Abstract
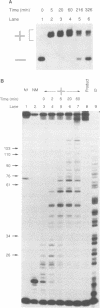
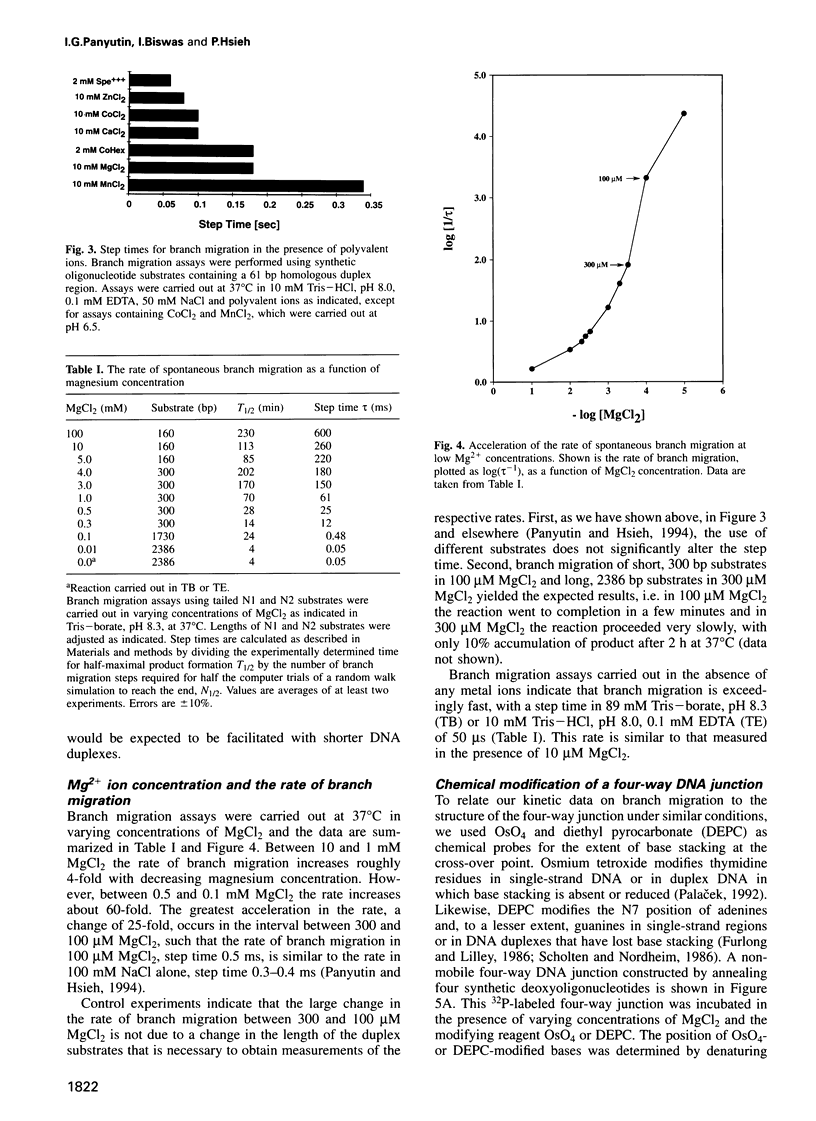
Branch migration of a DNA Holliday junction is a key step in genetic recombination that affects the extent of transfer of genetic information between homologous DNA sequences. We previously observed that the rate of spontaneous branch migration is exceedingly sensitive to metal ions and postulated that the structure of the cross-over point might be one critical determinant of the rate of branch migration. Other investigators have shown that in the presence of divalent metal ions like magnesium, the Holliday junction assumes a folded conformation in which base stacking is retained through the cross-over point. This base stacking is disrupted in the absence of magnesium. Here we measure the rate of branch migration as a function of Mg2+ concentration. The rate of branch migration increases dramatically at MgCl2 concentrations below 500 microM, with the steepest acceleration occurring between 300 and 100 microM MgCl2. This increase in the rate of branch migration coincides with the loss of base stacking in the four-way junction over this same interval of magnesium concentration, as measured by the susceptibility of junction residues to modification by osmium tetroxide and diethyl pyrocarbonate. We conclude that at physiological concentrations of intracellular Mg2+, base stacking in the Holliday junction constitutes one kinetic barrier to branch migration and that disruption of base stacking at the cross-over relieves this constraint.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Broker T. R., Lehman I. R. Branched DNA molecules: intermediates in T4 recombination. J Mol Biol. 1971 Aug 28;60(1):131–149. doi: 10.1016/0022-2836(71)90453-0. [DOI] [PubMed] [Google Scholar]
- Clegg R. M., Murchie A. I., Lilley D. M. The solution structure of the four-way DNA junction at low-salt conditions: a fluorescence resonance energy transfer analysis. Biophys J. 1994 Jan;66(1):99–109. doi: 10.1016/S0006-3495(94)80765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickie P., McFadden G., Morgan A. R. The site-specific cleavage of synthetic Holliday junction analogs and related branched DNA structures by bacteriophage T7 endonuclease I. J Biol Chem. 1987 Oct 25;262(30):14826–14836. [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988 Oct 7;55(1):79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Lilley D. M. The role of metal ions in the conformation of the four-way DNA junction. EMBO J. 1990 Feb;9(2):583–590. doi: 10.1002/j.1460-2075.1990.tb08146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubbs R. D., Maguire M. E. Magnesium as a regulatory cation: criteria and evaluation. Magnesium. 1987;6(3):113–127. [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Kohwi Y. Detection of non-B-DNA structures at specific sites in supercoiled plasmid DNA and chromatin with haloacetaldehyde and diethyl pyrocarbonate. Methods Enzymol. 1992;212:155–180. doi: 10.1016/0076-6879(92)12011-e. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994 Sep;58(3):401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy J. L., Broseta D., Guéron M. Proton exchange and base-pair kinetics of poly(rA).poly(rU) and poly(rI).poly(rC). J Mol Biol. 1985 Jul 5;184(1):165–178. doi: 10.1016/0022-2836(85)90050-6. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Clegg R. M. The structure of the four-way junction in DNA. Annu Rev Biophys Biomol Struct. 1993;22:299–328. doi: 10.1146/annurev.bb.22.060193.001503. [DOI] [PubMed] [Google Scholar]
- Lu M., Guo Q., Mueller J. E., Kemper B., Studier F. W., Seeman N. C., Kallenbach N. R. Characterization of a bimobile DNA junction. J Biol Chem. 1990 Oct 5;265(28):16778–16785. [PubMed] [Google Scholar]
- Lu M., Guo Q., Studier F. W., Kallenbach N. R. Resolution of branched DNA substrates by T7 endonuclease I and its inhibition. J Biol Chem. 1991 Feb 5;266(4):2531–2536. [PubMed] [Google Scholar]
- Møllegaard N. E., Murchie A. I., Lilley D. M., Nielsen P. E. Uranyl photoprobing of a four-way DNA junction: evidence for specific metal ion binding. EMBO J. 1994 Apr 1;13(7):1508–1513. doi: 10.1002/j.1460-2075.1994.tb06412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek E. Probing DNA structure with osmium tetroxide complexes in vitro. Methods Enzymol. 1992;212:139–155. doi: 10.1016/0076-6879(92)12010-n. [DOI] [PubMed] [Google Scholar]
- Panyutin I. G., Hsieh P. Formation of a single base mismatch impedes spontaneous DNA branch migration. J Mol Biol. 1993 Mar 20;230(2):413–424. doi: 10.1006/jmbi.1993.1159. [DOI] [PubMed] [Google Scholar]
- Panyutin I. G., Hsieh P. The kinetics of spontaneous DNA branch migration. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2021–2025. doi: 10.1073/pnas.91.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyutin I. G., Wells R. D. Nodule DNA in the (GA)37.(CT)37 insert in superhelical plasmids. J Biol Chem. 1992 Mar 15;267(8):5495–5501. [PubMed] [Google Scholar]
- Picksley S. M., Parsons C. A., Kemper B., West S. C. Cleavage specificity of bacteriophage T4 endonuclease VII and bacteriophage T7 endonuclease I on synthetic branch migratable Holliday junctions. J Mol Biol. 1990 Apr 20;212(4):723–735. doi: 10.1016/0022-2836(90)90233-C. [DOI] [PubMed] [Google Scholar]
- Pöhler J. R., Duckett D. R., Lilley D. M. Structure of four-way DNA junctions containing a nick in one strand. J Mol Biol. 1994 Apr 22;238(1):62–74. doi: 10.1006/jmbi.1994.1268. [DOI] [PubMed] [Google Scholar]
- Radding C. M., Beattie K. L., Holloman W. K., Wiegand R. C. Uptake of homologous single-stranded fragments by superhelical DNA. IV. Branch migration. J Mol Biol. 1977 Nov;116(4):825–839. doi: 10.1016/0022-2836(77)90273-x. [DOI] [PubMed] [Google Scholar]
- Sadowski P. D. Bacteriophage T7 endonuclease. I. Properties of the enzyme purified from T7 phage-infected Escherichia coli B. J Biol Chem. 1971 Jan 10;246(1):209–216. [PubMed] [Google Scholar]
- Scholten P. M., Nordheim A. Diethyl pyrocarbonate: a chemical probe for DNA cruciforms. Nucleic Acids Res. 1986 May 27;14(10):3981–3993. doi: 10.1093/nar/14.10.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Kallenbach N. R. DNA branched junctions. Annu Rev Biophys Biomol Struct. 1994;23:53–86. doi: 10.1146/annurev.bb.23.060194.000413. [DOI] [PubMed] [Google Scholar]
- Stark W. M., Boocock M. R., Sherratt D. J. Catalysis by site-specific recombinases. Trends Genet. 1992 Dec;8(12):432–439. [PubMed] [Google Scholar]
- Sullivan K. M., Lilley D. M. Influence of cation size and charge on the extrusion of a salt-dependent cruciform. J Mol Biol. 1987 Jan 20;193(2):397–404. doi: 10.1016/0022-2836(87)90227-0. [DOI] [PubMed] [Google Scholar]
- West S. C. The processing of recombination intermediates: mechanistic insights from studies of bacterial proteins. Cell. 1994 Jan 14;76(1):9–15. doi: 10.1016/0092-8674(94)90168-6. [DOI] [PubMed] [Google Scholar]
- de Massy B., Weisberg R. A., Studier F. W. Gene 3 endonuclease of bacteriophage T7 resolves conformationally branched structures in double-stranded DNA. J Mol Biol. 1987 Jan 20;193(2):359–376. doi: 10.1016/0022-2836(87)90224-5. [DOI] [PubMed] [Google Scholar]