Brassicaceae-specific divergent forms of RNA cap-binding proteins do not compete with the conserved form in translation initiation.
Abstract
Canonical translation initiation in eukaryotes begins with the Eukaryotic Initiation Factor 4F (eIF4F) complex, made up of eIF4E, which recognizes the 7-methylguanosine cap of messenger RNA, and eIF4G, which serves as a scaffold to recruit other translation initiation factors that ultimately assemble the 80S ribosome. Many eukaryotes have secondary EIF4E genes with divergent properties. The model plant Arabidopsis (Arabidopsis thaliana) encodes two such genes in tandem loci on chromosome 1, EIF4E1B (At1g29550) and EIF4E1C (At1g29590). This work identifies EIF4E1B/EIF4E1C-type genes as a Brassicaceae-specific diverged form of EIF4E. There is little evidence for EIF4E1C gene expression; however, the EIF4E1B gene appears to be expressed at low levels in most tissues, though microarray and RNA Sequencing data support enrichment in reproductive tissue. Purified recombinant eIF4E1b and eIF4E1c proteins retain cap-binding ability and form functional complexes in vitro with eIF4G. The eIF4E1b/eIF4E1c-type proteins support translation in yeast (Saccharomyces cerevisiae) but promote translation initiation in vitro at a lower rate compared with eIF4E. Findings from surface plasmon resonance studies indicate that eIF4E1b and eIF4E1c are unlikely to bind eIF4G in vivo when in competition with eIF4E. This study concludes that eIF4E1b/eIF4E1c-type proteins, although bona fide cap-binding proteins, have divergent properties and, based on apparent limited tissue distribution in Arabidopsis, should be considered functionally distinct from the canonical plant eIF4E involved in translation initiation.
Cap-dependent translation in eukaryotes begins with recognition of the 7-methylguanosine cap at the 5′ end of an mRNA by the translation initiation factor eIF4E, which forms the eIF4F complex with the scaffolding protein eIF4G. The binding of the RNA helicase eIF4A along with eIF4B promotes unwinding of mRNA secondary structure (Aitken and Lorsch, 2012). The eIF4F complex then serves to circularize mRNA by interaction of eIF4G with poly(A) binding protein and recruit the preinitiation complex through binding of eIF4G to eIF3 and eIF5, ultimately leading to the assembly of the 80S ribosome (Aitken and Lorsch, 2012). eIF4E is an attractive target for global regulation of translational activity through its position at the earliest step, mRNA cap recognition. In many organisms, eIF4E availability is regulated by 4E-binding proteins as well as phosphorylation and sumoylation (Jackson et al., 2010; Xu et al., 2010). However, plants appear to lack 4E-binding proteins, and the role of phosphorylation of eIF4E in translational control is less clear (Pierrat et al., 2007).
The eIF4E proteins generally thought to be involved in translation initiation are Class I eIF4E proteins (Joshi et al., 2005), of which two exist in flowering plants: eIF4E, which pairs with eIF4G to form the eIF4F complex, and the plant-specific isoform eIFiso4E, which pairs with eIFiso4G to form eIFiso4F (Mayberry et al., 2011; Patrick and Browning, 2012). Class I eIF4E family members have conserved Trp residues at positions equivalent to Trp-43 and Trp-56 of Homo sapiens eIF4E (Joshi et al., 2005), and the canonical members of this class, such as plant eIF4E and eIFiso4E, have the ability to promote translation through binding of mRNA cap structure and eIF4G (or eIFiso4G).
In some organisms, however, secondary Class I isoforms exist with expression patterns and functions divergent from the conserved eIF4E (Rhoads, 2009). Caenorhabditis elegans has four isoforms involved in differentiation between mono- and trimethylated mRNA caps (Keiper et al., 2000) and have specialized roles for regulation of certain sets of mRNAs, particularly in the germline (Amiri et al., 2001; Song et al., 2010). Trypanosoma brucei has four isoforms with varying ability to bind cap analog and eIF4G isoforms (Freire et al., 2011). Schizosaccharomyces pombe has a second eIF4E isoform, eIF4E2, which is nonessential under normal growth conditions, but accumulates in response to high temperatures (Ptushkina et al., 2001). It cannot, however, complement deletion of EIF4E1, and while it can bind capped mRNA and promote translation in vitro, it has reduced ability to bind an eIF4G-derived peptide.
Vertebrates encode a novel Class I isoform called EIF4E1B with oocyte-specific expression and functions (Evsikov and Marín de Evsikova, 2009). Zebrafish (Danio rerio) EIF4E1B, with expression limited to muscle and reproductive tissue, has conserved residues identified as necessary for binding cap analog and eIF4G, yet fails to bind either and cannot functionally complement deletion of yeast (Saccharomyces cerevisiae) eIF4E (Robalino et al., 2004). In Xenopus spp. oocytes, the eIF4E1b protein was found to bind eIF4E transporter and cytoplasmic polyadenylation element binding protein to form a translation-repressing complex (Minshall et al., 2007). Drosophila species have undergone extensive expansion of EIF4E-encoding loci to as many as seven different Class I eIF4E isoforms (Tettweiler et al., 2012). The seven EIF4E isoforms of Drosophila melanogaster are differentially expressed, with only five able to bind to eIF4G and complement deletion of yeast eIF4E (Hernández et al., 2005). The eIF4E-3 isoform of D. melanogaster was recently described as having a specific role in spermatogenesis (Hernández et al., 2012).
Upon completion of sequencing of the Arabidopsis (Arabidopsis thaliana) genome (Rhee et al., 2003), it was discovered that in addition to the conserved plant EIF4E (At4g18040) and EIFISO4E (At5g35620), there existed a tandem pair of genes of high sequence similarity on chromosome 1 that also encoded Class I eIF4E family proteins, EIF4E1B (At1g29550, also known as EIF4E3) and EIF4E1C (At1g29590, also known as EIF4E2). Published microarray and RNA Sequencing (RNA-Seq) data indicate little to no EIF4E1C gene expression; however, the EIF4E1B gene appears to be expressed at low levels in most tissues and enriched in tissues involved in reproduction. The protein sequences contain the residues predicted to be involved in regular eIF4E function but also showed some divergence at highly conserved residues of the canonical plant eIF4E. Genome sequencing data indicate that these genes are part of a divergent eIF4E clade specific to Brassicaceae.
The biochemical properties of the eIF4E1b and eIF4E1c proteins were investigated in this work, and it was found that while they can bind mRNA cap analog and eIF4G and support translation in yeast lacking eIF4E, their eIF4G-binding and translation initiation enhancing capabilities in vitro were less robust when compared with the conserved Arabidopsis eIF4E. In addition, it appears that these EIF4E1B-type genes cannot substitute for EIF4E or EIFISO4E in planta because deletion of both of these genes appears to be lethal. Taken together, these findings indicate the EIF4E1B-type genes represent a divergent eIF4E whose roles should be considered separately from the canonical eIF4E in plant translation initiation.
RESULTS AND DISCUSSION
In Silico Analysis
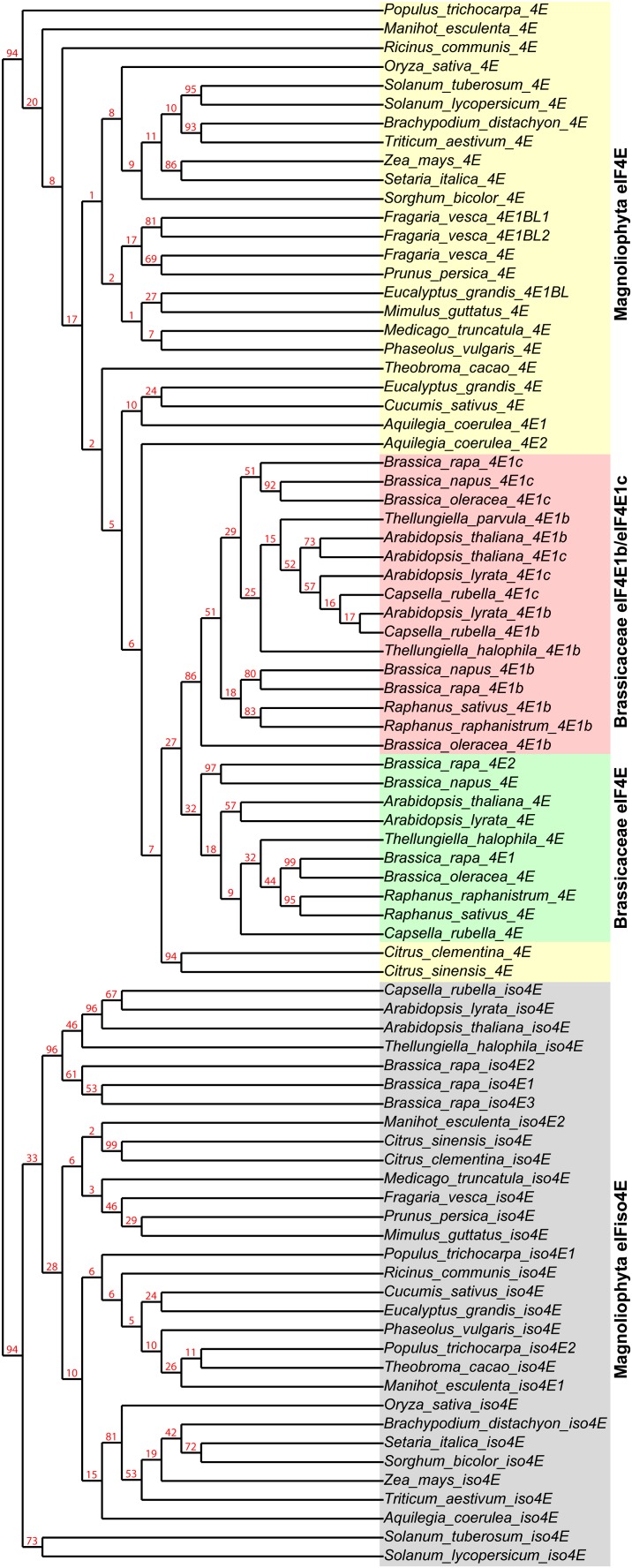
BLAST searches of available genomic and EST data using National Center for Biotechnology Information and Phytozome (Benson et al., 2012; Goodstein et al., 2012) find that EIF4E1B-type genes are present in close Arabidopsis relatives, including Capsella rubella and Brassica, Raphanus, and Thellungiella spp. However, there is no evidence of these genes outside of Brassicaceae, including the closest relative species sequenced, Carica papaya. It therefore appears that the EIF4E1B-type genes are the result of a Brassicaceae-specific gene duplication and specialization. The genomes of Eucalyptus grandis and Fragaria vesca also encode predicted divergent eIF4E protein forms (EIF4E1BL genes in Fig. 1), though it remains to be determined whether these genes are expressed or conserved in other related species.
Figure 1.
Cladogram of Brassicaceae eIF4E1b-like proteins in relation to the conserved eIF4E and eIFiso4E proteins of flowering plants. The Phylogeny.fr pipeline (Dereeper et al., 2008) was used for alignment and phylogenetic tree generation with alignment by MUSCLE and tree construction by PhyML using 500 bootstrap replicates.
The genomes of Arabidopsis, Arabidopsis lyrata, C. rubella, and Brassica rapa encode two EIF4E1B-type loci, called EIF4E1B and EIF4E1C, while Thellungiella halophila and Thellungiella parvula only have evidence for one copy of the gene. Alignment and phylogenetic tree construction of eIF4E and eIFiso4E sequences (Fig. 1) show that the eIF4E1b-type protein sequences cluster together, separately from the conserved eIF4E of flowering plants and from eIFiso4E, which diverged from eIF4E early in the flowering plant lineage (Patrick and Browning, 2012). In addition to completed and draft genomes, there is EST evidence of EIF4E1B and/or EIF4E1C in Brassica oleracea, Brassica napus, Raphanus raphanistrum, and Raphanus sativus, as well as Genome Survey Sequence support for the presence of an EIF4E1B-type gene in Sisymbrium irio (Supplemental Fig. S1).
Interestingly, the EIF4E1B and EIF4E1C genes of Arabidopsis and C. rubella are more closely conserved at the sequence level to each other than to EIF4E1B and EIF4E1C of Brassica spp. (Fig. 1), indicating that there has been recent duplication of EIF4E1B separately in each lineage. This is supported by the fact that while Arabidopsis and C. rubella. have EIF4E1B and EIF4E1C as a tandem duplication on one chromosome, B. rapa has EIF4E1B and EIF4E1C genes on separate chromosomes. Thellungiella spp., meanwhile, have only one copy of the gene.
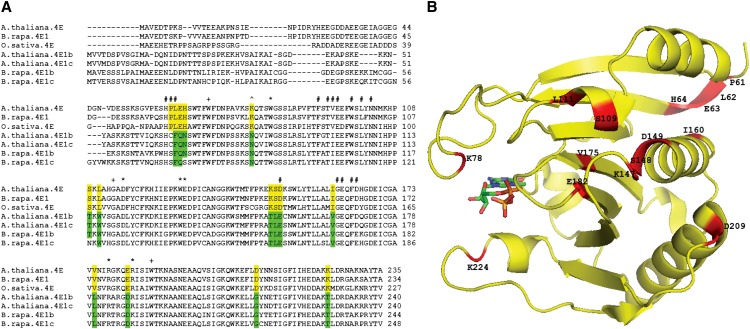
Fifteen residues within the protein have been identified as 90% conserved in flowering plant canonical eIF4E while consistently altered in eIF4E1b-type proteins; many of these residues are conserved as a specific divergent amino acid in eIF4E1b and eIF4E1c (Fig. 2; Supplemental Fig. S1). Residues that have been identified as being involved in cap binding from crystal structures of wheat (Triticum aestivum; Monzingo et al., 2007) and pea (Pisum sativum) eIF4E (Ashby et al., 2011) or by mutational analysis (Yeam et al., 2007; German-Retana et al., 2008) are well conserved in eIF4E1b-type proteins. One exception is the conserved positively charged residue at K78, which is predicted in the wheat eIF4E crystal structure to stabilize the negatively charged phosphate backbone of the cap structure. In eIF4E1b/eIF4E1c-type proteins, this residue is changed to Asn, which may weaken this interaction. However, mutation of this position in pea eIF4E had no effect on the ability to promote translation in yeast (Ashby et al., 2011).
Figure 2.
A, Representative flowering plant eIF4E and Brassica spp.-specific eIF4E1b/eIF4E1c sequences aligned by ClustalW2 (see Supplemental Fig. S1 for full alignment). Residues highlighted in yellow have 90% or greater identity in conserved flowering plant eIF4E sequences but are consistently altered in eIF4E1b-type sequences; residues highlighted in green are conserved divergent residues at these locations in eIF4E1b-type sequences. Residues marked # are conserved residues involved in interaction with eIF4G identified in yeast (Gross et al., 2003). Residues marked + are experimentally determined to be essential for cap binding (Yeam et al., 2007; German-Retana et al., 2008). Residues marked * are predicted to be involved in cap binding in plant eIF4E crystal structures (Monzingo et al., 2007; Ashby et al., 2011), and the residue marked ^ is predicted from the wheat eIF4E structure to form a salt bridge with the negatively charged phosphate backbone of the cap. B, Diverging residues in Arabidopsis eIF4E1b/eIF4E1c were modeled with PyMOL (DeLano, 2002) on the wheat eIF4E structure (Monzingo et al., 2007) as indicated.
Several mutations in eIF4E1b-type proteins occur at locations that are both well conserved between eIF4E of plants and mammals and predicted to be involved in eIF4E binding to eIF4G from a cocrystal structure of yeast eIF4E with a fragment of eIF4G (Gross et al., 2003). eIF4E residues P61, L62, and D149 (Fig. 2) are all predicted to be part of the eIF4G binding interface and are altered in eIF4E1b-type proteins. These changes appear to have altered the ability of eIF4E1b/eIF4E1c to interact with eIF4G compared with eIF4E (see below). While mutations in eIF4E that confer viral resistance are naturally occurring (Robaglia and Caranta, 2006) and directed mutagenesis has further identified residues conferring virus resistance (German-Retana et al., 2008; Ashby et al., 2011), the 15 conserved flowering plant eIF4E residues differing in eIF4E1b-type proteins do not overlap with these residues, with the exception of a K78 mutation, which confers virus resistance in pea (Ashby et al., 2011). Interestingly, transfer DNA (T-DNA) insertion mutants for EIF4E1B or EIF4E1C do not have any effect on turnip mosaic virus infection in Arabidopsis (Gallois et al., 2010). To date, neither EIF4E1B nor EIF4E1C has been reported to be a virus resistance gene in contrast to numerous reports of virus resistance attributed to EIF4E and EIFISO4E alleles (Wang and Krishnaswamy, 2012).
Analysis of homozygous T-DNA insertion lines for EIF4E1B (GK-874C07) or EIF4E1C (GK-361E12) do not show an obvious phenotype. Due to their close proximity on chromosome 1, it was not possible to obtain a double mutant even after screening more than 5,000 plants from a cross (data not shown).
EIF4E1B/EIF4E1C Expression
Due to their sequence similarity, EIF4E1B and EIF4E1C share a spot on many commonly used microarrays, limiting data as to whether either or both are expressed. However, RNA-Seq data can distinguish between the two genes and indicate that in flower tissue (Jiao and Meyerowitz, 2010; Niederhuth et al., 2013), shoot apical meristem (Torti et al., 2012), developing embryos (Nodine and Bartel, 2012), and the central cell of the female gamete (Schmid et al., 2012), EIF4E1B mRNA is expressed and associates with polysomes in flowers (Jiao and Meyerowitz, 2010); however, EIF4E1C mRNA was at much lower to undetectable levels in these tissues. In an analysis of 80 genomes released by the 1001 Genomes Project, EIF4E1C was predicted to be spontaneously deleted in 12 strains, suggesting that it is likely not providing any advantage to promote its retention in the genome (Cao et al., 2011).
Microarray data from the Arabidopsis eFP Browser (Winter et al., 2007) suggest that EIF4E1B is most highly expressed in developing flowers, while Genevestigator (Zimmermann et al., 2004) supports expression in shoots and reproductive tissue. EIF4E1B was identified as a gene up-regulated during pollen tube growth (Wang et al., 2008). Additionally, EIF4E1B was significantly enriched in pollen tubes grown by a semi in vivo method (Qin et al., 2009). EIF4E1B was identified as a sperm-enriched gene, while EIF4E was sperm depleted (Borges et al., 2008). In a microarray experiment investigating developing embryos, EIF4E1B was found to be expressed at high levels at the zygote stage of development relative to EIF4E, while EIF4E1C levels were near background levels (Xiang et al., 2011). Taken together, these findings may indicate a role for EIF4E1B in reproduction in Brassicaceae similar to that in Drosophila spp. or zebrafish.
eIFiso4E or eIF4E Is Required for Viability
The T-DNA knockout line for EIFISO4E (iso4e-1, Duprat et al., 2002) and the nonsense mutant for EIF4E (cucumovirus multiplication1 [cum1]; Yoshii et al., 2004) are viable and do not exhibit major developmental phenotypes individually. However, extensive attempts have been made by this laboratory to isolate an iso4e-1/cum1 double mutant without success. iso4e-1 plants heterozygous for cum1 are viable, but self-fertilized plants do not yield viable double mutants in the ratio expected for normal progeny (Table I). The defect appears to be embryo lethal, as nearly all planted seeds germinated and were successfully screened. Similarly, cum1 plants heterozygous for iso4E-1 do not yield viable double mutants (Supplemental Table S1). A lethal phenotype has also been reported with the iso4e/4e1 genotype, preventing the recovery of a double homozygous mutant (Callot and Gallois, 2014). These results suggest that EIF4E1B and EIF4E1C gene products are not sufficient to fulfill the necessary role for a canonical Class I eIF4E protein, either due to low or localized expression or loss of properties that contribute to translation initiation, such as binding eIF4G and/or mRNA cap structure.
Table I. Screening of iso4e-1 cum1/EIF4E progeny from self-fertilization.
One hundred seven seeds were planted on Murashige and Skoog agar plates, with 105 germinating and 103 successfully transplanted and screened. Recovery of iso4e-1 cum1/EIF4E was lower than expected (35%), and double homozygous mutant plants were not recovered.
| Genotype | No. with Genotype | Percentage of Total | Expected Mendelianb |
|---|---|---|---|
| iso4e-1 cum1/cum1 | 0/107 | 0 | 25% |
| iso4e-1 cum1/EIF4E | 37/107 | 35 | 50% |
| iso4e-1 EIF4E/EIF4E | 66/107 | 62 | 25% |
| Not screeneda | 4/107 | 4 | Not Applicable |
Seed did not germinate or died before screening. bExpected amounts if normal Mendelian inheritance.
Because EIF4E1B and EIF4E1C genes are products of a Brassicaceae-specific gene duplication, they may have specialized functions and/or lost the ability to function as cap-binding proteins. We sought to determine if eIF4E1b and eIF4E1c proteins are biochemically capable of performing the functions of cap-binding proteins in vitro, as well as in vivo using a yeast complementation system.
eIF4E1b and eIF4E1c Bind to m7GTP and Form Complexes with eIF4G
Arabidopsis eIF4E, eIF4E1b, and eIF4E1c were expressed in Escherichia coli and purified by affinity chromatography using 7-methyl guanosine triphosphate (m7GTP)-Sepharose (Fig. 3A). Both eIF4E1b and eIF4E1c were found to bind and elute from m7GTP-Sepharose in comparable yields to eIF4E (data not shown). eIF4E1b and eIF4E1c therefore seem to have biologically relevant cap-binding ability.
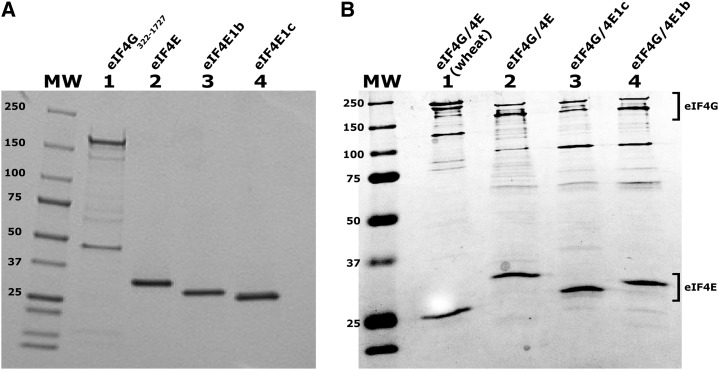
Figure 3.
PAGE analysis of purified proteins. A, Purified Arabidopsis eIF4G and eIF4E isoforms. Lane 1, eIF4G322–1727 (3 µg); lane 2, eIF4E (1.5 µg); lane 3, eIF4E1b (1.5 µg); and lane 4, eIF4E1c (1.5 µg), were separated by 12.5% SDS-PAGE and stained with Coomassie brilliant blue. B, Recombinant Arabidopsis eIF4F complexes from dicistronic constructs were expressed in E. coli and purified by m7GTP-Sepharose affinity and phosphocellulose chromatography. Lane 1, Wheat eIF4F (3 µg); lane 2, Arabidopsis eIF4G1–1727/eIF4E (3 µg); lane 3, eIF4G1–1727/eIF4E1c (3 µg); lane 4, eIF4G1–1727/eIF4E1b (3 µg). Proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. Note that the order of protein loading is different in A and B.
The initial selection of the coding sequence of Arabidopsis eIF4G (eIF4G322–1727) to express was made based on protein similarity to the N terminus of wheat eIF4G (Mayberry et al., 2007). However, subsequent peptide sequence data in the pep2pro database (http://fgcz-pep2pro.uzh.ch/index.php; Baerenfaller et al., 2011) suggested that upstream initiation codons are utilized. Because the precise initiation codon (or if there are multiple start sites, as occurs for mammalian eIF4G; [Coldwell et al., 2012]) is not known for Arabidopsis, the codon selected by The Arabidopsis Information Resource was also used to generate an expression construct for eIF4G1–1727, which includes all the peptides identified in the pep2pro database.
eIF4E, eIF4E1b, and eIF4E1c were coexpressed with Arabidopsis eIF4G322–1727, which retains the eIF4E binding site, to form eIF4F complexes that were purified by m7GTP-Sepharose affinity chromatography (Fig. 3B). eIF4E1b copurified with eIF4G322–1727 to form the eIF4F1b complex, and eIF4E1c copurified with eIF4G322–1727 to form the eIF4F1c complex. Both eIF4F1b and eIF4F1c purified with comparable yield to eIF4F, confirming that eIF4E1b and eIF4E1c were able to bind eIF4G and form a stable complex.
Purified eIF4E, eIF4E1b, and eIF4E1c were assayed by surface plasmon resonance (SPR) for their binding affinity to purified eIF4G322–1727 (Table II). The dissociation constant (KD) for eIF4E binding to eIF4G322–1727 was extremely tight, at 0.275 ± 0.002 nm. This finding is consistent with the measurement of the wheat eIF4G and eIF4E binding KD of 0.181 ± 0.002 nm (Mayberry et al., 2011). Surprisingly, eIF4E1b binding to eIF4G322–1727 was 1,640-fold weaker than eIF4E (451 ± 2 nm), while eIF4E1c binding was weaker still (970 ± 10 nm).
Table II. The binding affinity of purified Arabidopsis cap-binding proteins to eIF4G322–1727.
Affinities of the cap-binding proteins for eIF4G were measured by SPR.
| Cap-Binding Protein | KD |
|---|---|
| nm | |
| eIF4E1 | 0.275 ± 0.002 |
| eIF4E1b | 451 ± 2 |
| eIF4E1c | 970 ± 10 |
The eIF4E1b/eIF4E1c binding affinity to eIF4G322–1727 is lower than was observed in the wheat system for a mixed complex of eIFiso4E binding to eIF4G (14.3 ± 0.2 nm; Mayberry et al., 2011). It was previously shown that eIFiso4E was displaced from a mixed complex with wheat eIF4G by eIF4E, thus the correct binding partner is selectively favored (Mayberry et al., 2011). Based on the observed lower binding affinity, despite the ability of eIF4E1b or eIF4E1c to copurify with eIF4G322–1727 in vitro, it is unlikely either could form a complex with eIF4G in vivo in Arabidopsis unless eIF4E is absent (see Fig. 5 below).
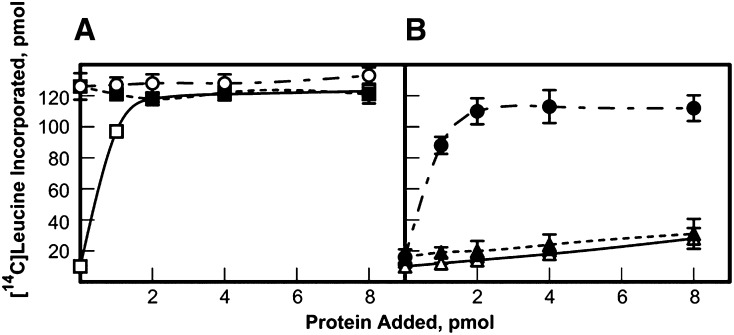
Figure 5.
Displacement of eIF4E1b from complex with eIF4G by eIF4E as measured by in vitro translation activity. A, The complex of eIF4G/eIF4E was presented with increasing amounts of eIF4E1b to determine if activity was reduced to the level of the eIF4G/eIF4E1b complex. White rectangle, Two picomoles of eIF4G titrated with eIF4E; black rectangle, 2 pmol of eIF4G/eIF4E titrated with additional eIF4E as indicated; and white circle, 2 pmol of eIF4G/eIF4E titrated with additional eIF4E1b as indicated. B, Alternatively, a mixed complex of eIF4G/eIF4E1b was presented with increasing amounts of eIF4E to determine if eIF4E1b could be displaced by eIF4E and form the more active eIF4G/eIF4E complex. The reaction mixture contained 4 pmol of barley α-amylase mRNA and 15 µL of a wheat germ S30 extract that had been depleted of eIF4F and eIFiso4F by passage over a m7GTP Sepharose column as described in “Materials and Methods.” White triangle, Two picomoles of eIF4G titrated with eIF4E1b as indicated; black triangle, 2 pmol of eIF4G/eIF4E1b titrated with additional eIF4E1b as indicated; black circle, 2 pmol of eIF4G/eIF4E1b titrated with additional eIF4E as indicated. Experiments were done in triplicate and averaged. The incorporation of [14C]Leu in the absence of any added factor was 10 pmol.
eIF4E1b and eIF4E1c Have Translation-Enhancing Activity in Vitro But Are Displaced by eIF4E
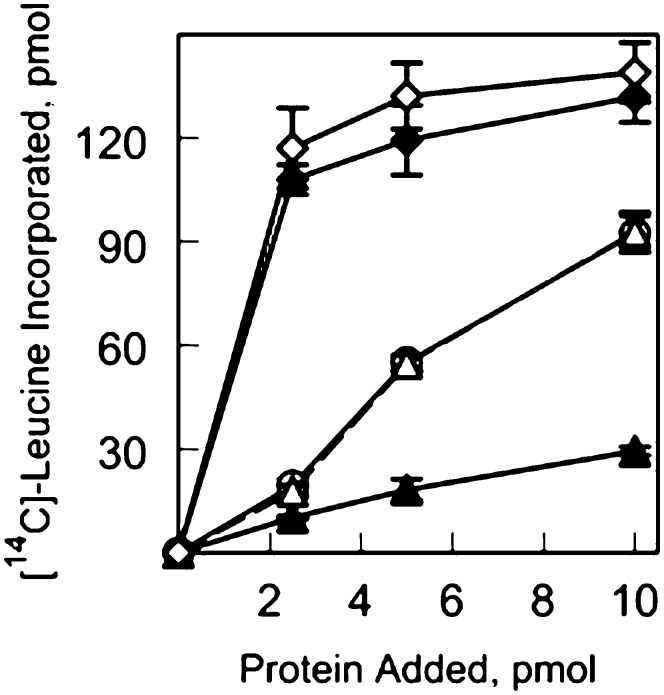
To measure the ability of eIF4E1b and eIF4E1c to function in the initiation of translation, in vitro translation assays in wheat germ S30 depleted of cap-binding complexes were carried out. Recombinant Arabidopsis eIF4G1–1727 was mixed with equimolar amounts of cap-binding proteins to form eIF4F complexes, and these were tested for their ability to translate mRNA compared to recombinant wheat eIF4F (Fig. 4). Arabidopsis eIF4G alone provided little stimulation of translation, while the conserved Arabidopsis eIF4F complex of eIF4G with eIF4E performed similarly to wheat eIF4F. eIF4G paired with either eIFE1b or eIF4E1c showed similar activity but required significantly higher concentrations of the complexes (approximately 5- to 10-fold) to approach the extent of stimulation of the eIF4F complex from either wheat or Arabidopsis.
Figure 4.
In vitro assay of eIF4E isoforms with eIF4G. Equal molar amounts of Arabidopsis eIF4G1–1727 and the respective eIF4E isoform were mixed and then added as indicated. The reaction mixture contained 5 pmol of barley (Hordeum vulgare) α-amylase mRNA and 15 µL of a wheat germ S30 extract that had been depleted of eIF4F and eIFiso4F by passage over a m7GTP Sepharose column as described in “Materials and Methods.” White diamond, Wheat eIF4F; black diamond, Arabidopsis eIF4G/eIF4E; white circle, Arabidopsis eIF4G/eIF4E1b; white triangle, Arabidopsis eIF4G/eIF4E1c; black triangle, Arabidopsis eIF4G. Note that curves for eIF4G/eIF4E1b and eIF4G/eIF4E1c overlap. Experiments were done in triplicate and averaged. The incorporation of [14C]Leu in the absence of any added factor was 5 pmol.
The contribution of eIF4E1b to in vitro translation was further examined with an assay placing eIF4E1b in competition with eIF4E for eIF4G to observe changes in activity of in vitro translation (Fig. 5). The complex of eIF4E/eIF4G was challenged with either additional eIF4E or eIF4E1b, and there was no significant change in the translational activity observed in either case (Fig. 5A); however, a complex of eIF4E1b/eIF4G, when challenged with eIF4E, clearly led to displacement of eIF4E1b to form the more stable and more translationally active complex of eIF4E/eIF4G (Fig. 5B). These results support the observed binding constants (Table II) and suggest that eIF4E must be absent for any complex to form in vivo between eIF4G and eIF4E1b/eIF4E1c. It is also evident that at least in vitro eIF4E1b does not act as a general translational repressor.
eIF4E1b and eIF4E1c Can Complement eIF4E Deletion in Yeast
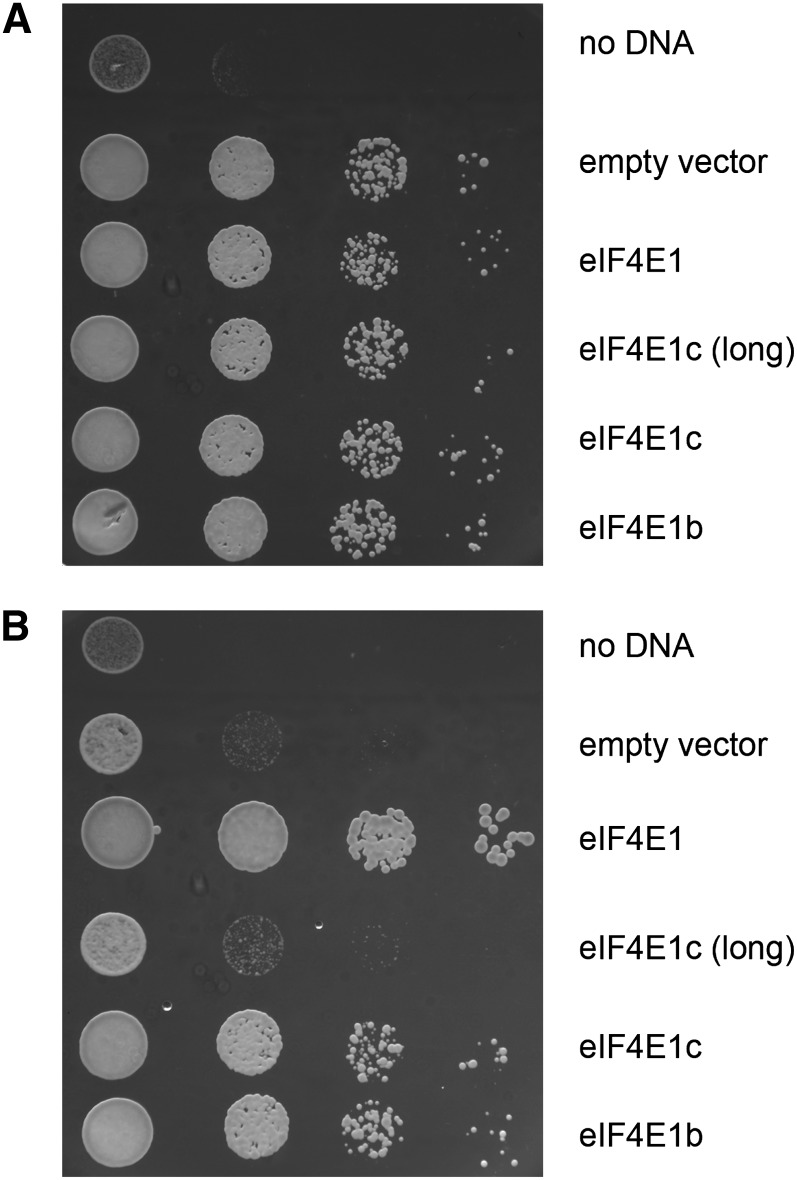
Arabidopsis eIF4E has previously been shown to be able to complement for an eIF4E deletion in yeast (Rodriguez et al., 1998). The in vitro data suggest that eIF4E1b and eIF4E1c are functional in that they bind to m7GTP-Sepharose and eIF4G. To further investigate the ability of eIF4E1b and eIF4E1c to function as bona fide cap-binding proteins, they were tested for their ability to complement eIF4E deletion in yeast. As shown in Figure 6, both EIF4E1B and EIF4E1C are able to substitute for yeast EIF4E gene deletion in vivo. This finding implies that eIF4E1b and eIF4E1c have biologically relevant ability to promote translation and, as shown in the in vitro experiments, retain sufficient cap-binding and eIF4G-binding properties in spite of their sequence differences from eIF4E. Thus, their inability to function in planta in a background lacking EIF4E and EIFISO4E suggests that their expression is highly localized or controlled.
Figure 6.
The ability of Arabidopsis eIF4E proteins to complement deletion of the eIF4E gene in yeast. Complementation was tested by introducing pG-1 plasmids for constitutive expression of Arabidopsis eIF4E genes into a yeast strain (T93C; Altmann et al., 1989) with eIF4E under control of a GAL promoter. Serial dilutions of midlog phase yeast were plated in 10-fold serial dilutions on SCM-Trp plates containing 2% Gal (A) or 2% Glc (B) and incubated at 30°C for 48 h. The experiment was performed in three biological replicates; representative results are shown.
A version of eIF4E1c protein with an additional N-terminal sequence predicted by The Arabidopsis Information Resource (Rhee et al., 2003) was also tested as EIF4E1C(long). This additional sequence is likely to be an artifact of gene assignment due to an incorrect prediction for the start site. The extra peptide sequence has no similarity to any known eIF4E peptide sequences and would be unique among the EIF4E1B-like genes as well as plant EIF4E genes. The EIF4E1C(long) gene was not able to complement the deletion of EIF4E in yeast, indicating the additional N-terminal amino acid residues may interfere with either cap recognition or eIF4G binding, preventing productive translation initiation. This is consistent with observations by our laboratory that some N- or C-terminal fusions of plant cap-binding proteins are not viable in vivo in yeast or in Arabidopsis (E. Levins, C. Tseng, and K. Browning, unpublished data).
CONCLUSION
Arabidopsis is the best model system for plant translation initiation currently available due to the availability of knockout lines of many translation initiation factors for in vivo study as well as the successful purification of many recombinant proteins for these factors. Arabidopsis and other members of the Brassicaceae family have noncanonical eIF4E related genes present in their genomes. There is little evidence for EIF4E1C gene expression, and the EIF4E1B gene is expressed at low levels in most tissues, though microarray and RNA-Seq data support enrichment in reproductive tissue. Unfortunately, AteIF4E antibody cross reacts poorly with eIF4E1b and eIF4E1c, so it is not possible to confirm that these proteins are actually produced in vivo (data not shown). The eIF4E1b and eIF4E1c are bona fide cap-binding proteins sufficient to promote translation initiation in yeast and display translation initiation activity in vitro. However, due to their low binding affinity for eIF4G relative to eIF4E and their low level of expression, it seems unlikely that these genes contribute substantially to translation initiation in most plant tissues. Increasing numbers of noncanonical eIF4E family members have been described in eukaryotes (Rhoads, 2009). As more plant genomes are sequenced, other events similar to the EIF4E duplication and divergence in Brassicaceae may be observed. E. grandis and F. vesca both encode apparent divergent EIF4E genes, though there is not yet available data to tell whether they are expressed or if the genes are conserved among close relatives. Given the data for poor interaction with eIF4G and its identification as a sperm-enriched gene, one might expect the role of eIF4E1b to be similar to what has been described in vertebrate oocytes: binding the 7-methylguanosine cap and excluding eIF4G binding to repress translation. The germline enrichment of Arabidopsis EIF4E1B seems in line with findings from vertebrates (Minshall et al., 2007), C. elegans (Amiri et al., 2001), and Drosophila spp. (Hernández et al., 2012) of specialized eIF4E isoforms with roles in reproductive tissue. However, the in vitro data from this work suggest that eIF4E1b does not contribute to translational repression in this manner, though mRNA-specific repression or interaction with other proteins cannot be ruled out. Arabidopsis EIF4E1B and EIF4E1C seem nonessential, as T-DNA insertion plants develop normally (data not shown); however, the strong conservation of the EIF4E1B-type genes within the Brassicaceae family implies that they provide some as-yet-unidentified contribution. In addition, crosses between EIF4E (cum1) and either EIF4E1B or EIF4E1C T-DNA lines do not have any observable phenotype or issues with reproduction (data not shown). Thus, although eIF4E1b and eIF4E1c appear to be able to function as cap-binding proteins in vitro and in yeast, it remains to be determined if EIF4E1C is even expressed in planta, and the actual levels of eIF4E1b protein expression and localization remain to be determined. Based on the large difference (approximately 1,600-fold) in binding affinity of eIF4E1b for eIF4G relative to eIF4E, it is unlikely eIF4E1b plays any role in canonical translation, but perhaps has a Brassicaceae-specific role in some tissues where eIF4E protein is not expressed and would allow eIF4E1b to interact with eIF4G or other proteins.
MATERIALS AND METHODS
In Silico Analysis
eIF4E and eIFiso4E gene sequences for alignment and analysis were collected from Phytozome (Goodstein et al., 2012) and BLAST searches to GenBank sequences (Benson et al., 2012). Alignment was performed by ClustalW2 (Larkin et al., 2007); residues defined as conserved in eIF4E were those with 90% or greater identity in the canonical coding from among the aligned sequences. The Phylogeny.fr pipeline (Dereeper et al., 2008) was used for alignment and phylogenetic tree generation with alignment by MUSCLE and tree construction by PhyML using 500 bootstrap replicates (see Supplemental Fig. S2).
eIF4E and eIFiso4E Cross
Mutant lines for eIF4E (cum1, a nonsense point mutation) and eIFiso4E (iso4E-1, Sainsbury Laboratory Arabidopsis Transformants library) have been previously described (Duprat et al., 2002; Yoshii et al., 2004). Crosses between these two lines were performed in both directions and the T2 progeny screened by PCR to identify wild-type, heterozygous, or double homozygous lines. iso4E-1 and cum1 lines were screened with primers as described in Supplemental Table S2.
Construction of eIF4E1, eIF4E1b, eIF4E1c, eIF4E1c(long), eIF4G322–1727, and eIF4G1–1727, and eIF4F, eIF4F1b, and eIF4F1c Expression Constructs
Initial attempts to express Arabidopsis (Arabidopsis thaliana) eIF4G protein from complementary DNA clones were unsuccessful. Using DNAWorks (Hoover and Lubkowski, 2002), Arabidopsis eIF4G322–1727, eIF4E1, eIF4E1b, eIF4E1c, and eIF4E1c(long) were designed with codon optimization for expression in Escherichia coli and assembled by overlap PCR of oligonucleotides (Supplemental Figs. S3-S8; Supplemental Tables S3-S7; Horton et al., 1989). Initial cloning of eIF4G322–1727 was into pCR-Blunt-II-TOPO (Invitrogen) followed by subcloning into pSB1AC3 (Shetty et al., 2008) and pET22b vectors (Novagen). eIF4E genes were cloned in one step. eIF4G322–1727 was cloned into pCR-Blunt-II-TOPO in four sections and then assembled in pSB1AC3. Full-length eIF4G1–1727 was created by cloning a synthetic DNA sequence (GenScript) to provide the missing N-terminal sequence to eIF4G322–1727; the restriction site used to ligate the synthetic DNA was then altered to match wild-type protein sequence by site-directed mutagenesis (Mutagenex). The pET22b eIF4G322–1727 vector was used to clone eIF4E1, eIF4E1b, and eIF4E1c genes at a site 3′ of the eIF4G coding region to create dicistronic plasmids for expression of eIF4F, eIF4F1b, and eIF4F1c complexes.
Purification of Recombinant Proteins
eIF4E proteins were expressed in BL21(DE3) E. coli and purified as previously described by m7GTP-Sepharose affinity chromatography (Mayberry et al., 2007). eIF4F complexes were expressed in Tuner(DE3) E. coli (Novagen) and purified as previously described for wheat (Triticum aestivum) eIFiso4F (Mayberry et al., 2007, 2011). eIF4G322–1727 and eIF4G1–1727 were expressed in Tuner(DE3) E. coli and purified as previously described for wheat eIF4G (Mayberry et al., 2007, 2011). Wheat eIF4A and eIF3 were purified as previously described (Lax et al., 1986; Mayberry et al., 2007).
In Vitro Translation Assay
Arabidopsis eIF4G and cap-binding proteins were assayed in an in vitro translation assay using wheat germ S30 extract that had been depleted of cap-binding proteins and complexes. Three 4-mL portions of m7GTP-Sepharose (GE Biosciences) were equilibrated in 20 mm HEPES, pH 7.6, 120 mm KAc, 5 mm MgAc2, 10% (v/v) glycerol, and 7.15 mm β-mercaptoethanol. A 2-mL aliquot of S30 extract was used to exchange the buffer from each of the three 4-mL portions of the m7GTP-Sepharose. Twenty-five milliliters of wheat germ S30 extract (Lax et al., 1986; Browning and Mayberry, 2006) was mixed for 15 min with 4 mL of m7GTP-Sepharose by rocking on ice; the supernatant was collected, and the process was repeated with the remaining two portions of 4 mL of m7GTP-Sepharose. The eIF4F/eIFiso4F-depleted S30 extract was aliquoted, flash frozen, and stored at –80°C.
The 50-μL translation assay reaction mixture contained 24 mm HEPES-KOH, pH 7.6, 2 mm MgAc2, 100 mm KAc, 30 mm KCl, 2.4 mm dithiothreitol, 0.1 mm spermine, 1 mm ATP, 0.2 mm GTP, 34 μm [14C]Leu, 50 μm 19 amino acids (−Leu), 7.8 μm creatine phosphate, 3 μg creatine kinase, 0.75 A260 units of yeast (Saccharomyces cerevisiae) tRNA, 15 μL of depleted S30 extract, 4 to 5 pmol barley (Hordeum vulgare) α-amylase mRNA, 10 µg of recombinant wheat eIF4A, 0.5 µg of recombinant wheat eIF4B, 6 µg of native wheat eIF3, and the indicated amounts of eIF4F, eIF4G, and/or cap-binding proteins. Incubation was for 30 min at 27°C, and the amount of [14C]Leu incorporated into protein was determined as previously described (Lax et al., 1986; Browning and Mayberry, 2006; Mayberry et al., 2007).
SPR Analysis
SPR (Biacore) experiments were carried out as described previously (Mayberry et al., 2011) at Biosensor Tools by Dr. David Myszka. Briefly, protein binding was measured at 25°C using a Biacore 2000 optical biosensor equipped with a CM4 sensor chip in running buffer (20 mm HEPES, 100 mm KCl, 1.5 mm tris[2-carboxyethyl]phosphine 0.1 mm EDTA, 100 μm m7GTP, 5% glycerol, 0.01% Tween 20, and 0.1 mg mL–1 bovine serum albumin, pH 7.6). eIF4G322–1727 was amine coupled at three surface densities (500, 1,370, and 4,430 resonance units). eIF4E1, eIF4E1b, and eIF4E1c proteins were tested for eIF4G binding in 3-fold dilution series performed in triplicate. For eIF4E1b and eIF4E1c, the highest concentration tested was 1.5 µm, and for eIF4E, it was 66.7 nm. Response data for each protein were fit globally to a 1:1 interaction model using Scrubber2 (Biologic Software) across the three eIF4G surface densities. Sample binding data are shown in Supplemental Figure S9.
Yeast Complementation of eIF4E
The yeast strain T93C (Altmann et al., 1989), containing a chromosomal deletion of eIF4E and a plasmid with eIF4E under control of a Gal promoter (eIF4E::LEU2 ura3 trp1 leu2 [pGal1-eIF4E URA3]), was transformed with pG-1 vectors (with an added NcoI site N terminus to BamHI in the cloning region) containing Arabidopsis eIF4E constructs. pG-1 provides constitutive gene expression and a TRP1 marker (Schena et al., 1991). Positive transformants were verified by plasmid reisolation and sequencing.
Yeast strains were grown overnight and then diluted to 0.1 optical density at A600, grown to midlog phase (approximately 0.3 optical density), and plated in serial dilutions on synthetic complete medium (SCM)-Trp plus 2% Gal and SCM-Trp plus 2% Glc. Plates were incubated at 30°C for 48 h and screened for ability of Arabidopsis eIF4E proteins to rescue yeast growth. This experiment was performed in triplicate.
Supplemental Data
The following figures are available in the online version of this article.
Supplemental Figure S1. ClustalW2 alignment of eIF4E genes of flowering plants.
Supplemental Figure S2. MUSCLE alignment of eIF4E/eIFiso4E for phylogeny construction (FASTA).
Supplemental Figure S3. eIF4E1 cloning.
Supplemental Figure S4. eIF4E1b cloning.
Supplemental Figure S5. eIF4E1c cloning.
Supplemental Figure S6. eIF4E1c(long) cloning.
Supplemental Figure S7. eIF4G322–1727 cloning.
Supplemental Figure S8. eIF4G1–321 segment.
Supplemental Figure S9. SPR curves for eIF4E, eIF4E1b, and eIF4E1c binding to eIF4G.
Supplemental Table S1. Screening of cum1 iso4e-1/EIFISO4E progeny from self-fertilization.
Supplemental Table S2. DNA oligonucleotides used for plant screening.
Supplemental Table S3. Oligonucleotides used for overlap PCR of eIF4E1.
Supplemental Table S4. Oligonucleotides used for overlap PCR of eIF4E1b.
Supplemental Table S5. Oligonucleotides used for overlap PCR of eIF4E1c.
Supplemental Table S6. Oligonucleotides used for overlap PCR of eIF4E1c(long).
Supplemental Table S7. Oligonucleotides used for overlap PCR of eIF4G.
Supplementary Material
Acknowledgments
We thank Monique Maldonado, Wing Tuet, and Travis Quintanilla for technical assistance.
Glossary
- RNA-Seq
RNA Sequencing
- T-DNA
transfer DNA
- m7GTP
7-methyl guanosine triphosphate
- SPR
surface plasmon resonance
- KD
dissociation constant
- SCM
synthetic complete medium
Footnotes
This work was supported by the National Science Foundation (grant nos. MCB1052530 and Arabidopsis 2010 S–0000335 to K.S.B.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aitken CE, Lorsch JR. (2012) A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 19: 568–576 [DOI] [PubMed] [Google Scholar]
- Altmann M, Sonenberg N, Trachsel H. (1989) Translation in Saccharomyces cerevisiae: initiation factor 4E-dependent cell-free system. Mol Cell Biol 9: 4467–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri A, Keiper BD, Kawasaki I, Fan Y, Kohara Y, Rhoads RE, Strome S. (2001) An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development 128: 3899–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby JA, Stevenson CE, Jarvis GE, Lawson DM, Maule AJ. (2011) Structure-based mutational analysis of eIF4E in relation to sbm1 resistance to pea seed-borne mosaic virus in pea. PLoS ONE 6: e15873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K, Hirsch-Hoffmann M, Svozil J, Hull R, Russenberger D, Bischof S, Lu Q, Gruissem W, Baginsky S. (2011) pep2pro: a new tool for comprehensive proteome data analysis to reveal information about organ-specific proteomes in Arabidopsis thaliana. Integr Biol DOI: 10.1039/C0IB00078G [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. (2012) GenBank. Nucleic Acids Res 40: D48–D53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD. (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol 148: 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning KS, Mayberry L. (2006) In vitro translation of plant viral RNA. Curr Protoc Microbiol Chapter 16: 1–13 [DOI] [PubMed] [Google Scholar]
- Callot C, Gallois JL. (2014) Pyramiding resistances based on translation initiation factors in Arabidopsis is impaired by male gametophyte lethality. Plant Signal Behav 9: e27940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Coldwell MJ, Sack U, Cowan JL, Barrett RM, Vlasak M, Sivakumaran K, Morley SJ. (2012) Multiple isoforms of the translation initiation factor eIF4GII are generated via use of alternative promoters, splice sites and a non-canonical initiation codon. Biochem J 448: 1–11 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C. (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J 32: 927–934 [DOI] [PubMed] [Google Scholar]
- Evsikov AV, Marín de Evsikova C. (2009) Evolutionary origin and phylogenetic analysis of the novel oocyte-specific eukaryotic translation initiation factor 4E in Tetrapoda. Dev Genes Evol 219: 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire ER, Dhalia R, Moura DM, da Costa Lima TD, Lima RP, Reis CR, Hughes K, Figueiredo RC, Standart N, Carrington M, et al. (2011) The four trypanosomatid eIF4E homologues fall into two separate groups, with distinct features in primary sequence and biological properties. Mol Biochem Parasitol 176: 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois JL, Charron C, Sánchez F, Pagny G, Houvenaghel MC, Moretti A, Ponz F, Revers F, Caranta C, German-Retana S. (2010) Single amino acid changes in the turnip mosaic virus viral genome-linked protein (VPg) confer virulence towards Arabidopsis thaliana mutants knocked out for eukaryotic initiation factors eIF(iso)4E and eIF(iso)4G. J Gen Virol 91: 288–293 [DOI] [PubMed] [Google Scholar]
- German-Retana S, Walter J, Doublet B, Roudet-Tavert G, Nicaise V, Lecampion C, Houvenaghel MC, Robaglia C, Michon T, Le Gall O. (2008) Mutational analysis of plant cap-binding protein eIF4E reveals key amino acids involved in biochemical functions and potyvirus infection. J Virol 82: 7601–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JEG, Wagner G. (2003) Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115: 739–750 [DOI] [PubMed] [Google Scholar]
- Hernández G, Altmann M, Sierra JM, Urlaub H, Diez del Corral R, Schwartz P, Rivera-Pomar R. (2005) Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila. Mech Dev 122: 529–543 [DOI] [PubMed] [Google Scholar]
- Hernández G, Han H, Gandin V, Fabian L, Ferreira T, Zuberek J, Sonenberg N, Brill JA, Lasko P. (2012) Eukaryotic initiation factor 4E-3 is essential for meiotic chromosome segregation, cytokinesis and male fertility in Drosophila. Development 139: 3211–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover DM, Lubkowski J. (2002) DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res 30: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68 [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Meyerowitz EM. (2010) Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol Syst Biol 6: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi B, Lee K, Maeder DL, Jagus R. (2005) Phylogenetic analysis of eIF4E-family members. BMC Evol Biol 5: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiper BD, Lamphear BJ, Deshpande AM, Jankowska-Anyszka M, Aamodt EJ, Blumenthal T, Rhoads RE. (2000) Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J Biol Chem 275: 10590–10596 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lax SR, Lauer SJ, Browning KS, Ravel JM. (1986) Purification and properties of protein synthesis initiation and elongation factors from wheat germ. Methods Enzymol 118: 109–128 [DOI] [PubMed] [Google Scholar]
- Mayberry LK, Allen ML, Nitka KR, Campbell L, Murphy PA, Browning KS. (2011) Plant cap-binding complexes eukaryotic initiation factors eIF4F and eIFISO4F: molecular specificity of subunit binding. J Biol Chem 286: 42566–42574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberry LK, Dennis MD, Leah Allen M, Ruud Nitka K, Murphy PA, Campbell L, Browning KS. (2007) Expression and purification of recombinant wheat translation initiation factors eIF1, eIF1A, eIF4A, eIF4B, eIF4F, eIF(iso)4F, and eIF5. Methods Enzymol 430: 397–408 [DOI] [PubMed] [Google Scholar]
- Minshall N, Reiter MH, Weil D, Standart N. (2007) CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J Biol Chem 282: 37389–37401 [DOI] [PubMed] [Google Scholar]
- Monzingo AF, Dhaliwal S, Dutt-Chaudhuri A, Lyon A, Sadow JH, Hoffman DW, Robertus JD, Browning KS. (2007) The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond. Plant Physiol 143: 1504–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhuth CE, Patharkar OR, Walker JC. (2013) Transcriptional profiling of the Arabidopsis abscission mutant hae hsl2 by RNA-Seq. BMC Genomics 14: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP. (2012) Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick RM, Browning KS. (2012) The eIF4F and eIFiso4F complexes of plants: an evolutionary perspective. Comp Funct Genomics 2012: 287814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrat OA, Mikitova V, Bush MS, Browning KS, Doonan JH. (2007) Control of protein translation by phosphorylation of the mRNA 5′-cap-binding complex. Biochem Soc Trans 35: 1634–1637 [DOI] [PubMed] [Google Scholar]
- Ptushkina M, Berthelot K, von der Haar T, Geffers L, Warwicker J, McCarthy JEG. (2001) A second eIF4E protein in Schizosaccharomyces pombe has distinct eIF4G-binding properties. Nucleic Acids Res 29: 4561–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R. (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al. (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads RE. (2009) eIF4E: new family members, new binding partners, new roles. J Biol Chem 284: 16711–16715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C, Caranta C. (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci 11: 40–45 [DOI] [PubMed] [Google Scholar]
- Robalino J, Joshi B, Fahrenkrug SC, Jagus R. (2004) Two zebrafish eIF4E family members are differentially expressed and functionally divergent. J Biol Chem 279: 10532–10541 [DOI] [PubMed] [Google Scholar]
- Rodriguez CM, Freire MA, Camilleri C, Robaglia C. (1998) The Arabidopsis thaliana cDNAs coding for eIF4E and eIF(iso)4E are not functionally equivalent for yeast complementation and are differentially expressed during plant development. Plant J 13: 465–473 [DOI] [PubMed] [Google Scholar]
- Schena M, Picard D, Yamamoto KR. (1991) Vectors for constitutive and inducible gene expression in yeast. Methods Enzymol 194: 389–398 [DOI] [PubMed] [Google Scholar]
- Schmid MW, Schmidt A, Klostermeier UC, Barann M, Rosenstiel P, Grossniklaus U. (2012) A powerful method for transcriptional profiling of specific cell types in eukaryotes: laser-assisted microdissection and RNA sequencing. PLoS ONE 7: e29685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty RP, Endy D, Knight TF., Jr (2008) Engineering BioBrick vectors from BioBrick parts. J Biol Eng 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song A, Labella S, Korneeva NL, Keiper BD, Aamodt EJ, Zetka M, Rhoads RE. (2010) A C. elegans eIF4E-family member upregulates translation at elevated temperatures of mRNAs encoding MSH-5 and other meiotic crossover proteins. J Cell Sci 123: 2228–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettweiler G, Kowanda M, Lasko P, Sonenberg N, Hernández G. (2012) The distribution of eIF4E-family members across Insecta. Comp Funct Genomics 2012: 960420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti S, Fornara F, Vincent C, Andrés F, Nordström K, Göbel U, Knoll D, Schoof H, Coupland G. (2012) Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24: 444–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Krishnaswamy S. (2012) Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol Plant Pathol 13: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH. (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol 148: 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D, Venglat P, Tibiche C, Yang H, Risseeuw E, Cao Y, Babic V, Cloutier M, Keller W, Wang E, et al. (2011) Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiol 156: 346–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Vatsyayan J, Gao C, Bakkenist CJ, Hu J. (2010) Sumoylation of eIF4E activates mRNA translation. EMBO Rep 11: 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeam I, Cavatorta JR, Ripoll DR, Kang BC, Jahn MM. (2007) Functional dissection of naturally occurring amino acid substitutions in eIF4E that confers recessive potyvirus resistance in plants. Plant Cell 19: 2913–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, Ishikawa M. (2004) The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J Virol 78: 6102–6111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.