In cold- or dehydration-stressed rice plants, up-regulation of genes related to starch degradation, sucrose metabolism, and the glyoxylate cycle results in the accumulation of sugars and with, abscisic acid signaling is inversely related to cytokinin signaling.
Abstract
Correlations between gene expression and metabolite/phytohormone levels under abiotic stress conditions have been reported for Arabidopsis (Arabidopsis thaliana). However, little is known about these correlations in rice (Oryza sativa ‘Nipponbare’), despite its importance as a model monocot. We performed an integrated analysis to clarify the relationships among cold- and dehydration-responsive metabolites, phytohormones, and gene transcription in rice. An integrated analysis of metabolites and gene expression indicated that several genes encoding enzymes involved in starch degradation, sucrose metabolism, and the glyoxylate cycle are up-regulated in rice plants exposed to cold or dehydration and that these changes are correlated with the accumulation of glucose (Glc), fructose, and sucrose. In particular, high expression levels of genes encoding isocitrate lyase and malate synthase in the glyoxylate cycle correlate with increased Glc levels in rice, but not in Arabidopsis, under dehydration conditions, indicating that the regulation of the glyoxylate cycle may be involved in Glc accumulation under dehydration conditions in rice but not Arabidopsis. An integrated analysis of phytohormones and gene transcripts revealed an inverse relationship between abscisic acid (ABA) signaling and cytokinin (CK) signaling under cold and dehydration stresses; these stresses increase ABA signaling and decrease CK signaling. High levels of Oryza sativa 9-cis-epoxycarotenoid dioxygenase transcripts correlate with ABA accumulation, and low levels of Cytochrome P450 (CYP) 735A transcripts correlate with decreased levels of a CK precursor in rice. This reduced expression of CYP735As occurs in rice but not Arabidopsis. Therefore, transcriptional regulation of CYP735As might be involved in regulating CK levels under cold and dehydration conditions in rice but not Arabidopsis.
Land plants must mount suitable responses to overcome the adverse effects of water stress caused by either drought or low-temperature conditions (Levitt, 1980). Among the external stresses, water stress is one of the most important limitations of crop productivity (Bartels and Sunkar, 2005). Discoveries of useful genes for molecular breeding using metabolomics and transcriptomics promise to facilitate the improvement of crop yields under water stress conditions. It is important to identify plant metabolites and transcripts that respond to water stress to determine the essential steps in molecular processes related to the effective adaptation of plants to stress conditions. Metabolomic and transcriptomic data have provided much information on the metabolite, phytohormone, and transcript networks that control plant growth and development. Studies using mass spectrometry (MS) -based metabolomic techniques have identified and characterized many stress-responsive metabolites (Guy et al., 2008; Saito and Matsuda, 2010; Urano et al., 2010; Obata and Fernie, 2012). Targeted analyses of phytohormones have been used to characterize signaling and metabolic networks (Müller et al., 2002; Chiwocha et al., 2003; Kojima et al., 2009). For example, abscisic acid (ABA) plays an essential role in the response to water stress (Yamaguchi-Shinozaki and Shinozaki, 2006; Hirayama and Shinozaki, 2007; Cutler et al., 2010), and an increase in endogenous ABA is required for certain water stress responses designated as ABA-dependent stress responses (Raghavendra et al., 2010; Weiner et al., 2010; Fujita et al., 2011). Cytokinins (CKs) play major roles in several developmental and physiological processes in plants (for example, in cell division, regulation of shoot growth, and stress responses; Mok et al., 2000; Sakakibara, 2006; Ha et al., 2012). Microarray-based transcriptomic analyses have identified many genes involved in the responses and tolerance to various stresses in several plant species. These genes encode metabolic enzymes, late embryogenesis-abundant (LEA) proteins, detoxification enzymes, chaperones, protein kinases, transcription factors, and other gene products (Kilian et al., 2007; Shinozaki and Yamaguchi-Shinozaki, 2007; Cutler et al., 2010; Maruyama et al., 2012).
Metabolite profiles have been reported for Arabidopsis (Arabidopsis thaliana) under dehydration conditions. The levels of raffinose family oligosaccharides, Pro, branched-chain amino acids (BCAAs), γ-aminobutyrate, saccharopine, and agmatine were reported to be significantly higher in dehydration-treated plants than in untreated plants (Urano et al., 2009; Skirycz et al.., 2010; Verslues and Juenger, 2011). Increased levels of several metabolites under dehydration conditions were correlated with the expression levels of their respective biosynthetic genes, many of which are regulated by endogenous ABA (Urano et al., 2009). Metabolite profiles have also been determined for crops exposed to dehydration conditions. The dehydration-induced changes in metabolites observed in Arabidopsis plants were similar to those reported for wheat (Triticum aestivum; Bowne et al., 2012), legumes (Sanchez et al., 2012), and maize (Zea mays; Sicher and Barnaby, 2012). Several research groups have profiled changes in metabolite levels in Arabidopsis after exposure to low temperatures. Metabolite analysis of natural accessions of Arabidopsis after cold acclimation revealed a clear correlation between the accumulation of several carbohydrate metabolites (e.g. Glc, Fru, and Suc) and cold stress tolerance (Cook et al., 2004; Hannah et al., 2006). Gray and Heath (2005) used Fourier transform ion cyclotron MS and a nontargeted metabolic fingerprinting approach to study the effects of cold acclimation on the metabolome. Gray and Heath (2005) found that a global reprogramming of metabolism occurs as a result of cold acclimation. An integrated analysis using transgenic Arabidopsis plants overexpressing DRE-binding protein 1A/C-Repeat binding factor3, which encodes a transcription factor that functions in cold stress responses, revealed that transcriptional regulation of the carbohydrate network is necessary for the accumulation of specific carbohydrates (e.g. Suc, galactinol, myoinositol, and raffinose). The accumulation of these carbohydrates may be important to improve tolerance to freezing stress in these transgenic plants (Cook et al., 2004; Maruyama et al., 2009).
Rice (Oryza sativa ‘Nipponbare’) is important not only as a major crop but also as a model monocot. In this study, we performed an integrated analysis of the metabolites, phytohormones, and gene transcripts in rice plants subjected to cold or dehydration treatments. Our aim was to comprehensively survey the molecular responses of rice to cold or dehydration stimuli. We used three types of MS systems: gas chromatography (GC) coupled with time-of-flight MS (GC-TOF-MS), capillary electrophoresis coupled with MS (CE-MS), and liquid chromatography coupled with MS (LC-MS). We identified and characterized representative cold- and dehydration-responsive metabolites and phytohormones. We also performed a transcriptome analysis using a rice oligonucleotide microarray. We analyzed metabolite-gene and phytohormone-gene correlations and identified several genes encoding metabolic enzymes that might play key roles in the responses of rice plants to cold or dehydration. We compared the roles of the identified rice genes with the roles of their counterparts in Arabidopsis under cold or dehydration conditions.
RESULTS
Metabolite Profiles of Rice Plants Exposed to Cold or Dehydration
We used GC-TOF-MS and CE-MS to determine the effects of seven environmental conditions on whole rice seedlings grown in soil for 2 weeks. The treatments were as follows: 1- or 2-d exposure to cold at 10°C, 2- or 3-d exposure to dehydration without watering, and no stress treatment (untreated for 1, 2, or 3 d). The GC-TOF-MS and CE-MS analyses identified 152 different metabolites according to their retention time indices and specific mass fragments (Supplemental Table S1). Relative to the levels in untreated plants, the levels of 38, 52, 24, and 91 metabolites were significantly increased in 1-d cold-treated plants, 2-d cold-treated plants, 2-d dehydration-treated plants, and 3-d dehydration-treated plants, respectively (Benjamini and Hochberg false discovery rate [FDR]: P < 0.05); the levels of 26, 12, 26, and eight metabolites, respectively, were significantly decreased in plants exposed to the same four stress conditions (FDR: P < 0.05).
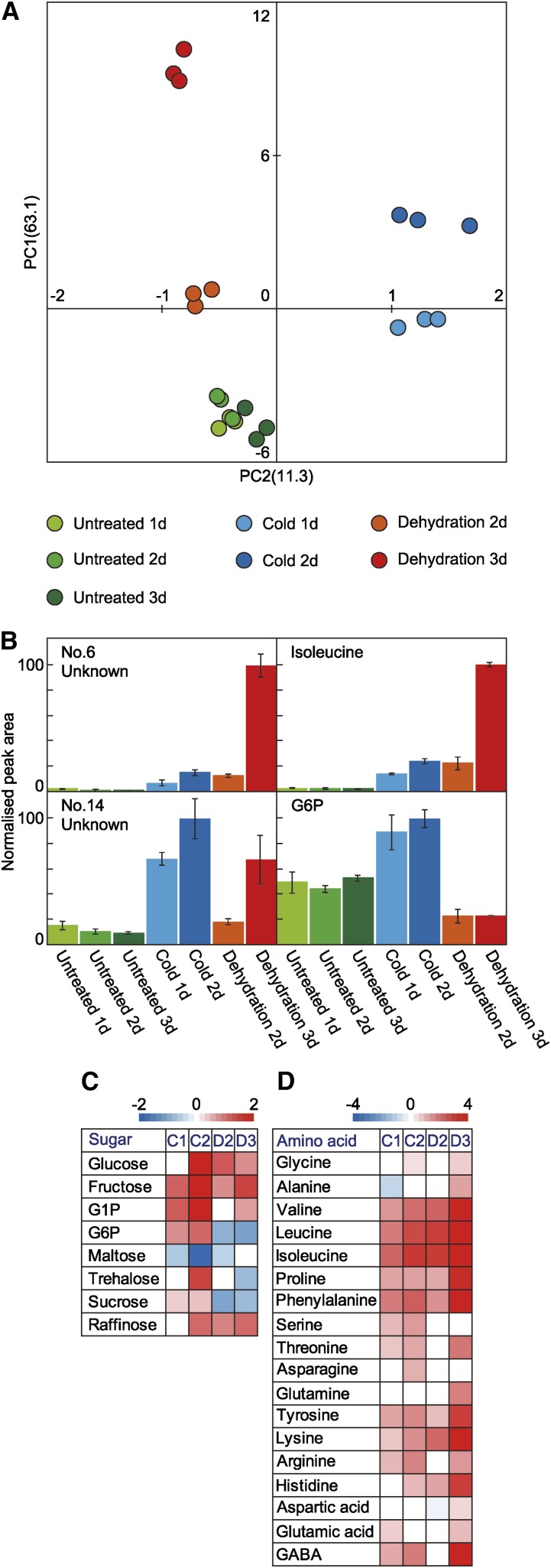
We used principal component analysis (PCA) to compare the metabolite profiles of plants subjected to seven treatments (Fig. 1A; Supplemental Tables S1–S5). The cumulative contribution ratio of the PCA was 74.4% up to the second principal component (PC2). The PCA analysis had two key features. The first principal component (PC1) reflected increases in the levels of metabolites in the rice plants subjected to seven treatments. The plants subjected to the 3-d dehydration treatment showed the highest PC1 value. Plants exposed to the 2-d cold treatment showed the second highest PC1 value. The PC1 values for the three classes of untreated plants were similar, and those values were the lowest among seven treatments. PC2 reflected differences in the variety of metabolites in plants among seven treatments. The PC2 values were positive for cold-treated plants and negative for dehydration-treated plants. The PC2 values for untreated plants were nearly zero. The results of the PCA indicated that the metabolite profiles of plants exposed to seven treatments were classified into three groups according to plant growth conditions: cold, dehydration, and untreated.
Figure 1.
Statistical analysis of 152 metabolites profiled in untreated rice plants and rice plants subjected to cold or dehydration treatments. Levels of 152 metabolites were measured in rice plants subjected to seven treatments: cold (10°C) for 1 d (Cold 1d) or 2 d (Cold 2d); dehydration (water withheld) for 2 d (Dehydration 2d) or 3 d (Dehydration 3d); and untreated for 1, 2, or 3 d (Untreated 1d, 2d, and 3d, respectively). A, PCA for metabolites. Values for y and x axes are values for PC1 and PC2, respectively. B, Representative metabolites showing the highest and second highest eigenvector values. In each case, maximum level of metabolite was set to 100. Values are means (n = 3 experiments); error bars show sd. C and D, Relative levels of representative sugars (C) and amino acids after cold or dehydration treatments. C1, Cold 1d; C2, cold 2d; D2, dehydration 2d; D3, dehydration, 3d; GABA, γ-aminobutyric acid; G1P, glucose 1-phosphate.
We selected representative metabolites showing the highest and second highest eigenvector values. Eigenvector values correspond to the coefficients of each PC. The levels of these metabolites are displayed as bar charts (Fig. 1B). The highest PC1 eigenvector value was for metabolite number 6 (unknown). The levels of metabolite number 6 were significantly higher in both cold-treated and dehydration-treated plants than in untreated plants. Ile had the second highest PC1 eigenvector value. The Ile levels were also significantly higher in both cold-treated and dehydration-treated plants than in untreated plants. The plants subjected to 3-d dehydration treatment showed the highest levels of metabolite number 6 and Ile. Metabolite number 14 (unknown) had the highest PC2 eigenvector value. The levels of metabolite number 14 were significantly higher in both cold-treated and dehydration-treated plants than in untreated plants. Glc-6-P (G6P) had the second highest PC2 eigenvector value. Cold-treated plants contained significantly higher levels of G6P and dehydration-treated plants contained significantly lower levels of G6P compared with the levels in untreated plants. The plants subjected to 2-d cold treatment showed the highest levels of metabolite number 14 and G6P.
The levels of several sugars and amino acids were higher in cold-treated and dehydration-treated plants than in untreated plants. We analyzed the levels of all detected sugars and amino acids and used heat maps to illustrate the log ratio of each metabolite (Fig. 1, C and D). The levels of Glc, Fru, and raffinose were significantly higher in cold-treated and dehydration-treated plants than in untreated plants. Compared with their levels in untreated plants, the levels of G6P, trehalose, and Suc were significantly higher in cold-treated plants and significantly lower in dehydration-treated plants. The levels of BCAAs (Val, Leu, and Ile) and Pro were also significantly higher in cold-treated and dehydration-treated plants than in untreated plants. The highest levels of these amino acids were in plants subjected to 3-d dehydration treatment.
Phytohormone Profiles of Rice Plants Exposed to Cold or Dehydration
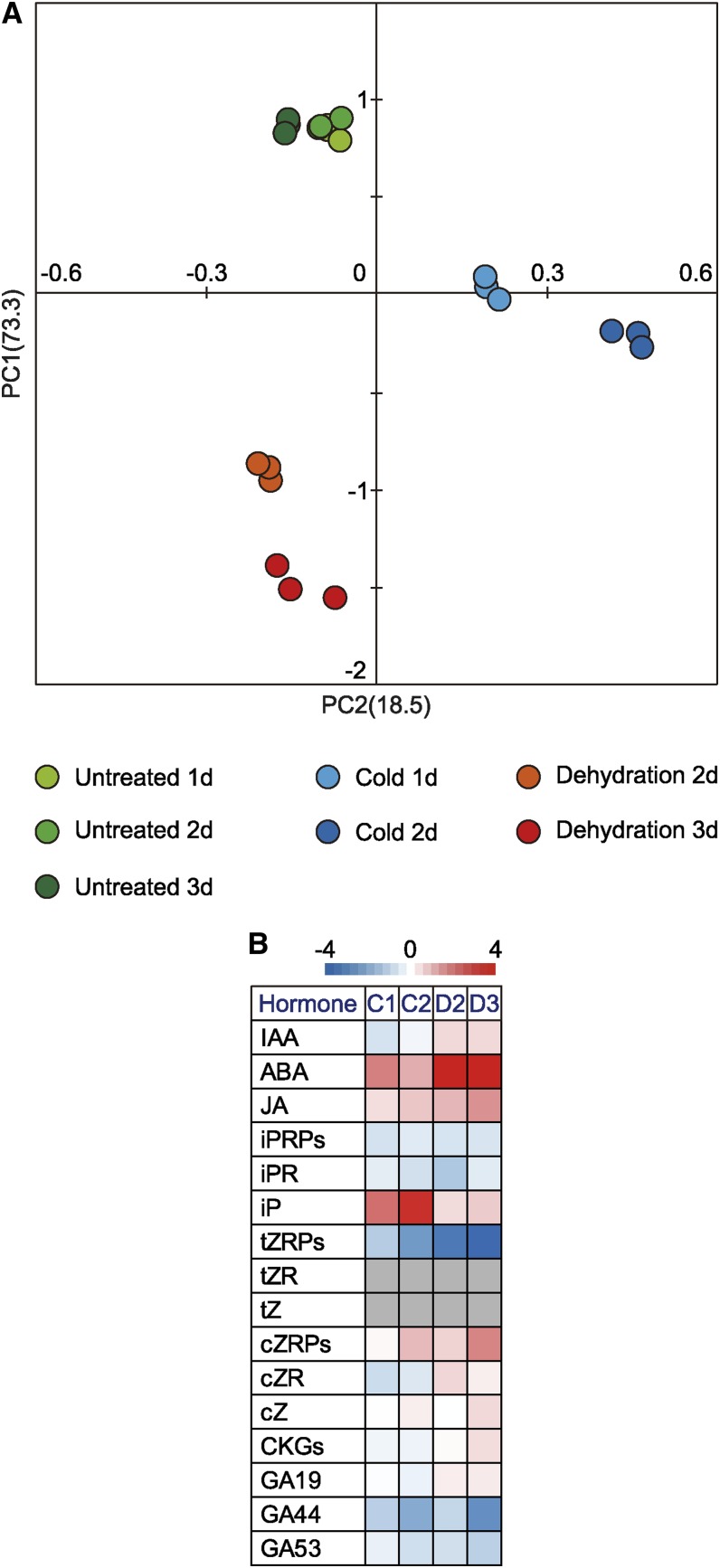
We used LC-MS to measure levels of phytohormones in plants subjected to seven different treatments and identified 14 phytohormones (Supplemental Table S6); PCA was used to compare the phytohormone profiles (Fig. 2A; Supplemental Tables S6–S10). The cumulative contribution ratio of the PCA was 91.8% up to PC2. Two key features emerged from the PCA. First, the PC1 value reflected decreases in the levels of phytohormones in plants subjected to seven different treatments. The lowest PC1 value was for plants subjected to 3-d dehydration treatment. Plants subjected to 2-d dehydration treatment showed the second lowest PC1 value. The PC1 values for the three classes of untreated plants were similar, and those values were the highest among all treatments. Second, the PC2 value reflected differences in the variety of phytohormones in plants under seven different environmental conditions. The PC2 values were positive for cold-treated plants and negative for dehydration-treated plants. The results of the PCA indicated that the phytohormone profiles of plants subjected to seven different treatments were classified into three groups according to plant growth conditions: cold, dehydration, and untreated.
Figure 2.
Statistical analysis of changes in profiles of rice phytohormone levels after exposure to cold or dehydration. A, PCA for phytohormones. Values for y and x axes are values for PC1 and PC2, respectively. B, Relative levels of representative phytohormones after cold or dehydration stress. A small gray square indicates that the phytohormone could not be detected. CKGs, Cytokinin glucosides; cZ, cis-zeatin; cZR, cis-zeatin riboside, cZRPs, cis-zeatin 5′-Ps; GA19, gibberellin A19; GA44, gibberellin A44; GA53, gibberellin A53; IAA, indoleacetic acid; iPR, isopentenyladenosine; iPRPs, N6-(Δ2-isopentenyl) adenine ribotides; JA, jasmonic acid; tZR, trans-zeatin riboside.
Next, we focused on representative phytohormones with the highest eigenvector values (Fig. 2B). The highest PC1 eigenvector value was for trans-zeatin riboside-5′-phosphatess (tZRPs). The level of tZRPs was significantly lower in both cold-treated and dehydration-treated plants than in untreated plants. The lowest level of tZRPs was in plants subjected to 3-d dehydration treatment. Isopentenyladenine (iP) had the highest PC2 eigenvector value. Compared with the level in untreated plants, the level of iP was significantly higher in cold-treated plants but only slightly higher in dehydration-treated plants. The highest level of iP was in plants subjected to 2-d cold treatment. ABA had the lowest PC2 eigenvector value; the ABA level was significantly higher in cold-treated and dehydration-treated plants than in untreated plants. The highest level of ABA was in plants exposed to the 3-d dehydration treatment.
Next, we compared rice plants with Arabidopsis in terms of the changes in phytohormone levels under cold or dehydration stress. In Arabidopsis, the ABA level in cold- and dehydration-treated plants was significantly higher than the level in untreated plants. Conversely, exposure of Arabidopsis plants to either cold or dehydration significantly decreased the levels of CKs, including iP and trans-zeatin (tZ). In Arabidopsis, the lowest levels of iP and tZ were in plants exposed to the dehydration treatment (Supplemental Fig. S1; Supplemental Table S11).
Transcriptomic Profiles of Rice Plants Exposed to Cold or Dehydration
Previously, we used oligonucleotide microarrays to identify cold- and dehydration-responsive genes in rice plants exposed to 1-d cold and 3-d dehydration treatments (Maruyama et al., 2012). The International Rice Genome Sequencing Project recently updated its rice genome sequence information, and the annotations for many rice genes have been revised in the rice annotation project database (http://rapdb.dna.affrc.go.jp/; Supplemental Fig. S2A). Given the update in the number of rice genes, we prepared a new, to our knowledge, oligonucleotide microarray for transcriptomic analyses. This array can detect 43,175 genes (Supplemental Fig. S2B). We used the microarray to identify additional cold- and dehydration-responsive genes in rice plants exposed to a 1- or 2-d cold treatment or a 2- or 3-d dehydration treatment. Overall, 3,576 (1-d cold treatment), 4,395 (2-d cold treatment), 2,089 (2-d dehydration treatment), and 5,927 (3-d dehydration treatment) genes were significantly up-regulated (FDR, P < 0.05; fold change, >2), and 3,147 (1-d cold treatment), 4,320 (2-d cold treatment), 1,678 (2-d dehydration treatment), and 6,184 (3-d dehydration treatment) genes were significantly down-regulated (FDR: P < 0.05; fold change: <0.5; Supplemental Tables S12–S15).
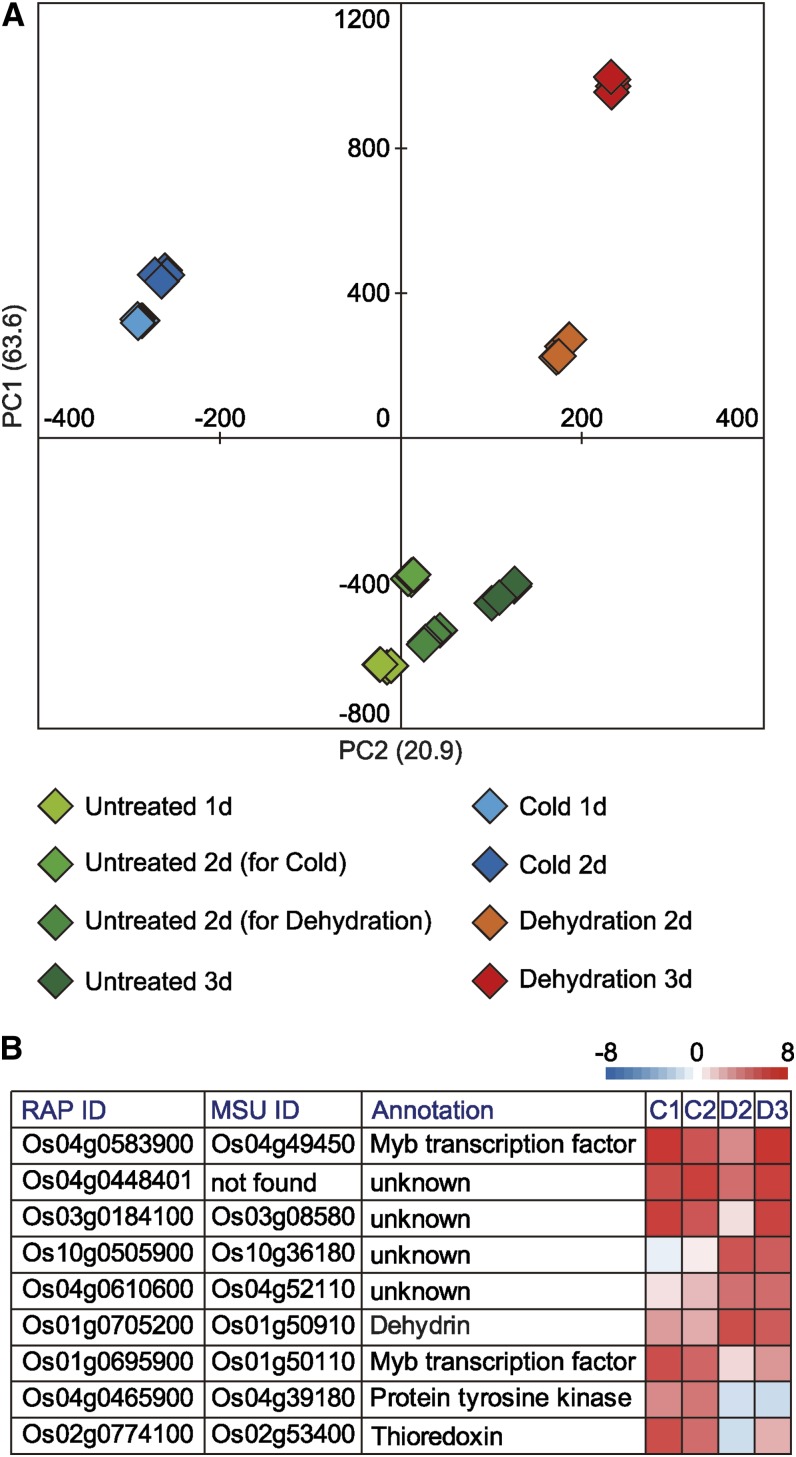
We used PCA to compare the transcript profiles among plants subjected to seven different treatments (Fig. 3A; Supplemental Tables S16–S19). Two features emerged from the PCA. First, the PC1 values reflected increased levels of transcripts in plants subjected to seven different treatments. The plants subjected to 3-d dehydration treatment showed the highest PC1 value. The PC1 values for the four classes of untreated plants were similar and represented the lowest values. Second, the PC2 value reflected differences in the types of transcripts that accumulated under the different treatments. The PC2 values were negative for cold-treated plants and positive for dehydration-treated plants. The PC2 values for untreated plants were nearly zero. The results of the PCA indicated that the transcript profiles of plants subjected to seven different treatments were classified into three groups according to plant growth conditions: cold, dehydration, and untreated.
Figure 3.
Statistical analysis of changes in the profiles of rice transcript levels after exposure to cold or dehydration. A, PCA for transcript data obtained from oligonucleotide microarrays. Values for y and x axes are values for PC1 and PC2, respectively. B, Representative cold- and dehydration-responsive genes identified using oligonucleotide microarrays. Genes shown are those genes with the first, second, and third highest eigenvector values. Heat maps illustrate transcript levels of representative cold- and dehydration-responsive genes. MSU, Michigan State University; RAP, The Rice Annotation Project.
We selected representative cold- or dehydration-responsive genes with the first, second, and third highest eigenvector values and displayed their transcript levels as heat maps (Fig. 3B). In the transcriptome profile, the highest PC1 eigenvector value was for Os04g0583900, which encodes an Myb-type transcription factor. Plants exposed to either cold or dehydration showed significantly increased Os04g0583900 transcript levels compared with the levels in untreated plants. The highest transcript level of this gene was in plants subjected to the 3-d dehydration treatment (Fig. 3B). The Os04g0448401 and Os03g0184100 transcripts showed the second and third highest PC1 eigenvector values, respectively. The levels of the Os04g0448401 and Os03g0184100 transcripts were also significantly higher in both cold- and dehydration-treated plants than in untreated plants (Fig. 3B). The Os10g0505900, Os04g0610600, and OsLEA14/water stress-induced (WSI) 18 (Os01g0705200) transcripts had the first, second, and third highest PC2 eigenvector values, respectively. Compared with their levels in untreated plants, the levels of these transcripts were significantly higher in dehydration-treated plants, and the levels in dehydration-treated plants were higher than those levels in cold-treated plants (Fig. 3B). The OsMYB4 (Os01g0695900), Os04g0465900, and Os02g0774100 transcripts had the first, second, and third lowest PC2 eigenvector values, respectively. The levels of these transcripts were significantly higher in cold-treated plants than in untreated plants, and the levels in cold-treated plants were higher than those levels in dehydration-treated plants (Fig. 3B).
Molecular Functions of Cold-Responsive and Dehydration-Responsive Genes in Rice
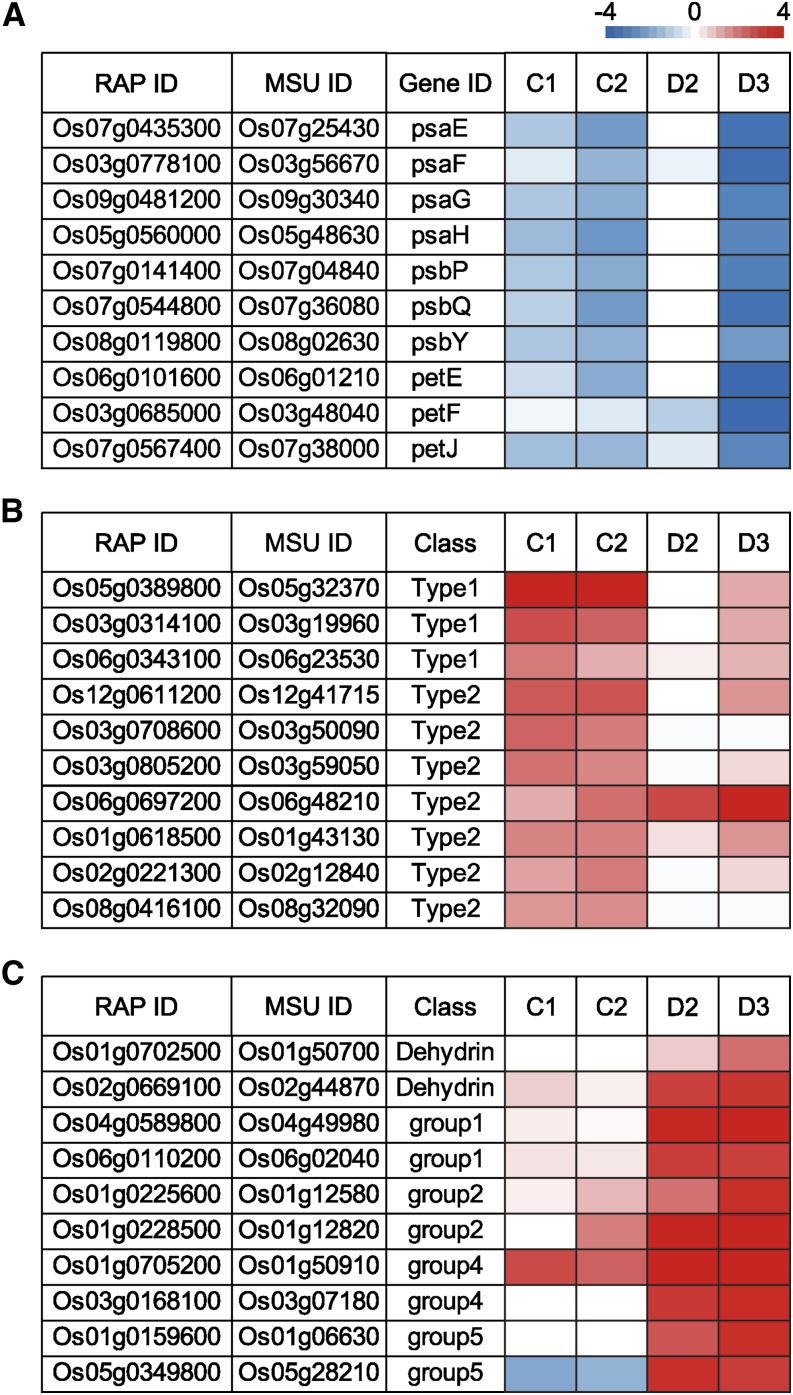
We used our in-house Gene Ontology database (Maruyama et al., 2012) to annotate the molecular functions of all identified cold- or dehydration-responsive genes (Supplemental Fig. S3). We selected the helicase gene family to represent genes up-regulated in plants subjected to cold treatments. Helicases are involved in RNA metabolism and may be important for increasing transcriptional and translational activities under cold stress conditions. The increase in the transcript abundance of helicase genes up-regulated under cold stress was 30.6% (1-d cold treatment) and 42.1% (2-d cold treatment). We selected the dehydrin/LEA gene family to represent genes up-regulated in plants subjected to a dehydration treatment. The increase in the transcript abundance of genes encoding dehydrin/LEA proteins up-regulated under dehydration stress was 26.0% (2-d dehydration treatment) and 32.9% (3-d dehydration treatment). We selected genes involved in photosynthesis to represent the genes down-regulated in both cold- and dehydration-treated plants. The decrease in the transcript abundance of photosynthesis-related genes down-regulated under 1-d cold treatment, 2-d cold treatment, and 3-d dehydration treatment was 31.6%, 44.7%, and 75.4%, respectively.
We focused on the cold- and dehydration-responsive genes in the three selected representative molecular classes (photosynthesis, helicases, and dehydrin/LEA proteins) and used heat maps to illustrate the log ratio of these cold- or dehydration-responsive genes (Fig. 4). Photosystem I subunit (psa) and photosystem II subunit (psb) encode subunits of PSI and PSII, respectively. psaE, psaF, psaG, psaH, psbP, psbQ, and psbY were significantly down-regulated in rice plants subjected to 1-d cold treatment, 2-d cold treatment, and 3-d dehydration treatment; photosystem and electron transport system E (plastocyanin), ferredoxin, and cytochrome c6 were also significantly down-regulated in rice plants subjected to these treatments. Compared with the expression levels in untreated plants, those levels in plants subjected to 2-d dehydration treatment were not significantly lower (Fig. 4A). Helicases are classified into three superfamilies: superfamily (SF) 1, SF2, and SF3 (Gorbalenya and Koonin, 1993). SF2 includes the DEAD/DEAH box helicases, which contain two conserved domains: DEADc (PF00270) and Helicase conserved C-terminal domains (PF00271). There are two types of DEAD/DEAH box helicases; only the type I protein has the helicase-associated domain 2 (PF04408). Representative members of DEAD/DEAH box helicases are shown in Figure 4B. The transcript levels of genes encoding several members of DEAD/DEAH box helicases were increased under cold stress. The levels of their corresponding transcripts in cold-treated plants were higher than those levels in dehydration-treated plants. The transcript level of Os06g0697200 was significantly higher in both cold- and dehydration-treated plants than in untreated plants and higher in dehydration- than cold-treated plants. Representative members of the dehydrin/LEA family are shown in Figure 4C. LEA proteins are classified into six groups in the Pfam database. Many LEA proteins were induced by dehydration. The transcript levels of genes encoding LEA proteins were higher in dehydration- than cold-treated plants. For example, whereas the transcript level of Oryza sativa early methionine (Os05g0349800) was significantly higher in dehydration-treated plants than in untreated plants, the level of this transcript was lower in cold-treated plants than in untreated plants. The transcript level of Os01g0705200 was significantly higher in both cold- and dehydration-treated plants than in untreated plants and higher in dehydration- than cold-treated plants.
Figure 4.
Molecular functions of cold-responsive and dehydration-responsive genes in rice plants. Heat maps of up-regulated (red) or down-regulated (blue) molecular functional classes indicate relative abundance (%; shading in boxes) in cold-treated (C1 and C2) and dehydration-treated (D2 and D3) plants. Heat maps illustrate the log ratios of cold- or dehydration-responsive genes in representative molecular classes of photosynthesis (A), helicases (B), and dehydrin/LEA proteins (C). psaE, PSI subunit IV; psaF, PSI subunit III; psaG, PSI subunit V; psaH, PSI subunit VI; psbP, PSII oxygen-evolving enhancer protein 2; psbQ, PSII oxygen-evolving enhancer protein 3; psbY, PSII PsbY protein.
Expressions of Genes Involved in Carbohydrate and Amino Acid Metabolism
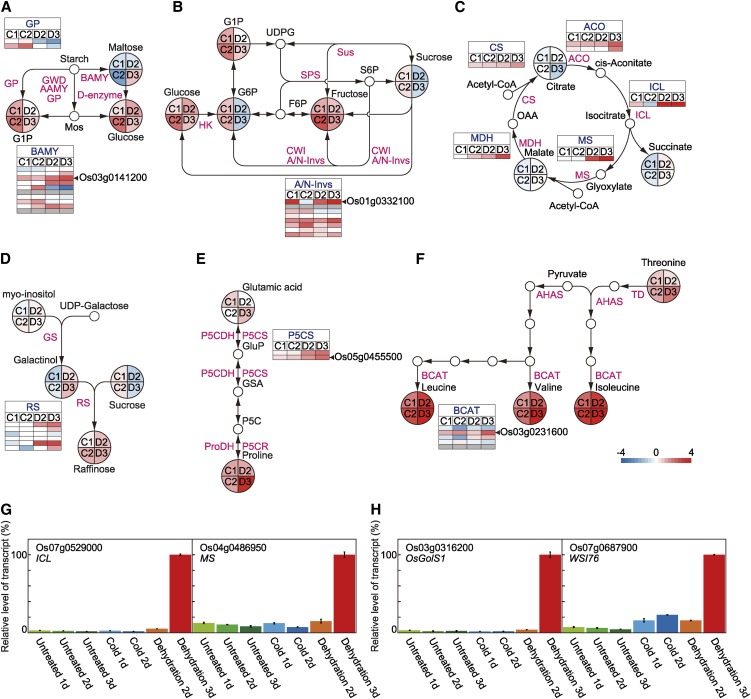
Our metabolome analyses revealed that the levels of monosaccharides were significantly higher in both cold- and dehydration-treated rice plants than in untreated plants (Fig. 1C). To predict candidate genes important for carbohydrate metabolism under cold or dehydration conditions, we screened cold- and dehydration-responsive genes for those encoding enzymes involved with starch degradation, Suc metabolism, gluconeogenesis, and the glyoxylate cycle (Fig. 5, A–D; Supplemental Figs. S4–S6). The abundances of several of the transcripts encoding starch degradation enzymes (e.g. α-amylase and β-amylase) were elevated in rice plants after exposure to cold or dehydration (Fig. 5A; Supplemental Fig. S4). In particular, the levels of the β-amylase (Os03g0141200) transcript were significantly up-regulated under both cold and dehydration conditions (Fig. 5A; Supplemental Fig. S4). The transcript level of the gene encoding α-glucanphosphorylase was increased under cold conditions but decreased under dehydration conditions compared with the transcript level in untreated plants (Fig. 5A; Supplemental Fig. S4). Of the genes associated with Suc metabolism, several encoding alkaline/neutral invertases showed increased transcript levels under both cold and dehydration conditions (Fig. 5B; Supplemental Fig. S5). Among the genes encoding alkaline/neutral invertases, the Os01g0332100 transcript was present at 10-fold higher levels in both cold-treated and dehydration-treated rice plants than in untreated plants (Fig. 5B; Supplemental Fig. S5). Gluconeogenesis is an important pathway for the synthesis of Glc from organic acids, and the glyoxylate cycle is an important source of the four-carbon compound succinate, which can enter the gluconeogenesis pathway. Phosphoenolpyruvate carboxykinase and Fru-1,6-bisphosphatase play key roles in gluconeogenesis (Rylott et al., 2003). Expressions of the genes encoding phosphoenolpyruvate carboxykinase and Fru-1,6-bisphosphatase did not increase markedly in either cold-treated or dehydration-treated rice plants (Supplemental Tables S12–S15). Conversely, the levels of transcripts encoding isocitrate lyase and malate synthase, both key enzymes in the glyoxylate cycle, increased significantly after dehydration (Fig. 5C; Supplemental Fig. S6). We confirmed the transcript levels of isocitrate lyase and malate synthase genes by quantitative real-time (qRT)-PCR. The highest levels of both transcripts were in plants subjected to the 3-d dehydration treatment (Fig. 5G). We also characterized the increases in raffinose, Pro, and BCAAs and screened cold- and/or dehydration-responsive genes for those genes involved in the biosynthesis of galactinol, raffinose, and BCAAs and metabolism of Pro (Fig. 5, D–F; Supplemental Figs. S7–S9). The levels of most raffinose synthase-related transcripts increased under dehydration conditions but decreased under cold conditions (Fig. 5D; Supplemental Fig. S7). The genes WSI76 (Os07g0687900) and Oryza sativa galactinol synthase1 (OsGolS1; Os03g0316200) encode galactinol synthases; however, probes corresponding to both of these genes were not included in our unique microarray slide. We analyzed the transcript levels of these genes using qRT-PCR. The WSI76 transcript level increased significantly under dehydration conditions and increased slightly under cold conditions. The OsGolS1 transcript level increased significantly under dehydration conditions but did not increase under cold conditions (Fig. 5H). The transcript level of OsP5CS (Os05g0455500), which encodes the Pro biosynthetic enzyme Δ1-pyrroline-5-carboxylate synthase, increased under dehydration conditions (Fig. 5E; Supplemental Fig. S8). Branched chain aminotransferase catalyzes the last step of BCAA biosynthesis. Expression of the Os03g0231600, a branched chain aminotransferase family gene, was induced by both cold and dehydration (Fig. 5F; Supplemental Fig. S9).
Figure 5.
Carbohydrate and amino acid metabolic pathways. A, Starch degradation. B, Suc metabolism. C, Glyoxylate cycle. D, Raffinose biosynthesis. E, Pro metabolism. F, BCAA biosynthesis. Heat maps illustrate transcript levels of representative cold- and dehydration-responsive genes. A small gray square indicates the absence of a probe for that gene in the oligonucleotide microarray. Heat maps illustrate accumulated levels of representative cold- and dehydration-responsive metabolites. G, Levels of transcripts for genes encoding isocitrate lyase and malate synthase determined by qRT-PCR. H, Transcript levels of OsGolS1 and WSI76 both encoding galactinol synthase. Mean values are shown (n = 3 experiments). Error bars indicate sd. AAMY, α-Amylase; ACO, aconitase hydratase; AHAS, acetolactate synthase; A/N-Invs, alkaline/neutral invertase; BAMY, β-amylase; BCAT, branched chain amino acid aminotransferase; CS, citrate synthase; CWI, apoplastic invertase; F6P, fructose 6-phosphate; GP, α-glucanphosphorylase; GSA, galactinol synthase A; GWD, glucan water dikinase; HK, hexokinase; ICL, isocitrate lyase; MDH, malate dehydrogenase; Mos, malto-oligosaccharides; MS, malate synthase; ProDH, Pro dehydrogenase; P5C, Δ1-pyrroline-5-carboxylate; P5CDH, Δ1-pyrroline-5-carboxylate dehydrogenase; P5CR, Δ1-pyrroline-5-carboxylate reductase; P5CS, Δ1-pyrroline-5-carboxylate synthetase; RS, raffinose synthase; SPS, Suc-P synthase; Sus, Suc synthase; S6P, sucrose-6-phosphate; TD, Thr dehydratase; UDPG, UDP-galactose.
Expression of Genes Involved in ABA and CK Metabolism
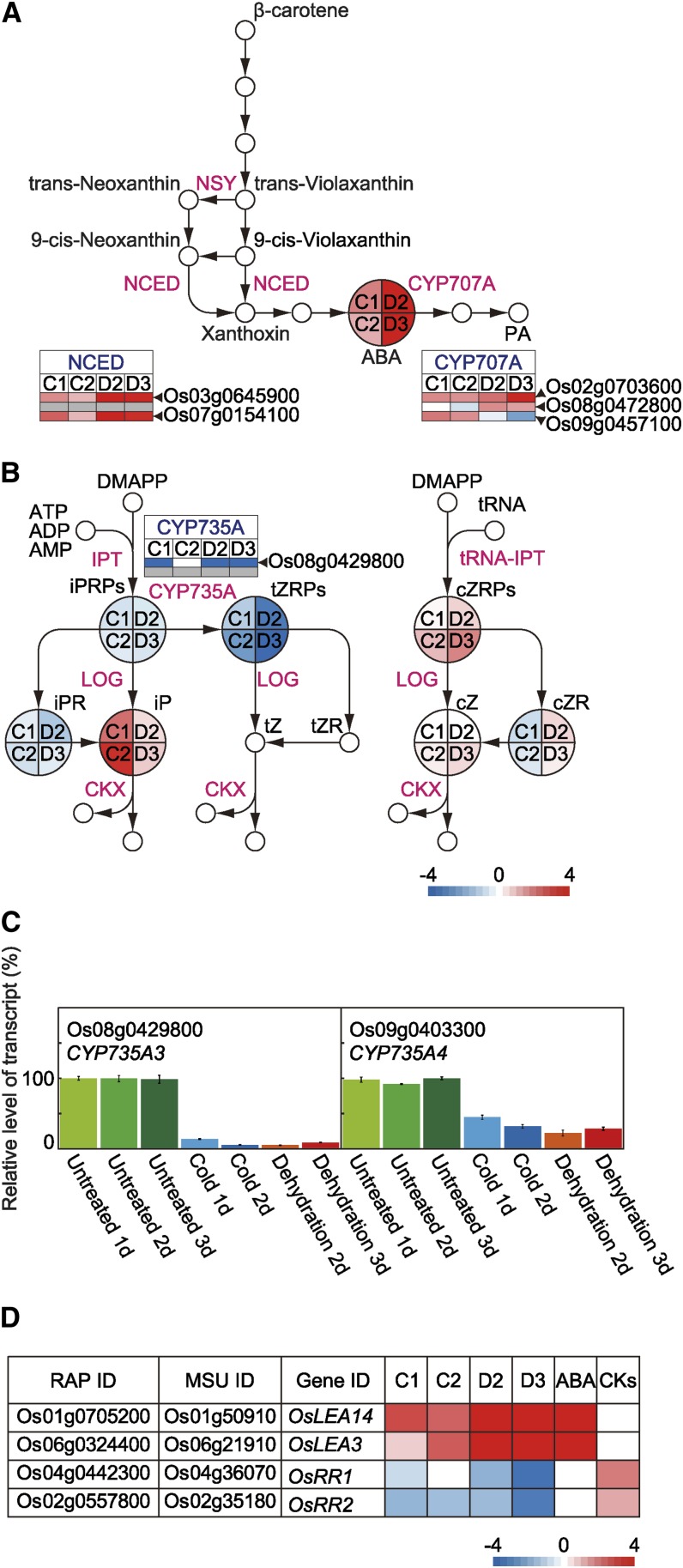
Our phytohormone analysis indicated that exposure to cold or dehydration increased ABA levels in rice plants. Cold treatment increased the level of iP, and cold and dehydration treatments decreased the level of tZRPs (Fig. 2B). We also investigated the expression of genes involved in ABA and CK biosyntheses under dehydration and cold conditions (Fig. 6, A–C; Supplemental Fig. S10). The enzyme 9-cis-epoxycarotenoid dioxygenase (NCED), which converts 9-cis-neoxanthin and 9-cis-violaxanthin into xanthoxin, plays a key role in ABA biosynthesis (Schwartz et al., 1997). Among the genes encoding NCED family members, OsNCED1 (Os03g0645900) and OsNCED3 (Os07g0154100) showed significantly increased transcript levels in both cold- and dehydration-treated plants (Fig. 6A; Supplemental Fig. S10). The major ABA catabolic pathway is thought to involve hydroxylation of ABA to yield 8′-hydroxy-ABA by cytochrome P450 (CYP) 707A followed by the spontaneous isomerization of 8′-hydroxy-ABA into phaseic acid (Krochko et al., 1998; Saito et al., 2004). The transcript level of ABA8OX1 (Os02g0703600), a CYP707A family member, was significantly higher in both cold- and dehydration-treated rice plants than in untreated plants (Fig. 6A). Compared with those levels in untreated plants, the levels of ABA8OX2 (Os08g0472800) and ABA8OX3 (Os09g0457100) transcripts were significantly higher in dehydration- and cold-treated plants, respectively (Fig. 6A; Supplemental Fig. S10). The genes CYP735A3 (Os08g0429800) and CYP735A4 (Os09g0403300) encode CK transhydroxylases. The transcript level of CYP735A3 was significantly lower in both cold- and dehydration-treated plants than in untreated plants (Fig. 6B; Supplemental Fig. S10). However, a probe corresponding to the CYP735A4 gene was not included in our unique microarray slide. We analyzed the transcript levels of CYP735A4 and CYP735A3 by qRT-PCR and found that both showed significant decreases under cold and dehydration conditions (Fig. 6C).
Figure 6.
Pathways for ABA and CK biosyntheses. A, ABA biosynthesis. B, CK biosynthesis. C, Levels of transcripts for genes encoding CYP735As determined by qRT-PCR. D, Expression of genes encoding LEAs and OsRRs. Heat maps illustrate transcript levels of representative cold- and dehydration-responsive genes. A small gray square denotes the absence of a probe for that gene in oligonucleotide microarray. We referred to http://ricexpro.dna.affrc.go.jp/ for expression of LEA and OsRR in ABA- or CK-treated rice plants. Heat maps illustrate accumulated levels of representative cold-responsive and dehydration-responsive phytohormones. CKX, Cytokinin oxidase/dehydrogenase; DMAPP, dimethylallyl diphosphate; IPT, adenosine P-isopentenyltransferase; LOG, lonely guy, cytokinin NMP phosphoribohydrolase; NSY, neoxanthin synthase; tRNA-IPT, tRNA isopentenyltransferase.
To clarify the relationship between the accumulation of phytohormones and phytohormone-responsive gene expression under cold or dehydration conditions, we analyzed the transcript levels of genes that are known to be ABA and CK responsive (Fig. 6D). Several genes for LEA proteins are known to be involved in abiotic stress tolerance in plants, and OsLEA14/WSI18 and OsLEA3 were reported to be ABA responsive (Moons et al., 1997; Oh et al., 2005). We found that OsLEA14/WSI18 and OsLEA3 were induced by cold and dehydration. The His-Asp phosphorelay system, which consists of a sensor His protein kinase, a His-containing phosphotransfer protein, and a response regulator, plays important roles in CK signaling. There are 13 type A response regulators in rice, all of which are up-regulated in response to CKs (Ito and Kurata, 2006; Jain et al., 2006; Hirose et al., 2007). However, we found that the CK-inducible marker genes Oryza sativa response regulator1 (OsRR1) and OsRR2 were down-regulated under cold conditions and dehydration conditions (Fig. 6D).
Next, we compared the transcriptional patterns of genes induced by ABA and CKs in rice plants with those patterns of their counterparts in Arabidopsis. We analyzed an Arabidopsis oligomicroarray and evaluated the transcript levels of the counterparts of the genes identified in rice in Arabidopsis plants. The ABA-inducible RD17 and RD29A genes were up-regulated in Arabidopsis after exposure to cold or dehydration. Conversely, the CK-inducible Arabidopsis response regulator5 (ARR5) and ARR6 genes were down-regulated by dehydration or ABA treatment (Supplemental Fig. S11; Supplemental Tables S20 and S21).
DISCUSSION
We performed an integrated analysis of the effects of cold or dehydration stress on the levels of metabolites, phytohormones, and gene transcripts in rice plants and characterized the events associated with changes in their levels. This analysis enabled us to identify important genes encoding metabolic enzymes for carbohydrates, amino acids, and phytohormones that are involved in the responses/adaptation of rice plants to cold or dehydration. In rice plants subjected to cold or dehydration treatments, the expression of several genes encoding enzymes involved in starch degradation, Suc metabolism, and the glyoxylate cycle increased dynamically, and these changes were correlated with the accumulation of Glc, Fru, and Suc (Figs. 1C and 5, A–C). We predicted that Os03g0141200, which encodes β-amylase, and Os01g0332100, which encodes an alkaline/neutral invertase, were important for starch degradation and Suc metabolism, respectively, in both cold- and dehydration-treated plants. Their transcript levels were more than 5-fold higher in both cold- and dehydration-treated plants than in untreated plants (Fig. 5, A and B; Supplemental Tables S11–S16). The high transcript levels of both of these genes were correlated with Glc accumulation. A similar relationship between high transcript levels of the counterparts of these genes and increased levels of Glc has been reported in cold- or dehydration-treated Arabidopsis plants (Kaplan and Guy, 2004; Kaplan et al., 2007; Maruyama et al., 2009).
Isocitrate lyase and malate synthase are key enzymes in the glyoxylate cycle, and the expression levels of their encoding genes play a role in activating gluconeogenesis during postgerminative growth and plant pathogenesis (Eastmond et al., 2000; Dunn et al., 2009). We predicted that isocitrate lyase (Os07g0529000) and malate synthase (Os04g0486950) would also play important roles under dehydration conditions in rice. We detected increased levels of isocitrate lyase and malate synthase transcripts in rice plants subjected to dehydration treatments (>20 times higher than those levels in untreated plants; Fig. 5, C and G). The high expression levels of these genes seemed to result in Glc accumulation (Figs. 1C and 5, C and G). A query of the gene expression database Genevestigator (Hruz et al., 2008) confirmed that the isocitrate lyase and malate synthase genes show increased expression under dehydration conditions in several rice cultivars. In contrast, expression of their counterparts in Arabidopsis plants does not increase after dehydration (Supplemental Fig. S12; Supplemental Table S21). These data imply that regulation of the glyoxylate cycle may be involved in Glc accumulation in response to dehydration in rice but not Arabidopsis.
The Arabidopsis galactinol synthase family contains eight genes. Among them, Arabidopsis thaliana galactinol synthase3 (AtGolS3) and AtGolS2 are significantly up-regulated in cold- and dehydration-treated Arabidopsis plants, respectively (Taji et al., 2002; Maruyama et al., 2009). Raffinose probably acts as an osmoprotectant to stabilize cellular membranes and a scavenger of reactive oxygen species to protect the photosynthetic complex in chloroplasts of Arabidopsis exposed to cold or dehydration (Nishizawa et al., 2008). Transgenic Arabidopsis plants overexpressing a gene encoding galactinol synthase accumulated more endogenous raffinose and exhibited improved dehydration tolerance relative to wild-type plants. Hence, genes for galactinol synthase likely play a key role in increasing levels of endogenous raffinose under cold or dehydration conditions in Arabidopsis (Taji et al., 2002). Our current analysis revealed comparable increases in the levels of raffinose after exposure of rice plants to cold or dehydration (Fig. 1C). In rice, galactinol synthase is encoded by only two genes: WSI76 and OsGolS1. The WSI76 transcript level increased significantly in plants exposed to dehydration but increased only slightly after exposure to cold. The OsGolS1 transcript level increased significantly after dehydration but not after cold treatment (Fig. 5H). These data suggest that the roles of galactinol synthase genes in raffinose accumulation under cold conditions differ between rice and Arabidopsis.
We identified tZRPs and iP as representative phytohormones involved in the responses of rice plants to cold or dehydration (Fig. 2). Regarding changes in CK levels, the level of tZRPs decreased significantly in rice plants subjected to cold or dehydration treatments, and tZ was not detected in any of the plants, regardless of the treatment. Conversely, the level of iP was significantly higher in cold-treated rice plants and slightly higher in dehydration-treated plants than in control plants (Figs. 2B and 6B). In rice, the transcript levels of CK-inducible genes encoding OsRR decreased after exposure to either cold or dehydration (Fig. 6C). These results suggest that the CK-dependent transcription pathway is inactive in both cold-treated and dehydration-treated rice plants and that an increased level of endogenous iP cannot activate the CK-dependent transcription pathway under cold or dehydration stress. We predicted that CYP735A3 (Os08g0429800) and CYP735A4 (Os09g0403300), both of which encode CK transhydroxylases, play important roles in rice plants under cold or dehydration stress (Fig. 6C); the low levels of both transcripts were correlated with decreased levels of tZRPs. The decreased levels of tZRPs associated with the down-regulation of CYP735A3 and CYP735A4 may be related to growth retardation of rice plants under cold or dehydration conditions. In Arabidopsis, the transcript levels of CYP735A1 (At5g38450) and CYP735A2 (At1g67110), which encode CK transhydroxylases, were unchanged in plants subjected to cold or dehydration treatments. However, the CK levels decreased significantly in Arabidopsis plants subjected to cold or dehydration treatments (Supplemental Fig. S13; Supplemental Tables S20 and S21). These data imply that the roles of the CK transhydroxylase in the decreased level of CKs under cold or dehydration conditions differ between Arabidopsis and rice. Transcriptional regulation of CYP735A genes might be involved in regulating CK levels under cold and dehydration conditions in rice but not Arabidopsis.
In Arabidopsis, five NCEDs (NCED2, NCED3, NCED5, NCED6, and NCED9) are probably involved in ABA biosynthesis (Iuchi et al., 2001; Tan et al., 2003). Among these proteins, NCED3 is the one most strongly induced by dehydration (Iuchi et al., 2001). Transgenic Arabidopsis plants overexpressing NCED3 show increased tolerance to dehydration, whereas the NCED3 knockout mutant shows enhanced sensitivity to dehydration, indicating that NCED3 plays a key role in ABA biosynthesis in Arabidopsis exposed to dehydration (Iuchi et al., 2001). Compared with the level in control plants, the level of ABA in dehydrated rice plants was significantly higher, whereas the level in cold-treated plants was only slightly higher (Fig. 2B). Rice harbors three OsNCED genes; of these genes, OsNCED1 and OsNCED3 showed expression levels that were correlated with the ABA level in both cold- and dehydration-treated plants (Figs. 2B and 6A). In addition, ABA-induced expression of LEA genes increased in rice plants exposed to cold or dehydration (Fig. 6D). These results imply that OsNCED1 and OsNCED3 are key candidates for ABA biosynthesis and that the ABA-dependent transcription pathway is active in rice plants exposed to cold or dehydration.
CKs and ABA are involved in regulating several processes in plant growth and development under various conditions, including water stress. Although CKs are considered to antagonize ABA in dehydration-stressed Arabidopsis (Nishiyama et al., 2011; Wang et al., 2011), little is known about the cross talk between CK signaling and ABA signaling during water stress in rice. Our integrated analysis of phytohormones and gene transcription indicated that there is an inverse relationship between CK signaling and ABA signaling in both rice and Arabidopsis plants exposed to dehydration stress (Figs. 2B and 6D; Supplemental Figs. S1 and S11). These results imply that the cross talk between CKs and ABA is fundamental to signal transduction in both rice and Arabidopsis under dehydration conditions and that this cross talk evolved during land colonization of plants before the divergence of monocots and dicots.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type rice (Oryza sativa ‘Nipponbare’) seedlings were grown in plastic pots filled with nutrient soil for 2 weeks under flooded lowland conditions under a 12-h-light (28°C)/12-h-dark (25°C) photoperiod (approximately 1,500 μmol photons m−2 s−1). The plants were then subjected to (1) cold treatment by transfer from 28°C to 10°C growing conditions for either 1 or 2 d or (2) dehydration treatment by growing them at 28°C without watering for either 2 d, until the soil moisture content was 18.7% (w/w; sd = 1.3%; 57.0% field capacity), or 3 d, until the soil moisture content was 15.6% (w/w; sd = 1.1%; 47.6% field capacity). Wild-type Arabidopsis (Arabidopsis thaliana) plants were grown in plastic pots filled with peat moss for 3 weeks (principal growth stage = 1.07–1.08) under a 16-h-light/8-h-dark photoperiod (50 ± 10 μmol photons m−2 s−1) at 22°C. The plants were then subjected to (1) a cold treatment by transfer from 28°C to 4°C with growth for 1 d or (2) a dehydration treatment by withholding water for 6 d at 28°C until the soil moisture content decreased to less than 10% (w/w; >47.2% field capacity).
GC-TOF-MS Analysis
Metabolites were extracted from the aerial parts of rice plants (100 mg per sample) with methanol. Extraction and derivatization were performed as described previously (Kusano et al., 2007). Metabolites were detected using a GC instrument (model 6890; Agilent Technologies) fitted with a 30-m DB-17ms column (0.25-mm i.d., 0.25-µm film; Agilent Technologies) coupled to a TOF mass spectrometer (Leco). Ribitol was used as the internal standard (Kusano et al., 2007). The reproducibility of GC-TOF-MS analysis was assessed with three biological replicates for each experiment.
CE-MS Analysis
Metabolites were extracted from aerial parts of rice plants (100 mg) with a chloroform:methanol solution (1:1) using a mixer and a 5-kD cutoff filter (Millipore). Extracted metabolites were detected using a CE-MS system consisting of an Agilent 1100 series MSD mass spectrometer, an Agilent 1100 series isocratic HPLC pump, a G1603A Agilent CE-MS adapter kit, and a G1607A Agilent CE-MS sprayer kit. Met sulphone was used as an internal standard. The reproducibility of CE-MS analysis was determined using three biological replicates in each experiment. Unknown metabolites were quantified using CE-MS.
LC-Coupled MS
The levels of phytohormones in aerial parts of rice plants (100 mg) were quantified as described previously (Kojima et al., 2009) using an LC-MS system (UPLC/Quattro Premier XE; Waters) fitted with an ODS column (1.7 μm, 2.1 × 100 mm, AQUITY-UPLC BEH-C 18; Waters). Reproducibility was assessed using three biological replicates in each experiment.
Microarray Analysis
Total RNAs were isolated from aerial parts of rice or Arabidopsis plants and labeled using Low RNA Input Linear Amplification/Labeling kit reagents (Agilent Technologies). Cy5-labeled complementary RNA experimental samples and Cy3-labeled complementary RNA control samples were hybridized to the microarray chips. Biological and technical (dye-swap) replicates of the sample sets were analyzed. After hybridization, microarray slides were scanned (scanner model G2505C with scan control software, version A.8.5.1; Agilent Technologies), and the data were analyzed using Feature Extraction software, version 10.10.1.1 (Agilent Technologies). Raw data were analyzed using GeneSpring GX software, version 12.0 (Agilent Technologies). Expression log ratios and the Benjamini and Hochberg FDR P values were calculated using GeneSpring GX (Maruyama et al., 2014). The microarray design and data were deposited at MIAMExpress (accession nos. E-MEXP-3863, E-MEXP-3864, E-MEXP-3865, E-MEXP-3862, E-MEXP-3999, and E-MEXP-4000).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Levels of ABA and CKs after cold or dehydration stress in Arabidopsis plants.
Supplemental Figure S2. Rice genes in the rice annotation project database and our unique oligonucleotide microarray.
Supplemental Figure S3. Molecular function of cold- and dehydration-responsive genes in rice plants.
Supplemental Figure S4. Effects of cold or dehydration on transcript levels of genes encoding starch degradation enzymes.
Supplemental Figure S5. Effects of cold or dehydration on transcript levels of genes encoding Suc metabolism enzymes.
Supplemental Figure S6. Effects of cold or dehydration on transcript levels of genes encoding glyoxylate cycle enzymes.
Supplemental Figure S7. Effects of cold or dehydration on transcript levels of genes encoding raffinose biosynthesis enzymes.
Supplemental Figure S8. Effects of cold or dehydration on transcript levels of genes encoding enzymes involved in Pro metabolism.
Supplemental Figure S9. Effects of cold or dehydration on transcript levels of genes encoding enzymes involved in BCAAs synthesis.
Supplemental Figure S10. Effects of cold or dehydration on transcript levels of representative cold- and dehydration-responsive genes involved in ABA and CK biosynthesis.
Supplemental Figure S11. Transcript levels of genes encoding rd17, rd29A, and ARRs.
Supplemental Figure S12. Glyoxylate cycle in Arabidopsis.
Supplemental Figure S13. CK biosynthesis in Arabidopsis.
Supplemental Table S1. Mean and sd values for metabolites.
Supplemental Table S2. Variance covariance matrix for metabolites.
Supplemental Table S3. Eigenvalues for metabolites.
Supplemental Table S4. Eigenvectors for metabolites.
Supplemental Table S5. PC scores for metabolites.
Supplemental Table S6. Mean and sd values for phytohormones in rice.
Supplemental Table S7. Variance covariance matrix for phytohormones.
Supplemental Table S8. Eigenvalues for phytohormones.
Supplemental Table S9. Eigenvectors for phytohormones.
Supplemental Table S10. Principal component scores for phytohormones.
Supplemental Table S11. Mean and sd for phytohormones in Arabidopsis.
Supplemental Table S12. Cold-treated rice plants (1 d).
Supplemental Table S13. Cold-treated rice plants (2 d).
Supplemental Table S14. Dehydration-treated rice plants (2 d).
Supplemental Table S15. Dehydration-treated rice plants (3 d).
Supplemental Table S16. Variance covariance matrix for transcripts.
Supplemental Table S17. Eigenvalues for transcripts.
Supplemental Table S18. Eigenvectors for transcripts.
Supplemental Table S19. PC scores for transcripts.
Supplemental Table S20. Cold-treated Arabidopsis plants (1 d).
Supplemental Table S21. Dehydration-treated Arabidopsis plants (6 d).
Supplementary Material
Acknowledgments
We thank the Rice Genome Resource Center at The National Institute of Agrobiological Sciences for use of the microarray analysis system and the technical support provided by Drs. Yoshiaki Nagamura and Ritsuko Motoyama. We thank Masami Toyoshima for skillful editorial assistance.
Glossary
- MS
mass spectrometry
- ABA
abscisic acid
- CK
cytokinin
- LEA
late-embryogenesis abundant
- BCAA
branched-chain amino acid
- GC
gas chromatography
- TOF
time-of-flight
- CE
capillary electrophoresis coupled with mass spectrometry
- LC
liquid chromatography coupled with mass spectrometry
- FDR
false discovery rate
- PCA
principal component analysis
- PC2
second principal component
- PC1
first principal component
- tZRP
trans-zeatin riboside-5′-phospate
- iP
isopentenyladenine
- tZ
trans-zeatin
- SF
superfamily
- NCED
9-cis-epoxycarotenoid dioxygenase
Footnotes
This work was supported by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (in part by Genomics for Agricultural Innovation, Development of Abiotic Stress Tolerant Crops by DREB Genes) and the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-Oriented Industry (BRAIN) of Japan. The Japan Advanced Plant Science Network provided funding for the hormone analysis.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Bartels D, Sunkar R. (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24: 23–58 [Google Scholar]
- Bowne JB, Erwin TA, Juttner J, Schnurbusch T, Langridge P, Bacic A, Roessner U. (2012) Drought responses of leaf tissues from wheat cultivars of differing drought tolerance at the metabolite level. Mol Plant 5: 418–429 [DOI] [PubMed] [Google Scholar]
- Chiwocha SD, Abrams SR, Ambrose SJ, Cutler AJ, Loewen M, Ross AR, Kermode AR. (2003) A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. Plant J 35: 405–417 [DOI] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF. (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 101: 15243–15248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dunn MF, Ramírez-Trujillo JA, Hernández-Lucas I. (2009) Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 155: 3166–3175 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Germain V, Lange PR, Bryce JH, Smith SM, Graham IA. (2000) Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc Natl Acad Sci USA 97: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124: 509–525 [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. (1993) Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol 3: 419–429 [Google Scholar]
- Gray GR, Heath D. (2005) A global reorganization of the metabolome in Arabidopsis during cold acclimation is revealed by metabolic fingerprinting. Physiol Plant 124: 236–248 [Google Scholar]
- Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK. (2008) Metabolomics of temperature stress. Physiol Plant 132: 220–235 [DOI] [PubMed] [Google Scholar]
- Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. (2012) Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci 17: 172–179 [DOI] [PubMed] [Google Scholar]
- Hannah MA, Wiese D, Freund S, Fiehn O, Heyer AG, Hincha DK. (2006) Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol 142: 98–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12: 343–351 [DOI] [PubMed] [Google Scholar]
- Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H. (2007) Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol 48: 523–539 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Kurata N. (2006) Identification and characterization of cytokinin-signalling gene families in rice. Gene 382: 57–65 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. (2006) Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL. (2004) β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol 135: 1674–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Sung DY, Zhao W, Popp M, Porat R, Guy CL. (2007) Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J 50: 967–981 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, Ashikari M, Ueguchi-Tanaka M, Matsuoka M, Suzuki K, et al. (2009) Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol 50: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krochko JE, Abrams GD, Loewen MK, Abrams SR, Cutler AJ. (1998) (+)-Abscisic acid 8′-hydroxylase is a cytochrome P450 monooxygenase. Plant Physiol 118: 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M, Fukushima A, Arita M, Jonsson P, Moritz T, Kobayashi M, Hayashi N, Tohge T, Saito K. (2007) Unbiased characterization of genotype-dependent metabolic regulations by metabolomic approach in Arabidopsis thaliana. BMC Syst Biol 1: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt J. (1980) Responses of Plants to Environmental Stress, Ed 2 Academic Press, New York [Google Scholar]
- Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano K, Fujita M, Yoshiwara K, Matsukura S, Morishita Y, et al. (2009) Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol 150: 1972–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Todaka D, Mizoi J, Yoshida T, Kidokoro S, Matsukura S, Takasaki H, Sakurai T, Yamamoto YY, Yoshiwara K, et al. (2012) Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res 19: 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. (2014) Gene expression profiling using DNA microarrays. In JJ Sanchez-Serrano, J Salinas, eds, Arabidopsis Protocols, Ed 3, Vol 1062 Humana Press, New York, pp 381–391 [DOI] [PubMed] [Google Scholar]
- Mok MC, Martin RC, Mok DWS. (2000) Cytokinins: biosynthesis, metabolism and perception. In Vitro Cell Dev Biol Plant 36: 102–107 [Google Scholar]
- Moons A, De Keyser A, Van Montagu M. (1997) A group 3 LEA cDNA of rice, responsive to abscisic acid, but not to jasmonic acid, shows variety-specific differences in salt stress response. Gene 191: 197–204 [DOI] [PubMed] [Google Scholar]
- Müller A, Düchting P, Weiler EW. (2002) A multiplex GC-MS/MS technique for the sensitive and quantitative single-run analysis of acidic phytohormones and related compounds, and its application to Arabidopsis thaliana. Planta 216: 44–56 [DOI] [PubMed] [Google Scholar]
- Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Kakimoto T, et al. (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S. (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Fernie AR. (2012) The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol Life Sci 69: 3225–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK. (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Gilday AD, Graham IA. (2003) The gluconeogenic enzyme phosphoenolpyruvate carboxykinase in Arabidopsis is essential for seedling establishment. Plant Physiol 131: 1834–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Matsuda F. (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61: 463–489 [DOI] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M. (2004) Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol 134: 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57: 431–449 [DOI] [PubMed] [Google Scholar]
- Sanchez DH, Schwabe F, Erban A, Udvardi MK, Kopka J. (2012) Comparative metabolomics of drought acclimation in model and forage legumes. Plant Cell Environ 35: 136–149 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Sicher RC, Barnaby JY. (2012) Impact of carbon dioxide enrichment on the responses of maize leaf transcripts and metabolites to water stress. Physiol Plant 144: 238–253 [DOI] [PubMed] [Google Scholar]
- Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R, Andriankaja M, Van Aken O, Van Breusegem F, Fernie AR, et al. (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR. (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Urano K, Kurihara Y, Seki M, Shinozaki K. (2010) ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr Opin Plant Biol 13: 132–138 [DOI] [PubMed] [Google Scholar]
- Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, Sakurai N, Suzuki H, Saito K, Shibata D, Kobayashi M, et al. (2009) Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J 57: 1065–1078 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Juenger TE. (2011) Drought, metabolites, and Arabidopsis natural variation: a promising combination for understanding adaptation to water-limited environments. Curr Opin Plant Biol 14: 240–245 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li L, Ye T, Zhao S, Liu Z, Feng YQ, Wu Y. (2011) Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J 68: 249–261 [DOI] [PubMed] [Google Scholar]
- Weiner JJ, Peterson FC, Volkman BF, Cutler SR. (2010) Structural and functional insights into core ABA signaling. Curr Opin Plant Biol 13: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.