Accurate regulation of callose metabolism during microsporogenesis is critical for plant male fertility.
Abstract
During angiosperm microsporogenesis, callose serves as a temporary wall to separate microsporocytes and newly formed microspores in the tetrad. Abnormal callose deposition and dissolution can lead to degeneration of developing microspores. However, genes and their regulation in callose metabolism during microsporogenesis still remain largely unclear. Here, we demonstrated that the Arabidopsis (Arabidopsis thaliana) CALLOSE DEFECTIVE MICROSPORE1 (CDM1) gene, encoding a tandem CCCH-type zinc finger protein, plays an important role in regulation of callose metabolism in male meiocytes and in integrity of newly formed microspores. First, quantitative reverse transcription PCR and in situ hybridization analyses showed that the CDM1 gene was highly expressed in meiocytes and the tapetum from anther stages 4 to 7. In addition, a transfer DNA insertional cdm1 mutant was completely male sterile. Moreover, light microscopy of anther sections revealed that microspores in the mutant anther were initiated, and then degenerated soon afterward with callose deposition defects, eventually leading to male sterility. Furthermore, transmission electron microscopy demonstrated that pollen exine formation was severely affected in the cdm1 mutant. Finally, we found that the cdm1 mutation affected the expression of callose synthesis genes (CALLOSE SYNTHASE5 and CALLOSE SYNTHASE12) and potential callase-related genes (A6 and MYB80), as well as three other putative β-1,3-glucanase genes. Therefore, we propose that the CDM1 gene regulates callose metabolism during microsporogenesis, thereby promoting Arabidopsis male fertility.
In flowering plants, male meiocytes undergo meiosis to generate microspores, eventually producing haploid gametes for double fertilization (Ma, 2005). At early meiosis, each meiocyte begins to synthesize a temporary wall mainly containing callose, which is deposited between the primary cell wall and the plasma membrane. Callose continues to be deposited through the whole meiosis, resulting in the enclosure of each newly formed microspore by a thick callose wall (McCormick, 2004). The major composition of callose is β-1,3-glucan, which consists of Glc residues with β-1,3-linkages (Ariizumi and Toriyama, 2011). The callose wall is subsequently, degraded by an enzyme mixture called callase, which is secreted by the tapetum and possesses β-1,3-glucanase (β-1,3-G) activities (Scott et al., 2004). Finally, the sibling microspores are released individually into the anther locule (Stieglitz, 1977). In addition to serving as a temporary envelope of newly formed microspores, callose also facilitates pollen wall formation (Ariizumi and Toriyama, 2011).
In most angiosperms, callose is synthesized by callose synthases from meiocytes (Scott et al., 2004). Reduced callose accumulation may lead to the abortion of developing microspores (Ariizumi and Toriyama, 2011). In the Arabidopsis (Arabidopsis thaliana) genome, there is a total of 12 callose synthase genes (CALLOSE SYNTHASE1 [CalS1] to CalS12; Hong et al., 2001). Among them, CalS5, CalS11, and CalS12 have been shown to be involved in callose synthesis during microsporogenesis (Dong et al., 2005; Enns et al., 2005; Nishikawa et al., 2005). CalS5 is responsible for callose deposition surrounding the meiocytes, tetrads, and microspores. On the other hand, CalS11 and CalS12 function redundantly in synthesizing callose between microspores in a tetrad. Double mutants of cals11/cals12 produce greatly reduced amounts of callose, leading to the degeneration of microspores (Dong et al., 2005; Enns et al., 2005; Nishikawa et al., 2005).
In addition to its synthesis, the proper timing of callose dissolution is also crucial for the formation of functional microspores. For example, premature or delayed callase activity leads to microspore abortion in petunia (Petunia hybrida; Frankel et al., 1969; Izhar and Frankel, 1971). Similarly, premature dissolution of callose walls during meiosis as a result of early expression of a β-1,3-G results in male sterility in transgenic tobacco (Nicotiana tabacum; Worrall et al., 1992). According to the enzymatic cleavage site, β-1,3-Gs have been classified as endoglucanases or exoglucanases (Zhang et al., 2007). In Lilium, it was shown that endo-β-1,3-Gs were responsible for callose wall degradation during microsporogenesis (Stieglitz, 1977). Although the Arabidopsis genome has >50 genes encoding β-1,3-Gs (Doxey et al., 2007), only the A6 gene was suggested to be a major component of the callase mixture secreted by the tapetum (Hird et al., 1993). Among known regulatory genes for anther development, only the AtMYB80 (formally AtMYB103) gene encoding a putative transcription factor was identified as a positive regulator of the A6 gene, affecting callose metabolism during microsporogenesis (Zhang et al., 2007). Therefore, the regulation of callase-related gene expression still remains largely unknown. In this study, we report that the Arabidopsis CALLOSE DEFECTIVE MICROSPORE1 (CDM1) gene, which encodes a tandem CCCH-domain zinc finger (TZF) protein, regulates callose metabolism during microsporogenesis and is required for male fertility.
RESULTS
The CDM1 Gene Expression Pattern
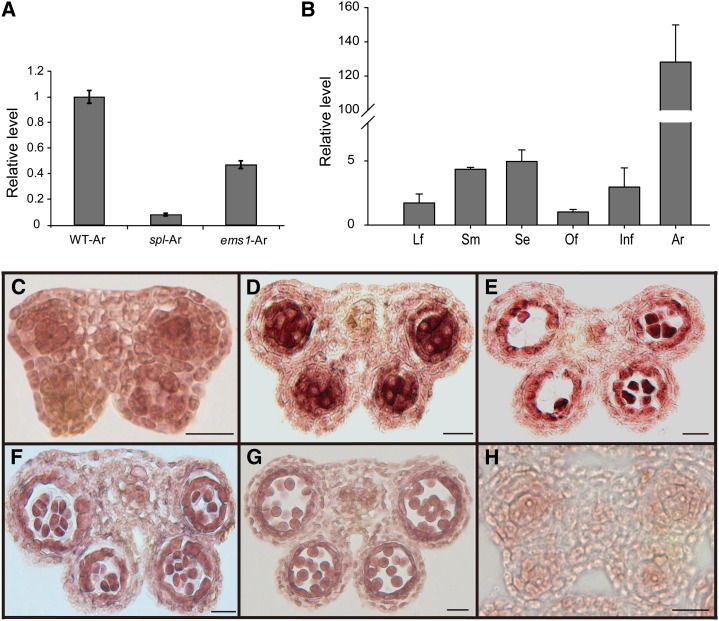
In Arabidopsis, SPOROCYTELESS (SPL) and EXCESS MICROSPOROCYTE1 (EMS1) are two important genes regulating microsporogenesis (Schiefthaler et al., 1999; Yang et al., 1999; Zhao et al., 2002). In an effort to identify new genes for anther and pollen development, we compared gene expression profiles between wild-type and spl and ems1 mutant anthers (Wijeratne et al., 2007). At1g68200 was among the genes that showed significantly less expression in the spl and the ems1 mutants than the wild-type anther, and this gene was named CDM1 because of the phenotype of a transfer DNA (T-DNA) insertional mutant (see below). To investigate the CDM1 gene expression pattern, we performed quantitative reverse transcription PCR (qRT-PCR). Our results confirmed the expression reductions in both spl and ems1 anthers (Fig. 1A). Although CDM1 expression was detectable in leaves, stems, young inflorescences, open flowers, and siliques at relatively low levels, its expression level in stage 4 to 7 anthers was ≥100-fold that of other tissues (Fig. 1B). Furthermore, RNA in situ hybridization with cross sections of wild-type floral buds showed that the CDM1 expression signal was low in precursors of meiocytes and the tapetum in stage 4 anthers (Fig. 1C), and then reached the highest level in stage 5 to 6 anthers, primarily in meiocytes and the tapetum (Fig. 1, D and E). At anther stage 7, the expression level remained relatively high in tetrads and the tapetum (Fig. 1F). Subsequently, it decreased after anther stage 8 (Fig. 1G). Therefore, the expression pattern of CDM1 strongly suggested that it functions in the tapetum and meiocytes, both crucial for microsporogenesis.
Figure 1.
The CDM1 expression pattern. A, Analysis of CDM1 expression in wild-type, spl, and ems1 anthers using qRT-PCR. B, Detection of CDM1 expression in various tissues using qRT-PCR. C–G, In situ hybridization of the CDM1 transcript with a CDM1 antisense probe in the wild type. Anthers at stages 4 (C), 5 (D), 6 (E), and 7 (F). CDM1 expression was greatly reduced (G). H, In situ hybridization of the CDM1 transcript with a CDM1 sense probe in a wild-type stage 5 anther. Only the background signal was detected. Ar, Anther; ems1-Ar, ems1 anther; Infl, young inflorescence; Lf, leaf; Of, open flower; Se, silique; Sm, stem; spl-Ar, spl anther. Bar = 20 μm in C–H.
The cdm1 Mutant Is Completely Male Sterile
The CDM1 gene has two exons and one intron (Supplemental Fig. S1A), encoding a predicted protein with 308 amino acids. CDM1 has two CCCH (C-X8-C-X5-C-X3-H) domains, which are separated by 18 amino acids (Supplemental Fig. S1B). To analyze CDM1 function genetically, we obtained a T-DNA insertion (SALK_065040; named cdm1) from the SIGnAL mutant collection (Alonso et al., 2003). Conventional sequencing of PCR fragments confirmed that the T-DNA was inserted in the second exon of CDM1 (Supplemental Fig. S1A). The T-DNA sequence was fused after the codon for the last amino acid residue of the first CCCH domain (Supplemental Fig. S1B), potentially producing a truncated protein lacking the second CCCH domain. The CDM1/cdm1 heterozygous plants showed normal development, whereas the cdm1/cdm1 homozygous mutants were completely sterile (see below). The progenies of a heterozygous plant segregated for sterile to normal phenotypes in an approximate 1:3 ratio (56:150 for mutant:normal), indicating that the mutant phenotype was caused by a single recessive nuclear mutation. Pollination of a cdm1 pistil with wild-type pollen resulted in full fertility, indicating that the mutant was female fertile. Reverse transcription PCR analysis from wild-type and cdm1 inflorescences revealed that the mRNA transcript (fa) spanning the T-DNA insertion was undetectable in cdm1, whereas a shortened one (fb) could be amplified (Supplemental Fig. S1, A and C), implying that the full-length mRNA is disrupted in cdm1.
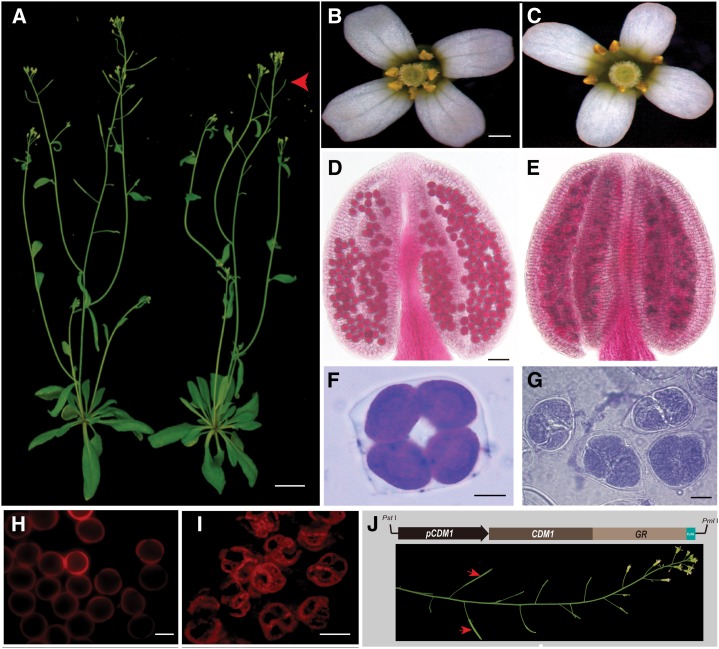
The cdm1 plants had vegetative growth similar to that of the wild type (Fig. 2A), but produced much shorter siliques than those of the wild type (Fig. 2A, red arrows). Further examination revealed that the cdm1 silique lacked any seeds (Supplemental Fig. S1D). Although wild-type and cdm1 flowers had similar sepals and petals (Fig. 2, B and C), no pollen grains were observed in cdm1 anthers (Fig. 2C), unlike wild-type anthers with plenty of pollen (Fig. 2B). Furthermore, Alexander staining showed that the wild-type anther produced many viable round pollen grains (Fig. 2D, in deep pink), but the cdm1 anther contained dead pollen grains (in blue) in clumps (Fig. 2E). Examination of tetrads using toluidine blue staining showed that the wild-type meiosis produced four microspores, each separately surrounded by a well-formed wall (Fig. 2F); however the cdm1 meiotic products were still attached to each other (Fig. 2G), implying that the wall biogenesis of newly formed microspores was affected. Visualization using 550-nm excitation indicated that the wild-type microspore wall was regular and round (Fig. 2H), but the degenerating microspores from the same meiocyte still remained attached in cdm1 (Fig. 2I). To verify that the mutant defects were caused by the cdm1 mutation, an inducible construct containing the CDM1 promoter driving a fusion of CDM1 with sequences encoding the glucocorticoid receptor (GR) domain was introduced into the mutant background. A PCR result further showed that, in the transgenic lines, the full CDM1 coding domain sequence was present in the cdm1 mutant background (Supplemental Fig. S1E). The fertility was restored after dexamethasone induction (Fig. 2J), confirming that the defect in the CDM1 gene was responsible for the mutant sterility.
Figure 2.
Phenotypes of wild-type, cdm1, and transgenic plants for functional rescue. A, A wild-type plant (left) and a cdm1 plant (right), showing no obvious differences in vegetative growth. However, the cdm1 plant produced short siliques without any seeds (red arrowhead). B, A wild-type flower. C, A cdm1 flower. D and E, Alexander staining. A wild-type anther showing viable pollen grains in deep pink (D). A cdm1 anther containing clusters of dead pollen grains in blue (E). F, A wild-type tetrad. G, cdm1 tetrads. H, Autofluorescence (red) of wild-type individual microspore wall under 550-nm excitation. I, Autofluorescence (red) of a cdm1 microspore wall under 550-nm excitation. J, The functional rescue of cdm1 using a construct carrying the CDM1 cDNA fused with GR sequences and driven by the CDM1 native promoter. Red arrows indicated fertile siliques with restored CDM1 function after GR induction. Bar = 20 mm in A; 500 μm in B and C; 50 μm in D and E; and 10 μm in F to K.
Delayed Meiotic Cytokinesis and Microspore Degeneration in cdm1
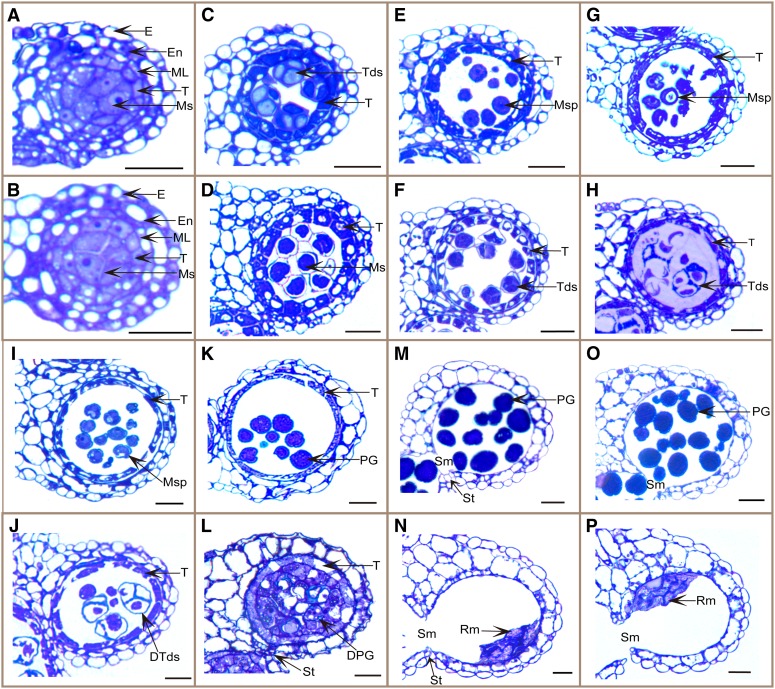
To examine the cdm1 mutant defects in more detail, semithin anther transverse sections were analyzed. The results showed that the cdm1 anther developed normally from anther stages 1 to 5, but began to show abnormal morphology starting at anther stage 6. At anther stage 6, the wild-type meiocyte had a thick callose wall (Fig. 3A), but the callose wall around cdm1 meiocytes seemed slightly thinner (Fig. 3B). In a stage 7 wild-type anther, the newly formed sibling microspores were well blanketed and completely separated by a thick callose wall, forming a tetrad (Fig. 3C). When the cdm1 anther reached the same size as (even slightly larger than) the wild-type stage 7 anther, somatic cell morphologies (e.g. the collapse of the middle layer) were similar to those of the wild-type cells; however, cdm1 meiocytes still did not complete cytokinesis and did not form tetrads with microspores (Fig. 3D), indicating that microspore formation was delayed. From anther stages 8 to 10, the callose wall was completely degraded in the wild type, releasing individual microspores to undergo pollen development in the anther locule (Fig. 3, E, G, and I). In a stage 8 cdm1 anther (according to anther size and somatic cells), although four cytoplasmic clusters were recognizable in some meiocytes (Fig. 3F), the callose wall between microspores was abnormally thin (Fig. 3C), suggesting that callose synthesis and/or deposition was severely impaired in cdm1. During the subsequent anther development in cdm1, sibling microspores remained attached (Fig. 3, H and J), possibly because of residual callose wall or other defects in microspore wall formation. In a stage 9 wild-type anther, microspores formed an exine wall and became vacuolated (Fig. 3G); they then continued to develop into pollen grains from anther stages 10 to 11 (Fig. 3, I and K). By contrast, cdm1 microspores degenerated (Fig. 3, H and J), leaving behind a mass of wall materials in the anther locule (Fig. 3L). At anther stage 12, mature wild-type pollen grains were formed (Fig. 3M), but cdm1 microspores were completely degenerated, with only remnants in the anther locule (Fig. 3N). At anther stage 13, stomium ruptured in the wild type, releasing pollen grains in a process called anther dehiscence (Fig. 3O). The cdm1 stomium also ruptured (Fig. 3P), indicating normal endothecium secondary cell wall thickening, which was required for stomium breakage. We also observed using semithin sections that tapetum morphology in the cdm1 mutant appeared normal, similar to that in the wild type (Supplemental Fig. S2).
Figure 3.
Comparison of wild-type and cdm1 anther development. Semithin anther sections were stained with toluidine blue, with one locule shown in each: the wild type (A, C, E, G, I, K, M, and O) and cdm1 (B, D, F, H, J, L, N, and P). A and B, At stage 6, five cell layers were presented and meiocytes underwent meiosis. C, At stage 7, tetrads were already formed. D, At stage 7, tetrads were not formed yet. E, At stage 8, individual microspores were released. F, At stage 8, meiocyte external wall was not degraded, and thinner than normal callose wall was formed between microspores. G, At stage 9, microspores became vacuolated. H, At stage 9, microspores in tetrads began to degenerate. I and J, At stage 10, tapetum degeneration was initiated in both the wild type and the cdm1 mutant. cdm1 microspores, which were undergoing degeneration, were still attached (J). K, At stage 11, pollen grains formed exine. L, At stage 11, microspores were almost completely degenerated, and masses of wall materials were found separately from the microspores in the locule. M and N, At stage 12, stomium was broken down. No pollen grains were generated, leaving remnants of microspores in the cdm1 mutant anther locule (N). O and P, At stage 13, stomium breakage allowed anther dehiscence and pollen grain release in the wild type (O). The cdm1 anther dehisced (P). DPG, Degenerating pollen grain; DTds, degenerating tetrad; E, epidermis; En, endothecium; ML, middle layer; Ms, meiocytes; MSp, microspore; PG, pollen grain; Rm, remnant of locule contents; Sm, septum; St, stomium; T, tapetum; Tds, tetrad. Bar = 20 μm.
Callose Dissolution during Microsporogenesis Was Defective in cdm1
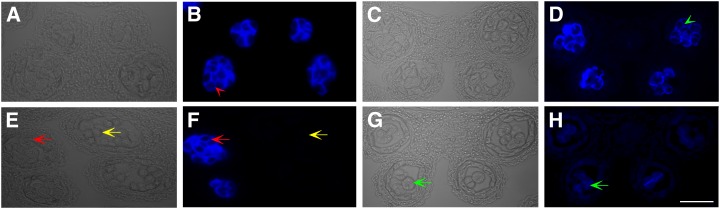
Callose metabolism during microsporogenesis was further examined using aniline blue staining in both wild-type and cdm1 anther sections. At anther stage 4, both wild-type and cdm1 meiocytes synthesized the callose wall (Supplemental Fig. S3, A–D), indicating that the initiation of callose synthesis was not obviously affected in cdm1. At anther stage 7, wild-type tetrads were well formed with a highly thickened callose wall surrounding each microspore (Fig. 4, A and B; Supplemental Fig. S3, E and F). However, in the cdm1 anther, although more callose was accumulated on meiocytes and few tetrads could be recognized, the callose between microspores was much thinner than that of the wild type (Fig. 4, C and D). Callose staining further revealed that individual cdm1 tetrads had less callose (Supplemental Fig. S3, G and H). These results suggested that callose synthesis and/or deposition between microspores was impaired in cdm1. Furthermore, callose was not detected on released microspores in the wild-type anther (Fig. 4, E and F, yellow arrows), indicating that callose was completely degraded when the microspore was released. Intriguingly, in a younger anther (still at the tetrad stage) of the same flower, microspores displayed strongly stained callose (Fig. 4, E and F, red arrows). This result suggested that callose dissolution was a rapid process in the wild type. However, in cdm1, residual callose was obviously observed even after the tetrad stage (Fig. 4, G and H, green arrows), resulting in the attachment between sibling microspores. Therefore, callose dissolution in cdm1 was incomplete or otherwise abnormal.
Figure 4.
Aniline blue staining analysis for callose in the wild type and cdm1. Analysis of staining in wild-type anther sections (A, B, E, and F) and cdm1 anther sections (C, D, G, and H), using bright-field microscopy (A, C, E, and G) and UV light (B, D, F, and H). A and B, A wild-type stage 7 anther, showing that each newly formed microspore was enveloped with a thick callose wall (red arrowhead in B). C and D, A cdm1 stage 7 anther. Although newly formed microspores were separated by callose (green arrow head in D), the callose wall was thinner than that formed in the wild type (red arrowhead in B). E and F, A wild-type stage 8 anther (yellow arrows), showing that the callose wall had been completely degraded, and individual microspores were released. In the same flower, the neighboring younger anther still remained at stage 7 (red arrows) with a strong signal for callose. G and H, A cdm1 stage 8 anther, showing residual callose between microspores (green arrows). Bar = 50 μm.
Exine Formation Was Disturbed in cdm1
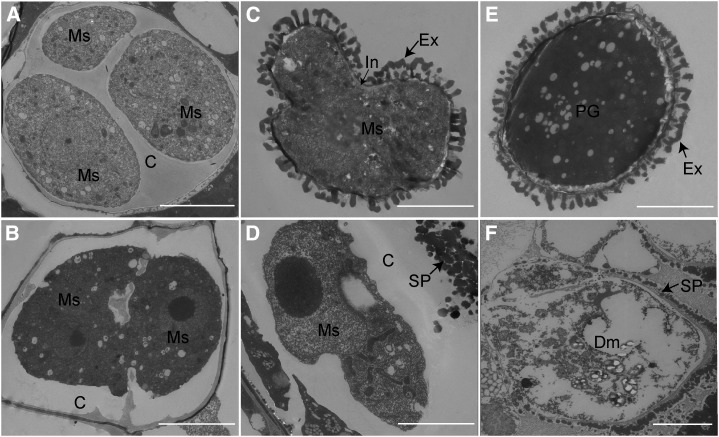
To further investigate cdm1 pollen defects, the ultrastructures of tetrads, microspores, and mature pollen grains were compared using transmission electron microscopy (TEM) between the wild type and cdm1. The wild-type tetrad was encased in a thick callose wall, which completely surrounded each microspore (three microspores are visible in this section; Fig. 5A). In the cdm1 anther, the callose wall surrounding the meiocyte (Fig. 5B) was similar to that of the wild-type meiocyte (Fig. 5A), but after cytokinesis, the callose wall could not completely form between newly formed adjacent microspores (two microspores were seen in the section; Fig. 5B). These results were consistent with the observations using aniline blue staining (Supplemental Fig. S3H). Later in development, wild-type microspores successfully formed the exine and intine portions of the pollen wall (Fig. 5C). However, there was not even primexine (a precursor of exine) outside the cdm1 microspore cell membrane (Fig. 5D). Spotted sporopollenin was blocked away from cell membrane by residue callose materials (Fig. 5D). The pollen wall was regularly assembled on the mature wild-type pollen (Fig. 5E), whereas the cdm1 microspore was degenerated, leaving spotted sporopollenin abnormally deposited outside the callose wall (Fig. 5F), indicating that pollen exine formation was defective in cdm1.
Figure 5.
TEM of the microspore wall in the wild type (A, C, and E) and cdm1 (B, D, and F). A, Callose formed a complete wall to fill between microspores in the tetrad. B, Poorly developed callose failed to completely fill spaces between microspores. C, Exine and intine were formed on the individual microspore. D, Cytoplasm had shrunk. The callose wall was still seen. Spotted sporopollenin was placed outside the callose wall. E, Mature exine and pollen coat were formed. F, Cytoplasm has degenerated. Spotted sporopollenin was randomly accumulated near the degenerating microspore. C, Callose; Dm, degenerated microspore; Ex, exine; In, intine; Ms, microspore; SP, spotted sporopollenin. Bar = 5 μm.
The Expression of CalS5 and CalS12 Was Down-Regulated in cdm1
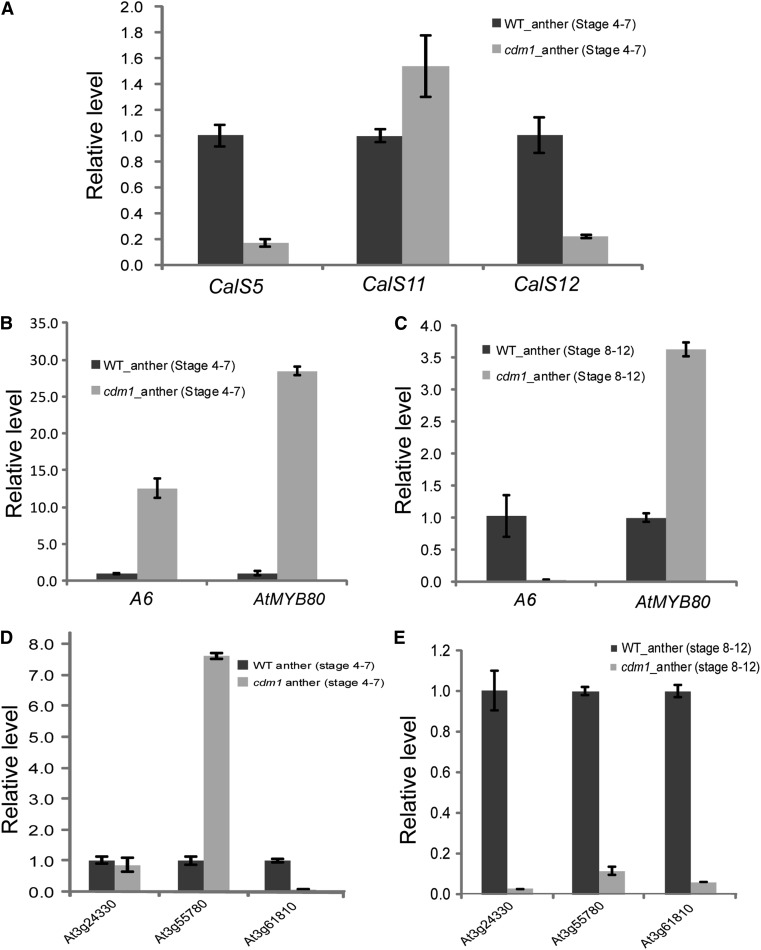
Because callose accumulation was reduced between cdm1 microspores (Fig. 5B; Supplemental Fig. S3H), we hypothesized that the expression of callose synthase genes near the time of microspore formation was possibly affected. To test this, we performed qRT-PCR for stage 4 to 7 anthers of the wild type and cdm1 (Fig. 6A). In cdm1 anthers, the expression level of CalS5 was dramatically reduced to only 20% of that in wild-type anthers. The expression of CalS12 was also decreased to approximately 20% of that in wild-type anthers, but the expression of CalS11 appeared normal in cdm1 anthers.
Figure 6.
qRT-PCR analyses of gene expression differences in the wild type and cdm1. A, Expression of CalS5, CalS11, and CalS12 in wild-type and cdm1 stage 4 to 7 anthers. B, Expression of A6 and AtMYB80 in wild-type and cdm1 stage 4 to 7 anthers. C, Expression of A6 and AtMYB80 in wild-type and cdm1 stage 8 to 12 anthers. D, Expression of three genes potentially encoding β-1,3-Gs in wild-type and cdm1 stage 4 to 7 anthers. E, Expression of three genes encoding putative β-1,3-Gs in wild-type and cdm1 stage 8 to 12 anthers. Arabidopsis ACTIN1 was used as a control. Error bars indicate sd. WT, Wild type.
A6 and AtMYB80 Expression Patterns Were Altered during Microsporogenesis in cdm1
Based on the observation of callose dissolution defects in cdm1 (Figs. 4H and 5D), we decided to examine the expression of A6 and AtMYB80 in wild-type and cdm1 anthers at different stages using qRT-PCR. Compared with the wild-type anther expression level, the A6 expression level in stage 4 to 7 cdm1 anthers was increased >10-fold (Fig. 6B), implying that the A6 gene was activated at an early stage in cdm1, at a time when its expression was low in the wild type. The expression of AtMYB80 in stage 4 to 7 cdm1 anthers was sharply elevated to approximately 30-fold of that in stage 4 to 7 wild-type anthers (Fig. 6B). Because MYB80 acts as a positive regulator of A6, the elevated MYB80 expression was consistent with the expression changes of A6 in cdm1. However, we further detected that the expression level of A6 in cdm1 stage 8 to 12 anthers was greatly reduced to a nearly undetectable level compared with that in wild-type stage 8 to 12 anthers (Fig. 6C). This suggested that cdm1 possibly lost callase activities much earlier than normal, finally leading to the failure to completely hydrolyze callose during pollen development. On the other hand, the expression of AtMYB80 in cdm1 stage 8 to 12 anthers was still slightly higher than that in wild-type stage 8 to 12 anthers (Fig. 6C), indicating that the remained AtMYB80 was not sufficient for A6 expression at these late stages of pollen development.
The Expression of Three Putative β-1,3-G Genes Was Altered in cdm1
We used Affymetrix ATH1 arrays to identify additional genes potentially regulated by CDM1 by comparing gene expression between cdm1 and wild-type young floral buds. Among 375 genes that showed differential expression (P < 0.05) with >1.5-fold changes between the wild type and cdm1, 185 genes were down-regulated by 1.5-fold to 7.10-fold (Supplemental Table S1), whereas 190 genes were up-regulated by 1.5-fold to 5.82-fold (Supplemental Table S2).
Interestingly, three genes (At3g24330, At3g55780, and At3g61810), encoding putative β-1,3-Gs, exhibited reduced expression levels ranging from 1.60-fold to 2.08-fold in cdm1 (Supplemental Table S3), consistent with qRT-PCR analyses (Supplemental Fig. S4A). Further examination of the expression levels of wild-type and cdm1 anthers at stages 4 to 7 and stages 8 to 12 revealed distinct patterns. In stage 4 to 7 anthers, the expression of At3g24330 was similar between the wild type and cdm1. The expression level of At3g55780 was up-regulated over 7-fold in cdm1; however, the At3g61810 expression was greatly reduced (Fig. 6D). In stage 8 to 12 anthers, all three genes showed a sharp decrease in expression levels (<10% remained) in cdm1 (Fig. 6E). These results strongly suggested that multiple β-1,3-Gs participated in callose dissolution during microsporogenesis, and the proper timing of different β-1,3-G activities was probably precisely regulated, further revealing the complexity of callose dissolution during pollen development. In addition, because the meiotic cytokinesis in the cdm1 mutant was delayed, it is also possible that some of the genes identified by microarray analysis were indirectly affected by the delayed meiotic cytokinesis.
The Expression of Pollen Developmental Genes Was Changed in cdm1
It was known that callose defects can affect pollen wall formation and pollen viability (Dong et al., 2005; Enns et al., 2005; Zhang et al., 2007). Thus, we examined the expression of MALE STERILITY2 (MS2), DEFECTIVE IN EXINE FORMATION1 (DEX1), NO EXINE FORMATION1 (NEF1), and FACELESS POLLEN1 (FLP1), which were involved in pollen exine formation and sporopollenin synthesis (Aarts et al., 1997; Paxson-Sowders et al., 2001; Ariizumi et al., 2003, 2004), in both the wild type and cdm1 by qRT-PCR. Our results indicated that the expression of DEX1 and NEF1 was not altered. The expression of MS2 in cdm1 was reduced to approximately 60% of the wild-type level. However, the expression of FLP1 was slightly up-regulated in cdm1 (Supplemental Fig. S4B). Furthermore, microarray analyses revealed that six other genes related to anther and pollen development showed differential expression levels in the wild type and cdm1 (Supplemental Table S3). Among them, IMPORTIN ALPHA ISOFORM8, BRIC-A-BRAC-TRAMTRACK-BROAD COMPLEX AND TRANSCRIPTIONAL ADAPTOR ZINC FINGER DOMAIN PROTEIN3, AtPV42a, ROXY2, and KOMPEITO were down-regulated in cdm1, with expression level changes ranging from 1.51-fold to 3.35-fold. However, SPERMIDINE HYDROXYCINNAMOYL TRANSFERASE expression was detected to be up-regulated to 3.32-fold in cdm1, as further supported by qRT-PCR results (Supplemental Fig. S4C). Therefore, the above data strongly suggested that various pollen development processes were influenced in cdm1.
On the other hand, genes known to be important for tapetum development were not dramatically affected. Our microarray analysis indicated that the ABORTED MICROSPORES (AMS) gene, which is important for early tapetum development, had expression values (±se) of 11.091 ± 0.004 in the wild type and 11.275 ± 0.0235 in cdm1. Another example is the late tapetum development gene MS188 (Zhu et al., 2011), which had expression values of 7.456 ± 0.102 in the wild type and 7.306 ± 0.105 in cdm1. These results are consistent with the normal appearance of the cdm1 tapetum layer mentioned earlier.
DISCUSSION
CDM1 Likely Regulates Expression of Genes for Both Callose Synthases and Callase
During microsporogenesis, the major role of callose is to serve as a temporary cell wall to separate the newly formed microspores and prevent their plasmic membrane from fusing together (Scott et al., 2004). Studies have shown that reduced accumulation of callose may result in microspore degeneration (Dong et al., 2005; Enns et al., 2005). Here, we found that the cdm1 mutant exhibited severe reduction of callose between newly formed microspores (Fig. 5B; Supplemental Fig. S3H), which is consistent with the decreased expression levels of CalS5 and CalS12 (Fig. 6A), suggesting that CDM1 might positively regulate CalS5 and CalS12 expression. Although CalS11 and CalS12 were suggested to function redundantly in synthesizing the callose wall between microspores in a tetrad (Enns et al., 2005), the fact that CalS11 expression was not affected (Fig. 6A) implied that CalS11 and CalS12 might have somewhat different functions and CalS11 alone is not sufficient for normal levels of callose synthesis between the microspores.
After the tetrad stage, callase digests the callose both at the exterior of the tetrad and between the microspores to release and separate individual microspores (Stieglitz and Stern, 1973; Scott et al., 2004). The accurate timing of callase activation is critical for normal microsporogenesis and pollen development. In tobacco, it was found that premature callose dissolution during meiosis affected subsequent pollen exine formation, resulting in male sterility (Worrall et al., 1992; Tsuchiya et al., 1995). In this study, we hypothesize that the increased expression of A6 and AtMYB80 in the stage 4 to 7 cdm1 anthers (from premeiotic to just postmeiotic stages) caused precociously activated callase, leading to premature callose dissolution. The concurrent increase in the expression of A6 and its positive regulator AtMYB80 strongly suggests that the increase in A6 expression was the consequence of the elevation in AtMYB80 expression. Therefore, CDM1 might be a repressor of A6 and AtMYB80 at anther stages 4 to 7 in the wild type. Alternatively, CDM1 might repress upstream factors of AtMYB80 to control the corresponding regulatory pathway.
Moreover, the severely reduced expression level of A6 in cdm1 anther stages 8 to 12 suggested that callase activity was impaired in cdm1 after the tetrad stage, providing an explanation for the presence of residual callose between microspores in cdm1. These results indicated that CDM1 is a critical factor affecting the temporal expression patterns of genes involved in callose metabolism at the right time and the right place during male reproductive development. In addition, it is also possible that the reduced expression levels of CalS5 and CalS12 were a result of feedback regulation caused by the earlier higher expression of callase-related genes.
Callose Dissolution during Pollen Development Is a Highly Complex Process
Although only the A6 gene was thought to be a potential major component of the callase enzyme mixture (Hird et al., 1993), Arabidopsis has >50 genes encoding putative β-1,3-Gs (Doxey et al., 2007), suggesting that the metabolism of β-1,3-glucan in plants is more complex than previously understood. We identified three other β-1,3-G genes (At3g24330, At3g55780, and At3g61810) showing different extents of reduced expression in cdm1, including two anther-specific ones (Supplemental Fig. S4A; Supplemental Table S3). The difference in their expression alteration at different anther stages (Fig. 6, D and E) suggested that they potentially function at different anther stages or at different steps of callose dissolution, possibly even in different subcellular compartments. The callase mixture includes endoglucanases and exoglucanases (Stieglitz and Stern, 1973; Scott et al., 2004). Endoglucanases cleave β-1,3-glucans into short-chain reducing sugars; exoglucanase hydrolysis releases a single Glc unit from the reducing ends of the substrate (Stieglitz, 1977; Zhang et al., 2007). A6 was suggested to possess endoglucanase activity (Hird et al., 1993). Based on the similarity of expression change patterns in cdm1 (Fig. 6, B–E), it is possible that At3g55780 acts in a way that is similar to that of A6. At3g24330 might play a role at a later stage. However, the reduced expression of At3g61810 at multiple stages suggested it possibly acts at different times during pollen development. Taken together, we concluded that multiple β-1,3-G genes participate in callose dissolution during microsporogenesis and pollen development, and the precise timing and place of their activities are critical. In addition, it is also possible that one or more of the above gene products possesses exoglucanase activity. Further genetic and biochemical studies are required to illuminate their biochemical and biological functions.
Callase is considered to be secreted by tapetum to separate newly formed microspores (Stieglitz and Stern, 1973; Stieglitz, 1977; Scott et al., 2004). However, recent RNA sequencing data from Arabidopsis male meiocytes detected several β-1,3-G genes (At3g55430, At2g01730, and At5g20390) expressed in these reproductive cells (Yang et al., 2011), suggesting that male meiocytes probably also synthesize β-1,3-Gs. Thus, it is possible that both meiocytes and the tapetum could synthesize β-1,3-Gs to degrade the callose wall.
Pollen Wall Formation Was Defective in cdm1
The mature pollen wall contains both the intine and exine layers, protecting pollen from harsh conditions (Ariizumi and Toriyama, 2011). During microspore and pollen development, the callose wall could guide exine formation (Scott et al., 2004). Therefore, in cdm1, the exine defect might be caused by the abnormal residual callose on the surface of developing pollen grains. We also noted that the expression of the MS2 gene, which is required for normal exine formation, was reduced in cdm1 (Supplemental Fig. S4B), suggesting that MS2 might act downstream of CDM1, providing another possible mechanism for cdm1 defects in exine formation. However, three other exine-related genes (DEX1, NEF1, and FLP1) had normal expression levels in cdm1, implying that at least some of the pollen wall materials might be normally produced in cdm1. In addition, TEM observation indicated that exine material was delivered outside the pollen callose wall, but could not be properly assembled (Fig. 5F), further revealing the exine defect in cdm1. Moreover, the expression changes of six other pollen viability genes in cdm1 suggested that multiple pollen developmental processes were potentially influenced.
Our in situ hybridization results showed that CDM1 also has a high level of expression in the wild-type tapetum, which suggests that it might play an important role in the tapetum. Therefore, we investigated whether tapetum development in the cdm1 mutant was affected as part of the phenotypic analyses. Our analyses using semithin sections (Supplemental Fig. S2) showed that the cdm1 tapetum morphology appeared normal and different from known mutants (e.g. dyt1 and ams) with defective tapetum (Sorensen et al., 2003; Zhang et al., 2006). In addition, our microarray analysis did not detect dramatic expression changes for genes known to be important for tapetum development, such as AMS and MS188. Therefore, it is likely that the cdm1 mutation did not affect the tapetum morphology or expression of key genes for tapetum development.
CDM1 Might Be an RNA-Binding Protein
CDM1 encodes a protein with TZF domains, each containing three Cys and one His that coordinate a zinc atom (Lai et al., 2000); this type of zinc finger protein is involved in various developmental processes and environmental responses in plants (Li and Thomas, 1998; Sun et al., 2007; Kim et al., 2008; Wang et al., 2008; Lin et al., 2011). TZF proteins were initially identified in animals with two highly similar C-X8-C-X5-C-X3-H motifs (X represents variable amino acids) that are separated by 18 amino acids between the carboxyl terminal H of the first zinc finger and the amino terminal C of the second zinc finger (Blackshear et al., 2005). In humans and mice, TZF proteins were found to bind to AU-rich elements of target mRNAs to reduce their stability (Carballo et al., 1998; Ramos et al., 2004; Blackshear et al., 2005; Stumpo et al., 2009). Arabidopsis has 68 CCCH-type zinc finger proteins (Wang et al., 2008); only two TZF proteins, CDM1 and AtC3H14, have the spacing features of a typical animal TZF motif (Wang et al., 2008; Pomeranz et al., 2010a). In addition to the above features, the sequence motifs (MM/TKTEL or RYKTEV) upstream of each finger in CDM1 and AtC3H14 are also similar to the conserved pattern R/KYKTEL in animals (Pomeranz et al., 2010a). Moreover, the subcellular localization of CDM1 was similar to that of the human TZF protein TRISTETRAPROLIN (Pomeranz et al., 2010b). Both of them were predominantly localized in specific cytoplasmic foci called processing bodies, which potentially process mRNA turnover and mediate translational repression (Pomeranz et al., 2010b). Based on the sequence and localization similarities between CDM1 and animal TZFs, we hypothesize that CDM1 might also function as an mRNA-binding protein to regulate the stability of its targets.
If CDM1 is an RNA binding protein, it could affect gene expression via different mechanisms. One possibility is that CDM1 could affect the stability of its target RNAs in the cytoplasm; another possibility is that it could regulate RNA processing in the nucleus, as suggested by recent studies of the Arabidopsis AtTZF1 protein and its rice (Oryza sativa) homolog. AtTZF1 is similar to CDM1 in having tandem CCCH zinc fingers and is predicted to be an RNA-binding protein; furthermore, AtTZF1 can shuttle between the nucleus and cytoplasm (Pomeranz et al., 2011), suggesting that it can function in both the cytoplasm and the nucleus. The rice homolog of AtTZF1, OsTZF1, was recently shown to be involved in stress response and leaf senescence and to have RNA-binding activity in vitro; furthermore, approximately 200 genes showed at least 2-fold expression changes in an OsTZF1 overexpression transgenic line compared with the wild type (Jan et al., 2013), indicating that a large number of genes could be affected by an RNA-binding protein in rice, similar to our data here for CDM1. If CDM1 can also enter the nucleus under certain conditions, it could also bind to RNAs in the nucleus and regulate their processing, such as splicing. Regardless of how CDM1 affects gene expression, it can regulate genes for callose metabolism either directly by binding to the mRNAs for those genes, or indirectly by binding to the mRNAs encoding regulatory proteins. For example, CDM1 could bind to one or more mRNAs that encode transcription factors, which then regulate callose metabolic genes in microsporogenesis and pollen development. Further RNA sequencing experiments to examine the RNAs bound to CDM1 could test these possibilities.
A Working Model for CDM1 Function during Meiosis and Pollen Development
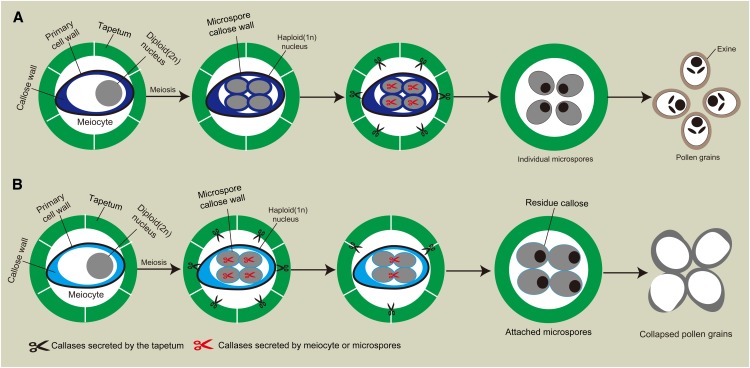
Taken together, we proposed a model to explain CDM1 function during microsporogenesis and pollen development (Fig. 7). In the wild type, at anther stages near the time of meiosis (stages 4 to 7), callose is synthesized and deposited initially around meiocytes and then between newly formed microspores in the tetrad (Fig. 7A). During this time, CDM1 represses the expression of genes involved in callose dissolution, including AtMYB80, A6, and others such as the above-mentioned three β-1,3-G genes, in both the tapetum and meiocytes probably at a posttranscriptional level. Thereafter, the expression of CDM1 is gradually reduced, leading to the higher expression levels of AtMYB80, A6, and others. The genes encoding β-1,3-G, including endoglucanases and exoglucanases, are expressed, releasing the callase mixture from the tapetum and meiocytes, to rapidly and completely degrade the callose wall and facilitate exine formation (Fig. 7A). However, in cdm1, the expression of AtMYB80, A6, and others are activated precociously (Fig. 6, B and D), somehow affecting the expression of CalS5 and CalS12 (Fig. 6A), leading to reduced callose accumulation between microspores (Fig. 7B). Furthermore, the disturbed expression of callase genes in cdm1 causes a lowering of activity after anther stage 8 via an unknown mechanism, failing to completely degrade callose between the microspores (Fig. 7B). The residual callose then blocks normal exine formation, causing defects and loss of viability of pollen grains.
Figure 7.
Proposed models for callose metabolism during microsporogenesis and pollen development in the wild type and cdm1. A, Callose metabolism during microsporogenesis in the wild-type anther. Normal callose synthesis and degradation are required for functional microspore formation. B, Callose metabolism during microsporogenesis in cdm1. The disturbance of callose metabolism leads to degeneration of developing microspores. Black scissors represent components of callase generated by the tapetum. Red scissors stand for potential components of callase from meiocytes. For easy visualization, only one meiocyte was shown in the anther locule.
MATERIALS AND METHODS
Plant Materials, Growth, and Phenotypic Analyses
Arabidopsis (Arabidopsis thaliana) plants used were of the Columbia 0 ecotype. Plants were grown under long-day conditions (16-h light/8-h dark) in a 22°C growth chamber. Floral images were obtained using a Nikon dissecting microscope with a digital camera (Optronics). Dissected tetrads were stained with 0.01% (w/v) toluidine blue. Pollen grains were stained with Alexander solution (Alexander, 1969) to detect pollen viability. Wild-type and mutant inflorescences were collected and fixed as described (Zhao et al., 2002). Floral buds were embedded in Spurr’s resin; semithin (0.5 μm) sections were prepared with an Ultracut E ultramicrotome (Leica Microsystems), stained with 0.05% (w/v) toluidine blue, and photographed under an Olympus BX-51 microscope. TEM was carried out as previously described (Li et al., 2004). Anther stages were referred as described (Sanders et al., 1999).
Complementation of the cdm1 Mutant
For functional complementation of the cdm1 mutant, a fusion containing an approximately 1.1-kb native promoter, the CDM1 coding sequence, and the sequence for the GR domain was constructed and subcloned into pCAMBIA1300 digested with PstI and PmlI using the In-Fusion HD Cloning System (Clontech; Fig. 2J). The CDM1/cdm1 plants verified by PCR were used for transformation with Agrobacterium tumefaciens GV3101 carrying the above plasmid by the floral dip method (Clough and Bent, 1998). The transformants were selected on plates containing 25 mg/L hygromycin in Murashige and Skoog medium (Sigma). After the selected cdm1 mutant plants commenced flowering, 30 mm dexamethasone in dimethyl sulfoxide was sprayed to their unopen flower buds to induce the expression of the fusion gene.
Reverse TranscriptionPCR and qRT-PCR
Plant tissues were collected and immediately frozen in liquid nitrogen. Anthers at approximately stages 4 to 7 or 8 to 12 were collected under a dissection microscope. Total RNA was extracted using the RNeasy Plant Kit (Qiagen). For gene expression analyses, 2 µg of total RNA was used for reverse transcription with the SuperScript II system (Invitrogen). qRT-PCR primers (Supplemental Table S4) were designed by GenScript Real-time PCR Primer Design with crossing exon junction first. PCR was performed for 30 cycles (quantitative PCR for 45 cycles; 95°C, 30 s; 60°C, 20 s; 72°C, 30 s).
RNA in Situ Hybridization
Nonradioactive RNA in situ hybridization was performed as previously described (Wijeratne et al., 2007). A gene-specific fragment (342 bp) of the CDM1 cDNA was amplified by oMC7545 and oMC7547 (Supplemental Table S4), and cloned into the pGEM-T easy vector (Promega) with the resulting plasmid named pMC3573. The CDM1 antisense and sense probes were synthesized using the linearized pMC3537 by digestion with, respectively, the enzymes SpeI and NcoI, as templates for labeling with digoxigenin using in vitro transcription. Floral sections were hybridized with the probes, and signals were detected with anti-digoxigenin antibodies conjugated with alkaline phosphatase and nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate (Roche).
Aniline Blue Staining for Callose
For callose staining, transverse anther sections and tetrads released from the anther were stained with 0.01% (w/v) aniline blue in 0.077 m phosphate buffer (pH 8.5; Regan and Moffatt, 1990) for 10 min at room temperature. They were visualized in a fluorescence microscope using a UV filter (Nikon).
Microarray Analysis
Wild-type and cdm1 young floral buds were used to isolate total RNA, with two biological replicates for each. Microarray analysis was performed using 500 ng of total RNA per sample as described in the Genechip Expression Analysis Technical Manual (Affymetrix). Microarray original data (*.cel files) were statistically analyzed using R software (http://www.r-project.org).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GSE55799.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The CDM1 gene and protein structures, and mRNA expression levels in the wild type and the T-DNA insertional mutant.
Supplemental Figure S2. Comparison of the wild type and cdm1 tapetum development.
Supplemental Figure S3. Aniline blue staining for callose during microsporogenesis in the wild type and cdm1.
Supplemental Figure S4. qRT-PCR analyses of different gene expression in the wild type and the cdm1 mutant young inflorescences.
Supplemental Table S1. Down-regulated genes with expression level changed >1.5-fold in the cdm1 mutant.
Supplemental Table S2. Up-regulated genes with expression level changed >1.5-fold in the cdm1 mutant.
Supplemental Table S3. The genes show differential expression levels between cdm1 and the wild type, involved in callose dissolution and anther and pollen development.
Supplemental Table S4. Primer sequences used in this study.
Supplementary Material
Glossary
- β-1,3-G
β-1,3-glucanase
- TZF
tandem CCCH-domain zinc finger
- T-DNA
transfer DNA
- qRT-PCR
quantitative reverse transcription-PCR
- TEM
transmission electron microscopy
Footnotes
This work was supported by grants from the Chinese Ministry of Science and Technology (2011CB944600 to H.M.), funds from Fudan University, Rijk Zwaan, and the Pennsylvania State University Department of Biology and Huck Institutes of the Life Sciences, and a start-up grant from Fudan University (to P.L.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, Kato T, Tabata S, Toriyama K. (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J 39: 170–181 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K. (2003) A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol Biol 53: 107–116 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K. (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Blackshear PJ, Phillips RS, Lai WS. (2005) Tandem CCCH zinc finger proteins in mRNA binding. In Iuchi S, Kuldell N, eds, Zinc Finger Proteins: From Atomic Contact to Cellular Function. Kluwer Academic/Plenum Publishers, New York, pp 80–90 [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. (1998) Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DP. (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J 42: 315–328 [DOI] [PubMed] [Google Scholar]
- Doxey AC, Yaish MW, Moffatt BA, Griffith M, McConkey BJ. (2007) Functional divergence in the Arabidopsis beta-1,3-glucanase gene family inferred by phylogenetic reconstruction of expression states. Mol Biol Evol 24: 1045–1055 [DOI] [PubMed] [Google Scholar]
- Enns LC, Kanaoka MM, Torii KU, Comai L, Okada K, Cleland RE. (2005) Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol Biol 58: 333–349 [DOI] [PubMed] [Google Scholar]
- Frankel R, Izhar S, Nitsan J. (1969) Timing of callase activity and cytoplasmic male sterility in Petunia. Biochem Genet 3: 451–455 [DOI] [PubMed] [Google Scholar]
- Hird DL, Worrall D, Hodge R, Smartt S, Paul W, Scott R. (1993) The anther-specific protein encoded by the Brassica napus and Arabidopsis thaliana A6 gene displays similarity to beta-1,3-glucanases. Plant J 4: 1023–1033 [DOI] [PubMed] [Google Scholar]
- Hong Z, Delauney AJ, Verma DP. (2001) A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell 13: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhar S, Frankel R. (1971) Mechanism of male sterility in Petunia: The relationship between pH, callase activity in the anthers, and the breakdown of the microsporogenesis. Theor Appl Genet 41: 104–108 [DOI] [PubMed] [Google Scholar]
- Jan A, Maruyama K, Todaka D, Kidokoro S, Abo M, Yoshimura E, Shinozaki K, Nakashima K, Yamaguchi-Shinozaki K. (2013) OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol 161: 1202–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G. (2008) SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20: 1260–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. (2000) Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem 275: 17827–17837 [DOI] [PubMed] [Google Scholar]
- Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B. (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Thomas TL. (1998) PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10: 383–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Pomeranz MC, Jikumaru Y, Kang SG, Hah C, Fujioka S, Kamiya Y, Jang JC. (2011) The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J 65: 253–268 [DOI] [PubMed] [Google Scholar]
- Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- McCormick S. (2004) Control of male gametophyte development. Plant Cell 16(Suppl): S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S-i, Zinkl GM, Swanson RJ, Maruyama D, Preuss D. (2005) Callose (beta-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxson-Sowders DM, Dodrill CH, Owen HA, Makaroff CA. (2001) DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- Pomeranz M, Lin PC, Finer J, Jang JC. (2010a) AtTZF gene family localizes to cytoplasmic foci. Plant Signal Behav 5: 190–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz M, Zhang L, Finer J, Jang JC. (2011) Can AtTZF1 act as a transcriptional activator or repressor in plants? Plant Signal Behav 6: 719–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz MC, Hah C, Lin PC, Kang SG, Finer JJ, Blackshear PJ, Jang JC. (2010b) The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol 152: 151–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos SB, Stumpo DJ, Kennington EA, Phillips RS, Bock CB, Ribeiro-Neto F, Blackshear PJ. (2004) The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development 131: 4883–4893 [DOI] [PubMed] [Google Scholar]
- Regan SM, Moffatt BA. (1990) Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu Y-C, Lee PY, Truong MT, Beals TP, Goldberg RB. (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K. (1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG. (2004) Stamen structure and function. Plant Cell 16(Suppl): S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H. (2003) The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J 33: 413–423 [DOI] [PubMed] [Google Scholar]
- Stieglitz H. (1977) Role of beta-1,3-glucanase in postmeiotic microspore release. Dev Biol 57: 87–97 [DOI] [PubMed] [Google Scholar]
- Stieglitz H, Stern H. (1973) Regulation of beta-1,3-glucanase activity in developing anthers of Lilium. Dev Biol 34: 169–173 [DOI] [PubMed] [Google Scholar]
- Stumpo DJ, Broxmeyer HE, Ward T, Cooper S, Hangoc G, Chung YJ, Shelley WC, Richfield EK, Ray MK, Yoder MC, et al. (2009) Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood 114: 2401–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Jiang H, Xu Y, Li H, Wu X, Xie Q, Li C. (2007) The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol 48: 1148–1158 [DOI] [PubMed] [Google Scholar]
- Tsuchiya T, Toriyama K, Yoshikawa M, Ejiri S, Hinata K. (1995) Tapetum-specific expression of the gene for an endo-beta-1,3-glucanase causes male sterility in transgenic tobacco. Plant Cell Physiol 36: 487–494 [DOI] [PubMed] [Google Scholar]
- Wang D, Guo Y, Wu C, Yang G, Li Y, Zheng C. (2008) Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne AJ, Zhang W, Sun Y, Liu W, Albert R, Zheng Z, Oppenheimer DG, Zhao D, Ma H. (2007) Differential gene expression in Arabidopsis wild-type and mutant anthers: insights into anther cell differentiation and regulatory networks. Plant J 52: 14–29 [DOI] [PubMed] [Google Scholar]
- Worrall D, Hird DL, Hodge R, Paul W, Draper J, Scott R. (1992) Premature dissolution of the microsporocyte callose wall causes male sterility in transgenic tobacco. Plant Cell 4: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Lu P, Wang Y, Ma H. (2011) The transcriptome landscape of Arabidopsis male meiocytes from high-throughput sequencing: the complexity and evolution of the meiotic process. Plant J 65: 503–516 [DOI] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V. (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H. (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhang ZB, Zhu J, Gao JF, Wang C, Li H, Li H, Zhang HQ, Zhang S, Wang DM, Wang QX, et al. (2007) Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J 52: 528–538 [DOI] [PubMed] [Google Scholar]
- Zhao DZ, Wang GF, Speal B, Ma H. (2002) The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 16: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Lou Y, Xu X, Yang ZN. (2011) A genetic pathway for tapetum development and function in Arabidopsis. J Integr Plant Biol 53: 892–900 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.