Disruption of a glucomannan synthase alters cellulose crystallinity and spatial distribution, yielding thinner adherent mucilage with increased density in seeds of Arabidopsis.
Abstract
Mannans are hemicellulosic polysaccharides that are considered to have both structural and storage functions in the plant cell wall. However, it is not yet known how mannans function in Arabidopsis (Arabidopsis thaliana) seed mucilage. In this study, CELLULOSE SYNTHASE-LIKE A2 (CSLA2; At5g22740) expression was observed in several seed tissues, including the epidermal cells of developing seed coats. Disruption of CSLA2 resulted in thinner adherent mucilage halos, although the total amount of the adherent mucilage did not change compared with the wild type. This suggested that the adherent mucilage in the mutant was more compact compared with that of the wild type. In accordance with the role of CSLA2 in glucomannan synthesis, csla2-1 mucilage contained 30% less mannosyl and glucosyl content than did the wild type. No appreciable changes in the composition, structure, or macromolecular properties were observed for nonmannan polysaccharides in mutant mucilage. Biochemical analysis revealed that cellulose crystallinity was substantially reduced in csla2-1 mucilage; this was supported by the removal of most mucilage cellulose through treatment of csla2-1 seeds with endo-β-glucanase. Mutation in CSLA2 also resulted in altered spatial distribution of cellulose and an absence of birefringent cellulose microfibrils within the adherent mucilage. As with the observed changes in crystalline cellulose, the spatial distribution of pectin was also modified in csla2-1 mucilage. Taken together, our results demonstrate that glucomannans synthesized by CSLA2 are involved in modulating the structure of adherent mucilage, potentially through altering cellulose organization and crystallization.
Mannan polysaccharides are a complex set of hemicellulosic cell wall polymers that are considered to have both structural and storage functions. Based on the particular chemical composition of the backbone and the side chains, mannan polysaccharides are classified into four types: pure mannan, glucomannan, galactomannan, and galactoglucomannan (Moreira and Filho, 2008; Wang et al., 2012; Pauly et al., 2013). Each of these polysaccharides is composed of a β-1,4-linked backbone containing Man or a combination of Glc and Man residues. In addition, the mannan backbone can be substituted with side chains of α-1,6-linked Gal residues. Mannan polysaccharides have been proposed to cross link with cellulose and other hemicelluloses via hydrogen bonds (Fry, 1986; Iiyama et al., 1994; Obel et al., 2007; Scheller and Ulvskov, 2010). Furthermore, it has been reported that heteromannans with different levels of substitution can interact with cellulose in diverse ways (Whitney et al., 1998). Together, these observations indicate the complexity of mannan polysaccharides in the context of cell wall architecture.
CELLULOSE SYNTHASE-LIKE A (CSLA) enzymes have been shown to have mannan synthase activity in vitro. These enzymes polymerize the β-1,4-linked backbone of mannans or glucomannans, depending on the substrates (GDP-Man and/or GDP-Glc) provided (Richmond and Somerville, 2000; Liepman et al., 2005, 2007; Pauly et al., 2013). In Arabidopsis (Arabidopsis thaliana), nine CSLA genes have been identified; different CSLAs are responsible for the synthesis of different mannan types (Liepman et al., 2005, 2007). CSLA7 has mannan synthase activity in vitro (Liepman et al., 2005) and has been shown to synthesize stem glucomannan in vivo (Goubet et al., 2009). Disrupting the CSLA7 gene results in defective pollen growth and embryo lethality phenotypes in Arabidopsis, indicating structural or signaling functions of mannan polysaccharides during plant embryo development (Goubet et al., 2003). A mutation in CSLA9 results in the inhibition of Agrobacterium tumefaciens-mediated root transformation in the rat4 mutant (Zhu et al., 2003). CSLA2, CSLA3, and CSLA9 are proposed to play nonredundant roles in the biosynthesis of stem glucomannans, although mutations in CSLA2, CSLA3, or CSLA9 have no effect on stem development or strength (Goubet et al., 2009). All of the Arabidopsis CSLA proteins have been shown to be involved in the biosynthesis of mannan polysaccharides in the plant cell wall (Liepman et al., 2005, 2007), although the precise physiological functions of only CSLA7 and CSLA9 have been conclusively demonstrated.
In Arabidopsis, when mature dry seeds are hydrated, gel-like mucilage is extruded to envelop the entire seed. Ruthenium red staining of Arabidopsis seeds reveals two different mucilage layers, termed the nonadherent and the adherent mucilage layers (Western et al., 2000; Macquet et al., 2007a). The outer, nonadherent mucilage is loosely attached and can be easily extracted by shaking seeds in water. Compositional and linkage analyses suggest that this layer is almost exclusively composed of unbranched rhamnogalacturonan I (RG-I) (>80% to 90%), with small amounts of branched RG-I, arabinoxylan, and high methylesterified homogalacturonan (HG). By contrast, the inner, adherent mucilage layer is tightly attached to the seed and can only be removed by strong acid or base treatment, or by enzymatic digestion (Macquet et al., 2007a; Huang et al., 2011; Walker et al., 2011). As with the nonadherent layer, adherent mucilage is also mainly composed of unbranched RG-I, but with small numbers of arabinan and galactan ramifications (Penfield et al., 2001; Willats et al., 2001; Dean et al., 2007; Macquet et al., 2007a, 2007b; Arsovski et al., 2009; Haughn and Western, 2012). There are also minor amounts of pectic HG in the adherent mucilage, with high methylesterified HG in the external domain compared with the internal domain of the adherent layer (Willats et al., 2001; Macquet et al., 2007a; Rautengarten et al., 2008; Sullivan et al., 2011; Saez-Aguayo et al., 2013). In addition, the adherent mucilage contains cellulose (Blake et al., 2006; Macquet et al., 2007a), which is entangled with RG-I and is thought to anchor the pectin-rich mucilage onto seeds (Macquet et al., 2007a; Harpaz-Saad et al., 2011, 2012; Mendu et al., 2011; Sullivan et al., 2011). As such, Arabidopsis seed mucilage is considered to be a useful model for investigating the biosynthesis of cell wall polysaccharides and how this process is regulated in vivo (Haughn and Western, 2012).
Screening for altered seed coat mucilage has led to the identification of several genes encoding enzymes that are involved in the biosynthesis or modification of mucilage components. RHAMNOSE SYNTHASE2/MUCILAGE-MODIFIED4 (MUM4) is responsible for the synthesis of UDP-l-Rha (Usadel et al., 2004; Western et al., 2004; Oka et al., 2007). The putative GALACTURONSYLTRANSFERASE11 can potentially synthesize mucilage RG-I or HG pectin from UDP-d-GalUA (Caffall et al., 2009). GALACTURONSYLTRANSFERASE-LIKE5 appears to function in the regulation of the final size of the mucilage RG-I (Kong et al., 2011, 2013). Mutant seeds defective in these genes display reduced thickness of the extruded mucilage layer compared with wild-type Arabidopsis seeds.
RG-I deposited in the apoplast of seed coat epidermal cells appears to be synthesized in a branched form that is subsequently modified by enzymes in the apoplast. MUM2 encodes a β-galactosidase that removes Gal residues from RG-I side chains (Dean et al., 2007; Macquet et al., 2007b). β-XYLOSIDASE1 encodes an α-l-arabinfuranosidase that removes Ara residues from RG-I side chains (Arsovski et al., 2009). Disruptions of these genes lead to defective hydration properties and affect the extrusion of mucilage. Furthermore, correct methylesterification of mucilage HG is also required for mucilage extrusion. HG is secreted into the wall in a high methylesterified form that can then be enzymatically demethylesterified by pectin methylesterases (PMEs; Bosch and Hepler, 2005). PECTIN METHYLESTERASE INHIBITOR6 (PMEI6) inhibits PME activities (Saez-Aguayo et al., 2013). The subtilisin-like Ser protease (SBT1.7) can activate other PME inhibitors, but not PMEI6 (Rautengarten et al., 2008; Saez-Aguayo et al., 2013). Disruption of either PMEI6 or SBT1.7 results in the delay of mucilage release.
Although cellulose is present at low levels in adherent mucilage, it plays an important adhesive role for the attachment of mucilage pectin to the seed coat epidermal cells. The orientation and amount of pectin associated with the cellulose network is largely determined by cellulose conformation properties (Macquet et al., 2007a; Haughn and Western, 2012). Previous studies have demonstrated that CELLULOSE SYNTHASE A5 (CESA5) is required for the production of seed mucilage cellulose and the adherent mucilage in the cesa5 mutant can be easily extracted with water (Harpaz-Saad et al., 2011, 2012; Mendu et al., 2011; Sullivan et al., 2011).
Despite all of these discoveries, large gaps remain in the current knowledge of the biosynthesis and functions of mucilage polysaccharides in seed coats. In this study, we show that CSLA2 is involved in the biosynthesis of mucilage glucomannan. Furthermore, we show that CSLA2 functions in the maintenance of the normal structure of the adherent mucilage layer through modifying the mucilage cellulose ultrastructure.
RESULTS
Expression Pattern of CSLA2
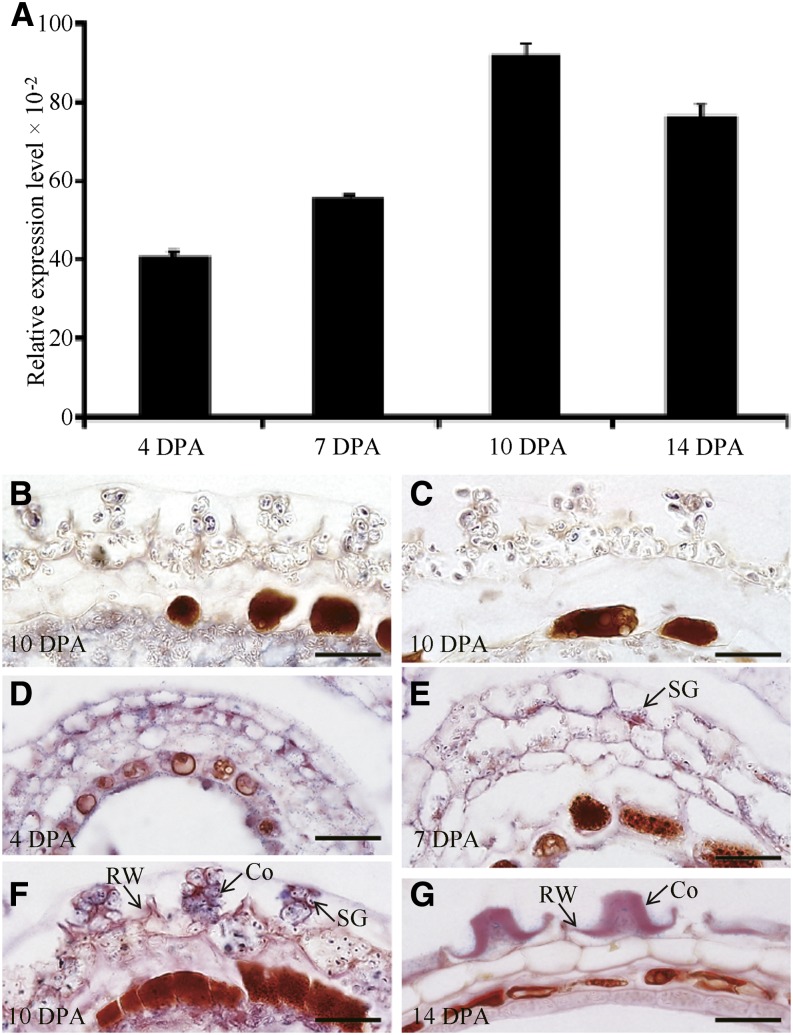
As shown in Supplemental Figure S1, RNA transcript abundance profiles of the CSLA2 (At5g22740) gene were detected in various vegetative and floral tissues examined from the Arabidopsis AtGenExpress dataset (Schmid et al., 2005). Further quantitative real-time (qRT)-PCR results were consistent with the initial expression profiling results (Liepman et al., 2007; Dean et al., 2011). In addition, transcriptome analyses using RNA extracted from laser-capture dissected seed coat tissue (Le et al., 2010; http://seedgenenetwork.net/arabidopsis) indicated that CSLA2 transcript levels are low in the seed coat during early embryogenesis, and that CSLA2 expression increases from the linear cotyledon stage onward (Supplemental Fig. S1A). According to a global expression dataset covering seed coat development, CSLA2 was significantly up-regulated at 11 DPA compared with 7 DPA (Dean et al., 2011). These results suggested its potential role during the formation of seed mucilage. Using qRT-PCR, we examined the expression pattern of CSLA2 in seeds at different developmental stages (4, 7, 10, and 14 DPA). As shown in Figure 1A, the CSLA2 gene was expressed throughout the seed differentiation process, and its maximum expression level was observed at 10 DPA with seeds at the bent cotyledon embryo stage. To detect the precise spatial expression of CSLA2 within seeds, we performed in situ hybridization to determine the specific cell type of the seed coat in which CSLA2 is expressed. The results demonstrated that CSLA2 expression was detected in all seed coat cell types, including the epidermal cells of the outer integument, where large quantities of mucilage are produced (Fig. 1, D–G). This result encouraged us to further investigate the potential functions of CSLA2 in modifying mucilage.
Figure 1.
Expression of CSLA2 in developing Arabidopsis seeds. A, qRT-PCR analysis of CSLA2 expression during seed development at the indicated time points. Error bars represent sd values (n = 3). B, In situ hybridization using a CSLA2-specific antisense probe to the csla2-1 seed coat. C, In situ hybridization using a CSLA2-specific sense probe to the wild-type seed coat. D to G, In situ hybridization using a CSLA2-specific antisense probe to the wild-type seed coat. Co, Columella; RW, Radial cell wall; SG, Starch granule. Bar = 20 μm.
The CSLA2 Is Targeted to the Golgi Apparatus
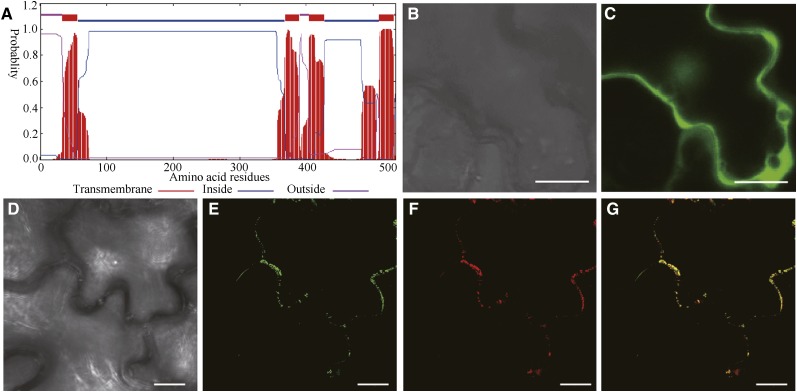
Sequence analysis using the TMHMM2.0 program for prediction of transmembrane helices in proteins predicted that CSLA2 contains four transmembrane helices, a cytoplasmic N terminus, a second cytoplasmic region located between the second and the third transmembrane helices, and a very short cytoplasmic C-terminal region (Fig. 2A). To confirm the actual subcellular location of CSLA2, a GFP-tagged CSLA2 (GFP-CSLA2) construct was cotransfected with the Golgi marker Man49-mCherry (Nelson et al., 2007) into tobacco (Nicotiana benthamiana) leaves. The examination of the epidermal cells of the infiltrated tobacco showed that the GFP control protein was distributed throughout the cytoplasm and the nucleus (Fig. 2C), whereas the GFP-CSLA2 signals displayed a punctate pattern (Fig. 2E). Colocalization experiments revealed that the GFP-CSLA2 signal was co-localized with the Man49-mCherry signal (Fig. 2G). The results demonstrated that CSLA2 is a Golgi-localized protein.
Figure 2.
Subcellular localization of fluorescence protein-tagged CSLA2. A, Sequence analysis using the TMHMM2.0 program for prediction of transmembrane helices of CSLA2 protein. B and C, Differential interference contrast (DIC) image (B) and the corresponding fluorescent signals (C) of tobacco leaf epidermal cells expressing GFP alone. D to G, DIC image (D) and the corresponding GFP-CSLA2 signals (E), mCherry-tagged Man 49 (F), and a merged image (G) of tobacco leaf epidermal cell expressing GFP-CSLA2 and the Golgi-localized mCherry-tagged Man 49. Note that the GFP-CSLA2 signals show a punctate pattern and co-localize with mCherry-tagged Man 49 signals. Bar = 20 μm in B and C; 10 μm in D to G.
csla2 Mutants Have Thinner But Denser Adherent Mucilage
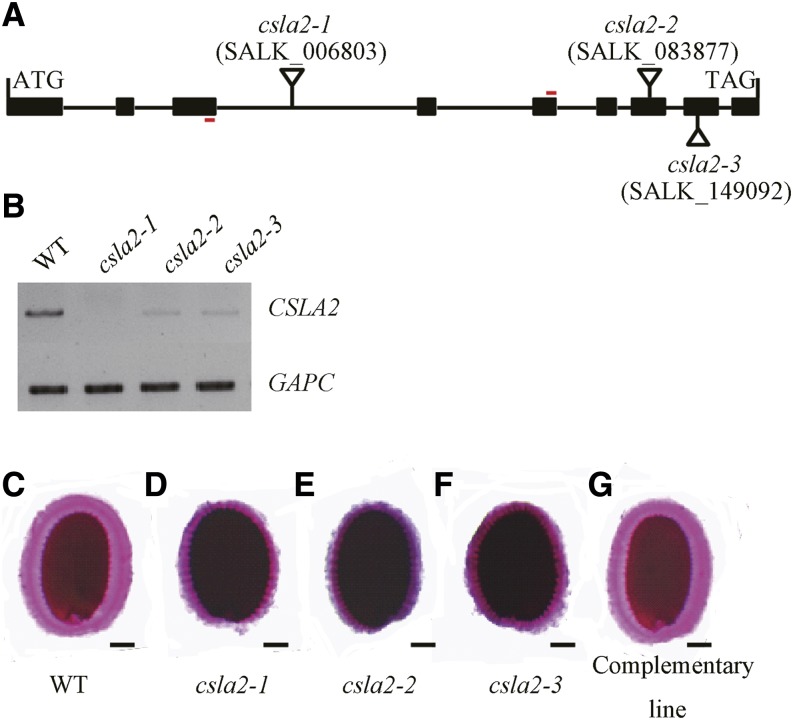
To extend our understanding of the functions of CSLA2, three independent homozygous transfer (T)-DNA (T-DNA) insertion lines, csla2-1 (SALK_006803), csla2-2 (SALK_083877), and csla2-3 (SALK_149092), were identified (Fig. 3A) and characterized. The csla2-1 line was completely null for the expression of CSLA2 as assessed by reverse transcription (RT)-PCR. Significant reductions in CSLA2 transcript expression were observed in the other two lines (csla2-2 and csla2-3; Fig. 3B). Ruthenium red staining of homozygous seeds showed that these three mutant alleles have similar mucilage defects, displaying thinner and darker mucilage halos compared with the wild type (Fig. 3, C–F). However, plants of the three mutant lines were morphologically indistinguishable from the wild type throughout development and reached similar sizes in their above-ground growth. The null mutant csla2-1 was primarily used for further analysis in subsequent experiments.
Figure 3.
CSLA2 is involved in maintaining the correct adherent mucilage structure. A, Schematic representation of the structure of CSLA2. The sites and orientation of insertion lines in csla2-1 (SALK_006803), csla2-2 (SALK_083877), and csla2-3 (SALK_149092) are indicated. Boxes and connecting lines represent exons and introns, respectively. Red bars indicate primers for RT-PCR amplification used in B. B, Expression of the CSLA2 gene as revealed by RT-PCR on siliques of wild-type (WT) and csla2 mutant plants. GAPC was amplified as the loading control. C to G, Arabidopsis seeds stained with ruthenium red after being shaken in water. C, Wild type. D to F, Three csla2 mutant alleles. G, Complemented csla2-1. Bar = 100 μm. [See online article for color version of this figure.]
To verify that the mutant phenotypes observed resulted from the knock-out of the CSLA2 gene, a wild-type copy of the CSLA2 gene driven by the Cauliflower mosaic virus 35S promoter was transformed into csla2-1 mutant plants. As shown in Figure 3G, the seed mucilage phenotypes of at least 18 independent transgenic lines were examined; all were restored to the wild-type characteristics, indicating that the phenotypes observed in csla2-1 did result from the mutation in the CSLA2 gene.
Mild shaking in water will remove the outer nonadherent mucilage layer, leaving the inner adherent mucilage layer attached to the seed surface (Western et al., 2000; Macquet et al., 2007a). Adherent mucilage mass and volume were measured to confirm the reduction of adherent mucilage thickness. Interestingly, no obvious difference was observed in adherent mucilage mass between wild-type and csla2-1 seeds, whereas the adherent mucilage volume of csla2-1 was substantially reduced 47% compared with the wild type (Table I). This reduction in volume implied a 92% increase in adherent mucilage density in csla2-1 compared with the wild type. These results illustrated that csla2-1 had thinner adherent mucilage with increased density.
Table I. Main parameters of adherent mucilage from wild-type and csla2-1 seeds.
The mass value is the average mass of adherent mucilage of 10,000 seeds of triplicate assays ± sd. The size value is the average size of adherent mucilage of 100 seeds of triplicate assays ± sd.
| Parameter | Mass | Size | Density |
|---|---|---|---|
| μg | mm3 | mg mL−1 | |
| Wild type | 1.78 ± 0.03 | 0.45 ± 0.05 | 3.92 ± 0.49 |
| csla2-1 | 1.77 ± 0.02 | 0.24 ± 0.05a | 7.53 ± 1.67a |
Significantly different from the wild type (P < 0.005).
Glucomannan Content Is Reduced in csla2-1 Mucilage
To determine whether the monosaccharide composition in the mucilage of csla2-1 seeds differed from that of the wild type, mucilage was extracted sequentially with distilled water, 0.2 n NaOH, and 2 n NaOH. These extracts were then hydrolyzed and their monosaccharide compositions were analyzed by HPLC. As shown in Table II, a significant decrease (approximately 30%) was observed for Man and Glc contents in both the nonadherent and 2 n NaOH-extracted mucilage from csla2-1 seeds. This result suggested a potential connection between the reduction of Man and the reduction of Glc. In the plant cell walls, Glc monomers link to each other to form cellulose, or link with Xyl/Man to form xyloglucan/glucomannan (Somerville, 2006; Sandhu et al., 2009; Pauly et al., 2013). Because there were no appreciable differences in the GalA, Rha, cellulosic Glc, Ara, or Xyl content in the extracted mucilage between wild-type and csla2-1 seeds (Table II), we hypothesized that the Glc reduction in the csla2-1 mutant might result from the loss of glucomannan in both the nonadherent and the adherent mucilage. In addition, Gal content decreased approximately 20% in 2 n NaOH-extracted mucilage of the mutant (Table II), suggesting that the reduced glucomannan might be substituted by Gal. The total amount of glucomannan-related sugars only accounted for a small percentage of the total sugar content in each extracted mucilage. Consequently, the reductions of Man and Glc were not reflected in the differences observed in csla2-1 across all three extractions (Table II). Considering the combined mucilage density and sugar composition results (Tables I and II), we deduce that the smaller adherent mucilage halo observed in the csla2-1 mutant is a result of compactness, not loss of mucilage.
Table II. Composition of sequentially extracted mucilage extracts from Arabidopsis wild-type and csla2-1 seeds.
The values are the average (mg g−1 dry seeds) of triplicate assays ± sd. ND, not detected.
| Sugar Composition | Wild Type | csla2-1 |
|---|---|---|
| Nonadherent mucilage polysaccharides | ||
| Man | 0.85 ± 0.03 | 0.59 ± 0.03a |
| Noncellulosic Glc | 0.24 ± 0.03 | 0.17 ± 0.01a |
| Gal | 0.18 ± 0.01 | 0.15 ± 0.04 |
| GalA | 8.34 ± 0.13 | 8.52 ± 0.21 |
| Rha | 7.32 ± 0.32 | 7.29 ± 0.06 |
| Ara | 0.06 ± 0.01 | 0.06 ± 0.00 |
| Xyl | 0.39 ± 0.03 | 0.41 ± 0.02 |
| Cellulosic Glc | ND | ND |
| Total sugars | 17.38 ± 0.56 | 17.19 ± 0.37 |
| 0.2 n NaOH-extracted mucilage polysaccharides | ||
| Man | 0.48 ± 0.01 | 0.43 ± 0.05 |
| Noncellulosic Glc | 0.52 ± 0.05 | 0.46 ± 0.01 |
| Gal | 0.41 ± 0.02 | 0.44 ± 0.09 |
| GalA | 2.87 ± 0.21 | 2.84 ± 0.07 |
| Rha | 2.05 ± 0.08 | 2.14 ± 0.12 |
| Ara | 0.25 ± 0.01 | 0.22 ± 0.04 |
| Xyl | 0.28 ± 0.04 | 0.26 ± 0.01 |
| Cellulosic Glc | ND | ND |
| Total sugars | 6.86 ± 0.42 | 6.79 ± 0.39 |
| 2 n NaOH-extracted mucilage polysaccharides | ||
| Man | 1.01 ± 0.02 | 0.71 ± 0.01a |
| Noncellulosic Glc | 1.03 ± 0.08 | 0.71 ± 0.05a |
| Gal | 0.60 ± 0.02 | 0.48 ± 0.01 |
| GalA | 4.99 ± 0.50 | 5.06 ± 0.13 |
| Rha | 3.44 ± 0.23 | 3.47 ± 0.09 |
| Ara | 0.43 ± 0.01 | 0.46 ± 0.02 |
| Xyl | 0.51 ± 0.03 | 0.53 ± 0.07 |
| Cellulosic Glc | 4.82 ± 0.37 | 4.77 ± 0.28 |
| Total sugars | 16.83 ± 1.26 | 16.19 ± 0.66 |
| Total mucilage polysaccharides | ||
| Total mucilage sugars | 41.07 ± 2.24 | 40.17 ± 1.42 |
| Total mucilage Man | 2.34 ± 0.06 | 1.73 ± 0.09a |
| Total mucilage noncellulosic Glc | 1.79 ± 0.16 | 1.34 ± 0.07a |
| Total mucilage cellulosic Glc | 4.82 ± 0.37 | 4.77 ± 0.28 |
| DM of mucilage pectin (%) | 34.33 ± 1.58 | 22.68 ± 1.72a |
Significantly different from the wild type (P < 0.005).
The degree of methylesterification (DM) impacts cell wall structure and mucilage adhesion, because unesterified HG regions can cross link via Ca2+ ions to form stronger gels (Voiniciuc et al., 2013). As shown in Table II, mucilage DM was determined to be reduced significantly in the csla2-1 mutant (approximately 12% reduction) compared with the wild type.
The Structure of the Mucilage Planar Surface Is Altered in csla2 Mutants
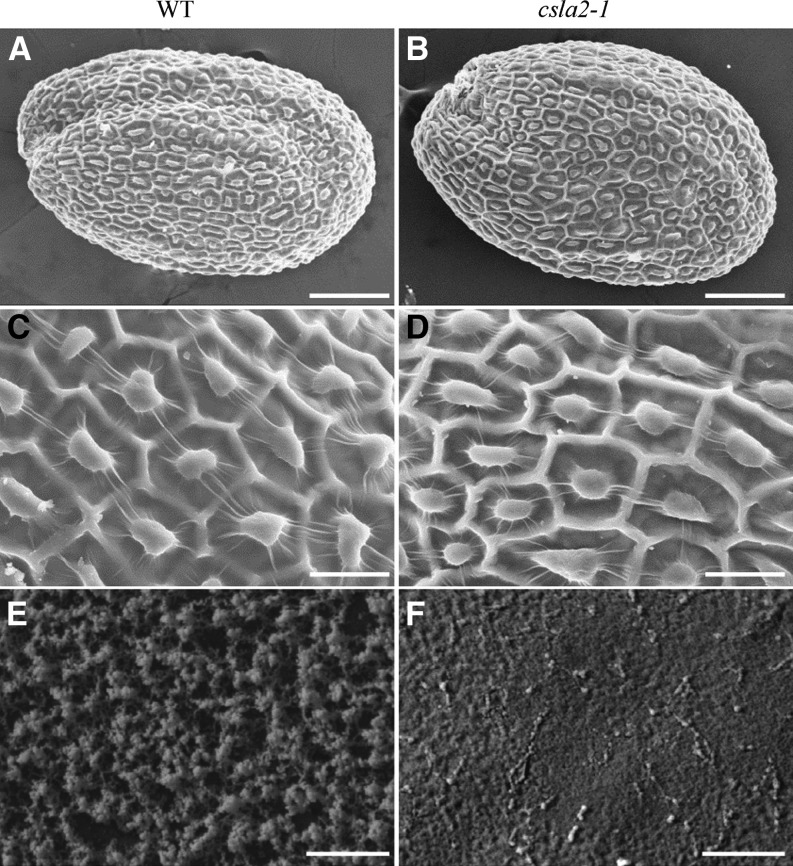
To determine whether mutation in CSLA2 affects mucilage structure or seed coat morphology during seed coat development and differentiation, cross sections of resin-embedded developing seeds were stained with toluidine blue O and examined. Comparison of csla2-1 and wild-type seed coats at different stages revealed no obvious differences in mucilage accumulation or in cell differentiation (Supplemental Fig. S2). As viewed by scanning electron microscopy (SEM), Arabidopsis seed coat epidermal cells appear as hexagonal epidermal cells outlined by reinforced radial cell walls with a volcano-shaped columella (Beeckman et al., 2000; Western et al., 2000; Stork et al., 2010). Inspection of mature dry seeds of the three mutant lines revealed no alteration in radial cell walls, columella, or mucilage pockets around the columella (Fig. 4, A–D; Supplemental Fig. S3A–F). When the outer nonadherent mucilage was removed by water extraction, cryo-SEM examination of the water-imbibed seeds showed that the mucilage surface seemed more compact in csla2-1 compared with that of the wild type (Fig. 4, E and F). This more compact mucilage surface phenotype was also observed in the other two mutant lines (csla2-2 and csla2-3; Supplemental Fig. S3, G–I).
Figure 4.
Analysis of dry and hydrated seeds by SEM. A to D, The surface morphology of dry mature Arabidopsis wild-type (WT; A and C) and csla2-1 mutant (B and D) seeds viewed with SEM. E and F, Cryo-SEM of mucilage extruded from dry mature Arabidopsis wild-type (E) and csla2-1 mutant (F) seeds hydrated in water. Bar = 100 μm in A and B; 25 μm in C and D; 5 μm in E and F.
The Biochemical Properties of Nonmannan Polysaccharides Are Not Altered in csla2-1 Mucilage
To investigate the structure of adherent mucilage in the wild type and the csla2-1 mutant, glycosyl linkage composition analysis was performed on the 2 n NaOH-extracted mucilage. Similar to the monosaccharide composition analysis (Table II), few changes were observed between the wild type and csla2-1 (Table III). The obvious differences observed were decreases in 4-linked Man (4-Manp), 4,6-linked Man (4,6-Manp), 4-linked Glc (4-Glcp), and 4,6-linked Glc (4,6-Glcp) in csla2-1 versus wild-type mucilage (Table III). Furthermore, the reduction of Man residues (4-Manp, 4,6-Manp) were in proportion to the reduction of Glc residues (4-Glcp, 4,6-Glcp), which further supported the putative glucomannan synthase activity for CSLA2. The level of t-Galp is also slightly decreased in csla2-1 mucilage.
Table III. Linkage analysis of 2 n NaOH-extracted mucilage from Arabidopsis wild-type and csla2-1 seeds.
The values are the average of two assays, with variance less than 5% for all samples. Results are given as the mean molar percentage. Values are scaled to monosaccharide analysis in Table II.
| Sugar and Linkage | Wild Type | csla2-1 |
|---|---|---|
| Man | ||
| t-Manp | 1.15 | 1.13 |
| 4-Manp | 4.26 | 2.86 |
| 4,6-Manp | 2.92 | 2.16 |
| Glc | ||
| t-Glcp | 0.16 | 0.14 |
| 4-Glcp | 5.13 | 3.87 |
| 6-Glcp | 0.76 | 0.54 |
| 2,4-Glcp | 0.50 | 0.51 |
| 3,4-Glcp | 0.77 | 0.62 |
| 4,6-Glcp | 1.16 | 0.47 |
| Gal | ||
| t-Galp | 2.39 | 1.87 |
| 3-Galp | 1.43 | 1.21 |
| 4-Galp | 1.13 | 1.08 |
| Rha | ||
| t-Rhap | 2.16 | 2.50 |
| 2-Rhap | 23.27 | 24.63 |
| 2,4-Rhap | 5.72 | 5.86 |
| GalA | ||
| t-GalAp | 1.68 | 1.91 |
| 4-GalAp | 34.19 | 35.95 |
| 2,4-GalAp | 1.91 | 2.40 |
| Ara | ||
| t-Araf | 1.67 | 1.78 |
| 3-Araf | 1.33 | 1.43 |
| 5-Araf | 1.25 | 1.57 |
| Xyl | ||
| t-Xylp | 0.69 | 0.84 |
| 4-Xylp | 3.38 | 3.42 |
| 2,4-Xylp | 0.49 | 0.62 |
| 3,4-Xylp | 0.50 | 0.63 |
To determine whether more subtle changes in the chemical structure of the mucilage occur in csla2-1 extracts, 28 monoclonal antibodies that recognize most major classes of cell wall polysaccharides were employed to probe the mucilage by ELISA. This antibody collection includes antibodies for pectin (RG-I, HG, arabinan, galactan, and arabinogalactan), hemicelluloses (xylan, xyloglucan, and heteromannan), and glycoproteins (extensin and arabinogalactan protein). As shown in Supplemental Figure S4, antibodies directed to the RG-I backbone, extensin, xylan, arabinan, and HG showed relatively strong binding to mucilage extracts, indicating the presence of these polymers in the mucilage. Other antibodies, including LM21 and CCRC-M170 directed to heteromannan (Marcus et al., 2010) and acetylated mannan (Zhang et al., 2014), respectively, showed very low binding to mucilage, confirming the low contents of these epitopes in mucilage. JIM5 and JIM7 can detect low methylesterified HG and high methylesterified HG (Clausen et al., 2003), respectively. Compared with the wild type, JIM5 displayed stronger epitope binding with the csla2-1 nonadherent mucilage. JIM7 showed weaker epitope binding in the csla2-1 nonadherent mucilage than in the wild-type mucilage, indicating a relative dearth of high methylesterified HG recognition in the mutant. This result further reinforced the notion that pectin DM was reduced in csla2-1 mucilage. No differential binding to wild-type and csla2-1 mucilage was observed for other antibodies.
To further examine whether the csla2-1 mucilage phenotype resulted from a change in the size of mucilage polysaccharides, the macromolecular parameters of the mucilage polysaccharides were examined using high-performance size exclusion chromatography (HPSEC) combined with refractive index detection. The elution profiles and macromolecular properties are listed in Supplemental Figure S5. Consistent elution profiles and macromolecular properties were observed for the sequential mucilage extracts from csla2-1 and wild-type seeds.
Crystalline Cellulose Content Is Reduced in csla2-1 Seeds
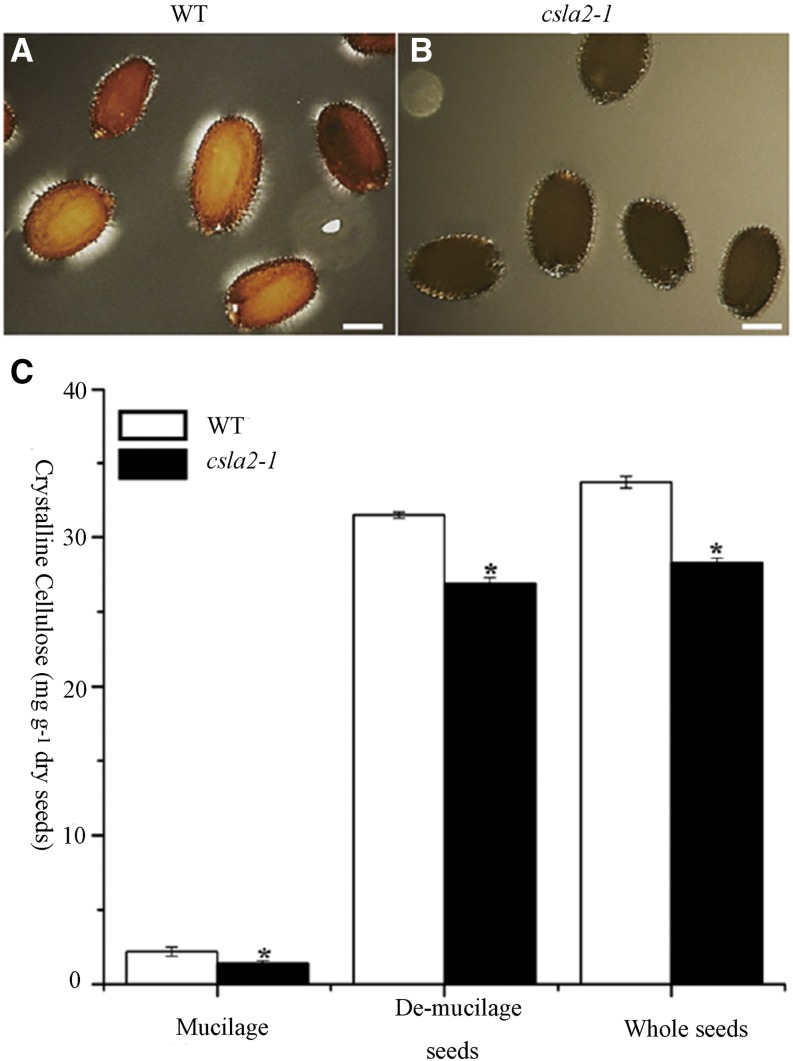
A previous report showed that highly ordered cellulose microfibrils can produce birefringence of polarized light within mucilage (Sullivan et al., 2011). As shown in Figure 5A, wild-type seeds have brighter regions with visible rays under polarized light, whereas the csla2-1 seeds only display bright spots on the edges of the seeds (Fig. 5B). This implied that the content of highly ordered cellulose microfibrils may be significantly reduced in the adherent mucilage of csla2-1. Subsequently, crystalline cellulose content in mucilage was calculated by subtracting crystalline cellulose content in demucilaged seeds from that in whole seeds, and it was found that csla2-1 mucilage had a 30% reduction in crystalline cellulose compared with the wild type (Fig. 5C).
Figure 5.
Crystalline cellulose content is reduced in the csla2-1 mutant. A and B, Observations of birefringence of polarized light by highly ordered cellulose microfibrils in adherent mucilage released from mature, imbibed Arabidopsis wild-type (WT; A) and csla2-1 (B) seeds. C, Crystalline cellulose content determined by the Updegraff (1969) method. Error bars represent sd (n = 3). Asterisks indicate a significant difference from the wild type (P < 0.005). Similar results were obtained in two biological repeats. Bar = 200 μm. [See online article for color version of this figure.]
The Deposition of Crystalline Cellulose Is Altered in csla2-1 Mucilage
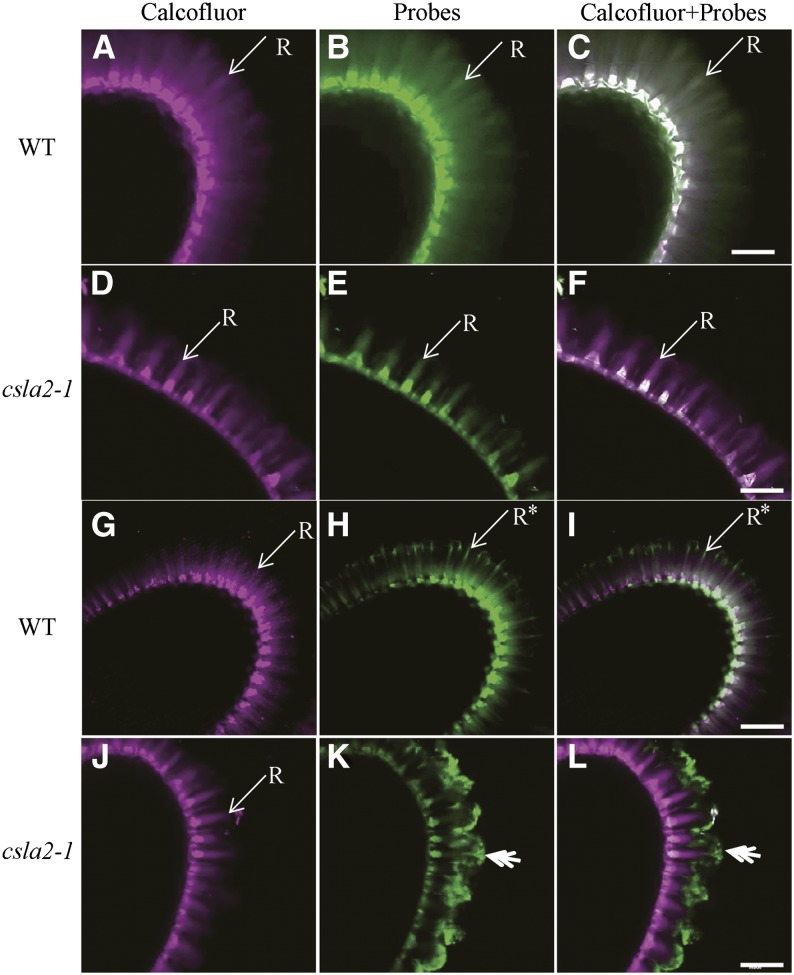
Heteromannan and crystalline cellulose was shown previously to be distributed in specific patterns within adherent mucilage (Blake et al., 2006; Sullivan et al., 2011; Lee et al., 2012). To further examine whether the spatial distribution of cellulose in adherent mucilage was modified in csla2-1, whole mount immunolabeling with probes that detect heteromanan and cellulose was carried out on mature dry seeds. Calcofluor White, which stains cellulose, callose, and other weakly substituted β-glycans, was used as the counter stain. Calcofluor White binding to wild-type mucilage displayed a combination of intense rays originating from the tops of the columella, and diffuse labeling between rays (Fig. 6, A and G; Sullivan et al., 2011). However, in the csla2-1 mutant, the mucilage rays emanating from the tops of the columella were observed as tufts and were shorter, and the diffuse labeling between rays was not obvious (Fig. 6, D and J). The LM21 antibody, which recognizes heteromannan specifically (Marcus et al., 2010), labeled adherent mucilage in a similar manner as Calcofluor White. The LM21 immunolabeling results between wild-type and csla2-1 seeds were similar to the Calcofluor White results (Fig. 6, B, C, E, and F).
Figure 6.
In situ localization of mannan and crystalline cellulose in adherent mucilage released from wild-type (WT) and csla2-1 seeds. A, D, G, and J, Staining of β-glycans with Calcofluor White. B and E, Indirect immunofluorescence detection of LM21 binding to heteromannan. C and F, Composite images of double labeling with calcofluor and LM21. H and K, Indirect immunofluorescence detection of His-tagged CBM3a with high affinity to crystalline cellulose. I and L, Composite images of double labeling with calcofluor and CBM3a. Asterisks indicate CBM3a labeling of mucilage not overlapped with calcofluor at the ends of the rays in the wild type. Double arrows indicate CBM3a binding of the external domain of the adherent mucilage-like mushroom-cap in csla2-1. R, Ray. Bar = 50 μm.
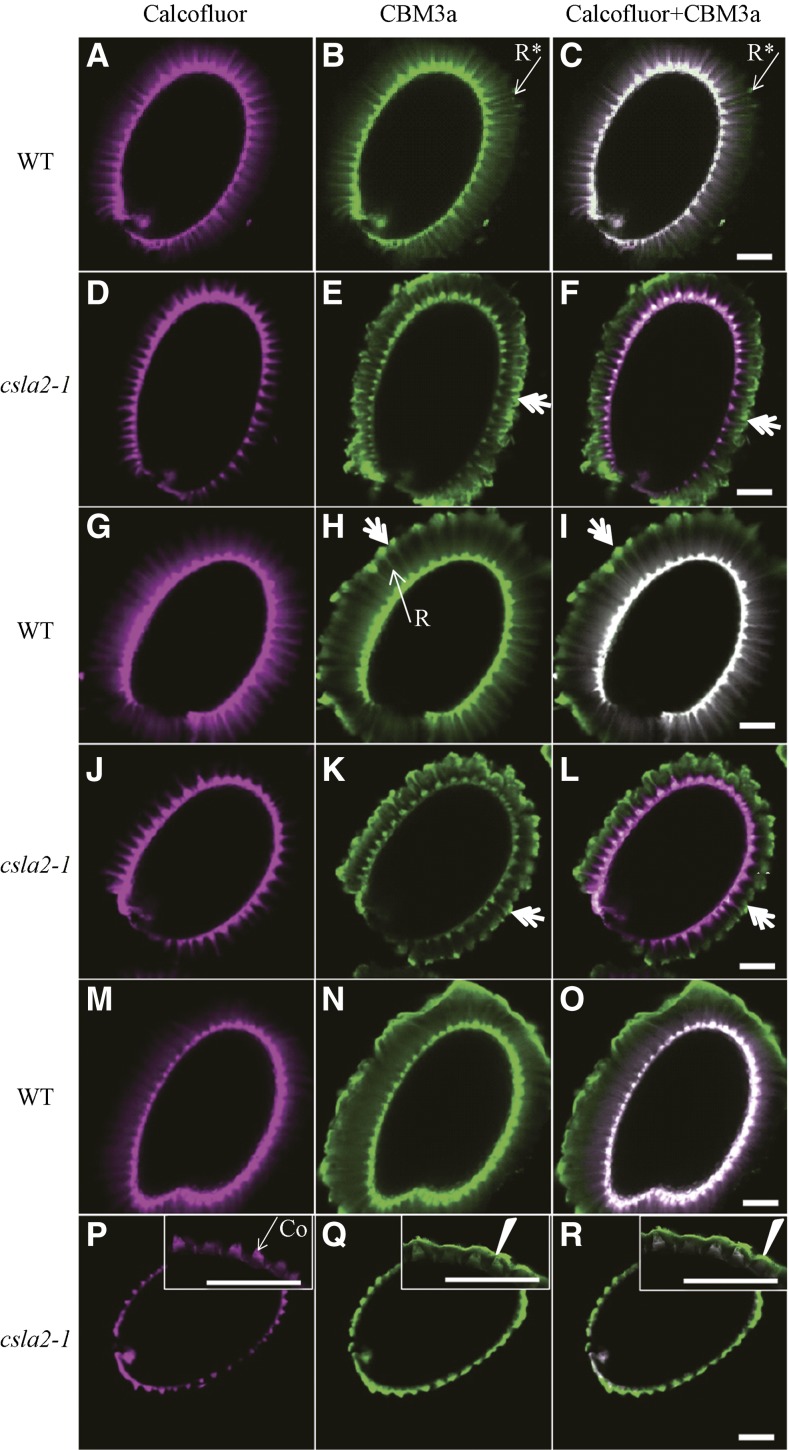
A carbohydrate-binding module with higher affinity to crystalline cellulose, designated CBM3a, was used to investigate the spatial distribution of cellulose (Blake et al., 2006). CBM3a labeling showed similar binding pattern with calcofluor staining in the wild type, but composite images of calcofluor staining and CBM3a labeling did not completely overlap; CBM3a labeling was stronger at the peripheral ends of cellulose rays (Figs. 6, G–I, and 7, A–C; Supplemental Fig. S6A). Nevertheless, in the csla2-1 mutant, CBM3a signals were prominently observed in the peripheral ends of cellulose rays, forming a “mushroom-cap” covering the columella; a CBM3a-type “mushroom-cap” was not observed by calcofluor staining. However, in the internal domain, CBM3a signals showed very weak binding along rays, and no diffuse labeling was observed between rays (Figs. 6, J–L, and 7, D–F; Supplemental Fig. S6B).
Figure 7.
Detection of crystalline cellulose with CBM3a in enzyme-treated Arabidopsis seed mucilage. A to F, Seeds without enzyme treatment were labeled as controls. G to L, Seeds were labeled after mannanase treatment. M to R, Seeds were labeled after EβG treatment. A, D, G, J, M, and P, Calcofluor labeling. B, E, H, K, N, and Q, CBM3a labeling. C, F, I, L, O, and R, Composite images of double labeling with Calcofluor and CBM3a. Asterisks indicate CBM3a labeling of mucilage not overlapped with Calcofluor at the ends of rays in the wild type. Double arrows indicate CBM3a binding of the external domain of the adherent mucilage-like mushroom-cap. Arrowheads indicate mucilage collapse in csla2-1. Co, Columella; R, Ray. Bar = 100 μm.
To test the potential role of mannan polysaccharides in modifying crystalline cellulose structure, Arabidopsis seeds were treated with endo-β-1,4-mannanase (EβM), which specifically degrades mannan polysaccharides. Ruthenium red staining of EβM-treated seeds was very similar to that of untreated samples, although the peripheral ends seemed slightly less compact in both wild-type and csla2-1 mutant mucilage (Supplemental Fig. S7, B and E). Notably, CBM3a labeling showed that treatment with EβM led to the formation of a mushroom-cap structure at the peripheral ends of wild-type adherent mucilage (Fig. 7, H and I), similar to the cap observed in csla2-1 seed mucilage (Fig. 6, K and L). However, this treatment had no effect on the binding of CBM3a in csla2-1 adherent mucilage (Fig. 7, K and L). These results suggest that the removal of mannan polysaccharides can alter the spatial distribution of cellulose in the external domain of adherent mucilage.
To confirm the conformational change of crystalline cellulose in csla2-1 adherent mucilage, endo-β-1,4-glucanase (EβG), which has no effect on fibrillar cellulose (Hurst et al., 1978; Takagaki et al., 2002), was applied prior to the ruthenium red and CBM3a labeling. Ruthenium red staining of wild-type seeds after EβG digestion was much lighter in color, but the mucilage structure remained (Supplemental Fig. S7C), and the remaining mucilage appeared to correspond to fibrillar cellulose, with less pectin. Compared with wild-type seeds without enzyme treatment, CBM3a binding was more diffuse, and the intense ray-like structure was less organized in the internal domain of the adherent mucilage (Fig. 7, N and O). In the external domain of EβG-treated wild-type mucilage, CBM3a binding was irregular, suggesting a potential role of less organized cellulose in maintaining the proper ray structure of mucilage (Fig. 7, N and O). For csla2-1 seeds digested with excess EβG, the structure of adherent mucilage had collapsed, with no ruthenium red staining on the mucilage and CBM3a signals covered the seeds tightly like a sheet, although the possibility cannot be excluded that these collapsed CBM3a signals may come from columella and primary cell wall (Fig. 7, P–R; Supplemental Fig. S7F). These findings indicated that csla2-1 adherent mucilage cellulose was more accessible to EβG treatment.
The Spatial Organization of Mucilage Pectin Is Altered in the csla2-1 Mutant
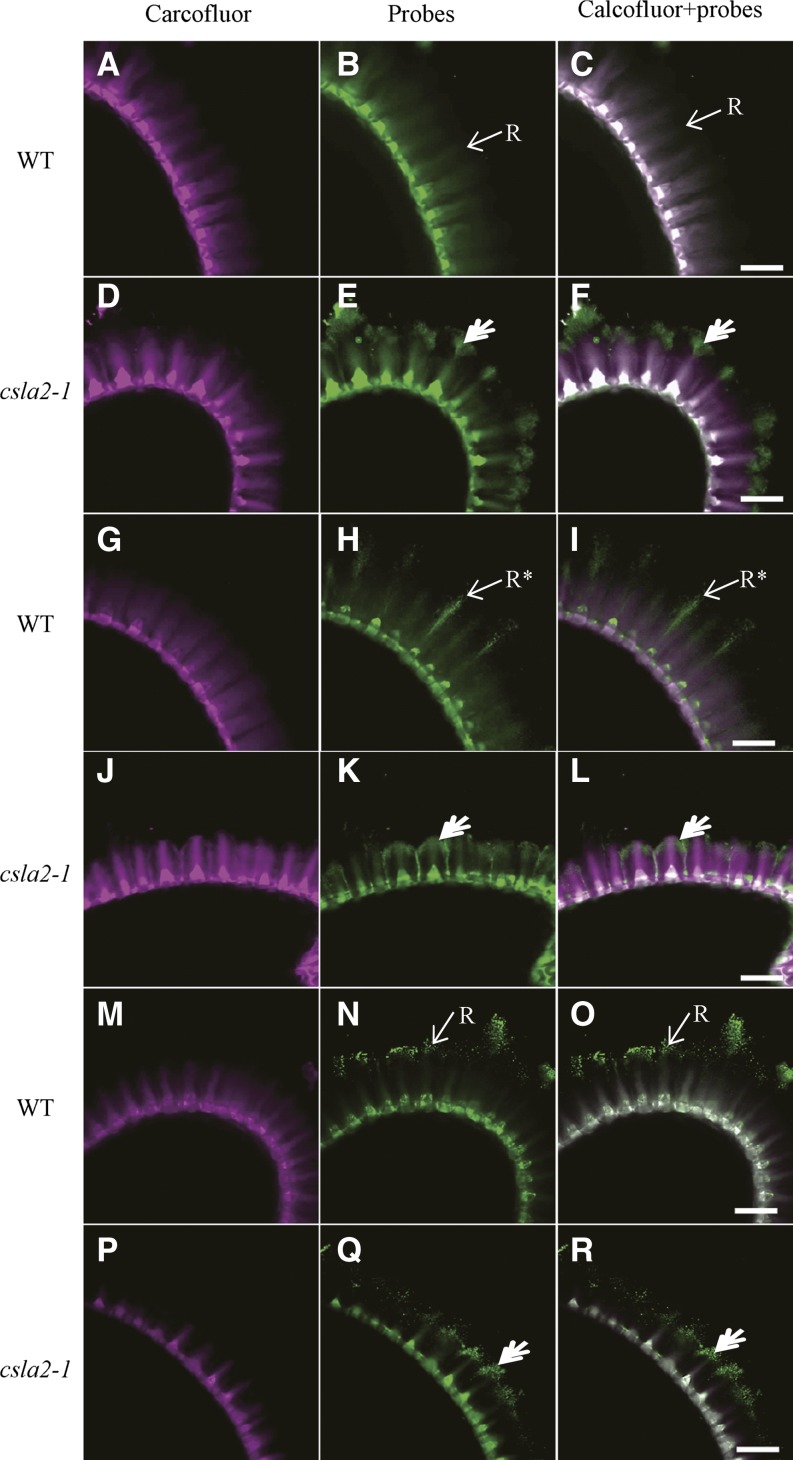
Pectins are major components of adherent mucilage and have been shown to be anchored to seeds by highly ordered crystalline cellulose (Harpaz-Saad et al., 2011, 2012; Mendu et al., 2011; Sullivan et al., 2011). Thus, the aberrant localization of cellulose in the csla2-1 mucilage suggests that pectin deposition might be compromised. CCRC-M14, JIM5, and JIM7 antibodies, specific to unbranched RG-I, low methylesterified HG, and high methylesterified HG, respectively (Clausen et al., 2003; Pattathil et al., 2010), were used to investigate if pectin was modified in response to the heteromannan defects and/or cellulose distribution changes in adherent mucilage. The CCRC-M14 antibody results showed the same binding pattern as Calcofluor White in the wild-type adherent mucilage (Fig. 8, A–C). However, labeling of unbranched RG-I with CCRC-M14 antibody in csla2-1 adherent mucilage indicated a similar binding pattern with CBM3a, except that labeling of the outer mushroom-cap was punctate (Fig. 8, D–F). JIM5 labeling showed strong signals of the rays originating from the columella in the wild type, but its labeling signals were punctate and denser, as if sprayed out from the top of columella (Fig. 8, G–I). In csla2-1 mucilage, JIM5 labeling of the rays bent to form an arc-shaped cap and labeling of the columella was localized in the central base (Fig. 8, J–L). As indicated in Figure 8, JIM7 labeled the external domain in dot patterns in the wild type, as previously observed (Macquet et al., 2007a). In csla2-1 mucilage, however, JIM7 labeling was decreased and displayed some aggregation (Fig. 8, Q and R). Overall, the in situ pectin epitope detection studies indicated that CSLA2 is involved in the spatial organization of mucilage pectin.
Figure 8.
Labeling of pectin in adherent mucilage released from wild type and csla2-1 seeds. A, D, G, J, M, and P, Calcofluor labeling. B and E, Indirect immunofluorescence detection of CCRC-M14 binding to unbranched RG-I. H and K, Indirect immunofluorescence detection of JIM5 binding to low methylesterified HG. N and Q, Indirect immunofluorescence detection of JIM7 binding to high methylesterified HG. C, F, I, L, O, and R, Composite images of double labeling with calcofluor and pectin probes. Asterisks indicate JIM5 labeling of mucilage not overlapped with calcofluor at the ends of rays in the wild type; double arrows indicate pectin probe binding to the external domain of the adherent mucilage-like mushroom-cap in csla2-1. R, Ray. Bar = 50 μm.
DISCUSSION
A screen exploiting new genes expressed in the Arabidopsis seed coat was used to identify a new mutant, clsa2, which displayed thinner adherent mucilage with increased density (Fig. 2, C–F; Tables I and II). CSLA2 has been demonstrated to have β-mannan/glucomannan synthase activity in vitro (Liepman et al., 2005, 2007), and mutation of CSLA2 decreases the mucilage glucomannan content in Arabidopsis (Tables II and III). In Arabidopsis seed mucilage, mannan polysaccharides represent less than 1% of the adherent mucilage, and their functions have not been characterized (Macquet et al., 2007a; Lee et al., 2012). Mutations identified in CSLA2 are likely to reduce glucomannan content in mucilage and thus affect interactions of glucomannan with other mucilage polysaccharides, and thereby modify the ultrastructure of mucilage. The role of CSLA2 in affecting the structure of adherent mucilage is confirmed by the observation of thinner adherent mucilage in three independent mutant lines and through molecular complementation of csla2-1 phenotypes with the CSLA2 CDS (Fig. 3, C–G).
CSLA2 Is Required for the Spatial Organization of Mucilage Cellulose
Several findings in this study lead to the conclusion that CSLA2 is required for the spatial organization of mucilage cellulose. First, the crystallinity of mucilage cellulose was clearly reduced in the csla2-1 mutant. Compared with the wild type, total cellulose Glc content was the same (Table II), but crystalline cellulose content was reduced by 30% in csla2-1 mucilage (Fig. 5C). In addition, csla2-1 is more sensitive to EβG, which specifically degrades noncrystalline cellulose glucans. csla2-1 mucilage was collapsed after EβG treatment (Fig. 7, P–R; Supplemental Fig. S7F). However, in the wild type, ruthenium red staining and CBM3a binding indicated that the mucilage cellulose remained (Fig. 7, M–O; Supplemental Fig. S7C). The reduction of cellulose crystallinity in csla2-1 mucilage could explain the sensitivity of csla2-1 mucilage to EβG, although it is also possible that the reduced glucomannan content allowed better access of this enzyme to mucilage cellulose. Second, the spatial distribution of crystalline cellulose was altered in the csla2-1 adherent mucilage. In the wild type, crystalline cellulose molecules detected with the CBM3a probe were abundant along the intense rays and diffuse between rays (Fig. 6, H and I; Supplemental Fig. S6A). In csla2-1, CBM3a labeling formed a mushroom-cap covering the whole seed, and was only weakly detected along the internal rays (Fig. 6, K and L; Supplemental Fig. S6B). Enzymatic removal of mannan polysaccharides revealed a similar mushroom-cap in the wild-type external domain of adherent mucilage (Fig. 7, H and I), suggesting that the loss of mannan polysaccharides in the external domain led to changes in the cellulose ultrastructure.
Heteromannans have been proposed to cross link cellulose by means of hydrogen bonds, and may be responsible for cellulose microfibril association in primary cell walls (Fry, 1986; Iiyama et al., 1994; Whitney et al., 1998; Obel et al., 2007). Whitney et al. (1998) have shown that glucomannan with different substitutions can form cross links with cellulose and change the crystallinity and ultrastructure of cellulose in vitro. Our data suggest that loss of glucomannans in csla2-1 adherent mucilage results in a reduction in cellulose crystallinity and heterogeneity of cellulose spatial distribution. The probable explanation is that glucomannans in adherent mucilage coat cellulose microfibrils to maintain cellulose association, or that glucomannans are entrapped into cellulose glucans to adopt the lowest free energy state with entropic contributions from cross links.
CSLA2 expression was detected in several seed tissues, including developing seed coat epidermal cells (Fig. 1), which produce large quantities of mucilage. CSLA2 expression was detected throughout seed development, and the peak of expression was observed at the bent cotyledon embryo stage (Fig. 1A). The timing and apparent peak of CSLA2 expression coincided with the expression of CESA5, which is responsible for the synthesis of mucilage cellulose (Sullivan et al., 2011). Although cellulose is synthesized at the plasma membrane and glucomannan is synthesized in the Golgi, both are deposited into and accumulated in the apoplast of the epidermal cells of the seed coat. During deposition, cellulose could be embedded in some of the existing matrix polysaccharides and then self-associate, or it could associate with other polymers to construct mucilage with unique structures (Cosgrove, 2005). Therefore, we hypothesize that glucomannan may be one such matrix polysaccharide and the loss of glucomannan may perturb the association between glucomannan and cellulose or the self-association of cellulose. However, precisely how glucomannans interact with cellulose and affect the association of cellulose requires further investigation.
Modulation of Mucilage Structure by Glucomannan
This study demonstrates that mutation in CSLA2 results in decreased width and increased density of adherent mucilage in Arabidopsis seeds. In some cases, notably in mutants affecting cellulose production within mucilage including the cellulose synthase subunit mutant cesa5, the fasciclin-like arbinogalactan protein mutant sos5, and the Leu-rich receptor-like kinase mutant fei2, reduced widths of adherent mucilage have been observed (Harpaz-Saad et al., 2011, 2012; Mendu et al., 2011; Sullivan et al., 2011). The reduction in the width of adherent mucilage in these three mutants may result from the partitioning of polysaccharide-derived sugars from the adherent layer to the nonadherent layer. In the case of the csla2-1 mutant, the mass and total sugar content of the adherent mucilage were similar with those in the wild type (Tables I and II), so the thinner adherent mucilage halo results from compactness (Fig. 3, C–F; Table I). This indicates the important function of glucomannan in modulating the ultrastructure of adherent mucilage.
Previous studies have shown that pectin in adherent mucilage is entangled with crystalline cellulose and that the orientation and amount of pectin associated with the cellulose network are largely determined by the properties of the crystalline cellulose (Macquet et al., 2007a; Harpaz-Saad et al., 2011, 2012; Mendu et al., 2011; Sullivan et al., 2011; Haughn and Western, 2012). In the csla2-1 mutant, crystalline cellulose in the external domain of adherent mucilage is not upright, but folded inside to cover the columella, forming a mushroom-like structure (Figs. 6, K and L, and 7, E and F; Supplemental Fig. S6B). The major components of adherent mucilage, RG-I and low methylesterified HG, which showed similar distributions as with crystalline cellulose in the csla2-1 mutant, formed caps on the surface of adherent mucilage (Fig. 8). One possible explanation for the compactness of the adherent mucilage in csla2-1 is that the crystalline cellulose properties are altered as a result of the loss of glucomannan, thus leading to the collapse of mucilage polymers (RG-I, HG, and cellulose) in the internal domain. Such a collapse could result in the observed reduction in width and the increased density of adherent mucilage in csla2-1.
Our biochemical analysis, antibody immunolabeling, and ELISA results indicated that pectin DM was decreased in csla2-1 mucilage (Figure 8, M–R; Table II; Supplemental Figure S4). The decrease in pectin DM has been reported in several mutants with disrupted cellulose synthesis or modified cellulose ultrastructure (Shedletzky et al., 1990; Encina et al., 2001; Manfield et al., 2004; Sullivan et al., 2011). It has been suggested that calcium-mediated cross linking of HG can compensate, to some extent, for the reduced load-bearing capacity of the disrupted cellulosic network. In the adherent mucilage of clsa2-1, both cellulose crystallinity and pectin DM are decreased. Consequently, the increased number of negatively charged carboxyl groups on partially methylesterified HG in csla2-1 could allow interactions through increased calcium cross links, and this would reinforce pectin structure and further rigidify adherent mucilage. Therefore, it seems likely that the glucomannan reduction in csla2-1 disrupted the adherent cellulosic network, thus leading to the modifications of pectin distribution and HG properties, which would also contribute to the compactness of csla2-1 adherent mucilage.
We have demonstrated that CSLA2 is involved in the biosynthesis of mucilage glucomannan, and our study has extended understanding of the function of glucomannans in structuring Arabidopsis mucilage. Our observations support the hypothesis that glucomannans interact with cellulose and participate in the organization and crystallization of mucilage cellulose, which is of great significance for understanding mucilage polymer cross links, assembly, and properties. Further investigations into the cross links of glucomannan with cellulose and other mucilage polymers may improve our understanding of the heteromannan structure-function relationship.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized, plated on Murashige and Skoog basal medium plates (Murashige and Skoog, 1962), stratified in darkness for 2 d at 4°C, and then germinated at 22°C under 16-h-light/8-h-dark conditions at 110 μmol m−2 s−1 irradiance. After 7 to 10 d, the seedlings were transferred to soil and grown in a growth chamber under the same environmental conditions.
Arabidopsis T-DNA insertion lines of csla2 (SALK_006803, SALK_083877, and SALK_149092) were obtained from the Salk T-DNA collection at the Arabidopsis Biological Resource Center. DNA was extracted by a simple sodium dodecylsulfate method and PCR was performed with primer sets to identify homozygous insertion lines.
RNA Isolation, RT-PCR, and qRT-PCR
Arabidopsis siliques were staged by marking flowers on the DPA and sampled at 4, 7, 10, and 14 DPA. Total RNA was isolated from these tissues using the cetyl-trimethyl-ammonium bromide method (Chang et al., 1993) with an additional DNase I (Promega) treatment to remove genomic DNA contamination. Total RNA was used for the synthesis of first-strand complementary (c)DNA (cDNA) using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). RT-PCR was performed with the cDNA and Extaq polymerase (Takara). GAPC was used as the reference gene for RT-PCR (Huang et al., 2011). The gene-specific primers used are listed in Supplemental Table S1. qRT-PCR was carried out using the LightCycler 480 Real-Time PCR System (Roche) with LightCycler 480 SYBR GreenIMaster (Roche). Data were analyzed using ACTIN2 as an internal control (Huang et al., 2011). All qRT-PCR reactions were performed in triplicate.
In Situ Hybridization
Preparation of sections of paraffin-embedded developing seeds and in situ hybridization were performed as previously described (Yu et al., 2013). Briefly, siliques were fixed in 4% paraformaldehyde in 0.1 m phosphate buffer (pH 7.0) overnight at 4°C, rinsed with phosphate buffer, and then dehydrated by passing through an ethanol and a xylene series. The xylene was gradually replaced by Paraplast Plus (Sigma) chips at 60°C. Finally, the samples were embedded in Paraplast Plus boxes and stored at 4°C before sectioning. The 307-bp fragment of CSLA2 was cloned into the pGM-T vector and sequenced. Probes were generated in vitro with digoxigenin-UTP using the SP6 or T7 RNA polymerase transcription kit (Roche). Serial sections of 8 μm were cut, placed on Superfrost Plus slides (Fisher), and dried for 2 d at 37°C. Sections were dewaxed with xylene and hydrated through an ethanol series (100% to 0%). The in situ hybridization procedure followed that of Mayer et al. (1998). Images were captured with the Olympus BX51 light microscope.
Subcellular Localization of the CSLA2 Protein
The complete open reading frame of CSLA2 was amplified from cDNA and the PCR fragment was ligated to the PGWC-T vector. The fully sequenced entry clone was recombined into the binary vector pEarleyGate104 using the Gateway cloning strategy, which allowed the production of a GFP:CSLA2 translational fusion under the control of the Cauliflower mosaic virus 35S promoter. The 35S::CSLA2-GFP fusion construct vector was introduced into Agrobacterium tumefaciens GV3101 by electroporation. The colocalization of fluorescent protein-tagged CSLA2 with Golgi markers was carried out in tobacco (Nicotiana benthamiana) leaf cells as previously described (Kong et al., 2011). Briefly, the 35S::CSLA2-GFP fusion construct was transfected into fully expanded leaves of tobacco plants, together with the CFP-tagged Golgi marker mCherry-tagged Man49. At 3 or 4 d after infection, the transfected leaf in the injected area was cut and examined using laser scanning confocal microscopy (FluoView FV1000; Olympus; Davis et al., 2010).
Cytochemical Staining
Mature dry seeds were stained for 30 min with 0.01% (m/v) ruthenium red dissolved in distilled water after being shaken in distilled water on a rotator at 150 rpm at 16°C for 1 h. Ruthenium red staining was observed with a microscope (SZX16; Olympus). Siliques at 4, 7, 10, and 14 DPA were fixed in 2.5% (w/v) glutaraldehyde in phosphate buffer at pH 7.0 overnight at 4°C. After being washed with phosphate buffer, samples were postfixed for 1 h in 1% (v/v) osmium tetraoxide in phosphate buffer, dehydrated in a series of washing steps with increasing ethanol concentrations, and embedded in Spurr’s resin. One-μm sections were stained for 5 min in 0.1% (w/v) toluidine blue O dissolved in phosphate-buffered saline (PBS) and viewed with a microscope (BX51; Olympus).
SEM
Dry seeds were mounted on stubs, coated with platinum in a Hitachi E1045 ion sputter, and examined with a Hitachi S4800 scanning electron microscope (Hitachi High-Technologies) at an accelerating voltage of 20 kV.
The cryo-SEM methods for seed mucilage were modified from a previous method (Voiniciuc et al., 2013). After being shaken in distilled water for 1 h, Arabidopsis seeds were quickly transferred to stubs topped with Tissue-Tek mounting medium (Sakura Finetek), and excess water was absorbed with small squares of filter paper. Once mounted on the stub, seeds were immediately frozen in liquid nitrogen and were directly transferred to the microscopy cold stage. After ice sublimation for 7 min at −95°C, seeds were sputter coated with gold for 2 min. Seeds were subsequently observed with a Hitachi S-3000N scanning electron microscope equipped with a Gatan ALTO 1000 cryo transfer system and cryo stage control, at an accelerating voltage of 15 kV.
Cell Wall Preparation
Soluble and adherent mucilage extracts were obtained as follows: 100 mg of dry seeds was extracted with distilled water (4 mL) with shaking on a rotator at 150 rpm for 1 h at 16°C. The resulting suspension was centrifuged at 8,000 rpm for 3 min. The supernatants were collected, and the seeds were rinsed twice with 1 mL of distilled water. The initial supernatants and subsequent wash volumes were then pooled and lyophilized to obtain nonadherent mucilage. After the water extraction treatment, seeds were treated sequentially with 0.2 n NaOH and 2 n NaOH at 16°C for 1 h and supernatants were collected after centrifugation at 8,000 rpm for 3 min. Extracts were then neutralized, dialyzed (Mr cutoff 3,500) three times against water, lyophilized, and then weighed.
For seed analysis, whole dry seeds and seeds lacking mucilage were frozen in liquid nitrogen and ground to a fine powder using mortars and pestles. Ten mg of ground seeds were destarched with a total starch assay kit (Megazyme) according to the manufacturer’s protocol, washed twice with 2 mL 80% (v/v) ethanol on a shaker at room temperature, and centrifuged at 5,000 rpm for 10 min. The ethanol was then removed and the pellet was washed sequentially with 95% (v/v) ethanol, 100% ethanol, and twice with 100% acetone. After the final acetone wash, samples were dried under a vacuum at 60°C to obtain the alcohol insoluble residue.
Determination of Adherent Mucilage Size and Mass
The volume of adherent mucilage was measured using a modified method for algal mucilage determination (Tien et al., 2002). Briefly, mature dry seeds were stained with ruthenium red (as described above) and measurements were performed by taking photographs of seeds and calibrated graticules using a microscope equipped with an automatic camera. An example of how measurements were taken from photographs is detailed in Supplemental Figure S8. The volume of a seed with or without mucilage was estimated using the formula for a spheroid (volume = 4/3 × 1/8 × length × width × depth). For this calculation, the measured width was also taken to represent depth (Riefler et al., 2006; Robert et al., 2008). Accordingly, the volume of adherent mucilage was determined by subtracting the volume of the seed from the total volume (seed + mucilage).
For the measurement of mucilage mass, 10,000 seeds were extracted with water (as described above), and the remaining seeds were then extracted with 2 n NaOH at 16°C for 1 h. The 2 n NaOH extract was collected, neutralized with acetic acid, dialyzed against distilled water, and lyophilized to obtain adherent mucilage. The adherent mucilage mass from 10,000 seeds was measured.
Compositional Analysis
Two mg of mucilage was hydrolyzed in 2 m trifluoroacetic acid (TFA) at 121°C for 2 h. TFA was removed by evaporation under vacuum. The hydrolysates were derivatized with 1-phenyl-3-methyl-5-pyrazolone and 0.3 m NaOH at 70°C for 30 min, extracted with chloroform for three times and then analyzed on a Hypersil ODS-2 C18 column (4.6 × 250 mm; Thermo Scientific) connected to a Waters HPLC System. The 1-phenyl-3-methyl-5-pyrazolone derivative (10 μL) was injected, eluted with 82% (v/v) phosphate buffer (0.1 M, pH 7.0) and 18% (v/v) acetonitrile at 1 ml/min, and monitored by UV A245. Monosaccharide standards used included l-Fuc, l-Rha, l-Ara, d-Gal, d-Glc, d-Xyl, d-Man, d-GalA, and d-GlcA.
For measurement of cellulosic Glc content, 10 mg of freeze-dried mucilage was hydrolyzed in 1 mL of 2 m TFA at 121°C for 2 h. After centrifugation at 10,000 rpm for 10 min, the pellets were dissolved in 0.2 mL of 72% (w/v) H2SO4 for 30 min at room temperature, followed by dilution with H2SO4 to 2 m with distilled water and boiling for 1 h. Cellulosic Glc content was then quantified colorimetrically using the phenol-sulfuric acid method (Saeman et al., 1954; Macquet et al., 2007a).
Biochemical Determination of Pectin DM
Mucilage for DM determination was extracted by vigorously shaking 10 mg of seeds with a vortex mixer at high speed in 300 μL of 40 mm EDTA for 1 h (Voiniciuc et al., 2013). The mucilage DM was determined from measurement of the methanol released by alkaline deesterification of extracts with 2 n NaOH for 1 h. After neutralization of extracts with 2 n HCl, methanol was oxidized with alcohol oxidase enzyme (Candida boidinii; Sigma-Aldrich) and quantified with a colorimetric method (Lionetti et al., 2007; Voiniciuc et al., 2013). The uronic acid content was determined by the m-hydroxydiphenyl method using GalA as the standard (Blumenkrantz and Asboe-Hansen, 1973). Mucilage pectin DM was calculated as the percentage molar ratio of methanol released after mucilage saponification to GalA.
Glycosyl Linkage Analysis
Monosaccharide-linkage analysis was performed by methylation of the mucilage with iodomethane in the presence of NaOH, followed by TFA hydrolysis (2 m at 120°C, 2 h), reduction with sodium borodeuteride (10 mg mL−1 in 1 m NaOH overnight at room temperature), and acetylation with acetic anhydride/TFA (100°C, 1 h). Partially methylated alditol acetates were dissolved in methylene chloride, separated on a SP-2330 column using a Agilent 7890A chromatograph, and detected by electron-impact ionization mass spectrometry with a 5975C mass selective detector (mass-to-charge ratio of 50 to 350). Temperature was held at 80°C during injection and ramping, the derivatives were separated in a temperature program of 80°C to 140°C at 20°C min−1, to 200°C at 2°C min−1, and then to 300°C at 30°C min−1, with a hold of 5 min at the higher temperature. Helium flow was 1 mL min−1, with splitless injection. Electron-impact mass spectrometry was performed at 70 eV and a source temperature of 230°C.
Crystalline Cellulose Observation and Content Determination
Dry seeds were imbibed in distilled water for 1 h, and were then mounted on a glass slide and observed with a Nikon Eclipse E600 POL microscope (Sullivan et al., 2011). For determination of crystalline cellulose content, 20 mg of alcohol insoluble residue was hydrolyzed in 2 mL of 2 m TFA at 121°C for 2 h. After centrifugation, the pellets were suspended in 2 mL of Updegraff reagent (acetic acid:nitric acid:water, 8:1:2 [v/v/v]) at room temperature for 30 min, and then boiled for 1 h (Updegraff, 1969). The crystalline cellulose pellets were then collected by centrifugation at 10,000 rpm for 10 min. The cellulose was then dissolved in 3 mL of 72% (w/v) H2SO4 for 30 min at room temperature. Crystalline cellulose amounts were quantified colorimetrically with the phenol-sulfuric acid method using a dehydration factor of 0.9 (Ge et al., 2012).
HPSEC
HPSEC was performed at room temperature on a system comprising Shodex OHpak SB-802 and SB-803 columns connected in series, using a differential refractive index detector (Wyatt Optilab rEX). The separation was carried out with 50 mm sodium nitrate buffer as the mobile phase at a constant flow rate of 1 mL min−1. Samples were solubilized in 50 mm sodium nitrate and filtered through disposable syringe filters (0.22 μm; Millipore) prior to injection. Samples were injected with a 0.5-mL loop in an autosampler. Data were analyzed using Astra software.
ELISA and Immunolabeling Procedures
ELISA was performed as previously described (Yu et al., 2011). In all cases, mucilage extractions were coated onto microtiter plates (3599; Costa) at 100 μg mL−1 concentrations. Coating solutions were removed, and 200 μL 3% (w/v) milk protein in phosphate-buffered saline (MP/PBS) was used to block the plates overnight at 4°C to prevent nonspecific binding in subsequent assays. Plates were washed and then 100 μL well−1 of the primary antibody at a 20-fold dilution in MP/PBS was added. After 2-h incubation at 37°C, plates were washed with PBS and wells were incubated with anti-rat/anti-mouse IgG coupled to horseradish peroxidase at a 1,000-fold dilution in MP/PBS for a further 2 h. After washing with PBS, antibody binding was determined by the addition of 150 μL well−1 of horseradish peroxidase-substrate (12 mL 0.1 m sodium acetate buffer pH 5.5, 200 μL tetramethylbenzidine, 18 μL 6% [v/v] hydrogen peroxide) prepared immediately before use. The reaction was stopped after 5 min by the addition of 50 μL well−1 of 1 n sulphuric acid. The absorbance was determined at 450 nm in a microplate reader.
Whole-seed immunolabeling was conducted according to a previously described method (Blake et al., 2006). For LM, JIM, and CCRC series antibodies, whole seeds were incubated first with 3% (w/v) MP/PBS, and then incubated in a 10-fold dilution of primary antibody in 3% (w/v) MP/PBS for 1.5 h. Seeds were then washed three times in PBS and incubated with a 200-fold dilution of AlexaFluor488-tagged donkey anti-rat IgG (for JIM and LM series) or anti-mouse IgG (for CCRC series) in MP/PBS in darkness for 1 h. To detect the carbohydrate-binding module CBM3a containing a His tag, seeds were incubated in MP/PBS containing 10 μg mL−1 CBM3a for 1.5 h. Seeds were then washed in PBS, incubated in a 100-fold dilution of mouse anti-His monoclonal antibody for 1 h, washed three time in PBS, and incubated in a 200-fold dilution of AlexaFluor488-tagged donkey anti-mouse IgG in MP/PBS in darkness for 1 h. After antibody or CBM3a labeling, the seeds were washed in PBS and counterstained for 5 min with Calcofluor White (Sigma-Aldrich). Seeds were imaged using a 405-and 488-nm laser on a FluoView FV1000 spectral confocal laser microscope (Macquet et al., 2007a). All confocal micrographs were processed with FV10-ASW application software. For enzyme treatment, 10 units of EβM (Cellvibrio japonicus; megazyme) or EβG (Aspergillus niger; Sigma-Aldrich) was used to treat 5 mg of seeds in 800 μL of distilled water for 90 min at 37°C. Seeds were then rinsed with water and stained with ruthenium red or immunolabeled with CBM3a.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NM_122180.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression profile of CSLA2.
Supplemental Figure S2. Sections of developing seeds stained with toluidine blue O.
Supplemental Figure S3. Analysis of dry and hydrated seeds by scanning electron microscopy.
Supplemental Figure S4. ELISAs of the mucilage extracted with H2O, 0.2 n NaOH, and 2 n NaOH from wild-type and csla2-1 seeds.
Supplemental Figure S5. Analysis of the mucilage extracted with H2O, 0.2 n NaOH, and 2 n NaOH from wild-type and csla2-1 seeds by size-exclusion chromatography combined with refractive index detection.
Supplemental Figure S6. 3D structure of Arabidopsis wild-type and csla2-1 adherent mucilage stained with Calcofluor and CBM3a.
Supplemental Figure S7. Ruthenium red staining of the adherent mucilage after digestion with enzymes.
Supplemental Figure S8. Illustration of how the measurements of mucilage and seed volumes of Arabidopsis seed were conducted.
Supplemental Table S1. Primer sets used in this study.
Supplementary Material
Acknowledgments
The authors thank Zhirui Wang (Complex Carbohydrate Research Center, University of Georgia) for excellent technical help in performing linkage analysis, Paul J. Knox (University of Leeds, U.K.) for providing cell wall probes, and Jay Chen and Olaf Czarnecki (Biosciences Division, Oak Ridge National Laboratory) for critical reading of the manuscript.
Glossary
- HG
homogalacturonan
- RG-I
rhamnogalacturonan I
- PME
pectin methylesterase
- qRT
quantitative real-time
- T-DNA
transfer DNA
- RT
reverse transcription
- DM
degree of methylesterification
- SEM
scanning electron microscopy
- HPSEC
high-performance size exclusion chromatography
- EβM
endo-β-1,4-mannanase
- EβG
endo-β-1,4-glucanase
- cDNA
complementaryDNA
- PBS
phosphate-buffered saline
- TFA
trifluoroacetic acid
- MP/PBS
milk protein in phosphate-buffered saline
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31070272 and 31370328), the National Science and Technology Support Program (grant no. 2013BAD22B01), the Youth Talent Plan of Chinese Academy of Agricultural Sciences (to Y.K.), and the Qingdao Municipal Science and Technology Plan (grant no. 11–2–4–8–(1)–jch).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Arsovski AA, Popma TM, Haughn GW, Carpita NC, McCann MC, Western TL. (2009) AtBXL1 encodes a bifunctional β-D-xylosidase/α-L-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol 150: 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T, De Rycke R, Viane R, Inzé D. (2000) Histological study of seed coat development in Arabidopsis thaliana. J Plant Res 113: 139–148 [Google Scholar]
- Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, Knox JP. (2006) Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem 281: 29321–29329 [DOI] [PubMed] [Google Scholar]
- Blumenkrantz N, Asboe-Hansen G. (1973) New method for quantitative determination of uronic acids. Anal Biochem 54: 484–489 [DOI] [PubMed] [Google Scholar]
- Bosch M, Hepler PK. (2005) Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 17: 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffall KH, Pattathil S, Phillips SE, Hahn MG, Mohnen D. (2009) Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol Plant 2: 1000–1014 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Clausen MH, Willats WGT, Knox JP. (2003) Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr Res 338: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Davis J, Brandizzi F, Liepman AH, Keegstra K. (2010) Arabidopsis mannan synthase CSLA9 and glucan synthase CSLC4 have opposite orientations in the Golgi membrane. Plant J 64: 1028–1037 [DOI] [PubMed] [Google Scholar]
- Dean G, Cao Y, Xiang D, Provart NJ, Ramsay L, Ahad A, White R, Selvaraj G, Datla R, Haughn G. (2011) Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol Plant 4: 1074–1091 [DOI] [PubMed] [Google Scholar]
- Dean GH, Zheng H, Tewari J, Huang J, Young DS, Hwang YT, Western TL, Carpita NC, McCann MC, Mansfield SD, et al. (2007) The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19: 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encina AE, Moral RM, Acebes JL, Álvarez JM. (2001) Characterization of cell walls in bean (Phaseolus vulgaris L.) callus cultures tolerant to dichlobenil. Plant Sci 160: 331–339 [DOI] [PubMed] [Google Scholar]
- Fry SC. (1986) Cross-linking of matrix polymers in the growing cell walls of angiosperms. Ann Rev Plant Physiol 37: 165–186 [Google Scholar]
- Ge X, Green VS, Zhang N, Sivakumar G, Xu J. (2012) Eastern gamagrass as an alternative cellulosic feedstock for bioethanol production. Process Biochem 47: 335–339 [Google Scholar]
- Goubet F, Barton CJ, Mortimer JC, Yu X, Zhang Z, Miles GP, Richens J, Liepman AH, Seffen K, Dupree P. (2009) Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J 60: 527–538 [DOI] [PubMed] [Google Scholar]
- Goubet F, Misrahi A, Park SK, Zhang Z, Twell D, Dupree P. (2003) AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol 131: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz-Saad S, McFarlane HE, Xu S, Divi UK, Forward B, Western TL, Kieber JJ. (2011) Cellulose synthesis via the FEI2 RLK/SOS5 pathway and cellulose synthase 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J 68: 941–953 [DOI] [PubMed] [Google Scholar]
- Harpaz-Saad S, Western TL, Kieber JJ. (2012) The FEI2-SOS5 pathway and CELLULOSE SYNTHASE 5 are required for cellulose biosynthesis in the Arabidopsis seed coat and affect pectin mucilage structure. Plant Signal Behav 7: 285–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn GW, Western TL. (2012) Arabidopsis seed coat mucilage is a specialized cell wall that can be used as a model for genetic analysis of plant cell wall structure and function. Front Plant Sci 3: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, DeBowles D, Esfandiari E, Dean G, Carpita NC, Haughn GW. (2011) The Arabidopsis transcription factor LUH/MUM1 is required for extrusion of seed coat mucilage. Plant Physiol 156: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst PL, Sullivan PA, Shepherd MG. (1978) Substrate specificity and mode of action of a cellulase from Aspergillus niger. Biochem J 169: 389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iiyama K, Lam TBT, Stone BA. (1994) Covalent cross-links in the cell wall. Plant Physiol 104: 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Zhou G, Abdeen AM, Schafhauser J, Richardson B, Atmodjo MA, Jung J, Wicker L, Mohnen D, Western T, et al. (2013) GALACTURONOSYLTRANSFERASE-LIKE5 is involved in the production of Arabidopsis seed coat mucilage. Plant Physiol 163: 1203–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Zhou G, Yin Y, Xu Y, Pattathil S, Hahn MG. (2011) Molecular analysis of a family of Arabidopsis genes related to galacturonosyltransferases. Plant Physiol 155: 1791–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJD, Dekkers BJW, Steinbrecher T, Walsh CT, Bacic A, Bentsink L, Leubner-Metzger G, Knox JP. (2012) Distinct cell wall architectures in seed endosperms in representatives of the Brassicaceae and Solanaceae. Plant Physiol 160: 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Nairn CJ, Willats WGT, Sørensen I, Roberts AW, Keegstra K. (2007) Functional genomic analysis supports conservation of function among cellulose synthase-like A gene family members and suggests diverse roles of mannans in plants. Plant Physiol 143: 1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102: 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D. (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Kronenberger J, Marion-Poll A, North HM. (2007a) In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol 48: 984–999 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Loudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM. (2007b) A naturally occurring mutation in an Arabidopsis accession affects a β-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield IW, Orfila C, McCartney L, Harholt J, Bernal AJ, Scheller HV, Gilmartin PM, Mikkelsen JD, Paul Knox J, Willats WGT. (2004) Novel cell wall architecture of isoxaben-habituated Arabidopsis suspension-cultured cells: global transcript profiling and cellular analysis. Plant J 40: 260–275 [DOI] [PubMed] [Google Scholar]
- Marcus SE, Blake AW, Benians TA, Lee KJD, Poyser C, Donaldson L, Leroux O, Rogowski A, Petersen HL, Boraston A, et al. (2010) Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J 64: 191–203 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Mendu V, Griffiths JS, Persson S, Stork J, Downie AB, Voiniciuc C, Haughn GW, DeBolt S. (2011) Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol 157: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira LRS, Filho EXF. (2008) An overview of mannan structure and mannan-degrading enzyme systems. Appl Microbiol Biotechnol 79: 165–178 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Obel N, Neumetzler L, Pauly M (2007) Hemicelluloses and cell expansion. In The Expanding Cell. Springer, Berlin, pp 57–88 [Google Scholar]
- Oka T, Nemoto T, Jigami Y. (2007) Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. J Biol Chem 282: 5389–5403 [DOI] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, et al. (2010) A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol 153: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Gille S, Liu L, Mansoori N, de Souza A, Schultink A, Xiong G. (2013) Hemicellulose biosynthesis. Planta 238: 627–642 [DOI] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW. (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13: 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T. (2008) A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J 54: 466–480 [DOI] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. (2000) The cellulose synthase superfamily. Plant Physiol 124: 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Noriega A, Tocino A, Cervantes E. (2008) Morphological analysis of seed shape in Arabidopsis thaliana reveals altered polarity in mutants of the ethylene signaling pathway. J Plant Physiol 165: 911–919 [DOI] [PubMed] [Google Scholar]
- Saeman JF, Moore WE, Mitchell RL, Millett MA. (1954) Techniques for the determination of pulp constituents by quantitiative paper chromatography. TAPPI J 37: 336–343 [Google Scholar]
- Saez-Aguayo S, Ralet MC, Berger A, Botran L, Ropartz D, Marion-Poll A, North HM. (2013) PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25: 308–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu APS, Randhawa GS, Dhugga KS. (2009) Plant cell wall matrix polysaccharide biosynthesis. Mol Plant 2: 840–850 [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Shedletzky E, Shmuel M, Delmer DP, Lamport DTA. (1990) Adaptation and growth of tomato cells on the herbicide 2,6-dichlorobenzonitrile leads to production of unique cell walls virtually lacking a cellulose-xyloglucan network. Plant Physiol 94: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Stork J, Harris D, Griffiths J, Williams B, Beisson F, Li-Beisson Y, Mendu V, Haughn G, Debolt S. (2010) CELLULOSE SYNTHASE9 serves a nonredundant role in secondary cell wall synthesis in Arabidopsis epidermal testa cells. Plant Physiol 153: 580–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Ralet MC, Berger A, Diatloff E, Bischoff V, Gonneau M, Marion-Poll A, North HM. (2011) CESA5 is required for the synthesis of cellulose with a role in structuring the adherent mucilage of Arabidopsis seeds. Plant Physiol 156: 1725–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki K, Iwafune M, Kakizaki I, Ishido K, Kato Y, Endo M. (2002) Cleavage of the xylosyl serine linkage between a core peptide and a glycosaminoglycan chain by cellulases. J Biol Chem 277: 18397–18403 [DOI] [PubMed] [Google Scholar]
- Tien CJ, Krivtsov V, Levado E, Sigee DC, White KN. (2002) Occurrence of cell-associated mucilage and soluble extracellular polysaccharides in Rostherne Mere and their possible significance. Hydrobiologia 485: 245–252 [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Usadel B, Kuschinsky AM, Rosso MG, Eckermann N, Pauly M. (2004) RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol 134: 286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Dean GH, Griffiths JS, Kirchsteiger K, Hwang YT, Gillett A, Dow G, Western TL, Estelle M, Haughn GW. (2013) Flying saucer1 is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in Arabidopsis seed mucilage. Plant Cell 25: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M, Tehseen M, Doblin MS, Pettolino FA, Wilson SM, Bacic A, Golz JF. (2011) The transcriptional regulator LEUNIG_HOMOLOG regulates mucilage release from the Arabidopsis testa. Plant Physiol 156: 46–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mortimer JC, Davis J, Dupree P, Keegstra K. (2012) Identification of an additional protein involved in mannan biosynthesis. Plant J 73: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Skinner DJ, Haughn GW. (2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Young DS, Dean GH, Tan WL, Samuels AL, Haughn GW. (2004) MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol 134: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SEC, Brigham JE, Darke AH, Reid JSG, Gidley MJ. (1998) Structural aspects of the interaction of mannan-based polysaccharides with bacterial cellulose. Carbohydr Res 307: 299–309 [Google Scholar]
- Willats WG, McCartney L, Knox JP. (2001) In-situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta 213: 37–44 [DOI] [PubMed] [Google Scholar]
- Yu L, Zhou YF, Knox JP. (2011) Ginseng root water-extracted pectic polysaccharides originate from secretory cavities. Planta 234: 487–499 [DOI] [PubMed] [Google Scholar]
- Yu Y, Hu R, Wang H, Cao Y, He G, Fu C, Zhou G. (2013) MlWRKY12, a novel Miscanthus transcription factor, participates in pith secondary cell wall formation and promotes flowering. Plant Sci 212: 1–9 [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogowski A, Zhao L, Hahn MG, Avci U, Knox JP, Gilbert HJ. (2014) Understanding how the complex molecular architecture of mannan-degrading hydrolases contributes to plant cell wall degradation. J Biol Chem 289: 2002–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Nam J, Carpita NC, Matthysse AG, Gelvin SB. (2003) Agrobacterium-mediated root transformation is inhibited by mutation of an Arabidopsis cellulose synthase-like gene. Plant Physiol 133: 1000–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.