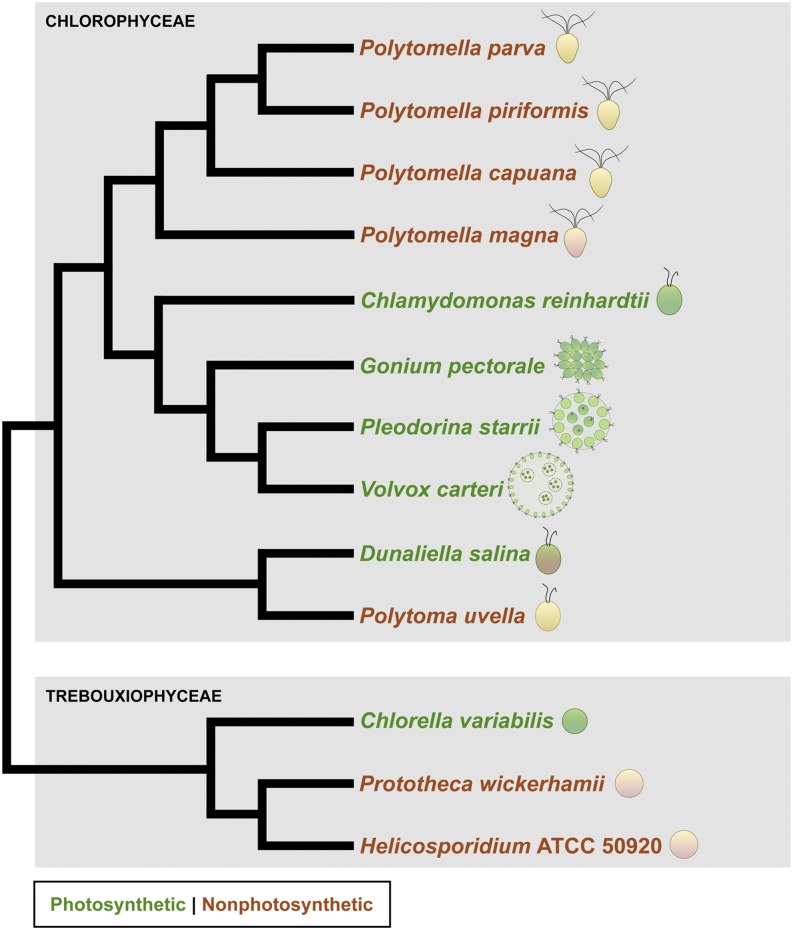
Genome and transcriptome sequencing of species from the nonphotosynthetic green algal genus Polytomella revealed no evidence of a plastid genome or associated gene expression system and presents an example of a primary plastid-bearing lineage without plastid DNA.
Abstract
Polytomella spp. are free-living, nonphotosynthetic green algae closely related to the model organism Chlamydomonas reinhardtii. Although colorless, Polytomella spp. have a plastid, but it is still unknown whether they harbor a plastid genome. We took a next generation sequencing approach, along with transcriptome sequencing, to search for a plastid genome and an associated gene expression system in Polytomella spp. Illumina sequencing of total DNA from four Polytomella spp. did not produce any recognizable plastid-derived reads but did generate a large number of mitochondrial DNA sequences. Transcriptomic analysis of Polytomella parva uncovered hundreds of putative nuclear-encoded, plastid-targeted proteins, which support the presence of plastid-based metabolic functions, similar to those observed in the plastids of other nonphotosynthetic algae. Conspicuously absent, however, were any plastid-targeted proteins involved in the expression, replication, or repair of plastid DNA. Based on these findings and earlier findings, we argue that the Polytomella genus represents the first well-supported example, to our knowledge, of a primary plastid-bearing lineage without a plastid genome.
The loss of photosynthesis has occurred in diverse eukaryotic lineages, including those with primary or secondary plastids (Keeling, 2010). Most of these lineages still retain a plastid (primary or secondary), which, although colorless, continues to perform crucial metabolic functions (Borza et al., 2005; Lim and McFadden, 2010). In all well-explored cases, with one potential exception (Fernández-Robledo et al., 2011), nonphotosynthetic plastids, like their photosynthetic counterparts, contain a genome (Gould et al., 2008; Krause, 2012). However, not surprisingly, it is highly reduced, having lost photosynthesis-related genes, and made up almost entirely of genes involved in plastid DNA (ptDNA) expression (Gould et al., 2008; Krause, 2012). Given the widespread preservation of ptDNA in diverse plants and algae, particularly those that are nonphotosynthetic, some have argued, often providing compelling evidence, that plastid gene expression is indispensable (Barbrook et al., 2006; Fleischmann et al., 2011; Alkatib et al., 2012a, 2012b).
Many nonphotosynthetic, plastid-bearing species are parasites (Lang-Unnasch et al., 1998; Gould et al., 2008), such as the malaria parasite Plasmodium falciparum (Wilson et al., 1996) as well as the green alga Helicosporidium sp., an insect gut parasite (de Koning and Keeling, 2006), and its close relative Prototheca wickerhamii (Borza et al., 2005), which causes protothecosis in humans. However, there are also strictly free-living species with colorless plastids, including all known members of the green algal genus Polytomella (Pringsheim, 1955), a monophyletic assemblage of unicellular freshwater flagellates closely related to the model photosynthetic organism Chlamydomonas reinhardtii (Nakada et al., 2008; Smith et al., 2013b; Fig. 1). Polytomella spp. are not to be confused with Polytoma spp., which are nonphotosynthetic chlamydomonadaleans that lost photosynthesis independently of Polytomella spp. and are polyphyletic (Nedelcu 2001; Nakada et al., 2008).
Figure 1.
Tree of chlorophycean and trebouxiophycean green algae with examples of species that have lost photosynthetic capabilities. Photosynthetic species are shown in green, and nonphotosynthetic species are shown in red. The cartoon beside the species name shows basic cellular structure (not to scale). Branching order is based on published phylogenetic analyses (Nakada et al., 2008; Smith et al., 2013b).
The plastid of Polytomella spp., which derives from a primary endosymbiosis, has been characterized by ultrastructural studies and typically contains copious amounts of starch (Moore et al., 1970; Brown et al., 1976). However, experiments searching for evidence of a plastid gene expression system within it have proven unsuccessful (Nedelcu et al., 1996; Nedelcu, 2001). No one, however, has addressed this issue with high-throughput sequencing and transcriptomic analysis. Here, we take precisely this approach to search for evidence of a genome and gene expression system in the plastid of Polytomella spp.
RESULTS AND DISCUSSION
An Absence of Plastid-Derived Sequencing Reads
We performed Illumina (HiSEquation 2000) sequencing of whole genomic DNA isolated from each of the four known Polytomella spp. lineages: Polytomella parva, Polytomella piriformis, Polytomella capuana, and Polytomella magna (Sammlung von Algenkulturen Goettingen, strains 63-3, 63-10, 63-5, and 63-9, respectively; Fig. 1). The resulting 225 million paired-end reads (length = 100 nucleotides; insert size ≈ 500 nucleotides) were scanned for organelle DNA sequences with BLAST (Altschul et al., 1990) and mapping-based methods using complete chlamydomonadalean plastid and mitochondrial genomes as search queries and reference sequences (Tables I and II; Supplemental Fig. S1). Mitochondrial DNA (mtDNA)-derived reads were easily detected, representing 5% of the raw data (Tables I and II). We could not, however, find any reads of obvious plastid origin. Some reads did map to the ptDNA reference sequences, but in all cases, they were shown to be false positives, corresponding, for example, to mitochondrial or nuclear ribosomal RNA (rRNA)-coding segments mapping to plastid rDNA. Similar results were observed from analyses of P. parva Illumina RNA sequencing (RNA-seq) reads (Table II).
Table I. Mapping Illumina DNA-seq reads to Polytomella spp. mitochondrial genomes.
| P. parva | P. piriformis | P. capuana | P. magna | |
|---|---|---|---|---|
| Mitochondrial genomea | ||||
| Approximate size (kb) | 16 | 16 | 13 | 28 |
| Gene number | 10 | 10 | 10 | 10 |
| Guanine + cytosine content (%) | 41 | 42 | 57 | 35 |
| GenBank accession no. | AY062933, AY062934 | GU108480, GU108481 | EF645804 | KC733827 |
| Illumina sequencing datab | ||||
| Total reads | 62.6 × 106 | 64.0 × 106 | 49.1 × 106 | 50.1 × 106 |
| Reads mapped to mtDNA | 2.3 × 106 | 2.6 × 106 | 1.3 × 106 | 3.4 × 106 |
| Perfect matches to mtDNA | 1.5 × 106 | 1.8 × 106 | 0.8 × 106 | 2.9 × 106 |
| Average mtDNA read coverage | 14,129 | 16,095 | 9,555 | 15,997 |
| mtDNA covered (%) | 100 | 100 | 100 | 100 |
The mtDNAs of P. parva and P. piriformis are fragmented into two chromosomes; genome architecture and mapping statistics include both chromosomes. Gene number includes all encoded proteins, rRNAs, and tRNAs; duplicate genes were counted only one time. bPaired-end reads (length = 100 nucleotides; insert size ≈ 500 nucleotides) were generated from total DNA on the Illumina HiSEquation 2000 sequencing system. Read mapping was performed with CLC Genomics Workbench version 6.0.4 using the following settings: mismatch cost = 2, insertion cost = 3, deletion cost = 3, length fraction = 0.5, similarity fraction = 0.5, and nonspecific match handling = map randomly.
Table II. Searching Polytomella spp. contigs for plastid DNA.
| Assembly and BLAST Results |
DNA-seq
a
|
RNA-seqa (P. parva) | |||
|---|---|---|---|---|---|
| P. parva | P. piriformis | P. capuana | P. magna | ||
| Illumina assembly summarya | |||||
| Contigs | 7,463 | 6,578 | 9,563 | 4,668 | 30,126 |
| N50 (nt) | 12,890 | 17,259 | 24,579 | 59,924 | 1,972 |
| Maximum scaffold length (nt) | 103,623 | 118,401 | 138,339 | 303,036 | 14,173 |
| Minimum scaffold length (nt) | 100 | 100 | 100 | 100 | 150 |
| Total assembly length (nt) | 47 × 106 | 47.4 × 106 | 80.3 × 106 | 68.2 × 106 | 40.2 × 106 |
| Fraction of reads assembled | 0.98 | 0.99 | 0.97 | 0.98 | 0.93 |
| BLASTn searchesb | |||||
| Mitochondrial rRNAs | Hit | Hit | Hit | Hit | Hit |
| Plastid rRNAs | No hit | No hit | No hit | No hit | No hit |
| Mitochondrial tRNAs | Hit | Hit | Hit | Hit | Hit |
| Plastid tRNAs | No hit | No hit | No hit | No hit | No hit |
| tBLASTx searchesb | |||||
| mtDNA protein-coding genes | Hit | Hit | Hit | Hit | Hit |
| ptDNA protein-coding genes | No hit | No hit | No hit | No hit | No hit |
| tBLASTn searchesb | |||||
| mtDNA-encoded proteins | Hit | Hit | Hit | Hit | Hit |
| ptDNA-encoded proteins | No hit | No hit | No hit | No hit | No hit |
Paired-end DNA-seq (length = 100 nucleotides; insert ≈ 500 nucleotides) and RNA-seq (length = 50 nucleotides; insert ≈ 400 nucleotides) data were generated from total Polytomella spp. DNA and RNA (standard Illumina TruSeq RNA library preparation) on the Illumina HiSEquation 2000 sequencing system. DNA reads were assembled with CLC Genomics Workbench version 6.0.4 using a word size of 20, a bubble size of 50, and paired-end scaffolding. RNA-seq assemblies were performed with ABySS by the National Center for Genome Research (details in “Materials and Methods”). bBLAST (Altschul et al., 1990) searches were performed with CLC Genomics Workbench using the following parameters: BLASTn with a word size of 11, match and mismatch scores of 1 and −3, respectively, and gap-cost values of 5 (existence) and 2 (extension); tBLASTx with a word size of 3 and the BLOSUM62 substitution matrix; and protein-translated nucleotide BLAST (tBLASTn) with a word size of 3, the BLOSUM62 substitution matrix, and gap-cost values of 11 (existence) and 1 (extension). In all cases, the filter low complexity regions option was switched off, and an expectation value of 10 was used. The following completely sequenced chlamydomonadalean mitochondrial and plastid genomes (GenBank accession numbers) were used as BLAST query sequences: C. reinhardtii (EU306622 and FJ423446), Gonium pectorale (AP012493 and AP012494), Pleodorina starrii (JX977845 and JX977846), Volvox carteri (GU048821 and GU084820), and Dunalielia salina (GQ250045 and GQ250046). The annotations (i.e. genes and exons) within each of these genomes were also individually searched against the Polytomella spp. assemblies. All annotated rRNA-, tRNA-, and protein-coding regions were considered. All contig regions with BLAST hits were subsequently searched against the National Center for Biotechnology Information (NCBI) nucleotide collection (nr/nt) database. Hits from the tBLASTx and tBLASTn analyses were also searched against the NCBI nonredundant protein sequences database using BLASTx (using the same parameters as the tBLASTn searches described above). Hits to NCBI were scanned for putative plastid-encoded genes. Any false positives, such as hits matching to mitochondrial- or nuclear-encoded genes, were removed and are not included. Query sequences matching to highly repetitive regions, such as poly(A) tracts, were also removed.
The Polytomella spp. Illumina DNA data were assembled, giving 4,668–9,563 contigs per species with an N50 statistic ranging from 12,890 (P. parva) to 59,924 nucleotides (P. magna) and an average assembly length of 6.1 × 107 nucleotides (Table II). We used nucleotide-nucleotide BLAST (BLASTn) and translated nucleotide-translated nucleotide BLAST (tBLASTx) to search the contigs for ptDNA and mtDNA sequences. Polytomella spp. mitochondrial genomes, despite their highly reduced gene contents, elevated rates of nucleotide substitution, fragmented architectures, and/or extreme nucleotide compositions (Table I; Smith et al., 2010), were readily recovered from the contig data when using chlamydomonadalean mtDNAs as BLAST queries (Table II; Supplemental Fig. S1). However, not one contig comprised of putative ptDNA could be identified through BLAST searches with chlamydomonadalean (or any other) plastid genomes, even when using low search stringencies and focusing on highly conserved genes (Table II). The only hits generated from the ptDNA queries were to contigs made up of mtDNA or nuclear genes of obvious nonplastid function, such as 16S or 18S rDNA. Contig analysis of P. parva RNA-seq assemblies gave the same findings (Table II).
No Nuclear-Encoded Proteins for Plastid Gene Expression
Transcriptome sequencing, assembly, and annotation of P. parva were performed by the National Center for Genome Resources in conjunction with the Microbial Eukaryote Transcriptome Initiative (http://marinemicroeukaryotes.org/). Approximately 24,000 P. parva protein-coding regions and their deduced amino acid sequences were predicted from 30,126 contigs (N50 = 1,970 nucleotides; Table II). Transcripts were annotated based on hidden Markov model and protein-protein BLAST (BLASTp) searches against protein and protein family databases, including Swiss-Prot, PhyloDB, TIGERFAMs, and Pfam, and then scanned for genes with plastid-related functions. The C. reinhardtii plastid proteome, which contains 996 experimentally chloroplast-localized proteins (Terashima et al., 2010, 2011), and all presumed genes for plastid-targeted proteins from the C. reinhardtii nuclear genome assembly (v5.3.1; Merchant et al., 2007) were also used to search the P. parva transcriptome for genes encoding plastid proteins.
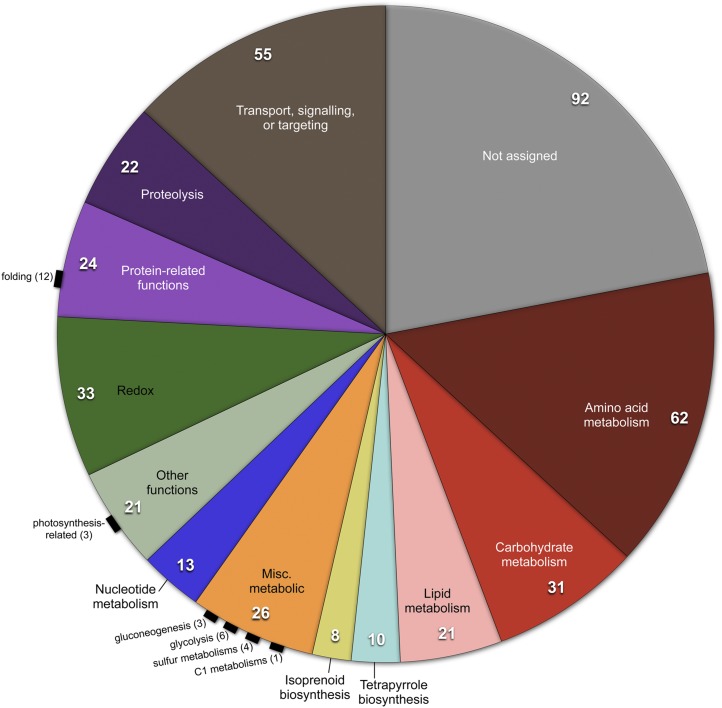
Altogether, we identified 418 P. parva transcripts that, given their annotation and/or similarity to C. reinhardtii homologs, encode possible plastid-targeted proteins (Supplemental Table S1). About one-half of these sequences give best hits to known plastid proteins in GenBank BLASTp searches and approximately one-third have an identifiable plastid transit peptide (Supplemental Table S1) based on TargetP analysis (Emanuelsson et al., 2007). A survey of these various proteins supports the hypothesis that the P. parva plastid carries out an array of functions (Fig. 2; Supplemental Table S1), paralleling those of other colorless plastids (de Koning and Keeling, 2004; Borza et al., 2005). Carbohydrate, lipid, and isoprenoid biosynthesis all seem to take place in the P. parva plastid along with amino acid, purine, and porphyrin biosynthesis as well as other diverse metabolic processes (Supplemental Table S1). Candidate plastid proteins with oxidation–reduction, transport, and proteolytic functions were also discovered along with those proteins involved in plastid biogenesis and sulfur metabolism. Conspicuously absent, however, were genes encoding proteins involved in the expression, replication, or repair of ptDNA (Supplemental Table S1). Not a single confirmed plastid-like ribosomal protein was found in the P. parva transcriptomic data. Mitochondrial-like ones, however, were abundant. There were also no clear traces of proteins associated with the regulation of plastid transcription or translation.
Figure 2.
Number of nuclear genes encoding potential plastid-targeted proteins in different functional categories determined from the P. parva transcriptome. The P. parva transcriptome is available from the Community Cyberinfrastructure for Advanced Microbial Ecology Research and Analysis database (http://camera.calit2.net/) under project ID MMETSP0052_2. Annotations were performed by the National Center for Genome Resources following the standard protocol of the Marine Microbial Transcriptome Initiative (http://marinemicroeukaryotes.org/). Annotated transcripts were scanned for plastid-like functions. The C. reinhardtii plastid proteome, which contains 996 experimentally chloroplast-localized proteins (Terashima et al., 2011), and all presumed plastid-targeted proteins from the C. reinhardtii nuclear genome assembly (version 5.3.1; downloaded from Phytozome version 9.1) were also used to search (with BLASTp) the P. parva transcriptome for putative plastid proteins. All hits to the P. parva transcriptome were collected. Duplicates and hits corresponding to well-known nuclear or mitochondrial proteins were removed; however, some nuclear or mitochondrial proteins still remain in the list, because they may represent proteins with dual functions (Terashima et al., 2011).
The Possibility of Plastid Genome Loss
The lack of any detectable plastid-derived sequencing reads and nuclear genes involved in ptDNA expression, replication, or repair from P. parva could be an indication that it and Polytomella spp. as a whole do not have a plastid genome, a hypothesis that is consistent with the inability to detect plastid rRNA or rDNA in northern blot (Nedelcu et al., 1996) and PCR experiments (Nedelcu, 2001) of P. parva. Next generation sequencing of total genomic DNA or RNA from plants and algae, including nonphotosynthetic ones, typically yields huge amounts of plastid-derived sequence data (Janouškovec et al., 2010; Smith and Keeling, 2012; McPherson et al., 2013) and has become the go-to method for plastid genomics. Thus, the absence of ptDNA reads from Illumina sequencing data of four different Polytomella spp. is perplexing as is the absence of an obvious plastid gene expression system from P. parva transcriptomic data, especially given that transcriptome sequencing of other colorless algae has consistently uncovered such a system (Bozdech et al., 2003; Borza et al., 2005; Wickett et al., 2011). Nuclear genes encoding proteins involved in the replication, repair, or expression of ptDNA represent a significant proportion (>7%) of the predicted C. reinhardtii plastid proteome (Terashima et al., 2011). If Polytomella spp. have a plastid genome, we would have expected to find an even greater proportion of proteins connected to these functions (because of the presumed loss of genes related to photosynthesis), but we found none (Supplemental Table S1).
Secondary plastids, which are derived from eukaryote–eukaryote endosymbioses (Keeling, 2010), are believed to have been lost, along with their genomes, from various eukaryotic lineages, including the apicomplexan Cryptosporidium parvum (Huang et al., 2004). Additionally, there is accumulating evidence to suggest that the oyster parasite Perkinsus marinus ‘Alveolata’ has a cryptic secondary plastid without DNA (Fernández-Robledo et al., 2011) and that some mitochondrion-derived organelles (i.e. hydrogenosomes and mitosomes) lack DNA (Hjort et al., 2010). However, there has not been an example of plastid genome loss or outright plastid loss within a primary plastid-bearing species, such as a green alga. Moreover, it has been consistently argued that a plastid gene expression system is essential in green plants, even under heterotrophic conditions (Fleischmann et al., 2011; Alkatib et al., 2012a, 2012b).
The plastid gene content among chlamydomonadalean algae is highly conserved (Smith et al., 2013a), making it easy to predict what genes the most recent green ancestor of Polytomella spp. had within its plastid genome (Fig. 3). When ignoring nonstandard open reading frames, 64 protein-coding genes are preserved among available chlamydomonadalean ptDNAs, 37 of which are directly or indirectly linked to photosynthesis and/or energy production (Fig. 3). The remaining 27 genes are all involved in plastid gene expression, with the exceptions of clpP (which encodes the proteolytic subunit of Clp protease) and the open reading frames ycf1 and ycf2 (also referred to as ftsH). Recent experiments showed that ycf1 encodes a vital component (Tic214) of the translocon at the inner membrane of chloroplasts (TIC) of Arabidopsis (Arabidopsis thaliana) and, thus, is essential for plant viability (Kikuchi et al., 2013). The same study, however, was unable to find direct homologs of ycf1 in glaucophytes, red algae, or extant cyanobacteria, and ptDNA sequencing has suggested that ycf1 is lost from the plastid genomes of certain grass (Poaceae) species (Guisinger et al., 2010); all of these findings cast doubts on the proposed universal function of the ycf1 product in protein import. Although their exact functions are sometimes debated, most available data indicate that clpP and ycf2 each encode a subunit for a distinct protease enzyme involved in plastid protein degradation (Sakamoto 2006; Wagner et al., 2012). Both genes are thought to be crucial for the survival of photosynthetic green plants (Majeran et al., 2000; Kuroda and Maliga, 2003), but their role in nonphotosynthetic taxa is less straightforward. For example, some viable nonphotosynthetic plant cell lines are missing clpP from their plastid genomes (Cahoon et al., 2003), and the colorless green alga Helicosporidium sp. is missing it as well (de Koning and Keeling, 2006). Also, the putative protein degradation processes carried out by the multisubunit proteases partially encoded by clpP and ycf2 are predicted to help in the transition of one plastid type to another (Adam and Clarke 2002; Sakamoto 2006), a process likely not required in colorless algae. Moreover, it has been suggested that ycf2 may be involved in plastid gene expression (Bock, 2007) and/or the linkage of ptDNA to proteins or membranes (Wicke et al., 2011), potentially eliminating its necessity in ptDNA-lacking species. Finally, there is evidence that ycf2 has been lost from the plastid genomes of Poaceae (Guisinger et al., 2010). We did not find obvious nuclear-located copies of clpP, ycf1, or ycf2 in the Illumina DNA assemblies of the four Polytomella spp. or the P. parva transcriptome. However, the nucleotide sequences of these three plastid-located genes, especially the latter two genes, are known to be poorly conserved and highly divergent among species (Wicke et al., 2011), which can make them very difficult to identify in some taxa.
Figure 3.
Venn diagram of plastid DNA-encoded genes in photosynthetic chlamydomonadalean algae and the nonphotosynthetic trebouxiophycean Helicosporidium sp. The chlamydomonadalean plastid genome (plastome) gene content (black line) is based on the following photosynthetic species: C. reinhardtii, Gonium pectorale, Pleodorina starrii, Volvox carteri, and Dunalielia salina (Fig. 1). A dashed line encloses the Helicosporidium sp. ptDNA genes. Genes numbered with black circles: (1) genes are nuclear encoded in C. reinhardtii and Polytomella spp.; (2) ycf1 in Arabidopsis (Arabidopsis thaliana) is known to encode a crucial component of the TIC complex required for plastid protein import (Kikuchi et al., 2013), and ycf2 is predicted to encode a subunit of protease enzyme involved in plastid protein degradation (Sakamoto 2006; Wagner et al., 2012) but may have a role in plastid gene expression, protein import, and/or the linkage of ptDNA to proteins or membranes (Bock, 2007; Wicke et al., 2011); and (3) like ycf2, data suggest that clpP encodes a subunit of a protease, which might be involved in the degradation of the chloroplast cytochrome b6f complex (Majeran et al., 2000) but may not be crucial for nonphotosynthetic plants and algae (Cahoon et al., 2003; de Koning and Keeling, 2006).
In certain land plants and algae, including the nonphotosynthetic green alga Helicosporidium sp., accD (a gene encoding the carboxyl transferase β subunit of acetyl-CoA carboxylase, which is essential for fatty acid biosynthesis) is located in the plastid genome (Fig. 3). In all explored chlamydomonadalean algae, however, accD is located in the nuclear genome and not the ptDNA (Smith et al., 2013a; Fig. 3). We were able to identify a putative nuclear-encoded copy of accD (and other ACCase subunits) in all four Polytomella spp., suggesting that they use the standard prokaryotic-type ACCase for fatty acid biosynthesis as found in other chlamydomonadalean algae and, therefore, do not require a plastid genome for accD.
Our evidence for the absence of a plastid gene expression system in Polytomella spp. challenges hypotheses about the essentiality of ptDNA and its associated expression in nonphotosynthetic taxa. Among the most compelling of these arguments is the essential tRNAs hypothesis (Barbrook et al., 2006). In plastid-containing species, with few exceptions (Oborník and Green, 2005), the first steps of heme biosynthesis, which involve the production of 5-aminolevulinate (ALA), occur in the plastid through the C5 (i.e. glutamate) pathway and use a plastid-encoded tRNA for Glu (Beale, 1999). This finding means that the plastid trnE, which encodes this tRNA, has a fundamental cellular role in both plastid protein production and heme biosynthesis. It is argued that heterotrophic plants and algae retain a plastid genome and its trnE for the sole purpose of maintaining a functional heme pathway (Barbrook et al., 2006). Our transcriptome analyses (Supplemental Table S1) confirm earlier data (Atteia et al., 2005) showing that heme biosynthesis does occur within the P. parva plastid and follows the typical five-carbon pathway found in other green plants. The ptDNA-located trnE is highly conserved among chlamydomonadalean algae (100% sequence identity among C. reinhardtii, Gonium pectorale, Pleodorina starrii, and Volvox carteri). However, as described above, we were unable to identify a plastid-like trnE gene in either the raw or assembled DNA sequencing (DNA-seq) or RNA-seq data. In the absence of a plastid genome and plastid trnE in Polytomella spp., the heme pathway could likely only be completed through the import of a nuclear tRNA for Glu into the plastid from the cytosol, because the Polytomella mitochondrial genome encodes only a single tRNA (an elongator tRNA for Met; Smith et al., 2010). Although plastid tRNA import has yet to be documented, it is thought to occur in various parasitic species (Wolfe et al., 1992; Alkatib et al., 2012a), such as the underground orchid Rhizanthella gardneri (Delannoy et al., 2011). Import of cytosolic tRNAs into mitochondria, however, is well-documented (Duchêne et al., 2009) and, in fact, must occur in the mitochondria of Polytomella spp. Some plastid-bearing species, such as apicomplexan parasites, are not reliant on a plastid trnE for heme production: they synthesize ALA in the mitochondrion using the Shemin (or succinyl-CoA) pathway and the enzyme ALA synthase (Oborník and Green, 2005). We did not find an ALA synthase enzyme in the P. parva transcriptome data or the DNA-seq data, implying that Polytomella spp. are not synthesizing heme through the Shemin pathway.
Another part of the essential tRNAs hypothesis centers around the potential use of plastid-derived tRNAs within mitochondria. Many mitochondrial genomes, including those of Polytomella spp., lack a gene for the initiator formylmethionyl-tRNA (tRNAfMet), which is required for the initiation of organelle translation and, therefore, gene expression. It has been hypothesized that for some species, such as P. falciparum and C. reinhardtii, tRNAfMet is imported into the mitochondrion from the plastid, thus, making the plastid genome unexpendable (Barbrook et al., 2006). Recently, however, it was shown that missing mitochondrial tRNAs in C. reinhardtii come from the cytosol and not the plastid and that the mitochondrial tRNAfMet is likely generated from a nuclear-encoded elongator methionyl-tRNA using the mitochondrial-targeted protein methionyl-tRNA formyltransferase (Vinogradova et al., 2009). We identified in the P. parva transcriptome a single nuclear-encoded, mitochondrial-like methionyl-tRNA formyltransferase with a putative mitochondrial transit peptide (accession no. MMETSP0052_2-20121109|21186, Community Cyberinfrastructure for Advanced Microbial Ecology Research and Analysis [CAMERA] database), suggesting that P. parva mitochondria are not reliant on a plastid genome for tRNAfMet.
The Search Will Go On
If Polytomella spp. have lost their plastid genome, the ultimate proof will come from an accumulation of data and experiments. The P. parva transcriptome is publically available from the CAMERA database (http://camera.calit2.net/; project ID MMETSP0052), and the DNA-seq reads from the four Polytomella spp. are deposited in GenBank (BioProject accession nos. PRJNA227032–PRJNA227034). We encourage researchers to use these data to help design additional experiments in search of a plastid genome. We have already attempted the obvious experiment of treating Polytomella spp. cells with the DNA-binding fluorescent dye Synergy Brands Green I, which readily visualizes both plastid and mitochondrial nucleotides in C. reinhardtii (Nishimura et al., 1998). Using this dye, we were able to detect nucleoids in P. parva that seem to be associated with highly reticulated mitochondrial structures, which are known to layer over its plastid (Moore et al., 1970). However, we were unable to eliminate the possibility that some of these signals might be located in the plastid.
Based on the evidence presented here, we argue that Polytomella spp. are the first example, to our knowledge, of a plastid-bearing lineage without a plastid genome. Why Polytomella spp. would have lost ptDNA when so many other heterotrophic lineages have kept it is puzzling. It may turn out that Polytomella spp. have a propensity for organelle genome reduction; their mitochondrial genomes are the smallest and have the most reduced gene content observed from any green plant (Smith et al., 2010). Perhaps the strongest evidence for the absence of a plastid genome would be the complete lack of genes encoding plastid-targeted, gene expression-related proteins in a complete nuclear genome sequence. This is certainly the next logical step in testing our hypothesis.
MATERIALS AND METHODS
Polytomella spp. strains were made axenic using the method of Mallet and Lee (2006), grown in darkness at 22°C in Polytomella spp. medium (Sheeler et al., 1968), and harvested in the late logarithmic growth phase (optical density at 750 nm ≈ 0.35). DNA library preparation and Illumina sequencing was performed by BGI Americas using total DNA isolated with the DNeasy Plant Mini Kit (Qiagen). Total P. parva RNA was isolated with the Qiagen RNeasy Plant Mini Kit and treated with Qiagen RNase-Free DNase. RNA library preparation, Illumina sequencing, assembly, and annotation were carried out by the National Center for Genome Resources following the standard protocols of the Marine Microbial Eukaryotic Transcriptome Project, which have been previously described (Curtis et al., 2012).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers PRJNA227032–PRJNA227034.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Mapping Polytomella spp. Illumina sequencing reads to chlamydomonadalean organelle genomes.
Supplemental Table S1. Putative nuclear-encoded, plastid targeted proteins from the P. parva transcriptome.
Note Added in Proof: While this manuscript was in review, Molina et al., 2014 (Molina J, Hazzouri KM, Nickrent D, Geisler M, Meyer RS, Pentony MM, Flowers JM, Pelser P, Barcelona J, Inovejas SA, et al [2014] Mol Biol Evol 31: 793\x{2013}803) published evidence, using similar approaches to those employed here, for the possible loss of the plastid genome in the primary plastid-bearing parasitic flowering plant Rafflesia lagascae.
Supplementary Material
Acknowledgments
We thank Martin Mallet for early work in search of the Polytomella spp. plastid genome, Jimeng Hua for help with nucleic acid isolation, Tudor Borza for preliminary analyses of the Polytomella parva transcriptome, and Stephen Whitefield for microscopy experiments.
Glossary
- ALA
5-aminolevulinate
- DNA-seq
DNA sequencing
- ptDNA
plastid DNA
- RNA-seq
RNA sequencing
- tRNAfMet
formylmethionyl-tRNA
Footnotes
This work was supported by Discovery Grants (to D.R.S. and R.W.L.) from the Natural Sciences and Engineering Research Council (NSERC) of Canada; transcriptomic data were generated by the National Center for Genome Resources as part of the Gordon and Betty Moore Foundation’s Marine Microbial Eukaryote Transcriptome Project.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adam Z, Clarke AK. (2002) Cutting edge of chloroplast proteolysis. Trends Plant Sci 7: 451–456 [DOI] [PubMed] [Google Scholar]
- Alkatib S, Fleischmann TT, Scharff LB, Bock R. (2012a) Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine. Nucleic Acids Res 40: 6713–6724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkatib S, Scharff LB, Rogalski M, Fleischmann TT, Matthes A, Seeger S, Schöttler MA, Ruf S, Bock R. (2012b) The contributions of wobbling and superwobbling to the reading of the genetic code. PLoS Genet 8: e1003076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Atteia A, van Lis R, Beale SI. (2005) Enzymes of the heme biosynthetic pathway in the nonphotosynthetic alga Polytomella sp. Eukaryot Cell 4: 2087–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. (2006) Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci 11: 101–108 [DOI] [PubMed] [Google Scholar]
- Beale SI. (1999) Enzymes of chlorophyll biosynthesis. Photosynth Res 60: 43–73 [Google Scholar]
- Bock R. (2007) Structure, function, and inheritance of plastid genomes. In Bock R, ed, Cell and Molecular Biology of Plastids, Topics in Current Genetics, Vol 19 Springer, Heidelberg, pp 29–63 [Google Scholar]
- Borza T, Popescu CE, Lee RW. (2005) Multiple metabolic roles for the nonphotosynthetic plastid of the green alga Prototheca wickerhamii. Eukaryot Cell 4: 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z, Llinás M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. (2003) The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol 1: E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DL, Massalski A, Patenaude R. (1976) Organization of the flagellar apparatus and associate cytoplasmic microtubules in the quadriflagellate alga Polytomella agilis. J Cell Biol 69: 106–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon AB, Cunningham KA, Stern DB. (2003) The plastid clpP gene may not be essential for plant cell viability. Plant Cell Physiol 44: 93–95 [DOI] [PubMed] [Google Scholar]
- Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, Arias MC, Ball SG, Gile GH, Hirakawa Y, et al. (2012) Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492: 59–65 [DOI] [PubMed] [Google Scholar]
- Delannoy E, Fujii S, Colas des Francs-Small C, Brundrett M, Small I. (2011) Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol 28: 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Keeling PJ. (2004) Nucleus-encoded genes for plastid-targeted proteins in Helicosporidium: functional diversity of a cryptic plastid in a parasitic alga. Eukaryot Cell 3: 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Keeling PJ. (2006) The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchêne AM, Pujol C, Maréchal-Drouard L. (2009) Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr Genet 55: 1–18 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Fernández-Robledo JA, Caler E, Matsuzaki M, Keeling PJ, Shanmugam D, Roos DS, Vasta GR. (2011) The search for the missing link: a relic plastid in Perkinsus? Int J Parasitol 41: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann TT, Scharff LB, Alkatib S, Hasdorf S, Schöttler MA, Bock R. (2011) Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23: 3137–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. (2008) Plastid evolution. Annu Rev Plant Biol 59: 491–517 [DOI] [PubMed] [Google Scholar]
- Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK. (2010) Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in poaceae. J Mol Evol 70: 149–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM. (2010) Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans R Soc Lond B Biol Sci 365: 713–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Mullapudi N, Lancto CA, Scott M, Abrahamsen MS, Kissinger JC. (2004) Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol 5: R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J, Horák A, Oborník M, Lukes J, Keeling PJ. (2010) A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci USA 107: 10949–10954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. (2010) The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci 365: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M. (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Krause K. (2012) Plastid genomes of parasitic plants: a trail of reductions and losses. In Bullerwell C, ed, Organelle Genetics, Springer, Berlin, pp 79–103 [Google Scholar]
- Kuroda H, Maliga P. (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425: 86–89 [DOI] [PubMed] [Google Scholar]
- Lang-Unnasch N, Reith ME, Munholland J, Barta JR. (1998) Plastids are widespread and ancient in parasites of the phylum Apicomplexa. Int J Parasitol 28: 1743–1754 [DOI] [PubMed] [Google Scholar]
- Lim L, McFadden GI. (2010) The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci 365: 749–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Wollman FA, Vallon O. (2000) Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b6f complex. Plant Cell 12: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet MA, Lee RW. (2006) Identification of three distinct Polytomella lineages based on mitochondrial DNA features. J Eukaryot Microbiol 53: 79–84 [DOI] [PubMed] [Google Scholar]
- McPherson H, van der Merwe M, Delaney SK, Edwards MA, Henry RJ, McIntosh E, Rymer PD, Milner ML, Siow J, Rossetto M. (2013) Capturing chloroplast variation for molecular ecology studies: a simple next generation sequencing approach applied to a rainforest tree. BMC Ecol 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J, Cantor MH, Sheeler P, Kahn W. (1970) The ultrastructure of Polytomella agilis. J Protozool 17: 671–676 [Google Scholar]
- Nakada T, Misawa K, Nozaki H. (2008) Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol Phylogenet Evol 48: 281–291 [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Spencer DF, Denovan-Wright EM, Lee RW. (1996) Discontinuous mitochondrial and chloroplast large subunit ribosomal RNAs among green algae: phylogenetic implications. J Phycol 32: 103–111 [Google Scholar]
- Nedelcu AM. (2001) Complex patterns of plastid 16S rRNA gene evolution in nonphotosynthetic green algae. J Mol Evol 53: 670–679 [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Higashiyama T, Suzuki L, Misumi O, Kuroiwa T. (1998) The biparental transmission of the mitochondrial genome in Chlamydomonas reinhardtii visualized in living cells. Eur J Cell Biol 77: 124–133 [DOI] [PubMed] [Google Scholar]
- Oborník M, Green BR. (2005) Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol Biol Evol 22: 2343–2353 [DOI] [PubMed] [Google Scholar]
- Pringsheim EG. (1955) The genus Polytomella. J Protozool 2: 137–145 [Google Scholar]
- Sakamoto W. (2006) Protein degradation machineries in plastids. Annu Rev Plant Biol 57: 599–621 [DOI] [PubMed] [Google Scholar]
- Sheeler P, Cantor M, Moore J. (1968) Growth characteristics of Polytomella agilis in batch culture. Life Sci 7: 289–293 [DOI] [PubMed] [Google Scholar]
- Smith DR, Hamaji T, Olson BJ, Durand PM, Ferris P, Michod RE, Featherston J, Nozaki H, Keeling PJ. (2013a) Organelle genome complexity scales positively with organism size in volvocine green algae. Mol Biol Evol 30: 793–797 [DOI] [PubMed] [Google Scholar]
- Smith DR, Hua J, Archibald JM, Lee RW. (2013b) Palindromic genes in the linear mitochondrial genome of the nonphotosynthetic green alga Polytomella magna. Genome Biol Evol 5: 1661–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Hua J, Lee RW. (2010) Evolution of linear mitochondrial DNA in three known lineages of Polytomella. Curr Genet 56: 427–438 [DOI] [PubMed] [Google Scholar]
- Smith DR, Keeling PJ. (2012) Twenty-fold difference in evolutionary rates between the mitochondrial and plastid genomes of species with secondary red plastids. J Eukaryot Microbiol 59: 181–184 [DOI] [PubMed] [Google Scholar]
- Terashima M, Specht M, Hippler M. (2011) The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr Genet 57: 151–168 [DOI] [PubMed] [Google Scholar]
- Terashima M, Specht M, Naumann B, Hippler M. (2010) Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol Cell Proteomics 9: 1514–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova E, Salinas T, Cognat V, Remacle C, Maréchal-Drouard L. (2009) Steady-state levels of imported tRNAs in Chlamydomonas mitochondria are correlated with both cytosolic and mitochondrial codon usages. Nucleic Acids Res 37: 1521–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R, Aigner H, Funk C. (2012) FtsH proteases located in the plant chloroplast. Physiol Plant 145: 203–214 [DOI] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76: 273–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJN, Honaas LAL, Wafula EKE, Das MM, Huang KK, Wu BB, Landherr LL, Timko MPM, Yoder JJ, Westwood JHJ, et al. (2011) Transcriptomes of the parasitic plant family Orobanchaceae reveal surprising conservation of chlorophyll synthesis. Curr Biol 21: 2098–2104 [DOI] [PubMed] [Google Scholar]
- Wilson RJM, Denny PW, Preiser PR, Rangachari K, Roberts K, Roy A, Whyte A, Strath M, Moore DJ, Moore PW, et al. (1996) Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol 261: 155–172 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. (1992) Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA 89: 10648–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.