Oil body dynamics reveal new roles for oleosins during seed development.
Abstract
Oil bodies (OBs) are seed-specific lipid storage organelles that allow the accumulation of neutral lipids that sustain plantlet development after the onset of germination. OBs are covered with specific proteins embedded in a single layer of phospholipids. Using fluorescent dyes and confocal microscopy, we monitored the dynamics of OBs in living Arabidopsis (Arabidopsis thaliana) embryos at different stages of development. Analyses were carried out with different genotypes: the wild type and three mutants affected in the accumulation of various oleosins (OLE1, OLE2, and OLE4), three major OB proteins. Image acquisition was followed by a detailed statistical analysis of OB size and distribution during seed development in the four dimensions (x, y, z, and t). Our results indicate that OB size increases sharply during seed maturation, in part by OB fusion, and then decreases until the end of the maturation process. In single, double, and triple mutant backgrounds, the size and spatial distribution of OBs are modified, affecting in turn the total lipid content, which suggests that the oleosins studied have specific functions in the dynamics of lipid accumulation.
The seed is a complex, specific structure that allows a quiescent plant embryo to cope with unfavorable germinating conditions and also permits dissemination of the species. To achieve these functions, seeds accumulate reserve compounds that will ensure the survival of the embryo and fuel the growth of the plantlet upon germination. Accumulation of lipids occurs in many eukaryotic cells and is a rather common means of storing carbon and energy. Lipid droplets (LDs) can be found in all eukaryotes, such as yeast (Saccharomyces cerevisiae; Leber et al., 1994), mammals (Murphy, 2001; Hodges and Wu, 2010), Caenorhabditis elegans (Zhang et al., 2010; Mak, 2012), Drosophila melanogaster (Beller et al., 2006, 2010), and plants (Hsieh and Huang, 2004), but also in prokaryotes (Wältermann et al., 2005). The basic structure of an LD is a core of neutral lipids covered by a phospholipid monolayer. LDs differ between species by the set of proteins covering their surface, the nature of the lipids stored, and their turnover. Nevertheless, they apparently always ensure the same function in the cell (i.e. energy storage; Murphy, 2012). In Brassicacea species such as Arabidopsis (Arabidopsis thaliana), seed reserves are mainly composed of carbohydrates, proteins, and lipids (Baud et al., 2002). The lipids are primarily stored as triacylglycerols (TAGs) in LDs, more commonly called oil bodies (OBs; Hsieh and Huang, 2004; Chapman et al., 2012; Chapman and Ohlrogge, 2012) of diameter 0.5 to 2 µm (Tzen et al., 1993).
The protein composition of seed OBs has been determined for several plant species, including Brassica napus (Katavic et al., 2006; Jolivet et al., 2009) and Arabidopsis (Jolivet et al., 2004; D’Andréa et al., 2007; Vermachova et al., 2011). In Arabidopsis, 10 proteins have been identified, and seed-specific oleosins represent up to 79% of the OB proteins (Jolivet et al., 2004; D’Andréa et al., 2007; Vermachova et al., 2011). Oleosins are rather small proteins of 18.5 to 21.2 kD with a specific and highly conserved central hydrophobic domain of 72 amino acid residues flanked by hydrophilic domains of variable size and amino acid composition (Qu and Huang, 1990; Tzen et al., 1990, 1992; Huang, 1996; Hsieh and Huang, 2004). It is generally agreed that oleosins cover the OB surface, with their central hydrophobic domain inserted in the TAG through the phospholipid layer (Tzen and Huang, 1992). Besides their structural function in OBs, oleosins may serve as docking stations for other proteins at its surface (Wilfling et al., 2013) and may participate in the biosynthesis and mobilization of plant oils (Parthibane et al., 2012a, 2012b). Oleosins are probably involved in OB stability (Leprince et al., 1998; Shimada et al., 2008) and in the regulation of OB repulsion (Heneen et al., 2008), preventing the coalescence of OBs into a single organelle (Schmidt and Herman, 2008). Nevertheless, the precise functions of oleosins in OB biogenesis and dynamics have not yet been established.
Global analysis of seed lipids can be performed using gas chromatography (Li et al., 2006), which allows the precise determination of both lipid content and fatty acid composition. Recently, direct organelle mass spectrometry has been used to visualize the lipid composition of cotton (Gossypium hirsutum) seed OBs (Horn et al., 2011). Nevertheless, in both cases, the methods are destructive. To observe lipid accumulation at the subcellular level, well-known nondestructive techniques for lipid visualization have been adapted to seeds. Third harmonic generation microscopy (Débarre et al., 2006) and label-free coherent anti-Stokes Raman scattering microscopy (Paar et al., 2012) allow dyeless observation of LDs but require very specific equipment. Magnetic resonance imaging enables topographic analysis of lipid distribution in cereal grains (Neuberger et al., 2008) and in submillimeter-sized seeds like those of tobacco (Nicotiana tabacum; Fuchs et al., 2013). Nevertheless, the use of fluorescent dyes such as Nile Red (Greenspan and Fowler, 1985), BODIPY (Pagano et al., 1991), or LipidTOX (Invitrogen) associated with confocal microscopy is also a powerful way to monitor LDs in living organisms.
Despite knowledge accumulated on this topic (Brasaemle and Wolins, 2012; Chapman et al., 2012), little is known about OB dynamics during seed maturation. In this article, we investigate this question by monitoring the evolution of OBs in living Arabidopsis embryos over time. This analysis showed a marked change in OB size at 9 to 10 d after flowering (DAF). We then examined single, double, and triple mutants of the major oleosins found in developing seeds (OLE1 [At4g25140], OLE2 [At5g40420], and OLE4 [At3g01570]; Jolivet et al., 2004). We analyzed the OB dynamics in these mutant backgrounds as if they would contain only these three proteins. We show that the lack of specific oleosins influences the dynamics and distribution of OBs during seed maturation, which in turn affects lipid accumulation. These results pave the way for analyzing specific functions of oleosins in the synthesis, growth, and evolution of OBs.
RESULTS
Oleosins and Storage Compound Accumulation
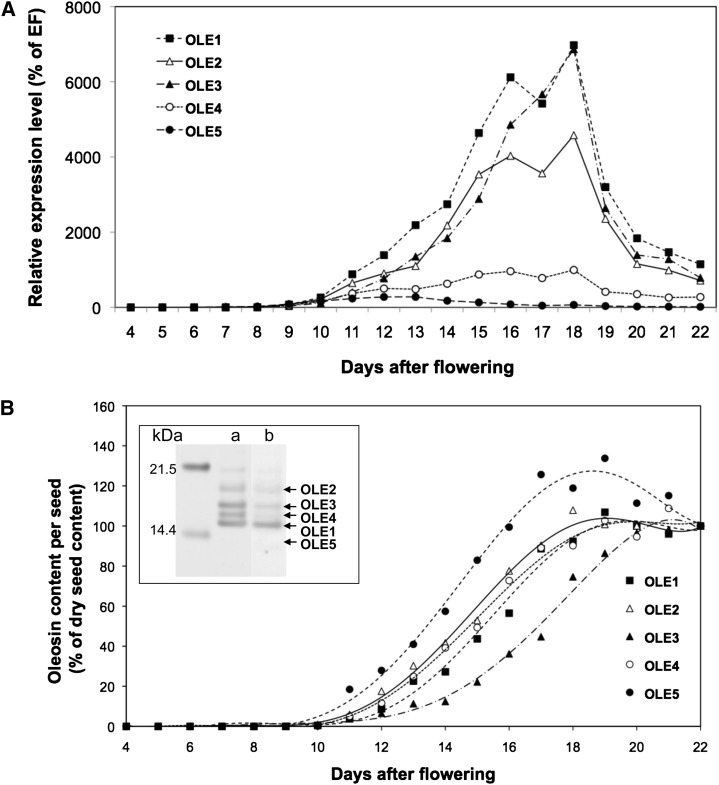
Several complementary techniques were used to study the spatiotemporal dynamics of oleosin expression. A quantitative reverse transcription (RT)-PCR approach first confirmed that oleosins were highly and specifically expressed in silique material. The relative expression profiles of the oleosins OLE1, OLE2, OLE3, OLE4, and OLE5 were investigated in siliques 4 to 22 DAF (Fig. 1A).
Figure 1.
Oleosin accumulation pattern during seed development. The abundance of oleosin mRNA (A) and oleosin protein accumulation (B) were determined in Arabidopsis siliques collected 7 to 22 DAF. Seeds were mature at 22 DAF. The abundance of oleosin mRNA was assessed by quantitative RT-PCR, and EF1αA1 was used as a control. Oleosin protein accumulation was measured by immunoblot using specific sera. Five seed-specific oleosins were quantified: OLE1 (black squares), OLE2 (white triangles), OLE3 (black triangles), OLE4 (white circles), and OLE5 (black circles). Luminescent signals from immunoblots, recorded using the LAS-3000 imaging system and quantified with the MultiGauge software, were within the linear range, allowing quantitative determination of oleosin content. For each oleosin, data were expressed as the percentage of the mature seed content of that oleosin, regardless of its relative amount among OB proteins. The inset in B shows OB proteins resolved by SDS-PAGE and stained with silver nitrate (column a) or Coomassie blue (column b) to illustrate the relative content of each oleosin in purified Arabidopsis OBs.
OLE1, OLE2, and OLE3 relative transcript levels were low during early silique development and increased sharply 9 to 15 DAF, corresponding to the onset of seed maturation; mRNA levels reached their maximal value at 16 to 18 DAF and then gradually decreased throughout the maturation phase. Transcript accumulation for OLE4 was much lower than for OLE1, OLE2, and OLE3 and also decreased at 18 DAF. In contrast, OLE5 showed low mRNA abundance, which decreased earlier than the other oleosins, starting 13 DAF (Fig. 1A).
Specific antibodies against each oleosin (D’Andréa et al., 2007) were used to investigate the temporal accumulation of oleosins by western blot (Fig. 1B). The content of each oleosin was determined quantitatively during seed development and expressed as a percentage of its mature seed content (Fig. 1B). In a manner compatible with the accumulation of their respective transcripts, OLE5 was the first protein detected, followed by OLE1, OLE2, and OLE4, and then OLE3. All oleosins accumulated during embryo development and were present in mature seeds. It is noteworthy that OLE5, although present in mature seeds, at maturity contained only three-fourths of its maximal level.
Another question was the potential redundancy between oleosins. To address this question, we obtained ole1, ole2, and ole4 mutants. Immunolocalization of the oleosins OLE1, OLE2, and OLE4 in their respective mutant backgrounds (Supplemental Fig. S1) confirmed that the ole1, ole2, and ole4 mutants used in this study are null mutants. Protein extracts from the wild type and the different single, double, and triple mutants were resolved by SDS-PAGE and stained with Coomassie blue (Supplemental Fig. S2A). Results of image analysis (Supplemental Fig. S2A) showed that deficiency in one or several oleosins was not compensated by an increase in the others. 12S and 2S storage proteins were also resolved (Supplemental Fig. S2A). While oleosin deficiency led in some cases to a reduced 12S and 2S storage protein content compared with the wild type (Supplemental Fig. S2B), with the exception of the ole1ole4 double mutant, the ratio between 12S and 2S storage proteins was not significantly affected (Supplemental Fig. S2C). In contrast, the total fatty acid content appeared more susceptible to a deficiency in one or more oleosins, as in all cases it was decreased compared with the wild type (Supplemental Fig. S2B).
OB Accumulation during Seed Formation and Germination
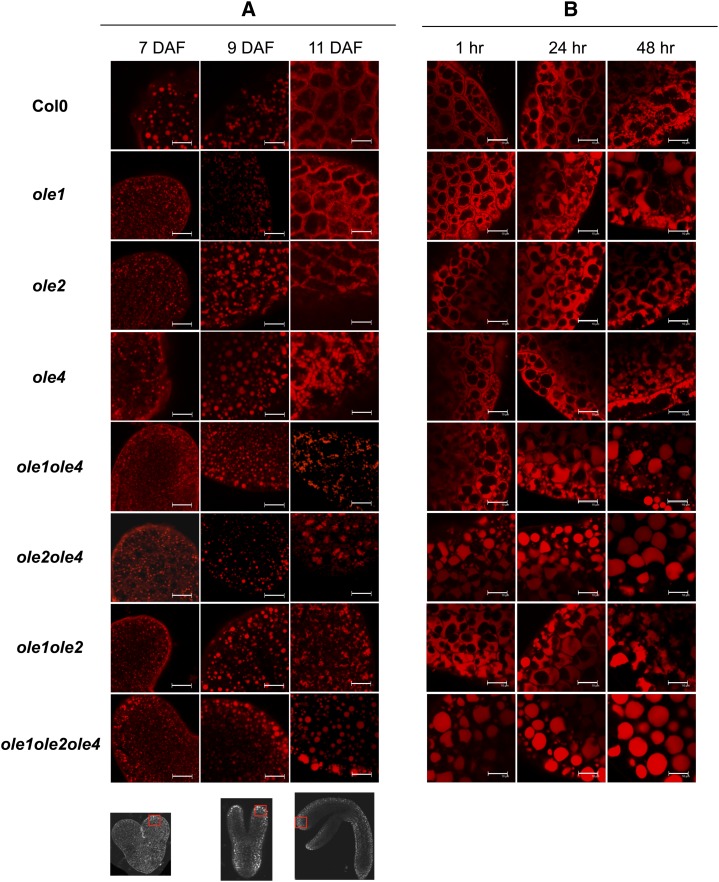
During wild-type seed development, at the early heart stage (7 DAF), OBs were scattered regularly in the embryo cells (Fig. 2A). The average estimated volume of OB (VOB) was 0.096 µm3 (after segmentation). This volume increased during embryo growth, with a marked increase after 9 DAF, at the transition between torpedo (9 DAF) and U-shaped (11 DAF) stages (Fig. 4A). At 11 DAF (Fig. 2A) the OBs decreased in size and were pushed to the edges of the cell, the cell center being occupied by protein bodies (PBs) and other intracellular components (nucleus, Golgi network, etc.). After 11 DAF, it was no longer possible to discriminate OBs with confocal microscopy, as they were too close together. The OB distribution during embryo development was not affected in the ole1, ole2, or ole4 single mutants (Fig. 2B). In double mutants, the OB accumulation pattern changed, especially after 9 DAF, when OBs displayed a different spatial distribution. In the ole1ole2ole4 mutant, the VOB continued to increase after 9 DAF, reaching an average of 2.5 µm3 at 11 DAF.
Figure 2.
Phenotypes of OBs stained with Nile Red in different genetic backgrounds during seed maturation (A) and imbibition (B). Red squares on the photographs of developing embryos at 7, 9, and 11 DAF (bottom) indicate the localization of image acquisition. Col0, Columbia wild type; hr, hours after imbibition start. Bars = 10 µm.
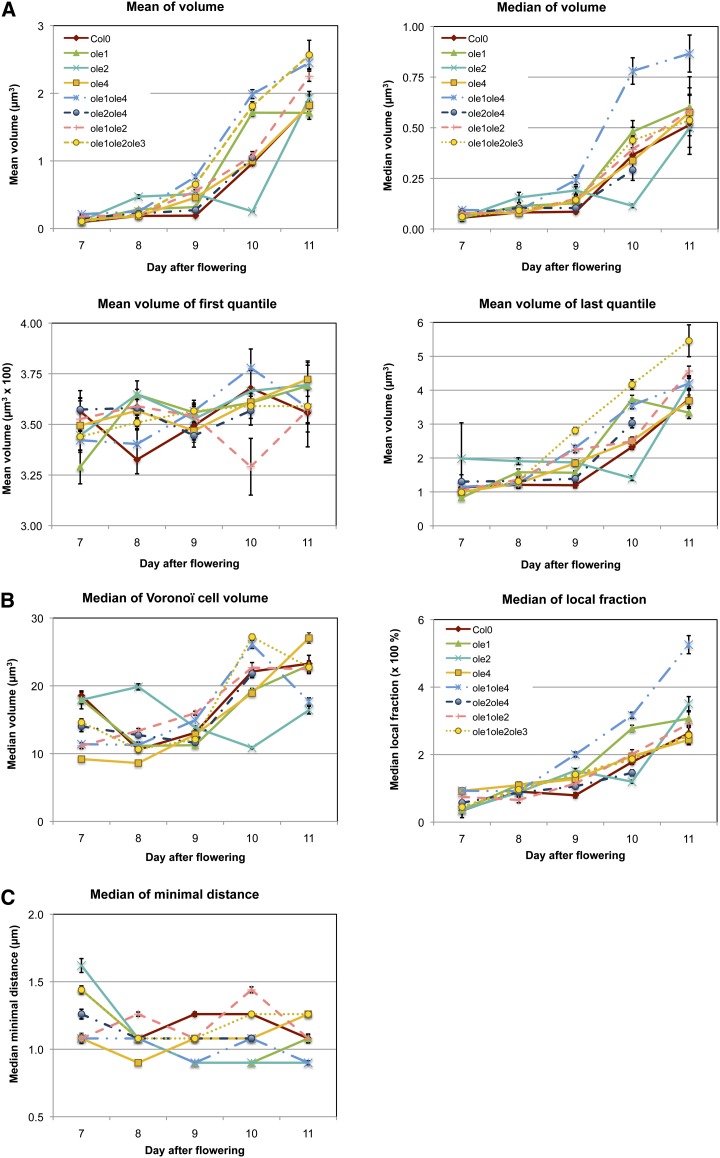
Figure 4.
Distribution of volume (A), spatial disposition (B), and minimal distance (C) of OBs during seed development of oleosin mutants. Col0, Columbia wild type.
During germination, after a 1-h imbibition of dry mature seeds, rehydration was not complete, only minor changes in the transcriptome were observed (Preston et al., 2009), and the OBs were still located on the edges of the cell. They were of very small size, and individual OBs could not be distinguished in the single mutants. After 24 h of imbibition, the OBs began to increase in size and reduce in number. After 48 h of imbibition, OBs began to be distinguishable (Fig. 2B). In the double and triple mutants, the OB distribution was affected. In 1-h imbibed seeds, compared with the wild type and single mutants, the ole2ole4 mutant contained larger OBs, and those of ole1ole4 and ole1ole2 were less compacted. The ole2ole4 OBs remained voluminous from the dry stage to 48 h after imbibition. In the ole1ole2ole4 mutant, the average VOB was rather constant (between 8 and 9 µm3) from 1-h imbibed seeds until 48 h after imbibition (Fig. 2B). In the case of large OBs, the PBs were pushed to the sides of cells by the OBs (Supplemental Fig. S3), indicating a structural flexibility of the PBs when confronted with increased VOB.
Characterizing OB Dynamics
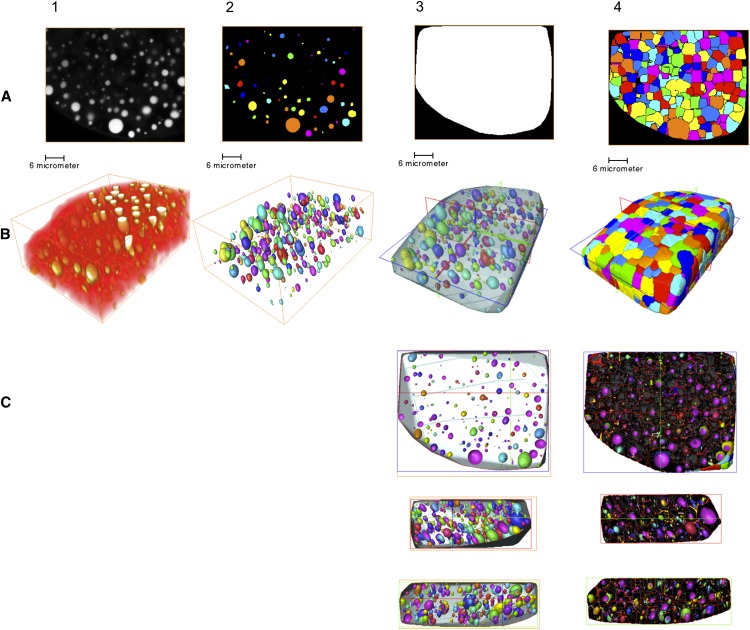
We wanted to address the distribution of OBs in the plant cells where OBs are normally distributed. OB dynamics in the wild type and oleosin mutant lines could be studied most easily if both OBs and cell walls of living cells could be stained. Unfortunately, as Nile Red was not compatible with cell wall stains, no fluorescent dye allowed discriminating OBs from cell walls in living embryos (Fig. 3, A1 and B1). Consequently, individual plant cells could not be detected, and we used estimators of the distance between each OB and its neighbors to answer the question of distribution in the three-dimensional (3D) space. We have segmented the OB (Fig. 3, A2 and B2) and defined a volume containing all the OBs (the convex envelope; Fig. 3, A3, B3, and C3). This 3D space was thus divided independently of the cellular segmentation in regions using Voronoï diagrams (Fig. 3, A4, B4, and C4). The distance to the next neighbor or the minimal distance between OBs (Fig. 4C) was computed, and the Voronoï cell corresponded to the junction of all the points at half the minimal distance between the surface of two OBs. Therefore, the outer surface of all the Voronoï cells defined a global volume (the convex envelope) that resembled the surface of the organ considered (embryo cotyledon or hypocotyl) without the outer plant cell walls (Fig. 3, B3 and B4).
Figure 3.
Sequential steps of image processing and 3D volume rendering. This example is taken from a 10-DAF ole1ole2ole4 sample image with a resolution of 512 × 512 × 512 pixels. A, Segmentation: crude acquired images (1), segmented OBs (2), convex envelope (3), and Voronoï cell processing (4). B, 3D volume rendering of the image stack for each step. C, Transverse section and orthoslice of the 3D reconstruction showing individual OBs (3) and individual OBs with the shape of the Voronoï cells (4).
Use of the Voronoï cells allowed calculating the “local fraction” (ϕloc) estimator (i.e. the ratio of volumes between a given OB and the Voronoï cell containing it), and therefore estimating densities of OBs and compaction processes, independently of the plant cell walls (Fig. 4B). To evaluate the spatial distribution of OBs during seed development, we designed a pipeline based on morphological operators for the quantitative analysis and characterization of OB size and organization. The pipeline was implemented in a 3D imaging and analysis software, Avizo-Fire (Burlington). It processed laser scanning microscope image stacks of OBs, yielding a mathematical representation of segmented OB images built on watershed-based techniques. Three estimators were used: the VOB, the ϕloc, and the distance of each OB to its nearest neighbor (Fig. 4). These estimators are detailed in “Materials and Methods.” Because the population of OBs contains small and large OBs, and the growing process is expected to be continuous between small and large OBs, we used different representations of the statistical distribution of the OB populations: the mean of volume, the median of volume, and the mean of the first and last quantiles (Fig. 4A).
To go further in the analysis of OB size and dynamics in the different genotypes, a linear mathematical model was developed (Trigui et al., 2012), and a statistical analysis of it using a quantile regression (Trigui et al., 2013) was performed (Table I), which confirmed that the VOB increased significantly as a function of time (DAF) regardless of the genotype (P < 2 × 10−10). The maximum rate of increase occurred between 9 and 10 DAF, as the log(VOB) was increased by approximately 0.35 µm3 with respect to the intercept (7 DAF; fitted value = −1.120 ± 0.009; Supplemental Table S1). Focusing first on OBs lacking OLE1, OLE2, and OLE4 proteins (ole1ole2ole4 mutant), the increase of VOB (Fig. 4A, mean of volume) indicated a fusion process, even without the effect of the studied oleosin. Adding OLE4 oleosin (ole1ole2 mutant) resulted in a decrease of the VOB compared with the ole1ole2ole4 mutant (Fig. 4A; fitted value = −0.02 ± 0.008; Supplemental Table S1). Similarly, OBs containing only OLE1 oleosin (ole2ole4 mutant) were smaller than OBs of ole1ole2ole4 mutant (Fig. 4A; fitted value = −0.091 ± 0.009; Supplemental Table S1). In addition, OLE1 acted more efficiently then OLE4 in decreasing VOB (ole2ole4 compared with ole1ole2; Fig. 4A; Supplemental Table S1). On the other hand, the effect of OLE2 was shown to increase the VOB (fitted value = 0.156 ± 0.007; Supplemental Table S1): the VOB in the ole1ole4 mutant is greater than that of ole1ole2ole4 (Fig. 4A). The mutant (ole2) with OLE1 and OLE4 together exhibited a reduction of the mean VOB compared with ole1ole2ole4, but their interaction seemed to be negligible (P = 0.031; Supplemental Table S1), suggesting a simple additive effect. The minimal VOB was obtained by this mutant background (Fig. 4A). However, the interaction between OLE2 and OLE4 was significant (fitted value = −0.127 ± 0.012; Supplemental Table S1) and acted by reducing the VOB even if OLE2 acted to increase VOB. The common presence of OLE2 with OLE1 (in mutant ole4) led to a reduction of the VOB when compared with the mutant ole1ole2ole4. This reduction was even larger than when it was present with OLE4 (mutant ole1). This result is in agreement with OLE1 being more effective than OLE4 in preventing OB fusion.
Table I. Quantile regression analysis.
A variance analysis using a quantile regression statistical model was performed on the mathematical model (see “Materials and Methods”) developed to study the effect of oleosins, the developmental stage, and the interactions of all these factors on VOB. Fitted estimate values of VOB, se, and P are given for factors affecting VOB. Intercept was 7 DAF.
| Sample | τ = 0.25 |
τ = 0.5 |
τ = 0.75 |
|||
|---|---|---|---|---|---|---|
| Estimate ± se | P | Estimate ± se | P | Estimate ± se | P | |
| Intercept | −1.479 ± 0.016 | 0.000 | −1.186 ± 0.010 | 0.000 | 0.828 ± 0.012 | 0.000 |
| 8 DAF | 0.130 ± 0.010 | 0.000 | 0.191 ± 0.010 | 0.000 | 0.226 ± 0.013 | 0.000 |
| 9 DAF | 0.246 ± 0.009 | 0.000 | 0.355 ± 0.010 | 0.000 | 0.469 ± 0.012 | 0.000 |
| 10 DAF | 0.538 ± 0.011 | 0.000 | 0.733 ± 0.011 | 0.000 | 0.902 ± 0.013 | 0.000 |
| 11 DAF | 0.722 ± 0.014 | 0.000 | 0.950 ± 0.012 | 0.000 | 1.114 ± 0.014 | 0.000 |
| OLE1 | −0.058 ± 0.011 | 0.000 | −0.056 ± 0.011 | 0.000 | −0.104 ± 0.013 | 0.000 |
| OLE2 | 0.137 ± 0.011 | 0.000 | 0.195 ± 0.011 | 0.000 | 0.210 ± 0.011 | 0.000 |
| OLE4 | −0.037 ± 0.011 | 0.001 | −0.004 ± 0.011 | 0.712 | 0.008 ± 0.012 | 0.490 |
| OLE1:OLE4 | −0.038 ± 0.016 | 0.020 | −0.066 ± 0.017 | 0.000 | −0.029 ± 0.020 | 0.134 |
| OLE2:OLE4 | −0.089 ± 0.017 | 0.000 | −0.156 ± 0.017 | 0.000 | −0.190 ± 0.017 | 0.000 |
| OLE1:OLE2 | −0.139 ± 0.018 | 0.000 | −0.168 ± 0.017 | 0.000 | −0.126 ± 0.018 | 0.000 |
| OLE1:OLE2:OLE4 | 0.138 ± 0.026 | 0.000 | 0.179 ± 0.026 | 0.000 | 0.106 ± 0.028 | 0.000 |
Finally, when considering the wild type, the VOB obtained by the contribution of OLE1, OLE2, and OLE4 was reduced compared with the ole1ole2ole4 mutant, and its effect was significant (fitted value = 0.126 ± 0.019; Supplemental Table S1). The presence of these oleosins on the surface of OBs modulates the fusion process by both contributing to and preventing OB growth.
Analysis of the evolution of the Voronoï cell volume showed a global decrease of this volume between 7 and 8 DAF, then an increase during maturation from 8 DAF on, except for the ole2 mutant (Fig. 4B). At the beginning of embryo development, fusion events would be rather scarce, while small OBs started filling the cell, thus limiting the space occupied by each OB. As OB density increased, OBs would fuse to optimize cell space, as shown by an increase in the volume of the Voronoï cell (Fig. 4B). The minimal distance between OBs decreased over time, indicating an increase in OB density (Fig. 4C), but the evolution of the curve was different for the ole2 mutant, which exhibited a low distance between OBs, which was confirmed by the presence of OB aggregates at 11 DAF (Fig. 2A; Supplemental Fig. S4).
Similar to the volume, the ϕloc decreased significantly as a function of time (DAF; Fig. 4B; Supplemental Table S1; P < 2 × 10−10) but more linearly between 8 and 11 DAF [log(ϕloc) increased by approximately 1.5 per time step]. While the effects of OLE1 and OLE4 contributed to a low decrease (fitted value = −0.078 ± 0.007 and −0.053 ± 0.006, respectively; Supplemental Table S1) of the ϕloc, OLE2 contributed to its increase (fitted value = 0.155 ± 0.006; Supplemental Table S1). The double and triple interactions involving OLE2 (OLE2:OLE4, OLE1:OLE2, and OLE1:OLE2:OLE4) were shown to have a synergistic effect on the VOB (P = 7.81−9; Supplemental Table S1).
OB Fusion
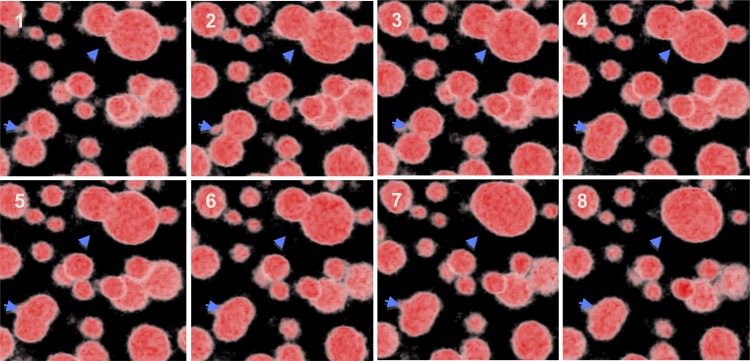
In order to monitor OB behavior, 3D time-lapse live-cell imaging of developing embryos was performed. No specific OB movement of large amplitude was detected. Nevertheless, long acquisition of immobilized embryos at early torpedo stage showed fusions of OBs of similar or different sizes, resulting in larger OBs (Fig. 5; Supplemental Video S1). In a wild-type embryo, a typical fusion event was completed in about 15 min.
Figure 5.
Fusion of OBs. OBs in living torpedo (10 DAF) Columbia embryos immobilized in low-melting agarose were stained with Nile Red and observed with a confocal laser scanning microscope during 1 h (one z-stack each 2 min). The time series images presented were extracted each 7.5 min. Image stacks were processed with the 3D medical image-processing software OsiriX for volume rendering.
DISCUSSION
The processes of OB synthesis and dynamics are still largely unknown (Robenek et al., 2006; Ploegh, 2007; Walther and Farese, 2009; Yang et al., 2012). Concerning OB synthesis, the currently favored model is that they originate from discrete regions of the endoplasmic reticulum (ER) where enzymes for neutral lipid biosynthetic machinery are located (Lacey et al., 1999; Martin and Parton, 2006; Shockey et al., 2006). However, no exact mechanism has been described so far that can explain, for example, why the phospholipid composition of the half membrane enclosing OBs is different from that of the cytosolic leaflet of the ER membrane (Tauchi-Sato et al., 2002).
For OB dynamics and, especially, OB maturation, several proposals have been made (Yang et al., 2012). First, small OBs would bud off the ER at sites specialized in TAG biosynthesis and would then expand by recruiting enzymes of this biosynthetic pathway (McFie et al., 2011; Wilfling et al., 2013). Second, OBs would remain tethered to the ER, filling with neutral lipids until an optimal size is reached, allowing their release into the cytosol. Third, large OBs could also result from the fusion or coalescence of smaller ones (Gong et al., 2011; Ariotti et al., 2012). These different mechanisms may depend on the type of LD, on cell type, and on the neutral lipids accumulated (Cheng et al., 2009; Thiam et al., 2013). In addition to the biophysics at play, the protein complement of OBs is consequential (Krahmer et al., 2013; Thiam et al., 2013), and specific proteins presumably play specific roles. In the case of seed OBs, the role of the conserved family of oleosins is still not clear (Chapman et al., 2012; Murphy, 2012). The analysis presented here was made on living material and represents a comprehensive analysis of OB dynamics in developing embryos, without any treatment but Nile Red staining for the visualization of neutral lipids. After a size increase, at least in part by fusion, during embryo development, OB size decreases during maturation and reaches a size smaller than the optical resolution of standard confocal microscopes after 12 d of seed development. We did not address the question of de novo OB formation itself but, rather, OB dynamics after they have been released from the ER. Thus, our study was mainly based on image analysis and focused on both the first half of seed development and very early seed imbibition, the latter to provide an image of the very last phase of seed development. We have chosen to model OB population dynamics to (1) better estimate the volumes of the OBs in the developing embryos and (2) estimate the relationships between OBs in a 3D space and evaluate their relationships with their neighbors. This time-course analysis differs from previous studies based on mature seed OBs and thus offers a new perspective on the subcellular events linked to OB growth. Finally, it allowed drawing conclusions and new hypotheses about the role of oleosins in OB dynamics.
Five of the 16 members of the Arabidopsis oleosin family are specifically expressed in seeds (Kim et al., 2002; Jolivet et al., 2004; D’Andréa et al., 2007; Liu et al., 2012). In this study, we focused on the most abundant oleosins, OLE1, OLE2, and OLE4. At the time, only a leaky mutant (i.e. with some protein accumulated in seeds) was available for OLE3 (Shimada et al., 2008), which was not useful for evaluation of its role in OB growth. No mutant was available for OLE5, which exhibits a different expression pattern, both in terms of intensity and timing of expression, when compared with other members of the family (Fig. 1), although OLE2 and OLE5 originated from a duplication of OLE3 and OLE1, respectively (Liu et al., 2012). We focused our study on single, double, and triple oleosin mutants and excluded the use of antisense or RNA interference approaches to reduce the risk of cross effects between oleosins. Lack of oleosin was associated in previous studies with delayed germination (Siloto et al., 2006) or lower freezing tolerance (Shimada et al., 2008). We have investigated the germination capabilities of the mutants already characterized and available in the frame of this study, and in our culture conditions no specific delay in germination was observed on freshly harvested seed batches.
OBs Exhibit Volume Variations during Seed Development
We have constructed a treatment pipeline that allowed processing of the image data and transforming them into computational objects after segmentation. Doing so, we have eliminated the possible errors of manual diameter measurements, and we have increased the size of the population of measured OBs. Furthermore, we worked with measured volumes and not with extracted diameters, thus making the analysis much more robust. During early seed maturation in wild-type seeds, OB mean volume increased sharply and then reached a maximum before decreasing until the end of the maturation to sizes below the resolution of conventional confocal microscopes. At the same time, at first the distance between OBs grows and then it decreases, thus increasing the compaction of OBs. We have shown in planta that the increase in OB size is at least partly due to OB fusions and, thus, not only from additional cargo originating from the ER, as suggested previously (Walther and Farese, 2009). We have shown that either the size of OBs or the distance between them is disturbed in oleosin mutants, and in particular, the lack of OLE2 decreased the minimal distance between OBs. This is consistent with the median size of the Voronoï volumes, which is an indicator of the spatial distribution between an OB and all the other OBs in the vicinity, which remains at the same level between 9 and 11 DAF when compared with the control. From these data, we can conclude that one of the putative roles of the oleosin OLE2 could be to maintain the distance between OBs, limiting too-close contacts and fusions. Nevertheless, the ole2 mutant showed aggregates of smaller sized OBs that are not fused (Fig. 2A; Supplemental Fig. S4), thus indicating that the role of OLE2 is not to prevent fusions.
The decrease in OB size during early seed maturation (11 DAF) is more surprising. At the end of seed maturation, when the seed has reached the desiccation stage, the OBs are highly compacted and small (Mansfield and Briarty, 1992). Nevertheless, we have shown an increase followed by a decrease in OB size early during maturation. This variation in OB size is probably not due to an increase in intracellular space constraints, since the cell content in OBs is far from its maximal value. Furthermore, the process is not due to oil content decrease by lipid breakdown, as occurs during late maturation (Baud et al., 2002) and seed germination, since at 11 d after pollination (U-shaped embryo), the oil production rate and the oil content increased sharply. One may hypothesize a discrepancy between the rates of accumulation of oil and oleosins, limiting transiently the quantity of OBs (or half membrane) available to absorb the increase in oil production. From a physical point of view, the optimization of intracellular space by compaction is more efficient with very small OBs but demands much more membrane surface. Such a feedback adaptation of OB production to the rate of oil synthesis may take a few days. The mechanical process underlying this OB size variation is also a matter of debate (Walther and Farese, 2009; Yang et al., 2012). Fission or budding processes could be involved, via the action of specialized proteins docked to the OB surface, although we have not been able to detect such events to date. The fact that embryos are detached from their mother plant when observed should be considered, since the lipid precursor flux is thus modified and the observation of fission events is perhaps just not possible using these techniques. Alternatively, OB size reduction could be obtained by filling an OB from another through intracellular cargos or direct filling. Lastly, large OBs could reattach to the ER to pour their TAG content in newly formed OBs. Whatever the process involved, oleosins play a role, since this size variation disappears in oleosin mutants.
Mutants for Specific Oleosins Show Different Dynamics of OB Growth and Affect Storage Compound Accumulation
Changes in OB size and distribution did perturb the flux of lipid accumulation, since a small but significant decrease in total fatty acids was evidenced in the different mutant backgrounds. The quantity of 12S and 2S storage proteins accumulated was also modified in some mutants, but with the exception of the ole1ole4 background, the ratio between those proteins was not significantly affected. Finally, lack of one or more oleosins seems to have a low impact on storage protein accumulation in the seed. Of note is that the shape of the PBs in the ole1ole2ole4 mutant is completely different from that of the control. In wild-type cells, PBs are found in the center of the cells and are surrounded with small OBs, whereas when the mean VOB increases in the ole1ole2ole4 triple mutant to form one or several very large OBs in the cell, PBs are pushed to the side of the cell with a clearly different deposition profile (Supplemental Fig. S3), as observed previously in an ole1 mutant (Siloto et al., 2006). This certainly indicates a relative plasticity of PBs and, contrarily, that increased OB density can perturb the distribution of accumulated proteins.
Oleosins OLE1, OLE2, and OLE4 Have Different Roles
On the basis of the data accumulated and the statistical analysis performed, we propose that oleosins are not only structural proteins necessary for covering the OB half membrane but also have specific roles in OB dynamics during seed development. In the light of our analyses, five rules can be drawn: (1) fusions can occur between OBs; (2) oleosins OLE1 and OLE4 limit OB fusion; (3) oleosin OLE2 promotes OB fusion; (4) rule 2 is stronger than rule 3; (5) oleosin OLE1 is more efficient in preventing OB fusion than oleosin OLE4.
These five simple rules are sufficient to explain the phenotypes observed in OB distribution or size in the mutant backgrounds. Rule 1 was directly drawn from observations in the wild type as well as the mutant genotypes. Rule 2 is rather straightforward: when lacking oleosins, OBs increased in volume, leading to very large OBs in the cell, pushing aside the PBs and other cell contents (Supplemental Fig. S3). Rule 3 addresses a new function specific to oleosin OLE2. Thus, from these data, one can associate oleosins with specific OB behavior during oil accumulation. Rule 4 defines the fact that if OBs are not properly covered with oleosins, they will fuse in spite of factors keeping them separate, probably in order to optimize the oleosin-surface ratio.
The main model for OB biogenesis relies on the budding of LDs from the ER followed by the fusion of small OBs to form larger ones (Murphy and Vance, 1999; Siloto et al., 2006; Guo et al., 2009), although the mechanisms underlying the delivery of the newly synthesized TAGs to the OB remain unclear. While we did not study these early stages of OB biogenesis, our data clearly showed that OB growth can occur by fusion. Nile Red staining does not allow discrimination between two possibilities: (1) the fusion of two OBs with redistribution of the oleosins and phospholipids from donor to acceptor, and (2) a net transfer of storage lipids from smaller OBs to larger ones without redistribution.
In the case of the depletion of one or more oleosins, fusion is promoted, thus likely maintaining an average number of oleosin molecules per surface unit of OB. Recent data suggest that this view of OBs is probably too simple to explain OB fusions and fragmentations (fissions), organelle interactions, or the transport needed for the dynamic variations of VOB (Walther and Farese, 2009; Thiam et al., 2013). In animals, the net transfer of storage lipids from smaller LDs to larger ones without redistribution of LD proteins from donor to acceptor has been shown (Gong et al., 2011). Also, the synthesis of TAGs can be directly coupled with the formation of LDs that share common features with plant OBs, thus indicating a growth process independent of fusions with existing LDs or with the ER (Wilfling et al., 2013). In that case, one can hypothesize that the role of the oleosins on the surface of the OB is not only to ensure the stability of the organelle (role of OLE1 and OLE4) or the distance between organelles (role of OLE2) but probably also the docking of enzymes of the lipogenic pathway, such as members of the TAG synthesis machinery (Wilfling et al., 2013), that will allow direct filling of the OB. These docking properties could be carried out by OLE1, OLE2, or OLE4 in addition to their structural functions but also by other members of the family, such as OLE5 or OLE3. The fact that OLE1 and OLE5, as well as OLE2 and OLE3, derive from duplications suggests that these protein may have similar functions. Moreover, other proteins found in the protein complement of the OB, such as steroleosins, caleosins, and seed- and microspore-specific oleosins, also could be involved in the docking process.
In the future, we aim to use these rules to further characterize the importance of the different oleosins in different processes, including the movement of OBs, the measurement of the fusion rate, and the movement of oleosins on the surface of OBs. This will pave the way for the construction and improvement of a mathematical model of OB dynamics that would improve our understanding of the roles of OBs in addition to being the central player in the accumulation of oil.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana), accession Columbia and oleosin-deficient mutants, were surface sterilized and stratified for 48 h at 4°C in the dark. They were then germinated on full-strength Gamborg B5 medium, pH 5.6 (Duchefa Biochemie; G0210.0050), containing 0.8% (w/v) agar, 1% (w/v) Suc, and the appropriate selection agent in a growth cabinet (16-h-light/8-h-dark photoperiod at 150 μmol m−2 s−1 ; 15°C night/20°C day temperature). After 14 d, the plantlets were transferred to compost, grown in a greenhouse under the following conditions (13 h of light, diurnal temperature of 25°C, and nocturnal temperature of 17°C), and irrigated twice per week with mineral nutrient solution. To harvest seeds at defined developmental stages, individual flowers were tagged on the day of opening, and then opened flowers and developing siliques were counted daily. Materials used for RNA and protein extractions and lipid analysis were frozen in liquid nitrogen immediately after harvest and stored at −80°C prior to analysis. Weight determinations of seed samples were realized on an M2P balance (Sartorius).
Oleosin-Deficient Mutants
Seeds of ole1 (SM_3_29875; At4g25140), ole2 (SALK 072403; At5g40420), and ole4 (SM_20767; At3g01570) were obtained from the Nottingham Arabidopsis Stock Centre (NASC). The double (ole1ole2, ole2ole4, and ole1ole4) and triple (ole1ole2ole4) mutants were generated by crosses between single mutants.
Fatty Acid Content and Composition
For total fatty acid quantity and composition analyses by gas chromatography of the corresponding fatty acyl methyl esters, pools of 20 individual seeds were placed in a glass reaction tube prior to methanol:sulfuric acid:toluene (100:2.5:30, v/v/v) with the addition of C17:0 fatty acid as an internal standard (Li et al., 2006). After derivatization for 1.5 h at 95°C, fatty acid methyl esters were extracted in hexane, separated by gas chromatography on a 30-m × 0.53-mm EC-WAX column (Alltech Associates), and quantified using a flame ionization detector. The gas chromatograph was programmed for an initial temperature of 160°C for 1 min, followed by an increase of 20°C min−1 to 190°C and a ramp of 4°C min−1 to 230°C, with a 9-min hold of the final temperature.
Imaging
Developing seeds were dissected from siliques for each stage of development of interest. Seeds were spread on a glass slide and incubated with Nile Red (Sigma-Aldrich), a neutral lipid stain, at 2 µg mL−1 in a 60% (v/v) glycerol solution. Embryos were removed from the seed teguments by gently pressing seeds between the slide and lamella and observed after 30 min of incubation in the dark. For germinating seeds, mature seeds were placed on wet Whatman paper, stratified for 2 d at 4°C, and then placed in a growth cabinet (16-h-light/8-h-dark photoperiod at 150 μmol m−2 s−1 ; 15°C night/20°C day temperature) for 1, 24, or 48 h. Seed teguments were manually removed, and the embryos were incubated with Nile Red as described for developing seeds.
Images of dissected Arabidopsis embryos were acquired using an inverted LEICA SP2-AOBS spectral confocal laser microscope (Leica Microsystems) using an HCX PL APO CS 40 × 1.25 objective. Fluorescence labeling from Nile Red was observed with a 488-nm light wavelength generated by an argon laser and an emission band of 550 to 650 nm. Each image consists of the maximum projection of 90 to 100 optical sections. Each section is the average of two scans conducted at the resolution of 512 × 512 pixels with a spatial resolution of 0.09 µm × 0.09 µm × 0.16 µm in the x, y, z referential.
For 3D time-lapse live-cell imaging, developing embryos were stained with Nile Red and mounted in 0.008% (w/v) low-gelling-point agarose in 50% (v/v) glycerol in a glass-bottom dish. Images were acquired using an inverted LEICA tandem SP5-AOBS spectral confocal laser microscope (Leica Microsystems) using an HCX PL APO CS 40 × 1.25 objective. Fluorescence labeling from Nile Red was observed with a 488-nm light wavelength generated by an argon laser and an emission band of 550 to 650 nm. Each image consists of the maximum projection of 100 optical sections, and images were acquired every 2 min over a 12-h period. Each section is the average of eight scans conducted at the resolution of 512 × 512 pixels with a spatial resolution of 0.09 µm × 0.09 µm × 0.16 µm in the x, y, z referential. Image stacks were processed with OsiriX, a medical image-processing software for 3D rendering
Immunocytochemistry
Siliques of different developmental stages were fixed in 4% (w/v) paraformaldehyde in phosphate-buffered saline as described previously (Macquet et al., 2007). Sections of 8 µm were incubated with the primary antibody raised against oleosins (D’Andréa et al., 2007) and secondary antibody (anti-Alexa Fluor 488; Molecular Probes, Invitrogen). Samples were visualized with an inverted LEICA SP2-AOBS spectral confocal laser microscope (Leica Microsystems) equipped with an argon laser.
Quantitative PCR
Frozen tissues were ground in liquid nitrogen, and total RNA was extracted using the Sigma-Aldrich GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich). RNA extraction of dry seeds was performed as described (Suzuki et al., 2004). An on-column DNase treatment was performed using the RNase-free DNase 1 (Qiagen). For RT-PCR studies, the DNA-free RNA was converted into first-strand complementary DNA (cDNA) using the SuperScript II RT preamplification system for first-strand cDNA synthesis (Invitrogen) with oligo(dT)22.
Quantitative PCR was performed with a LightCycler using SYBR Green mix (Roche). Specific primers for OLE1, OLE2, OLE3, OLE4, and OLE5 were designed using the LightCycler probe design software version 3.5 (for oligonucleotide sequences, see Supplemental Table S2) and tested for efficiency rates as well as sensibility to dilutions of cDNA samples. EF1αA1 was used as a control. The parameters for quantitative PCR were as follows: (1) initial denaturation at 95°C for 8 min; (2) 45 cycles of 94°C for 10 s, 55°C for 10 s, and 72°C for 15 s, with a temperature transition rate of 20°C s−1; and (3) determination of the fusion curve at 94°C for 10 s, 65°C for 30 s, and an increase to 94°C, with a temperature transition rate of 0.1°C s−1.
Quantitative Western Blots
Oleosin contents in Arabidopsis developing siliques and mature seeds were measured by western blot using specific sera against the five seed-specific oleosins, OLE1, OLE2, OLE3, OLE4, and OLE5 (D’Andréa et al., 2007). Total protein extracts were prepared by grinding siliques (7–19 DAF) or seeds isolated from siliques (20 and 21 DAF) in 2× Laemmli buffer (125 mm Tris-HCl, pH 6.8, 4% (v/v) SDS, and 20% (v/v) glycerol) using a glass-glass conical tissue grinder at the ratio of one silique per 100 µL. After centrifugation, protein extracts were separated on 4–12% (w/v) NuPAGE gels using MOPS SDS running buffer (Novex, Life Technologies). Proteins were transferred onto polyvinylidene difluoride membranes (Immobilon-P 0.45 µm; Millipore) in NuPAGE transfer buffer (Life Technologies). Saturation and incubation with antibodies were carried out according to D’Andréa et al. (2007). Luminescent signals from western blots were recorded using a LAS-3000 luminescent image analyzer (Fujifilm) and quantified with the MultiGauge software (Fujifilm). Mature dry seed extract was used as a standard to determine the linear relation between luminescent signal and oleosin content. Using this linear relation, the amount of oleosin in developing seeds was determined from the luminescent signal (provided that this signal was within the linear range) and expressed as the percentage of mature seed content.
Quantitative Analyses of OB Size Distribution and Dispersion
A total of 180 3D stack images were first denoised using Nd Safir (Boulanger et al., 2010) before being treated within the pipeline. First, the extraction of the volumes from individualized OBs through the pipeline is based on the watershed procedure dedicated to the separation of touching objects (Soille, 2003). The case of nearest OBs is quite abundant in the image data we analyzed, resulting in overlapped OB segmentation when considering a simple threshold (Supplemental Fig. S5, A, B, and F). The distance transform assigns a value to each pixel on an object in the threshold image corresponding to its distance from the object border. The complement of this result (Supplemental Fig. S5, C and G) was transformed into its topographical representation, forming the catchment basins. The new separated objects corresponding to the OBs are then labeled with a specific color (Supplemental Fig. S5, D and H). The VOB was obtained by counting the number of voxels on an object and multiplying by the elementary volume of the voxel.
The ϕloc estimator is based on the concept of the Voronoï diagram. The Voronoï cells were obtained by dividing the space containing OBs into cells so that each point of a given cell is equidistant from any OB surface. The ϕloc is so defined by:
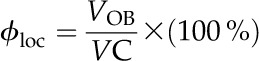 |
where VC is the volume of the Voronoï cell.
In order to determine the boundaries of the sample, and to avoid overestimation of OB Voronoï cells on the edge of the sample, we used the convex envelope (Fig. 3, A3, B3, and C3). Intuitively, the convex envelope is defined by the smallest convex polygon that contains all the elements T of a finite set E, where T represents a lipid droplet and E represents the set of lipid droplets. The corrected Voronoï cells are then computed with respect to the convex envelope (Fig. 3, A4, B4, and C4).
The VC is determined from labeled components as explained previously for VOB. Finally, extracting the minimal value across the boundaries of each Voronoï cell leads to the middistance to the nearest neighbor (Supplemental Fig. S6).
In order to study the influence of each oleosin, the day factor, and their interactions on both VOB and VC, a mathematical model was developed (Trigui et al., 2012), and a variance analysis of this model was performed using a quantile regression (Trigui et al., 2013).
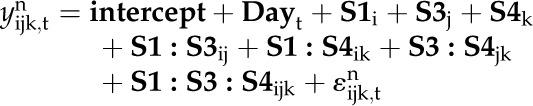 |
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Validation of antibody specificity on sections of developing siliques of mutants affected in one oleosin.
Supplemental Figure S2. Effect of oleosin(s) deficiency on protein content.
Supplemental Figure S3. 3D reconstruction of storage lipid and protein accumulation in the wild-type and ole1ole2 mutant backgrounds in 1-h imbibed seeds.
Supplemental Figure S4. Distribution of OBs at 12 DAF in wild-type and ole2 embryos.
Supplemental Figure S5. Segmentation steps of OBs using the watershed method.
Supplemental Figure S6. Determination of the Voronoï cells and the distance to the next neighbor estimator steps.
Supplemental Table S1. Adjusted parameters for the quantile regression statistical analysis of VOB and ϕloc factors.
Supplemental Table S2. Sequences of quantitative RT-PCR primers used in this study.
Supplemental Video S1. 3D time-lapse live-cell imaging of wild-type developing embryos stained with Nile Red.
Supplementary Material
Acknowledgments
We thank Thierry Chardot for many helpful suggestions and interesting discussions, Mark Tepfer for critical reading of the manuscript and kind correction of the English, Michael Anjuere for taking care of the plants, and Johanne Thévenin, David Rodrigues, and Manon Payre for technical help.
Glossary
- LD
lipid droplet
- TAG
triacylglycerol
- OB
oil body
- DAF
days after flowering
- RT
reverse transcription
- PB
protein body
- 3D
three-dimensional
- VOB
volume of oil bodies
- ϕloc
local fraction
- ER
endoplasmic reticulum
- cDNA
complementary DNA
- VC
volume of the Voronoï cell
Footnotes
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ariotti N, Murphy S, Hamilton NA, Wu L, Green K, Schieber NL, Li P, Martin S, Parton RG. (2012) Postlipolytic insulin-dependent remodeling of micro lipid droplets in adipocytes. Mol Biol Cell 23: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C. (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Beller M, Bulankina AV, Hsiao HH, Urlaub H, Jäckle H, Kühnlein RP. (2010) PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab 12: 521–532 [DOI] [PubMed] [Google Scholar]
- Beller M, Riedel D, Jänsch L, Dieterich G, Wehland J, Jäckle H, Kühnlein RP. (2006) Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics 5: 1082–1094 [DOI] [PubMed] [Google Scholar]
- Boulanger J, Kervrann C, Bouthemy P, Elbau P, Sibarita JB, Salamero J. (2010) Patch-based nonlocal functional for denoising fluorescence microscopy image sequences. IEEE Trans Med Imaging 29: 442–454 [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Wolins NE. (2012) Packaging of fat: an evolving model of lipid droplet assembly and expansion. J Biol Chem 287: 2273–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Dyer JM, Mullen RT. (2012) Biogenesis and functions of lipid droplets in plants. J Lipid Res 53: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Ohlrogge JB. (2012) Compartmentation of triacylglycerol accumulation in plants. J Biol Chem 287: 2288–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Fujita A, Ohsaki Y, Suzuki M, Shinohara Y, Fujimoto T. (2009) Quantitative electron microscopy shows uniform incorporation of triglycerides into existing lipid droplets. Histochem Cell Biol 132: 281–291 [DOI] [PubMed] [Google Scholar]
- D’Andréa S, Jolivet P, Boulard C, Larré C, Froissard M, Chardot T. (2007) Selective one-step extraction of Arabidopsis thaliana seed oleosins using organic solvents. J Agric Food Chem 55: 10008–10015 [DOI] [PubMed] [Google Scholar]
- Débarre D, Supatto W, Pena AM, Fabre A, Tordjmann T, Combettes L, Schanne-Klein MC, Beaurepaire E. (2006) Imaging lipid bodies in cells and tissues using third-harmonic generation microscopy. Nat Methods 3: 47–53 [DOI] [PubMed] [Google Scholar]
- Fuchs J, Neuberger T, Rolletschek H, Schiebold S, Nguyen TH, Borisjuk N, Börner A, Melkus G, Jakob P, Borisjuk L. (2013) A noninvasive platform for imaging and quantifying oil storage in submillimeter tobacco seed. Plant Physiol 161: 583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Sun Z, Wu L, Xu W, Schieber N, Xu D, Shui G, Yang H, Parton RG, Li P. (2011) Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J Cell Biol 195: 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan P, Fowler SD. (1985) Spectrofluorometric studies of the lipid probe, Nile red. J Lipid Res 26: 781–789 [PubMed] [Google Scholar]
- Guo Y, Cordes KR, Farese RV, Jr, Walther TC. (2009) Lipid droplets at a glance. J Cell Sci 122: 749–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneen WK, Karlsson G, Brismar K, Gummeson PO, Marttila S, Leonova S, Carlsson AS, Bafor M, Banas A, Mattsson B, et al (2008) Fusion of oil bodies in endosperm of oat grains. Planta 228: 589–599 [DOI] [PubMed] [Google Scholar]
- Hodges BDM, Wu CC. (2010) Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J Lipid Res 51: 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Ledbetter NR, James CN, Hoffman WD, Case CR, Verbeck GF, Chapman KD. (2011) Visualization of lipid droplet composition by direct organelle mass spectrometry. J Biol Chem 286: 3298–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K, Huang AHC. (2004) Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiol 136: 3427–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AH. (1996) Oleosins and oil bodies in seeds and other organs. Plant Physiol 110: 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolivet P, Boulard C, Bellamy A, Larré C, Barre M, Rogniaux H, d’Andréa S, Chardot T, Nesi N. (2009) Protein composition of oil bodies from mature Brassica napus seeds. Proteomics 9: 3268–3284 [DOI] [PubMed] [Google Scholar]
- Jolivet P, Roux E, D’Andrea S, Davanture M, Negroni L, Zivy M, Chardot T. (2004) Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 42: 501–509 [DOI] [PubMed] [Google Scholar]
- Katavic V, Agrawal GK, Hajduch M, Harris SL, Thelen JJ. (2006) Protein and lipid composition analysis of oil bodies from two Brassica napus cultivars. Proteomics 6: 4586–4598 [DOI] [PubMed] [Google Scholar]
- Kim HU, Hsieh K, Ratnayake C, Huang AHC. (2002) A novel group of oleosins is present inside the pollen of Arabidopsis. J Biol Chem 277: 22677–22684 [DOI] [PubMed] [Google Scholar]
- Krahmer N, Hilger M, Kory N, Wilfling F, Stoehr G, Mann M, Farese RV, Jr, Walther TC. (2013) Protein correlation profiles identify lipid droplet proteins with high confidence. Mol Cell Proteomics 12: 1115–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey DJ, Beaudoin F, Dempsey CE, Shewry PR, Napier JA. (1999) The accumulation of triacylglycerols within the endoplasmic reticulum of developing seeds of Helianthus annuus. Plant J 17: 397–405 [Google Scholar]
- Leber R, Zinser E, Zellnig G, Paltauf F, Daum G. (1994) Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast 10: 1421–1428 [DOI] [PubMed] [Google Scholar]
- Leprince O, van Aelst AC, Pritchard HW, Murphy DJ. (1998) Oleosins prevent oil-body coalescence during seed imbibition as suggested by a low-temperature scanning electron microscope study of desiccation-tolerant and -sensitive oilseeds. Planta 204: 109–119 [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J. (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Liu Q, Sun Y, Su W, Yang J, Liu X, Wang Y, Wang F, Li H, Li X. (2012) Species-specific size expansion and molecular evolution of the oleosins in angiosperms. Gene 509: 247–257 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Kronenberger J, Marion-Poll A, North HM. (2007) In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol 48: 984–999 [DOI] [PubMed] [Google Scholar]
- Mak HY. (2012) Lipid droplets as fat storage organelles in Caenorhabditis elegans. J Lipid Res 53: 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. (1992) Cotyledon cell development in Arabidopsis thaliana during reserve deposition. Can J Bot 70: 151–164 [Google Scholar]
- Martin S, Parton RG. (2006) Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol 7: 373–378 [DOI] [PubMed] [Google Scholar]
- McFie PJ, Banman SL, Kary S, Stone SJ. (2011) Murine diacylglycerol acyltransferase-2 (DGAT2) can catalyze triacylglycerol synthesis and promote lipid droplet formation independent of its localization to the endoplasmic reticulum. J Biol Chem 286: 28235–28246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ. (2001) The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res 40: 325–438 [DOI] [PubMed] [Google Scholar]
- Murphy DJ. (2012) The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249: 541–585 [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Vance J. (1999) Mechanisms of lipid-body formation. Trends Biochem Sci 24: 109–115 [DOI] [PubMed] [Google Scholar]
- Neuberger T, Sreenivasulu N, Rokitta M, Rolletschek H, Göbel C, Rutten T, Radchuk V, Feussner I, Wobus U, Jakob P, et al (2008) Quantitative imaging of oil storage in developing crop seeds. Plant Biotechnol J 6: 31–45 [DOI] [PubMed] [Google Scholar]
- Paar M, Jüngst C, Steiner NA, Magnes C, Sinner F, Kolb D, Lass A, Zimmermann R, Zumbusch A, Kohlwein SD, et al (2012) Remodeling of lipid droplets during lipolysis and growth in adipocytes. J Biol Chem 287: 11164–11173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano RE, Martin OC, Kang HC, Haugland RP. (1991) A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J Cell Biol 113: 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthibane V, Iyappan R, Vijayakumar A, Venkateshwari V, Rajasekharan R. (2012a) Serine/threonine/tyrosine protein kinase phosphorylates oleosin, a regulator of lipid metabolic functions. Plant Physiol 159: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthibane V, Rajakumari S, Venkateshwari V, Iyappan R, Rajasekharan R. (2012b) Oleosin is bifunctional enzyme that has both monoacylglycerol acyltransferase and phospholipase activities. J Biol Chem 287: 1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploegh HL. (2007) A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature 448: 435–438 [DOI] [PubMed] [Google Scholar]
- Preston J, Tatematsu K, Kanno Y, Hobo T, Kimura M, Jikumaru Y, Yano R, Kamiya Y, Nambara E. (2009) Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: a comparative study on dormant and non-dormant accessions. Plant Cell Physiol 50: 1786–1800 [DOI] [PubMed] [Google Scholar]
- Qu RD, Huang AH. (1990) Oleosin KD 18 on the surface of oil bodies in maize: genomic and cDNA sequences and the deduced protein structure. J Biol Chem 265: 2238–2243 [PubMed] [Google Scholar]
- Robenek H, Hofnagel O, Buers I, Robenek MJ, Troyer D, Severs NJ. (2006) Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J Cell Sci 119: 4215–4224 [DOI] [PubMed] [Google Scholar]
- Schmidt MA, Herman EM. (2008) Suppression of soybean oleosin produces micro-oil bodies that aggregate into oil body/ER complexes. Mol Plant 1: 910–924 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I. (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM. (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18: 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soille P (2003) Morphological Image Analysis: Principles and Applications, Ed 2. Springer-Verlag, Berlin [Google Scholar]
- Suzuki Y, Kawazu T, Koyama H. (2004) RNA isolation from siliques, dry seeds, and other tissues of Arabidopsis thaliana. Biotechniques 37: 542–544 [DOI] [PubMed] [Google Scholar]
- Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. (2002) The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem 277: 44507–44512 [DOI] [PubMed] [Google Scholar]
- Thiam AR, Farese RV, Jr, Walther TC. (2013) The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol 14: 775–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigui G, Laroche B, Miquel M, Dubreucq B, Trubuil A. (2012) The dynamics of oil bodies in A. thaliana seeds: a mathematical model of biogenesis and coalescence. World Academy of Science, Engineering, and Technology 64: 763–768 [Google Scholar]
- Trigui G, Miquel M, Dubreucq B, David O, Trubuil A. (2013) Analysis of factors affecting the growth of oil bodies in A. thaliana seeds: comparison of ordinary least squares and quantile regression. Tampere International Center for Signal Processing 63: 98–101 [Google Scholar]
- Tzen J, Cao Y, Laurent P, Ratnayake C, Huang A. (1993) Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol 101: 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzen JT, Huang AH. (1992) Surface structure and properties of plant seed oil bodies. J Cell Biol 117: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzen JT, Lai YK, Chan KL, Huang AH. (1990) Oleosin isoforms of high and low molecular weights are present in the oil bodies of diverse seed species. Plant Physiol 94: 1282–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzen JT, Lie GC, Huang AH. (1992) Characterization of the charged components and their topology on the surface of plant seed oil bodies. J Biol Chem 267: 15626–15634 [PubMed] [Google Scholar]
- Vermachova M, Purkrtova Z, Santrucek J, Jolivet P, Chardot T, Kodicek M. (2011) New protein isoforms identified within Arabidopsis thaliana seed oil bodies combining chymotrypsin/trypsin digestion and peptide fragmentation analysis. Proteomics 11: 3430–3434 [DOI] [PubMed] [Google Scholar]
- Wältermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla HJ, Kalscheuer R, Stöveken T, von Landenberg P, et al (2005) Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol Microbiol 55: 750–763 [DOI] [PubMed] [Google Scholar]
- Walther TC, Farese RV., Jr (2009) The life of lipid droplets. Biochim Biophys Acta 1791: 459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Fröhlich F, et al. (2013) Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell 24: 384–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Galea A, Sytnyk V, Crossley M. (2012) Controlling the size of lipid droplets: lipid and protein factors. Curr Opin Cell Biol 24: 509–516 [DOI] [PubMed] [Google Scholar]
- Zhang SO, Box AC, Xu N, Le Men J, Yu J, Guo F, Trimble R, Mak HY. (2010) Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proc Natl Acad Sci USA 107: 4640–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.