LODMQM = 4.16; 229B, LODMQM = 3.02) SO3 bones (Figure 5, K and L). Moreover, the same cofactors were selected for both sides (131C, 223C) with the exception of marker 26A, which was identified on the right side only. No epistatic interactions between cofactors were observed on either side (Figure 5, K and L).
Figure 5.
The area of third suborbital bone (SO3) demonstrates a symmetric genetic basis. The third suborbital bone (dashed box in A and B) was scored on the right (A, C, and E) and left sides (B, D, and F), as demonstrated using specimen 159 from our Asty12 pedigree. The total area of this bone (dashed outline of an exemplary bone composed of two fragments in C–F) harbored a genetic basis. Two markers, 55B and 229B, demonstrated LOD values >3.0 on both the right (G) and left (I) sides of the head. Effect plots revealed an intermediate dominant effect for 55B and a cave dominant effect for marker 229B, on both right and left sides of the head (H and J). MQM analyses identified the same two QTL associated with SO3 bone area on the left and right sides (55B, 229B). Three cofactors were found for the right side (26A, 131C, and 223C) (K), only two of which were present on the left side (131C and 223C) (L). No epistatic interactions between cofactors were observed. Bars: 3 mm in A and B (11×) and 1 mm in C and D (20×). In G and I, black line shows marker regression, green line shows HK, and blue line shows EM mapping methods, respectively.
SO4 area demonstrated partial genetic symmetry (Figure 6). The same genetic marker, 214F, is associated with area changes on both the left and right sides (Figure 6, G and I), although the LOD value is higher on the left side. Two additional genetic markers, 222E and 229B, are associated with SO4 area differences on the left side only (Figure 6I). Interestingly, the effect plot for the symmetric marker 214F demonstrates intermediate dominance on the right side (Figure 6H), but dominance for the cave allele on the left side (Figure 6J). Marker 222E demonstrates a weak dominant effect for the surface allele, while marker 229B demonstrates intermediate dominance (Figure 6J). Interestingly, the phenotypic polarity is different for the three markers: cave alleles for 222E and 214F are associated with smaller SO4 area, while cave alleles for 229B are associated with a larger SO4 area (Figure 6, H and J). MQM analyses supported the notion of partial genetic symmetry for SO4 area (Figure 6, K and L), identifying the same genetic marker (229B) on both right (LODMQM = 3.12) and left sides (LODMQM = 3.81). However, only one cofactor was selected for the right side (223C; Figure 6K) compared to three cofactors that were selected for the left side (202E, NYU53, and 223C; Figure 6L).
SO5 area is associated with the same linkage group (26) on both the left and right sides of the cranium (Figure 7). The effect plots for identified markers on the right (216C, NYU53) and left sides (2B) are nearly identical, demonstrating dominance at the cave locus (Figure 7, I and J). In both cases, the phenotypic polarity is the same—cave alleles are associated with a smaller SO5 area. One marker, 222E, harbored a LOD value just below our 3.0 threshold on the left side (Figure 7H). The same marker on the right side demonstrated an insignificant LOD value (Figure 7G). MQM analyses identified different results for this trait compared to one-scan analyses. On the right side, marker 214F on LG26 demonstrated an association with SO5 area (LODMQM = 2.29; Figure 7K). However, on the left side, a different marker on LG27 (229B) demonstrated an association with SO5 area (LODMQM = 2.88; Figure 7L). Two cofactors were identified on the right side (NYU53 and 233D); however, no cofactors were selected on the left side.
We performed additional analyses of area for the SO2, SO3, and SO5 bones based on residual values following normalization for standard length of each specimen. Interestingly, these results identified both symmetric and asymmetric QTL. For instance, a symmetric genetic signal was detected for right- and left-sided SO2 area at marker 227A (LODMR = 5.56 and 5.69 for the right and left sides, respectively). However, one marginally significant QTL (106C; LODMR = 4.0) was detected only on the right side (Table 3). SO2 area effect plots again revealed a larger bone associated with the homozygous cave genotype (“CC”).
Residual values for the SO3 bony area yielded similarly mixed results. Both 26A and 136B were identified for both the left and right sides; however, two additional markers (203F and 6A) were detected on the left side only. QTL effect plots from this analysis yielded contrasting results. Markers 136B, 203F, and 6A were all associated with larger values in the cave homozygous condition; however, marker 26A was associated with a smaller area. These results may explain, in part, the discrepancy between our results and those of Yamamoto et al. (2003), particularly if the causative gene(s) associated with marker 26A exerts a stronger effect at the earlier developmental stages evaluated in their study. At present, however, the gene(s) mediating this potential effect remains unknown. Finally, residual values for the SO5 bone also demonstrated asymmetry since significant QTL were strictly detected on the left side (marker 2B; LODMR = 3.61). Interestingly, this marker demonstrated a smaller value associated with the homozygous cave condition.
QTL associated with craniofacial phenotypes map to syntenic regions of the D. rerio genome
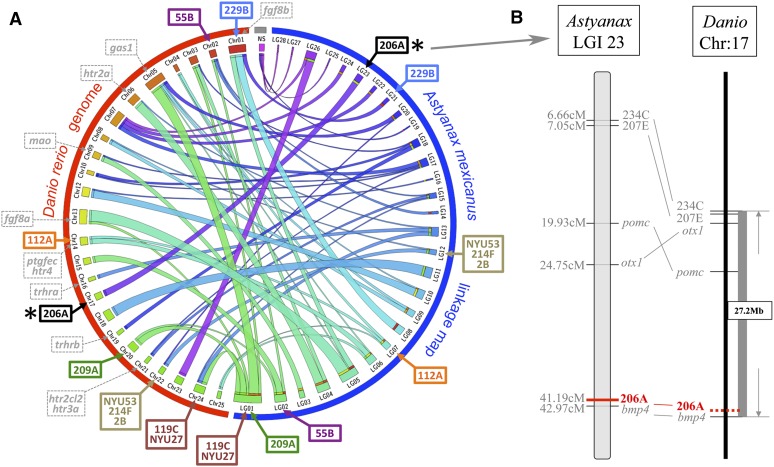
At present, there is not a fully annotated genome sequence available for A. mexicanus. Therefore, as a step toward nominating candidate genes linked to the QTL we detected, we analyzed the positions of microsatellite loci on an integrated linkage map that is anchored to the D. rerio genome (Figure 8). This integrated map represents a comprehensive analysis of multiple cave × surface fish crosses and therefore is a proxy for the genomic positions of several markers (Gross et al. 2008). Three analyses have identified numerous syntenic blocks shared between Astyanax and D. rerio (Gross et al. 2008, 2013; O’Quin et al. 2013). These blocks were identified through sequence similarity (BLAST hit) queries of noncoding sequences flanking polymorphic microsatellites or SNP markers used in RAD-seq studies.
Figure 8.
A syntenic analysis of the position of genomic markers anchored to the Danio rerio genome reveals bmp4 as a possible candidate gene mediating SO3 fragmentation. A Circos representation of synteny between an integrated Astyanax linkage map (blue hemicircle in A) and the D. rerio genome (red hemicircle in A) reveals significant stretches of shared synteny between these teleost species. Few candidate genes selected from prior studies (gray dashed markers, A) of genetic asymmetry overlap with linked microsatellite markers identified from this study. Marker 229B (blue) is associated with SO1+2 fusion and SO2–4 bone areas and is predicted to reside near the gene fgf8b in D. rerio. Additionally, marker 206A (black, asterisk in A), which is associated with SO3 fragmentation, maps near the bmp4 locus (B). This may indicate a potential role for this gene in mediating the asymmetric SO3 bone fragmentation phenotype in Astyanax cavefish. NS, no observed synteny between a whole linkage group and any particular Danio chromosome.
The microsatellite marker 119C on linkage group 1 (Figure 8A), linked to right-sided SO1+2 fusion and SO2 area, lies at approximate position 24.2 Mb on chromosome 24, ∼8.8 cM away from the gene musculin (msc). The microsatellite marker NYU27, which is linked to left-sided SO2 area, is positioned on chromosome 24 in D. rerio (Figure 8A). Marker NYU27 is ∼3 cM away from 119C and is estimated to reside near the same syntenic block on chromosome 24 in Danio. Marker 229B is linked to a number of the traits we analyzed, including right-sided SO1+2 fusion, symmetric SO2 bony area, symmetric SO3 bony area, and symmetric SO4 bony area phenotypes (Figure 8A). This marker is predicted to be ∼0.83 cM away from the gene alpha-A-crystallin (crystaa) within a 38.4-Mb syntenic block of Danio chromosome 1.
The microsatellite marker 209A (Figure 8A), linked to symmetric SO2 bony area, resides in a syntenic block on chromosome 20 in D. rerio, at approximate position 55.9 Mb. Microsatellite marker 55B, which is linked to the symmetric SO3 bony area phenotype, comprises a short (∼0.1 Mb) syntenic region on Danio chromosome 2 along with the marker 135C (Figure 8A). Marker 206A, on integrated linkage group (LGI) 23, which produces the peak LOD score for right-sided SO3 fragmentation, is predicted to reside within a 27.2-Mb syntenic block of Danio chromosome 17 (Figure 8, A and B). This marker is located ∼1.78 cM away from bone morphogenetic protein 4 (bmp4). Marker NYU53, also linked to right-sided SO3 fragmentation and right-sided SO5 area, was not evaluated in our prior publication. However, this marker is ∼2 cM away from marker 216C in our current linkage map, which is anchored to a syntenic block of Danio chromosome 22 (Figure 8A).
Microsatellite marker 214F, linked to symmetric SO4 bony area, resides near a block of synteny with Danio chromosome 22 (Figure 8A). The marker 112A, linked to left-sided SO4+5, fusion maps between two genetic markers (224C, ccng1), both of which are anchored to Danio chromosome 14 (Figure 8A). Marker 2B, linked to symmetric SO5 bony area, maps near a syntenic region shared between Astyanax LGI 12 and chromosome 22 (Figure 8A).
Discussion
Genetic asymmetry in the cavefish craniofacial complex
Here we demonstrated genetic asymmetry in SO3 fragment number and fusion of the SO1+2 and SO4+5 bones. Prior work has established a role for fluctuating asymmetry in the generation of numerous aberrant morphologies, including wing vein patterns in Drosophila (Klingenberg and Mcintyre 1998), male sexual ornaments in swallows (Møller 1990), and mandibular bone landmarks in mice (Leamy 1993). This phenomenon is frequently attributed to developmental instability (Klingenberg and McIntyre 1998) caused by any of a number of factors such as environmental stress (Palmer 1994), interspecific biotic stresses such as pathogens (Watson and Thornhill 1994), intraspecific biotic stress such as competition (Manning and Chamberlain 1993), or genetic stress occurring as a result of hybridization (Clarke et al. 1986; Hochwender and Fritz 1999; Bourguet 2000; Dongen 2006), which leads to random-sided morphological aberrations. Some of the craniofacial changes we report here may be fluctuating asymmetrically. In our mapping cross, all of the craniofacial traits for which we observed a genetic effect appear with roughly equal frequency on both sides of the head (Figure S1). Interestingly however, the genetic effects for three of these craniofacial traits were detected only on one side. We observed a genetic effect on strictly the right side for two traits (SO3 fragment number and SO1+2 fusion) and a genetic effect on strictly the left side for one trait (SO4+5 fusion).
A few prior studies have reported the genetic basis of craniofacial asymmetry. Leamy et al. (1997) evaluated directional asymmetry in a cross of Large (LG/J) and Small (SM/J) inbred mouse strains and discovered 16 significant QTL associated with nine mandibular characters (Leamy et al. 1997). Portuguese water dogs commonly develop hip dysplasia, which arises from both heritable and environmental factors (Chase et al. 2004). Chase et al. (2004) evaluated the genetic basis for this trait through quantitative autoradiographic measurements of hip joint laxity of the hind legs. They discovered two QTL, one affecting the right hip (explaining 16% of the heritable variation) and one affecting the left hip (explaining 14% of the heritable variation). There was significantly greater laxity for the left vs. the right hip, and this polarity was conserved in 80% of the dogs they evaluated. This directional asymmetry could be linked to behavioral or physiological preferences (Chase et al. 2004). For instance, a genetically influenced behavior (“right-” or “left-footedness”) may lead to undue joint strain and result in a more severe phenotype on one (but not the other) side of the animal.
Across many generations, this form of heritable variation may lead to more significant morphological asymmetries [e.g., claw size in fiddler crabs (Morgan 1923; Leamy et al. 1997)]. A fascinating example of morphological asymmetry has been described in cichlids. The Lake Tanganyikan scale-eating fish, Perissodus microlepis, harbors two morphotypes that differ in the (left–right) direction of their mouth opening (Hori 1993). These fish attack and feed on the scales of other living fish based on their handedness, which was originally reported as a simple Mendelian trait (Hori 1993). Recently, Stewart and Albertson (2010) identified a jaw laterality locus segregating in an F2 cross of two herbivorous species from Lake Malawi based on length variation of the retroarticular processes of the jaw. The patterns of inheritance for this were consistent with the genetic effects predicted by earlier studies (Hori 1993; Hori et al. 2007). The ratio of right- and left-handed morphs within the population is determined by a mechanism of frequency-dependent natural selection, mediated by the alertness of the prey (Hori 1993). Thus, natural selection appears to have favored the evolution of these distinct jaw asymmetries to facilitate predation success (Stewart and Albertson 2010). Within the wider context of the Perissodini tribe, these asymmetries likely arise in response to shifts in habitat and predation strategy (Stewart and Albertson 2010).
It is difficult to imagine how such “handedness” could be adaptive in blind Mexican cavefish. However, cranial neuromasts (which mediate responses to water movements) densely populate the SO3 bone (Yoshizawa et al. 2012), which demonstrated the most severe fragmentation phenotype in our hybrid pedigree (Figure 1). Perhaps increased cranial neuromast density leads to bone fragmentation by interfering with normal condensation and ossification of the underlying cranial mesenchyme (Stensiö 1947; Lekander 1949; Reno 1969). If so, a consistent directional asymmetry affecting cranial neuromast number might also be predicted. At present, a connection between increased neuromast numbers and severity of fragmentation, or an attendant directional asymmetry in cranial neuromast number in natural populations, remains unknown.
Novel craniofacial QTL in the context of past studies
Several of the markers identified from our analysis overlapped with QTL positions identified by Protas et al. (2007) (Table 1). For instance, four microsatellites—234B, NYU14, NYU27, and 229B—colocalized with map locations for previously identified eye and lens QTL. In addition, two genomic markers (119C, 206A) mapped to positions associated with melanophore number, one marker (55B) mapped to a linkage group position associated with taste bud number, one marker (112A) is also associated with the maxilla, and two markers (229B, 2B) mapped to a region associated with tooth phenotypes (Protas et al. 2007). We did not observe colocalization of our craniofacial traits with the SO3 width trait discovered by Protas et al. (2008). However, one of the six SO3 width QTL reported by Protas et al. (2008) maps to marker 203E, which neighbors marker 55B. SO3 area phenotypes also map with a peak LOD score to this marker (Protas et al. 2008). Thus, this marker may broadly indicate a genetic basis for bone size differences between morphotypes, and our scoring methods may have redundantly reported the same underlying phenotypic difference.
Of the 11 craniofacial phenotypes that yielded QTL in our study, only four linked markers failed to map to regions associated with previously identified troglomorphic phenotypes. This may indicate that certain regions of the cavefish genome, populated with multiple different genes, have undergone accelerated evolution through the process of cave adaptation (Yokoyama and Yokoyama 1990). Alternatively, the craniofacial QTL identified, along with previously discovered QTL, may be mediated by the same pleiotropic genes (Protas et al. 2008) or be closely linked to the causative gene(s) controlling other cave-associated traits. Future studies evaluating a larger pedigree along with a higher-resolution physical map of the genome will enable us to discern between these two possibilities.
Yamamoto et al. (2003) demonstrated that eye removal results in both dependent and independent effects on the craniofacial complex. One trait concluded to be independent of eye removal was the number of SO3 bony fragments (Yamamoto et al. 2003). Our results support this conclusion; the two SO3 fragment number QTL we identified (linked markers 206A and NYU53) do not overlap with the positions of any eye or lens QTL (Protas et al. 2008). However, 206A maps to a previously discovered QTL for melanophore number (Protas et al. 2007). Moreover, the genetic effect we discovered was detected only on the right side, implying a genetic asymmetry in SO3 bone fragmentation. Yamamoto et al. (2003) hypothesized that SO3 area may be reduced as an indirect consequence of olfactory sense expansion in cavefish. In this study, we measured the area of the external opening to the olfactory epithelium to determine whether nares expansion is associated with SO3 fragmentation, but found no QTL associated with this trait.
Interestingly, effect plots for SO3 bone QTL revealed homozygous cave alleles are associated with a larger SO3 bone area (Figure 5, H and J). In contrast, Yamamoto et al. (2003) observed that the size of the circumorbital bones correlated with presence of the eye in their experimental studies; i.e., eyeless experimental subjects had smaller circumorbital bones (Yamamoto et al. 2003). Our differing results could be attributed to at least two explanations. First, in experimental manipulations, bone growth may be critically reliant upon the developing eye. However, under nonexperimental conditions, the genetic basis for SO3 bone size increase may be fundamentally distinct from that for eye loss in cavefish. Second, different-aged specimens were evaluated in Yamamoto et al. (2003) compared to the present study. We therefore wanted to determine whether larger-sized bones were observed, in part, due to allometric size increases in this bone that may be revealed by the older specimens used in our study.
A regression analysis of the individuals within our pedigree indicated positive relationships between bone area measurements (for both the right and left sides of the cranium) and standard length (Table 3). The SO2, SO3, and SO5 bones are all positively associated with standard length in our hybrid pedigree, with R2 values ranging from 0.37 to 0.75, suggesting that larger-sized specimens harbor larger circumorbital bones. We further investigated this relationship by scanning for QTL based on residual values obtained for each of our circumorbital bone measurements. This was carried out following correction for standard length of each specimen (Materials and Methods). Significant results were obtained only for the SO2, the SO3, and the left SO5 bony area measures (Table 3).
These residuals analyses allowed us to dissect apart the genetic effects associated with suborbital bone areas that are not indirectly influenced by the overall size of each specimen. For example, larger fish harbor larger bones, and therefore QTL identified using raw values alone may indirectly report associations based on overall size differences. Indeed, for the SO2 and SO3 bones (Table 3), our residuals analyses identified new QTL (i.e., on different linkage groups) compared to those detected using raw, uncorrected values. Interestingly, however, these genetic effects were largely symmetric, consistent with our analysis of raw area measurements. Thus, normalization based on body size provided us with additional sensitivity to detect relevant genetic loci involved in complex craniofacial features evolving in cave-dwelling fish. These results further underscore the complex genetic regulation of craniofacial diversity in this species.
Potential genes affecting bone fragmentation and fusion
Leamy et al. (1997) observed QTL associated with directional asymmetry in mandibular dimensions evaluated in a cross of two inbred mouse strains. The authors speculated that directional asymmetry might be governed by genes encoding hormones, growth factors, and/or hormone receptors (Leamy et al. 1997, 1998). They noted the positions of three candidate genes, including growth arrest-specific 1 (gas1) on mouse chromosome 13 and thyrotropin-releasing hormone receptor (trhr) and platelet derived growth factor, endothelial cell (ptgfec) on mouse chromosome 15 (Leamy et al. 1997). A subsequent study identified QTL associated with fluctuating asymmetry, many of which localized to similar chromosomal regions to those identified for directional asymmetry (Leamy et al. 1998). Our preliminary data do not indicate that any of these genes reside close to the QTL identified in our study, with the exception of the genes ptgfec and htr4, which reside near marker 112A, and bmp4, which maps close to marker 206A. Overall, this observation may indicate that the causative genetic lesions associated with directional asymmetry are divergent in our model system or may be governed by one or more paralogous genes in Astyanax, whose homologous genomic positions differ significantly from the genomic position of these genes in mice.
One prospective candidate gene, bmp4, plays a key role in the evolution and craniofacial development of a broad set of species (Schubert et al. 2000; Terai et al. 2002; Trainor et al. 2003). For instance, bmp4 is associated with adaptive jaw shape differences in cichlid fish (Albertson and Kocher 2006) as well as the depth and breadth of upper beaks in Darwin’s finches (Abzhanov et al. 2004). Alongside its role in craniofacial evolution and development, bmp4 has also been implicated in laterality of the heart and gut (Monteiro et al. 2008). This wider role in establishing left–right asymmetry in other organ systems during development renders bmp4 a particularly attractive candidate molecule for mediating asymmetric phenotypes in our system.
While the genes controlling suborbital bone fragmentation in cavefish remain unknown, bmp4 may play a critical role in this abnormal process. The SO3 dermal bone in cavefish develops from condensations of mesenchymal cells derived from the embryonic cranial neural crest (Northcutt and Gans 1983; Gross and Hanken 2008). Therefore, the origin of directional asymmetry in SO3 bone fragmentation may ultimately reflect aberrant migration, proliferation, or survival of the osteogenic cranial neural crest (in a directionally biased manner) during embryonic development.
Another prospective candidate gene mediating craniofacial asymmetry in our system is fgf8, which influences laterality in the zebrafish and mouse craniofacial skeletons (Albertson and Yelick 2005, 2007; Griffin et al. 2012). Specifically, zebrafish juveniles with attenuated fgf8 activity demonstrate consistent left–right asymmetry in craniofacial structures (Albertson and Yelick 2005). Using the transgenic aceti282a/fgf8 heterozygous zebrafish line, Albertson and Yelick (2007) further demonstrated a role for fgf8 in mediating a variety of abnormalities, including facial asymmetry, altered craniofacial geometry and sutural pattern, and ectopic bone formation. These abnormalities, which resemble certain human disorders (such as craniosynostosis and hemifacial microsomia), may originate from increased bone metabolism observed in the aceti282a/fgf8 transgenic line (Albertson and Yelick 2007). A study of fgf8 attenuation in mice, combined with gain-of-function mouse–chicken chimeric studies, suggests a deeply conserved role for fgf8 in craniofacial development and symmetry. Griffin et al. (2012) recently demonstrated a key role for fgf8 in midfacial integration and development of the optic capsule. Thus, fgf8 is an excellent candidate gene for future analyses in cavefish given that fgf8 signaling is a likely target for evolutionary selective pressures (Griffin et al. 2012). The gene fgf8b is predicted to reside near marker 229B, associated with SO1+2 fusion in the present study (Figure 8).
Alternative explanations for genetic asymmetry
Serotonergic signaling has been implicated in consistent left–right asymmetry in vertebrate embryos (Fukumoto et al. 2005; Levin et al. 2006). Serotonin is a neurotransmitter molecule with critical functions in a variety of processes, including physiology (Fuller 1992), cognition (Canli and Lesch 2007), and circadian rhythms (Moore and Klein 1974). In Xenopus embryos, proper serotonergic signaling through the 5-HTR3, 5-HTR4 receptors and the metabolism of monoamine oxidase (MAO) is essential for establishment of asymmetric gene expression and organ development (Fukumoto et al. 2005). Serotonin is functional very early in development through maternal deposition, demonstrates dynamic spatial localization, and exerts its action as early as the four-cell stage (Fukumoto et al. 2005). Fukumoto et al. (2005) did not evaluate craniofacial bone formation, which occurs later in development. However, prior studies in mice have indicated serotonergic signaling through the 5-HT2B receptor is critical for craniofacial morphogenesis (Choi et al. 1997). Thus, altered expression patterns of serotonin, its receptors, or related enzymatic pathways (e.g., through MAO) may have similarly evolved in the cavefish lineage. Our syntenic analysis does not indicate that these particular receptors map near the critical region of our QTL (Figure 8); however, we cannot rule out the role of a paralogous gene or other member of the serotonergic pathway in causing one or more directionally symmetric phenotypes in cavefish.
Another possibility is the asymmetric activation of a microRNA locus (He and Hannon 2004). In Caenorhabditis elegans, a functional asymmetry is generated during development through the two-step activation of lsy-6 in a postmitotic pair of sensory neurons (Chang et al. 2004). lsy-6 is first “primed” in the precursor of the left (but not right-sided) neuronal cell and then boosted to higher expression levels several divisions later (Chang et al. 2004). The downstream consequence of functionally relevant levels of expression leads to asymmetric functional identity, based on the expression of distinct guanylate cyclase receptors (Chang et al. 2004). In cave-dwelling vertebrates, such altered patterns of expression for molecules leading to directionally asymmetric craniofacial phenotypes may similarly be at play. These changes might have arisen randomly through drift or as an indirect consequence of selection for another unrelated function.
The evolutionary mechanism of regressive craniofacial trait evolution
The craniofacial traits we describe here may not evolve as a (direct or indirect) consequence of selection pressure. In the complete darkness of the cave, there are relaxed selection pressures associated with visually mediated traits (Borowsky and Wilkens 2002). In the above-ground environment, craniofacial symmetry may be under intense sexual selection, as an indicator of mate quality (Rhodes et al. 2001). Conversely, in the subterranean environment perhaps total darkness and loss of vision lead to a relaxation of this selective constraint, resulting in the asymmetries we observed in this hybrid cross.
Here we provide further evidence that organismal development can be influenced differentially on the left and right sides, leading to asymmetric phenotypes. Such asymmetries, being controlled by particular loci, may provide a substrate upon which natural selection can act to elaborate phenotypic differences along the left–right axis of the organism. We also demonstrate here that certain craniofacial QTL do not colocalize with previously identified cave-associated traits, while other QTL are associated with a variety of traits such as vision and pigmentation (Table 1). It remains unclear whether the latter result indicates pleiotropy or the close physical linkage of the causative genes for the phenotypes evaluated in this study. Future studies directed toward identifying the precise genetic lesion(s) explaining these traits will enable functional analyses. Functional validation will, in turn, clarify whether multiple phenotypes are affected by gain or loss of encoded gene products. Ultimately, the evolutionary mechanism (selection vs. drift) driving craniofacial changes in cavefish may differ, depending on the trait.
Supplementary Material
Acknowledgments
We thank the members of the Gross laboratory for helpful discussions. We especially thank Meredith Protas, Richard Borowsky, and Cliff Tabin for providing the cleared-and-stained specimens evaluated in this study. K. Dougherty and L. Bruns assisted with phenotypic measurements during an earlier phase of this project. We also thank Krista Nichols and two anonymous reviewers for providing helpful suggestions on an earlier draft of this manuscript. This project was supported by National Institutes of Health (National Institute of Dental and Craniofacial Research) grant DE022403 (to J.B.G.).
Footnotes
Communicating editor: K. M. Nichols
Literature Cited
- Abzhanov A., Protas M., Grant B. R., Grant P. R., Tabin C. J., 2004. Bmp4 and morphological variation of beaks in Darwin’s finches. Science 305: 1462–1465. [DOI] [PubMed] [Google Scholar]
- Abzhanov A., Kuo W. P., Hartmann C., Grant B. R., Grant P. R., et al. , 2006. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature 442: 563–567. [DOI] [PubMed] [Google Scholar]
- Albertson R. C., Kocher T. D., 2006. Genetic and developmental basis of cichlid trophic diversity. Heredity 97: 211–221. [DOI] [PubMed] [Google Scholar]
- Albertson R. C., Yelick P. C., 2005. Roles for fgf8 signaling in left–right patterning of the visceral organs and craniofacial skeleton. Dev. Biol. 283: 310–321. [DOI] [PubMed] [Google Scholar]
- Albertson R. C., Yelick P. C., 2007. Fgf8 haploinsufficiency results in distinct craniofacial defects in adult zebrafish. Dev. Biol. 306: 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson R. C., Streelman J. T., Kocher T. D., 2003. Genetic basis of adaptive shape differences in the cichlid head. J. Hered. 94: 291–301. [DOI] [PubMed] [Google Scholar]
- Alvaréz, J., 1946 Revisión del género Anoptichthys con descipción de una especie nueva (Pisces, Characidae). An. Esc. Nac. Cien. Biol. México 4: 263–282. [Google Scholar]
- Alvaréz J., 1947. Descripción de Anoptichthys hubbsi caracindo ciego de La Cueva de Los Sabinos. S. L. P. Rev. Soc. Mexicana Hist. Nat. 8: 215–219. [Google Scholar]
- Anyonge W., Baker A., 2006. Craniofacial morphology and feeding behavior in Canis dirus, the extinct Pleistocene dire wolf. J. Zool. 269: 309–316. [Google Scholar]
- Arends D., Prins P., Jansen R. C., Broman K. W., 2010. R/qtl: high-throughput multiple QTL mapping. Bioinformatics 26: 2990–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif S., Aguirre W. E., Bell M. A., 2009. Evolutionary diversification of opercle shape in Cook Inlet threespine stickleback. Biol. J. Linn. Soc. Lond. 97: 832–844. [Google Scholar]
- Beavis W. D., 1998. QTL analyses: power, precision, and accuracy, pp. 145–162 in Molecular Dissection of Complex Traits, edited by Paterson A. H. CRC Press, Boca Raton, FL. [Google Scholar]
- Borowsky R., Wilkens H., 2002. Mapping a cave fish genome: polygenic systems and regressive evolution. J. Hered. 93: 19–21. [DOI] [PubMed] [Google Scholar]
- Bourguet D., 2000. Fluctuating asymmetry and fitness in Drosophila melanogaster. J. Evol. Biol. 13: 515–521. [Google Scholar]
- Bradic M., Beerli P., García-de León F. J., Esquivel-Bobadilla S., Borowsky R. L., 2012. Gene flow and population structure in the Mexican blind cavefish complex (Astyanax mexicanus). BMC Evol. Biol. 12: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Speed T. P., 1999. A review of methods for identifying QTLs in experimental crosses. Stat. Mol. Biol. 1999: 114–142. [Google Scholar]
- Broman K. W., Wu H., Sen Ś., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Broman K. W., Sen Ś., 2009. A Guide to QTL Mapping with R/qtl. Springer-Verlag, New York, USA [Google Scholar]
- Canli T., Lesch K., 2007. Long story short: the serotonin transporter in emotion regulation and social cognition. Nat. Neurosci. 10: 1103–1109. [DOI] [PubMed] [Google Scholar]
- Chang S., Johnston R. J., Frøkjær-Jensen C., Lockery S., Hobert O., 2004. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430: 785–789. [DOI] [PubMed] [Google Scholar]
- Chase K., Lawler D. F., Adler F. R., Ostrander E. A., Lark K. G., 2004. Bilaterally asymmetric effects of quantitative trait loci (QTLs): QTLs that affect laxity in the right vs. left coxofemoral (hip) joints of the dog (Canis familiaris). Am. J. Med. Genet. A 124: 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D., Ward S. J., Messaddeq N., Launay J., Maroteaux L., 1997. 5–HT2B receptor-mediated serotonin morphogenetic functions in mouse cranial neural crest and myocardiac cells. Development 124: 1745–1755. [DOI] [PubMed] [Google Scholar]
- Clarke G. M., Brand G. W., Whitten M. J., 1986. Fluctuating asymmetry: a technique for measuring developmental stress caused by inbreeding. Aust. J. Biol. Sci. 39: 145–154. [Google Scholar]
- Cooper W. J., Parsons K., McIntyre A., Kern B., McGee-Moore A., et al. , 2010. Bentho-pelagic divergence of cichlid feeding architecture was prodigious and consistent during multiple adaptive radiations within African rift-lakes. PLoS ONE 5: e9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongen S. V., 2006. Fluctuating asymmetry and developmental instability in evolutionary biology: past, present and future. J. Evol. Biol. 19: 1727–1743. [DOI] [PubMed] [Google Scholar]
- Fukumoto T., Kema I. P., Levin M., 2005. Serotonin signaling is a very early step in patterning of the left-right axis in chick and frog embryos. Curr. Biol. 15: 794–803. [DOI] [PubMed] [Google Scholar]
- Fuller R. W., 1992. The involvement of serotonin in regulation of pituitary-adrenocortical function. Front. Neuroendocrinol. 13: 250–270. [PubMed] [Google Scholar]
- Griffin J. N., Compagnucci C., Hu D., Fish J., Klein O., et al. , 2012. Fgf8 dosage determines midfacial integration and polarity within the nasal and optic capsules. Dev. Biol. 374: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J., Wilkens H., 2013. Albinism in phylogenetically and geographically distinct populations of Astyanax cavefish arises through the same loss-of-function Oca2 allele. Heredity 111: 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. B., 2012. The complex origin of Astyanax cavefish. BMC Evol. Biol. 12: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. B., Hanken J., 2008. Review of fate-mapping studies of osteogenic cranial neural crest in vertebrates. Dev. Biol. 317: 389–400. [DOI] [PubMed] [Google Scholar]
- Gross J. B., Protas M., Conrad M., Scheid P. E., Vidal O., et al. , 2008. Synteny and candidate gene prediction using an anchored linkage map of Astyanax mexicanus. Proc. Natl. Acad. Sci. USA 105: 20106–20111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. B., Furterer A., Carlson B. M., Stahl B. A., 2013. An integrated transcriptome-wide analysis of cave and surface dwelling Astyanax mexicanus. PLoS ONE 8: e55659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley C. S., Knott S. A., 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324. [DOI] [PubMed] [Google Scholar]
- He L., Hannon G. J., 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5: 522–531. [DOI] [PubMed] [Google Scholar]
- Hinaux H., Pottin K., Chalhoub H., Père S., Elipot Y., et al. , 2011. A developmental staging table for Astyanax mexicanus surface fish and Pachón cavefish. Zebrafish 8: 155–165. [DOI] [PubMed] [Google Scholar]
- Hochwender C. G., Fritz R. S., 1999. Fluctuating asymmetry in a Salix hybrid system: the importance of genetic vs. environmental causes. Evolution 53: 408–416. [DOI] [PubMed] [Google Scholar]
- Hori M., 1993. Frequency-dependent natural selection in the handedness of scale-eating cichlid fish. Science 260: 216–219. [DOI] [PubMed] [Google Scholar]
- Hori M., Ochi H., Kohda M., 2007. Inheritance pattern of lateral dimorphism in two cichlids (a scale eater, Perissodus microlepis, and an herbivore, Neolamprologus moorii) in Lake Tanganyika. Zoolog. Sci. 24: 486–492. [DOI] [PubMed] [Google Scholar]
- Jeffery W. R., 2009. Regressive evolution in Astyanax cavefish. Annu. Rev. Genet. 43: 25–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jheon A., Schneider R., 2009. The cells that fill the bill: neural crest and the evolution of craniofacial development. J. Dent. Res. 88: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavalco K. F., Almeida-Toledo L. F., 2007. Molecular cytogenetics of blind Mexican tetra and comments on the karyotypic characteristics of genus Astyanax (Teleostei, Characidae). Zebrafish 4: 103–111. [DOI] [PubMed] [Google Scholar]
- Kearsey M., Hyne V., 1994. QTL analysis: a simple ‘marker-regression’ approach. Theor. Appl. Genet. 89: 698–702. [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ullmann B., Walker C., Wilson C., Currey M., et al. , 2005. Evolution and development of facial bone morphology in threespine sticklebacks. Proc. Natl. Acad. Sci. USA 102: 5791–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Cresko W. A., Phillips P. C., Ullmann B., Currey M., et al. , 2012a Independent axes of genetic variation and parallel evolutionary divergence of opercle bone shape in threespine stickleback. Evolution 66: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Hohenlohe P. A., Ullmann B., Currey M., Cresko W. A., 2012b Developmental dissociation in morphological evolution of the stickleback opercle. Evol. Dev. 14: 326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus S., Mendoza J. C., Liew J. H., Plath M., Meier R., et al. , 2013. Rapid evolution of troglomorphic characters suggests selection rather than neutral mutation as a driver of eye reduction in cave crabs. Biol. Lett. 9: 20121098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg C. P., McIntyre G. S., 1998. Geometric morphometrics of developmental instability: analyzing patterns of fluctuating asymmetry with Procrustes methods. Evolution 52: 1363–1375. [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol İ., Connors J., Gascoyne R. et al, 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy L., 1993. Morphological integration of fluctuating asymmetry in the mouse mandible. Genetica 89: 139–153. [Google Scholar]
- Leamy L. J., Routman E. J., Cheverud J. M., 1997. A search for quantitative trait loci affecting asymmetry of mandibular characters in mice. Evolution 51: 957–969. [DOI] [PubMed] [Google Scholar]
- Leamy L. J., Routman E. J., Cheverud J. M., 1998. Quantitative trait loci for fluctuating asymmetry of discrete skeletal characters in mice. Heredity 80: 509–518. [DOI] [PubMed] [Google Scholar]
- Lekander B., 1949. The sensory line system and the canal bones in the head of some Ostariophysi. Acta Zool. 30: 1–131. [Google Scholar]
- Levin M., Buznikov G. A., Lauder J. M., 2006. Of minds and embryos: left-right asymmetry and the serotonergic controls of pre-neural morphogenesis. Dev. Neurosci. 28: 171–185. [DOI] [PubMed] [Google Scholar]
- Mallarino R., Grant P. R., Grant B. R., Herrel A., Kuo W. P., et al. , 2011. Two developmental modules establish 3D beak-shape variation in Darwin’s finches. Proc. Natl. Acad. Sci. USA 108: 4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning J. T., Chamberlain A. T., 1993. Fluctuating asymmetry, sexual selection and canine teeth in primates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 251: 83–87. [DOI] [PubMed] [Google Scholar]
- Martínez O., Curnow R. N., 1992. Estimating the locations and the sizes of the effects of quantitative trait loci using flanking markers. Theor. Appl. Genet. 85: 480–488. [DOI] [PubMed] [Google Scholar]
- Mitchell R. W., Russell W. H., Elliott W. R., 1977. Mexican Eyeless Characin Fishes, Genus Astyanax: Environment, Distribution, and Evolution. Texas Tech Press, Lubbock, TX. [Google Scholar]
- Møller A. P., 1990. Fluctuating asymmetry in male sexual ornaments may reliably reveal male quality. Anim. Behav. 40: 1185–1187. [Google Scholar]
- Monteiro R., van Dinther M., Bakkers J., Wilkinson R., Patient R., et al. , 2008. Two novel type II receptors mediate BMP signalling and are required to establish left-right asymmetry in zebrafish. Dev. Biol. 315: 55–71. [DOI] [PubMed] [Google Scholar]
- Montgomery J. C., Coombs S., Baker C. F., 2001. The mechanosensory lateral line system of the hypogean form of Astyanax fasciatus. Environ. Biol. Fishes 62: 87–96. [Google Scholar]
- Moore R. Y., Klein D. C., 1974. Visual pathways and the central neural control of a circadian rhythm in pineal serotonin N-acetyltransferase activity. Brain Res. 71: 17–33. [DOI] [PubMed] [Google Scholar]
- Morgan T. H., 1923. The development of asymmetry in the fiddler crab. Am. Nat. 57: 269–273. [Google Scholar]
- Northcutt R. G., Gans C., 1983. The genesis of neural crest and epidermal placodes: a reinterpretation of vertebrate origins. Q. Rev. Biol. 58: 1–28. [DOI] [PubMed] [Google Scholar]
- O’Quin K. E., Yoshizawa M., Doshi P., Jeffery W. R., 2013. Quantitative genetic analysis of retinal degeneration in the blind cavefish Astyanax mexicanus. PLoS ONE 8: e57281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, A. R., 1994 Fluctuating asymmetry analyses: a primer, pp. 335–364 in Developmental Instability: Its Origins and Evolutionary Implications. Kluwer Academic, Dordrecht, Netherlands. [Google Scholar]
- Parnell N. F., Hulsey C. D., Streelman J. T., 2012. The genetic basis of a complex functional system. Evolution 66: 3352–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouilly M., Miranda G., 2003. Morphology and reproduction of the cavefish Trichomycterus chaberti and the related epigean Trichomycterus cf. barbouri. J. Fish Biol. 63: 490–505. [Google Scholar]
- Protas M., Conrad M., Gross J. B., Tabin C., Borowsky R., 2007. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr. Biol. 17: 452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas M., Tabansky I., Conrad M., Gross J. B., Vidal O., et al. , 2008. Multi-trait evolution in a cave fish, Astyanax mexicanus. Evol. Dev. 10: 196–209. [DOI] [PubMed] [Google Scholar]
- Protas M. E., Hersey C., Kochanek D., Zhou Y., Wilkens H., et al. , 2006. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat. Genet. 38: 107–111. [DOI] [PubMed] [Google Scholar]
- Reno H. W., 1969. Cephalic lateral-line systems of the cyprinid genus Hybopsis. Copeia 4: 736–773. [Google Scholar]
- Rhodes G., Zebrowitz L. A., Clark A., Kalick S. M., Hightower A., et al. , 2001. Do facial averageness and symmetry signal health? Evol. Hum. Behav. 22: 31–46. [DOI] [PubMed] [Google Scholar]
- Şadoğlu P., 1957a Mendelian inheritance in the hybrids between the Mexican blind cave fishes and their overground ancestor. Verh. Dtsch. Zool. Ges. Graz. 1957: 432–439. [Google Scholar]
- Şadoğlu P., 1957b A Mendelian gene for albinism in natural cave fish. Cell. Mol. Life Sci. 13: 394. [Google Scholar]
- Schubert M., Holland L. Z., Panopoulou G. D., Lehrach H., Holland N. D., 2000. Characterization of amphioxus AmphiWnt8: insights into the evolution of patterning of the embryonic dorsoventral axis. Evol. Dev. 2: 85–92. [DOI] [PubMed] [Google Scholar]
- Smith K. K., 2006. Craniofacial development in marsupial mammals: developmental origins of evolutionary change. Dev. Dyn. 235: 1181–1193. [DOI] [PubMed] [Google Scholar]
- Stensiö, E. A., 1947 The Sensory Lines and Dermal Bones in the Cheek in Fishes and Amphibians. Almqvist & Wiksell, Stockholm. [Google Scholar]
- Stewart T. A., Albertson R. C., 2010. Evolution of a unique predatory feeding apparatus: functional anatomy, development and a genetic locus for jaw laterality in Lake Tanganyika scale-eating cichlids. BMC Biol. 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker U., Bernatchez L., Wilkens H., 2003. Genetic divergence between cave and surface populations of Astyanax in Mexico (Characidae, Teleostei). Mol. Ecol. 12: 699–710. [DOI] [PubMed] [Google Scholar]
- Streelman J., Peichel C. L., Parichy D., 2007. Developmental genetics of adaptation in fishes: the case for novelty. Annu. Rev. Ecol. Evol. Syst. 38: 655–681. [Google Scholar]
- Streelman J. T., Albertson R. C., 2006. Evolution of novelty in the cichlid dentition. J. Exp. Zool. B Mol. Dev. Evol. 306: 216–226. [DOI] [PubMed] [Google Scholar]
- Terai Y., Morikawa N., Okada N., 2002. The evolution of the pro-domain of bone morphogenetic protein 4 (Bmp4) in an explosively speciated lineage of East African cichlid fishes. Mol. Biol. Evol. 19: 1628–1632. [DOI] [PubMed] [Google Scholar]
- Trainor, P. A., K. R. Melton, and M. Manzanares, 2003 Origins and plasticity of neural crest cells and their roles in jaw and craniofacial evolution. Int. J. Dev. Biol. 47: 541–553. [PubMed]
- Watson P. J., Thornhill R., 1994. Fluctuating asymmetry and sexual selection. Trends Ecol. Evol. 9: 21–25. [DOI] [PubMed] [Google Scholar]
- Wilkens H., 1971. Genetic interpretation of regressive evolutionary processes: studies on hybrid eyes of two Astyanax populations (Characidae, Pisces). Evolution 25: 530–544. [DOI] [PubMed] [Google Scholar]
- Wilkens H., 1988. Evolution and genetics of epigean and cave Astyanax fasciatus (Characidae, Pisces): support for the neutral mutation theory, pp. 271–367 in Evolutionary Biology, edited by Hecht M. K., Wallace B. Plenum, New York. [Google Scholar]
- Wilkens H., 2001. Convergent adaptations to cave life in the Rhamdia laticauda catfish group (Pimelodidae, Teleostei). Environ. Biol. Fishes 62: 251–261. [Google Scholar]
- Wilkens H., 2010. Genes, modules and the evolution of cave fish. Heredity 105: 413–422. [DOI] [PubMed] [Google Scholar]
- Xu S., 2010. An expectation–maximization algorithm for the Lasso estimation of quantitative trait locus effects. Heredity 105: 483–494. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Espinasa L., Stock D. W., Jeffery W. R., 2003. Development and evolution of craniofacial patterning is mediated by eye-dependent and -independent processes in the cavefish Astyanax. Evol. Dev. 5: 435–446. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Byerly M. S., Jackman W. R., Jeffery W. R., 2009. Pleiotropic functions of embryonic sonic hedgehog expression link jaw and taste bud amplification with eye loss during cavefish evolution. Dev. Biol. 330: 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R., Yokoyama S., 1990. Isolation, DNA sequence and evolution of a color visual pigment gene of the blind cave fish Astyanax fasciatus. Vision Res. 30: 807–816. [DOI] [PubMed] [Google Scholar]
- Yoshizawa M., Goricki S., Soares D., Jeffery W. R., 2010. Evolution of a behavioral shift mediated by superficial neuromasts helps cavefish find food in darkness. Curr. Biol. 20: 1631–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M., Yamamoto Y., O’Quin K. E., Jeffery W. R., 2012. Evolution of an adaptive behavior and its sensory receptors promotes eye regression in blind cavefish. BMC Biol. 10: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.