Members of the GRF family are conserved transcriptional regulators in both monocot and dicot plants.
Abstract
KNOTTED1-LIKE HOMEOBOX (KNOX) genes are important regulators of meristem function, and a complex network of transcription factors ensures tight control of their expression. Here, we show that members of the GROWTH-REGULATING FACTOR (GRF) family act as players in this network. A yeast (Saccharomyces cerevisiae) one-hybrid screen with the upstream sequence of the KNOX gene Oskn2 from rice (Oryza sativa) resulted in isolation of OsGRF3 and OsGRF10. Specific binding to a region in the untranslated leader sequence of Oskn2 was confirmed by yeast and in vitro binding assays. ProOskn2:β-glucuronidase reporter expression was down-regulated by OsGRF3 and OsGRF10 in vivo, suggesting that these proteins function as transcriptional repressors. Likewise, we found that the GRF protein BGRF1 from barley (Hordeum vulgare) could act as a repressor on an intron sequence in the KNOX gene Hooded/Barley Knotted3 (Bkn3) and that AtGRF4, AtGRF5, and AtGRF6 from Arabidopsis (Arabidopsis thaliana) could repress KNOTTED-LIKE FROM ARABIDOPSIS THALIANA2 (KNAT2) promoter activity. OsGRF overexpression phenotypes in rice were consistent with aberrant meristematic activity, showing reduced formation of tillers and internodes and extensive adventitious root/shoot formation on nodes. These effects were associated with down-regulation of endogenous Oskn2 expression by OsGRF3. Conversely, RNA interference silencing of OsGRF3, OsGRF4, and OsGRF5 resulted in dwarfism, delayed growth and inflorescence formation, and up-regulation of Oskn2. These data demonstrate conserved interactions between the GRF and KNOX families of transcription factors in both monocot and dicot plants.
KNOTTED1-LIKE HOMEOBOX (KNOX) class I homeobox genes play an essential role in the development and maintenance of the shoot apical and floral meristems (Hake et al., 1995, 2004; Endrizzi et al., 1996; Hake, 1996; Reiser et al., 2000; Brand et al., 2002; Hake and Ori, 2002; Ito et al., 2002; Tsuda et al., 2011). KNOX proteins contribute to the regulation of meristem maintenance by regulating the production of GAs. Different KNOX genes have been shown to inhibit GA biosynthesis in the shoot apical meristem (SAM) through down-regulation of the key biosynthetic gene GA20 oxidase (Kusaba et al., 1998; Tanaka-Ueguchi et al., 1998; Sakamoto et al., 2001; Hay et al., 2002; Rosin et al., 2003) or by controlling the level of GA2 oxidase1 that degrades GA (Bolduc and Hake, 2009). Down-regulation of KNOX gene expression at the flanks of the SAM is thought to permit biosynthesis of GA and consequently to result in organized cell proliferation and determination of cell fate (Sakamoto et al., 2001).
A large body of evidence suggests that the precise regulation of KNOX activity is central to the determination of organ versus meristem identity in a wide range of plant species (Hake et al., 1995, 2004; Hake, 1996; Reiser et al., 2000). In the monocot plants rice (Oryza sativa) and maize (Zea mays), misexpression of KNOX genes has a profound effect on the blade-sheath boundary in the leaves (Freeling and Hake, 1985). In Arabidopsis (Arabidopsis thaliana), KNOX misexpression also affects leaf formation, leading to serrations and the formation of ectopic meristems in the sinuses (Chuck et al., 1996; Long et al., 1996; Dean et al., 2004; Kuijt et al., 2004). A loss-of-function mutation of BREVIPEDICELLUS (BP) in Arabidopsis causes defects in stem elongation due to a lower number of cell divisions and defects in the differentiation and elongation of epidermal and cortical cells (Venglat et al., 2002). The phenotypes of shoot meristemless (stm) alleles range from a reduced SAM size to a complete lack of the meristem and fused cotyledons (Endrizzi et al., 1996; Venglat et al., 2002).
KNOX gene expression is regulated at multiple levels to prevent misexpression in leaves and leaf primordia (Hibara et al., 2002; Kumaran et al., 2002; Kim et al., 2003b; Lin et al., 2003; Kuijt et al., 2004). Several negative regulators of KNOX gene expression have been identified because their loss-of-function phenotypes resemble the phenotypes of KNOX overexpressors. MYB domain transcription factors are such negative regulators conserved between maize (rough sheath2), Antirrhinum majus (PHANTASTICA), and Arabidopsis (ASYMETRIC LEAVES1 [AS1]; Waites et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000, 2002). In Arabidopsis, a network of negative interactors provides a mechanism to distinguish between founder cells and stem cells in the SAM. AS1 plays a central role in this network, repressing the activity of BP and KNOTTED-LIKE FROM ARABIDOPSIS THALIANA2 (KNAT2) and being negatively regulated itself by STM (Byrne et al., 2002). AS1 forms a complex with AS2, a member of the LATERAL ORGAN BOUNDARY DOMAIN (LBD) family, to create a hairpin in the promoters of BP and KNAT2 that blocks their transcription (Guo et al., 2008). Other negative regulators of KNOX gene expression include members of BLADE ON PETIOLE, YABBY, and BEL1-like gene families (Kumaran et al., 2002; Ha et al., 2003; Kumar et al., 2007). Positive regulators of KNOX gene expression have also been described, such as genes of the CUP-SHAPED COTYLEDON (CUC) and the LBD gene families, with members CUC1, CUC2, and JAGGED LATERAL ORGANS (JLO) genes (Hibara et al., 2002; Borghi et al., 2007). The expression patterns of CUC1, CUC2, and STM overlap during early embryogenesis in the region of the organizing SAM (Aida et al., 1999; Takada et al., 2001). Overexpression of CUC1 leads to ectopic expression of STM, BP, KNAT2, and KNAT6 in cotyledons (Hibara et al., 2002). The microRNA miR164A regulates the extent of serrations by regulating the expression of CUC2 in the leaf sinuses (Nikovics et al., 2006). Misexpression of JLO increases the expression of STM and KNAT1 in leaves, whereas the jlo-D mutant has small lobed leaves resembling the bp mutant phenotype (Borghi et al., 2007).
Previously, we addressed the transcriptional control of a rice KNOX gene, Oskn2, showing that its promoter sequence is sufficient to mediate the initial down-regulation in developing organ primordia (Postma-Haarsma et al., 2002). Here, we used the Oskn2 promoter as a bait sequence in a yeast (Saccharomyces cerevisiae) one-hybrid screen aimed at the identification of novel upstream regulators. This screen resulted in the isolation of two members of the GROWTH-REGULATING FACTOR (GRF) family, OsGRF3 and OsGRF10, both interacting specifically with the Oskn2 promoter region. The first member of the rice GRF family, named OsGRF1, was described by van der Knaap et al. (2000) and isolated in a search for genes that are differentially expressed in the intercalary meristem of deepwater rice internodes in response to GA. Two highly conserved regions, the QLQ and WRC domains, are distinctive characteristics of the GRF family members in both monocot and dicot species (van der Knaap et al., 2000; Kim et al., 2003a; Choi et al., 2004; Zhang et al., 2008; Osnato et al., 2010). In this report, we provide evidence of functional links between GRF proteins and transcriptional regulation of KNOX genes. We show that OsGRF3 and OsGRF10 repress Oskn2 promoter activity in planta. Likewise, a GRF protein from barley (Hordeum vulgare) named BGRF1 (Osnato et al., 2010) was found to act as a repressor on an intron sequence in the barley KNOX gene Hooded/Bkn3, and three Arabidopsis GRF proteins could repress KNAT2 promoter activity. These data support the proposition that repressor activity on KNOX genes is a conserved function of members of the GRF family in monocot and dicot plant species.
RESULTS
Specific Interaction of OsGRF3 and OsGRF10 Proteins with the Oskn2 Promoter in Yeast
To identify novel genes involved in the regulation of the rice KNOX class I gene Oskn2, we performed a yeast one-hybrid screen. A rice zygote-derived embryonic complementary DNA (cDNA) expression library in pACTII vector was used as prey, and HIS3 reporter gene constructs containing promoter fragments of 662 and 1,264 bp upstream of the ATG of Oskn2 were used as bait. In three independent rounds, we screened a total of 900,000 yeast transformants with the short bait construct (ProOskn2S:HIS3) and 700,000 transformants with the long bait (ProOskn2L:HIS3). This resulted in 18 positive clones, which, after sequence analysis, all showed homology to the rice gene encoding OsGRF1, the founding member of the GRF family of putative transcription factors (van der Knaap et al., 2000). The clones obtained represented two genes, designated OsGRF3 (two clones with the short bait) and OsGRF10 (14 clones with the short bait and two clones with the long bait). By comparison with an OsGRF3 EST sequence (GenBank accession no. AU182732) and TIGR gene model 12004m.10015, we derived that both OsGRF3 clones from our screen encoded a partial OsGRF3 sequence lacking 15 amino acids at the N-terminal end. This partial sequence was in frame with the Gal4p activation domain (AD) sequence in the pACTII vector. To obtain a full-length OsGRF3 clone, we rescreened the library using the short bait construct and performed colony PCR on positive yeast transformants. This resulted in six clones encoding the full-length OsGRF3 sequence of 384 amino acids in frame with the Gal4 AD. The OsGRF10 clones from our screen were also in frame with the Gal4 AD and corresponded to two different splice variants of the OsGRF10 gene, one fitting with TIGR gene model 12002m.33821 (nine clones) and the other fitting with gene model 12002m.09594 (five clones). The two OsGRF10 splice variants are 209 and 211 amino acids long and differ only at the C-terminal end. The cDNA encoding the 211 amino acid variant was used in this study.
The OsGRF3 and OsGRF10 clones in pACTII were retransformed to the ProOskn2:HIS3 reporter strains and were shown in a titration experiment to grow on media containing up to 25 mm of 3-aminotriazole (3-AT; a competitive inhibitor of the His-3p reporter protein), thereby confirming activation of the ProOskn2:HIS3 reporter constructs (data not shown). To determine the DNA-binding specificity of the OsGRF3 and OsGRF10 factors, we used yeast strains containing HIS3 reporter gene constructs with different fragments of the 5′ regulatory sequences of two other rice KNOX class I genes, Oskn1 and Oskn3. As shown in Supplemental Figure S1 and summarized in Table I, factors OsGRF3 and OsGRF10 were not able to activate these other reporter constructs, indicating that their interaction with the Oskn2 promoter is specific.
Table I. Yeast one-hybrid assays with OsGRF3 and OsGRF10 expression constructs and KNOX promoter HIS3 constructs.
The plasmids pACTII-OsGRF3 and pACTII-OsGRF10 transformed into different yeast strains were scored for growth on medium containing His or on medium supplemented with 10 mm 3-AT (without His). The pACTIIa empty vector was used as negative control. YPO101 contains a HIS3 reporter preceded by only a TATA box and also serves as control. Growth of colonies on plates with 3-AT indicates specific activation of the expression of the ProOskn:HIS3 constructs by the GRF expression constructs. Results are based on Supplemental Figure S1.
Conserved Binding of Rice and Arabidopsis GRF Proteins to the Oskn2 Promoter
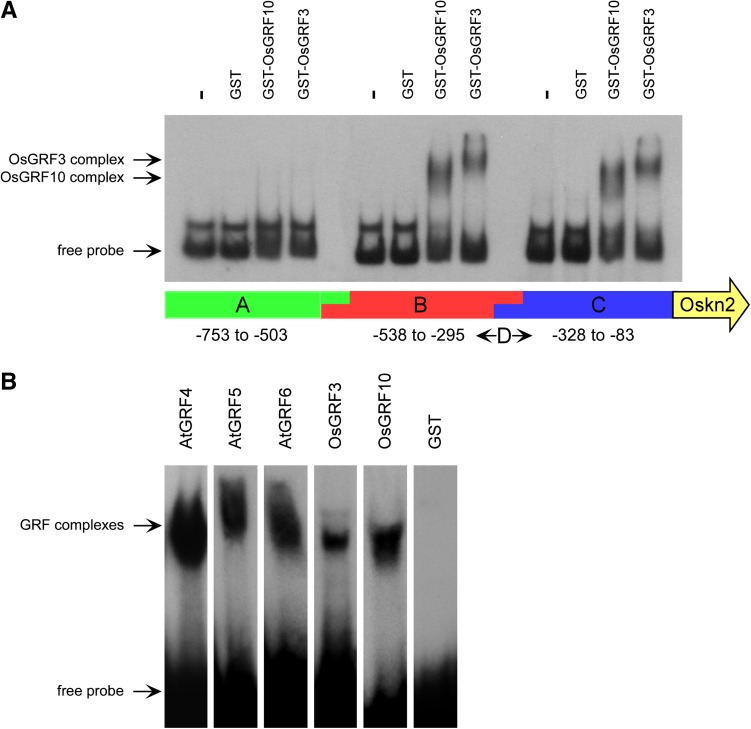
To map the region where OsGRF3 and OsGRF10 proteins interact with the Oskn2 promoter, we divided the 662-bp promoter fragment used in the screen into three subfragments (A, B, and C) of 230 bp, with overlaps of 10 and 16 bp (Fig. 1A). Reporter constructs of these subfragments were integrated into the yeast genome. OsGRF3 and OsGRF10 could not activate the reporter with subfragment A but were able to activate both the reporters with fragments B and C (Table II; Supplemental Fig. S2). Based on this result, a fourth reporter strain was constructed, containing a 34-bp promoter fragment (D) overlapping with the distal end of subfragment B and the proximal end of subfragment C (Fig. 1A). Both OsGRF3 and OsGRF10 were also able to activate the reporter construct with subfragment D (Table II; Supplemental Fig. S2).
Figure 1.
In vitro interactions of GST-OsGRF and GST-AtGRF proteins with Oskn2 promoter fragments in EMSAs. A, Subfragments A, B, and C of the Oskn2 promoter were tested for interaction with GST, GST-OsGRF3, and GST-OsGRF10 proteins. Bottom, Schematic organization of the Oskn2 promoter in subfragments A to D. B, Complex formation of GST-AtGRF4, GST-AtGRF5, GST-AtGRF6, GST-OsGRF3, and GST-OsGRF10 proteins with Oskn2 promoter fragment D. Similar to the yeast assays (Supplemental Figs. S1–S3), the GRF proteins interacted with Oskn2 promoter subfragments B, C, and D. GST was used as a control protein and shows no binding.
Table II. Interaction of OsGRF3 and OsGRF10 proteins with Oskn2 promoter subfragments A, B, C, and D.
Constructs pACTII-OsGRF3, pACTII-OsGRF10, and pACTIIa were analyzed in yeast strains Y187::ProOskn2-A, Y187::ProOskn2-B, Y187::ProOskn2-C, and Y187::ProOskn2-D on medium containing His or supplemented with 10 mm 3-AT (and without His). The locations of the Oskn2 promoter subfragments are schematically indicated in Figure 1A. Results are based on Supplemental Figure S2.
To investigate whether subfragment D contains a conserved GRF binding site, we tested three distantly related GRF proteins from Arabidopsis, AtGRF4, AtGRF5, and AtGRF6 (Kim et al., 2003a). Like OsGRF3 and OsGRF10, all three Arabidopsis GRF proteins in vector pACTII could activate the reporter with subfragment D, while none of them activated the reporter with subfragment A (Table III; Supplemental Fig. S3). AtGRF4, AtGRF5, and AtGRF6 were also able to activate the reporter with subfragment B, but only AtGRF5 could additionally activate the reporter construct with subfragment C (Table III; Supplemental Fig. S3).
Table III. Interaction of OsGRF3 and Arabidopsis GRF proteins with the Oskn2 promoter and subfragment D.
Yeast strains Y187::ProOskn2-662 and Y187::ProOskn2-D and control strain YPO101 harboring pACTII, pACTII-AtGRF4, pACTII-AtGRF5, pACTII-AtGRF6, and pACTII-OsGRF3 were examined for growth on medium containing His or medium supplemented with 10 mm 3-AT (without His). All GRF constructs show growth on 3-AT containing medium when grown in strains with constructs ProOskn2-662, ProOskn2-B, or ProOskn2-D but not in the control strain YPO101 that contains a promoterless HIS3 reporter. ProOskn2-C is only activated by pACTII-AtGRF5. Results are based on Supplemental Figure S3.
| Yeast Strain | pACTII |
pACTII-AtGRF4 |
pACTII-AtGRF5 |
pACTII-AtGRF6 |
pACTII-OsGRF3 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| His | 3-AT | His | 3-AT | His | 3-AT | His | 3-AT | His | 3-AT | |
| ProOskn2-A | + | − | + | − | + | − | + | − | + | − |
| ProOskn2-B | + | − | + | + | + | + | + | + | + | + |
| ProOskn2-C | + | − | + | − | + | + | + | − | + | − |
| ProOskn2-D | + | − | + | + | + | + | + | + | + | + |
| ProOskn2-662 | + | − | + | + | + | + | + | + | + | + |
| YPO101 | + | − | + | − | + | − | + | − | + | − |
Binding of OsGRF3 and OsGRF10 proteins to subfragments of the Oskn2 promoter was confirmed in vitro using glutathione S-transferase (GST) fusion proteins in electrophoretic mobility shift assays (EMSAs; Fig. 1). A DNA-protein complex was observed for the subfragments B, C, and D, but not for subfragment A (Fig. 1, A and B), fully consistent with the results obtained in yeast. GST fusion proteins of Arabidopsis AtGRF4, AtGRF5, and AtGRF6 were also able to bind to subfragment D, whereas GST control protein was not. Based on the combined results of yeast one-hybrid and EMSA analysis, we conclude that specificity for a DNA-binding site within the 34-bp sequence from Oskn2 promoter subfragment D is conserved at least among the five tested members of the GRF family in monocot and dicot plant species. Subfragments B, C, and D are located between positions –538 and –83 upstream of where we assume the start codon of Oskn2 is located (as indicated in GenBank accession no. AF323785). Based on comparison of the genomic sequence of Oskn2 (UniGene Os.4162) with 41 EST clones (of which GenBank accession no. CI740419 is the longest at the 5′ end), we define the putative mRNA transcription start at position –605. Therefore, OsGRF3 and OsGRF10 appear to interact with the 5′-untranslated region (UTR) of Oskn2.
Furthermore, analysis of deletion constructs of OsGRF10 showed that binding to subfragment D requires both the conserved QLQ and WRC domains described by van der Knaap et al. (2000; Table IV; Supplemental Fig. S2C). The QLQ domain was proposed to be involved in protein-protein interactions based on homology with the Switch/Sucrose Nonfermentable (SWI2/SNF2) chromatin remodeling complex in yeast (van der Knaap et al., 2000). The WRC domain contains a nuclear localization signal and a CCCH (C3H)-type zinc-finger motif, consistent with a function in DNA binding and transcriptional control (van der Knaap et al., 2000). Our results are in line with results of Osnato et al. (2010), who showed that deletion of the C3H zinc-finger motif of the WRC domain abolished DNA binding in vitro. However, this does not exclude the possibility that other domains of GRF factors also contribute to DNA binding.
Table IV. Analysis of DNA-binding properties of the QLQ and WRC domains from OsGRF10 in yeast one-hybrid screens.
Yeast strains Y187::ProOskn2-662 and Y187::ProOskn2-D harboring pACTII-QLQ, pACTII-AtGRF4WRC, or pACTII-QLQ-WRC were examined for growth on medium containing His or medium supplemented with 10 mm 3-AT (without His). Individual QLQ and WRC domains cannot activate the ProOskn2:HIS3 reporters when fused to the Gal4p AD in pACTIIa, whereas the combination QLQ-WRC can. Results are based on Supplemental Figure S2C.
Recently, Kim et al. (2012) identified a binding site for Arabidopsis GRF7 with the core sequence TGTCAGG. We checked the Oskn2 promoter for homology and found that the subfragments binding OsGRF3 and OsGRF10 (B, C, and D) are enriched for the motif CAG, which is contained within this core sequence. Subfragment D contains a cluster of seven CAG repeats or of its reverse complementary sequence CTG. Subfragment B has 14 CAG or CTG repeats (without the overlapping subfragment D), of which most are clustered in three regions with three or four repeats. Subfragment C also has 14 CAG or CTG repeats (without the overlapping subfragment D), of which one-half are in one cluster. By contrast, subfragment A, which has the same length as B and C, did not show any GRF binding in the yeast assays or the EMSAs and has only three CAG or CTG repeats, which are not clustered. Furthermore, the 305-bp fragment from the Bkn3 intron that is bound by BGRF1 has 14 CAG or CTG repeats, including one cluster of four repeats and two clusters of two repeats (Osnato et al., 2010). Taken together, the data suggest that GRF binding is associated with the presence of CAG or CTG repeats.
Expression Patterns of OsGRF3 and OsGRF10 Are Partially Overlapping with Oskn2
As shown above, yeast one-hybrid and in vitro binding studies have demonstrated DNA binding of GRF proteins to the Oskn2 promoter. Consistent with a possible transcription factor function, GFP tagging resulted in nuclear localization upon transient expression in onion (Allium cepa) cells (Supplemental Fig. S4A). Next, we compared expression patterns of the GRF genes in relation to their predicted target gene, Oskn2. To this end, we analyzed mRNA samples from 13 different tissues by reverse transcription (RT)-PCR (Fig. 2). OsGRF3 was broadly expressed in all tissues tested, except in roots. By contrast, OsGRF10 was only expressed in embryos, seedlings, immature rachis, and mature nodes but not in other tissues from mature plants, which is also confirmed by promoter GUS analysis (Supplemental Fig. S4). In several tissues, both Oskn2 and the GRF genes were found to be expressed, but Oskn2 was also expressed in roots where the GRF genes were not expressed. On the other hand, Oskn2 expression was absent in the region just above the SAM, which consist of leaf sheath and young leaves rolled upon each other, where OsGRF3 and OsGRF10 were both expressed.
Figure 2.
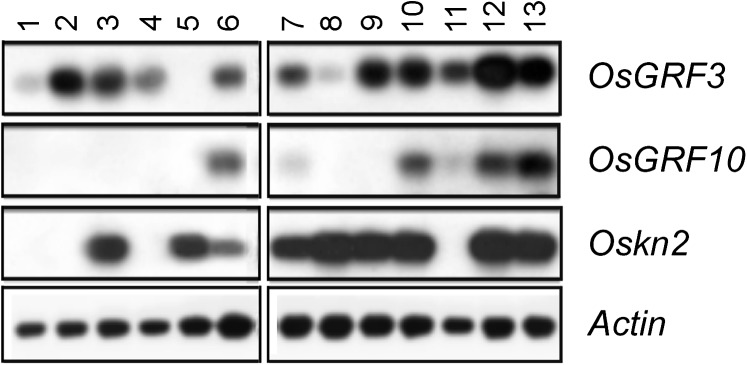
Analysis of tissue-specific expression of OsGRF3, OsGRF10, and Oskn2 by RT-PCR. Expression levels of OsGRF3 and OsGRF10 in 13 tissues compared with expression levels of Oskn2 and Actin by RT-PCR. 1, 2, 5, 6, and 11, Tissues from 3-week-old tissue-cultured seedlings, and; 3, 4, 7–9, 10, 12, and 13, tissues from mature greenhouse-grown plants. Leaf blade (1), leaf sheath (2), ligule-auricle region (3), leaves (4), roots (5), basal 0.5 cm of the seedling including the SAM region (6), immature rachis (7), immature spikelets (8), internodes (9), nodes (10), leaf sheath and blade 1.5 cm above the SAM (11), embryos at 6 to 8 d after pollination (12), and mature embryos (13).
Ectopic Expression of OsGRF3 and OsGRF10 in Rice Reduces Tillering and Induces Ectopic Root and Shoot Formation on Nodes
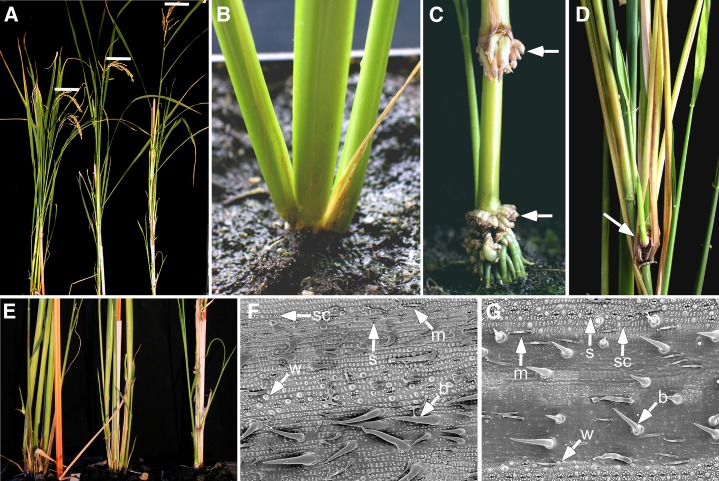
To study the possible function of the GRF factors in regulation of KNOX genes, we generated rice plants in which the Cauliflower mosaic virus (CaMV) 35S promoter drives overexpression of the full-length cDNAs of OsGRF3 (two independent lines) and OsGRF10 (seven independent lines) or a partial OsGRF3 cDNA (OsGRF3s) from the original yeast screen, expressing a shorter protein from an internal ATG (five independent lines). Overexpressor lines (T0 generation) of all three constructs showed similar phenotypical alterations, although effects of the longer OsGRF3 cDNA were stronger than effects of the shorter version and the effects of OsGRF10. The T1 generation of OsGRF3s, OsGRF3, and OsGRF10 overexpressors segregated into three classes, showing severe, mild, or no phenotypical changes. PCR analysis confirmed the presence of the transgene in the plants with mild and severe phenotypes and its absence in plants without phenotypical changes (data not shown). Normally, the stem of a rice plant produces a large number of tillers, reaching around 50 or even more at the maximum tiller stage under field conditions. The tillers arising on a main stem are called primary tillers. Secondary tillers can develop from the axils of leaves. T1 progeny of all independent overexpressor lines contained plants with a mild phenotype, forming three to four tillers (n = 16, 3.4 ± 0.8) in the time that wild-type plants produced four to six (n = 14, 5.1 ± 1.1). Furthermore, they formed adventitious roots on the nodes of tillers (Fig. 3C), which was not observed in the wild type (Fig. 3B). The severe class of GRF overexpressors (T1 progeny of two OsGRF3 lines, four OsGRF3s lines, and two OsGRF10 lines) consisted of slender plants with only one or two long tillers (Fig. 3, A and E), which had swollen nodes, showing extensive formation of adventitious roots and bundles of ectopic shoots (Fig. 3D). Increased tiller length was due to an increase in internode number, whereas internode lengths were not significantly increased (Supplemental Fig. S5). Inflorescence formation of the overexpressors was delayed approximately 2 weeks (OsGRF3s and OsGRF10 lines) up to 3 months (OsGRF3 lines), and the most severely affected plants (OsGRF3 lines) senesced when inflorescences were still enclosed within the leaf sheath. A region with a distinctive epidermal structure was noticed at the base of leaf blades of the overexpressors. This phenotype, affecting only the epidermis but not the internal leaf structure, was most apparent in OsGRF3 overexpressors. Scanning electron microscopy (SEM) analysis showed that the epidermis had formed fewer specialized epidermal structures such as silica knobs, bristles, and macro hairs and that the distance between stomata was decreased (Fig. 3, F and G). This phenotype was distinct from that of Oskn2 overexpressors or other rice KNOX gene overexpressors in which sectors showing sheath-like epidermal and internal leaf organization were displaced over the leaf blade area (Matsuoka et al., 1993; Postma-Haarsma et al., 2002). In conclusion, OsGRF3 and OsGRF10 overexpression inhibited the formation of new tillers, while increasing the formation of nodes and adventitious roots and shoots within the primary tiller(s). Additionally, OsGRF3 and OsGRF10 overexpression affected the epidermal structure in basal sectors of the leaf blade.
Figure 3.
Phenotypes of greenhouse-grown rice plants ectopically expressing OsGRF3 or OsGRF10. A and E, Comparison of plant length and number of tillers formed by wild-type plants (left) and OsGRF10 overexpressors with a mild (middle) and severe (right) phenotype. Bars in A mark the top of the first panicle. B, Basal part of a wild-type plant. C, Basal part of an OsGRF3 overexpressor showing ectopic root formation on the nodes, indicated with arrows. D, Severe OsGRF3-overexpressing plant showing ectopic shoot and root formation on the nodes, indicated with an arrow. F and G, SEM images of the adaxial epidermis of leaf blades of control plants (F) or plants ectopically expressing OsGRF3 (G). B, Bristle; M, macrohair; S, stomata; sc, silica cell; W, warts.
KNOX Gene Expression Is Down-Regulated by GRF Overexpression
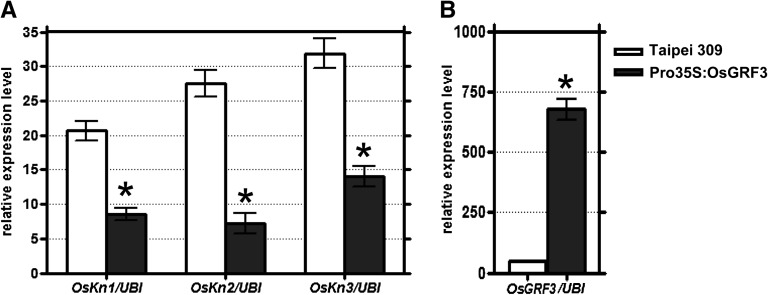
The expression of KNOX genes was analyzed in T2 seedlings of OsGRF3 overexpressors. This analysis could not be performed for OsGRF10 overexpressors because the T1 generation did not produce seeds. Down-regulation of Oskn1, Oskn2, and Oskn3 expression was observed by quantitative PCR (qPCR) analysis on RNA samples from SAM-containing (bottom) stem segments of OsGRF3 overexpressing seedlings (Fig. 4). Taken together with our DNA binding studies, these data strongly suggest that OsGRF3 acts as transcriptional repressor of Oskn2 and that Oskn1 and Oskn3 may be down-regulated by an indirect mechanism, because GRF proteins only showed a direct interaction with the Oskn2 promoter.
Figure 4.
Down-regulation of KNOX expression in OsGRF3 overexpressors. A, Oskn1, Oskn2, and Oskn3 expression levels were determined in SAM tissue of 2-week-old wild-type cv Taipei 309 seedlings and OsGRF3 overexpressor seedlings (Pro-35S:OsGRF3). B, OsGRF3 expression levels in SAM tissues are shown as verification of the overexpression of this gene. Total RNA for qPCR analysis was extracted from a 1-cm region of the stem containing the SAM of a pool of five plants. The OsUbi gene was used to normalize the data. qPCR was performed in triplicate, and mean values with sd are shown. Two independent biological replicates were performed with similar results. The data were analyzed with an unpaired Student’s t test. Asterisks indicate significant differences (P < 0.05) compared with the untransformed controls.
KNOX Gene Expression Is Up-Regulated by RNA Interference (RNAi)-Mediated OsGRF Silencing
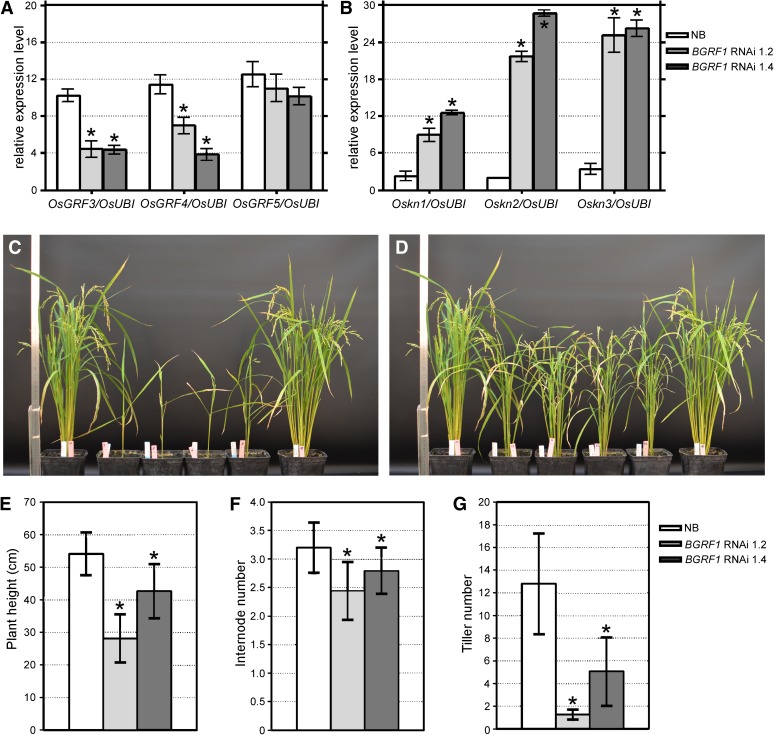
Because no suitable transfer DNA (T-DNA) or transposon insertion mutants for OsGRF3 or OsGRF10 were available, an RNAi approach was set up. For this, a construct was used based on the WRC motif from a barley GRF gene (BGRF1) that shares 93%, 85%, and 73% identity, respectively, with OsGRF3, OsGRF4, and OsGRF5, which are, according to a phylogeny reconstruction (Supplemental Figs. S6–S9), all in subclass C (Zhang et al., 2008). Different T0 lines exhibiting OsGRF gene silencing were selected (lines 1.2 and 1.4), self-pollinated, and amplified to T1 and T2 generations. T2 RNAi rice plants were analyzed by qPCR to assess the expression levels of endogenous GRF genes. As shown in Figure 5A, the expression of OsGRF3 and OsGRF4 was greatly reduced in the lines examined, whereas the expression of OsGRF5 was less affected likely due to the lower sequence similarity with the sequence of BGRF1 (Supplemental Figs. S6 and S8). Silencing of rice GRF genes caused developmental alterations, including reduced plant height (Fig. 5, C and D), which were strongest in line 1.2. The average plant height of the cv Nipponbare controls was 54.1 cm, which dropped to 28.1 and 44.2 cm for lines 1.2 and 1.4 (Fig. 5E), respectively. This phenomenon is largely caused by reduced internode length, because the average internode number (on the master tiller) stayed with 3.2, the same in line 1.4, and dropped only slightly to 2.4 in line 1.2 (Fig. 5F), but is not statistically significant. A strong effect was found on tiller number, which dropped from an average of 12.8 for the wild type to 1.3 and 5.4 for lines 1.2 and 1.4 (Fig. 5G), respectively. These phenotypic effects were accompanied by up-regulation of Oskn1, Oskn2, and Oskn3 gene expression, as revealed by qPCR analyses on a seedling sample corresponding to a 1-cm region of leaf blade and sheath comprising the SAM (Fig. 5B). These data are consistent with the GRF overexpression analysis, which resulted in down-regulation of the same KNOX genes.
Figure 5.
Down-regulation of GRF genes in rice results in up-regulation of KNOX genes and affects plant architecture. A and B, Relative expression levels of endogenous OsGRF3, OsGRF4, and OsGRF5 (A) and Oskn1, Oskn2, and Oskn3 genes (B) in BGRF1 RNAi lines 1.2 and 1.4 (T2 generation), respectively. Total RNA was extracted from a pool of leaves of five 10-d-old seedlings. The OsUbi gene was used to normalize data, and cv Nipponbare was used as wild-type control. qPCR was performed in triplicate, and mean values with sd are shown. Two independent biological replicates were performed with similar results. The data were analyzed with an unpaired Student’s t test. Asterisks indicate significant differences (P < 0.05) compared with the untransformed controls. C and D, Phenotypes of greenhouse-grown rice plants at anthesis stage expressing the BGRF1 RNAi construct. Line 1.2 (C) has a stronger phenotype than line 1.4 (D). The control cv Nipponbare plants are at the left and right, and four T2 plants are in the middle. E to G, The BGRF1 RNAi plants were phenotyped for plant height (E), internode number on the master tiller (F), and tiller number (G). The mean values with sd of two lines (n = 15 and n = 29 plants for lines 1.2 and 1.4, respectively) are compared with wild-type cv Nipponbare (NB; n = 5), and the data were analyzed using ANOVA followed by Bonferroni corrections. Asterisks indicate significant differences (P < 0.05) compared with the untransformed controls.
OsGRF3 and OsGRF10 Repress ProOsKn2:GUS Expression in Rice Calli
To provide further evidence for the regulatory function of the GRF proteins, we set up a transient expression system based on agroinoculation with Agrobacterium tumefaciens strains transferring T-DNA constructs expressing either OsGRF3 or OsGRF10. These T-DNA constructs were transferred to embryonic rice calli harboring construct ProOskn2:GUS. In this construct, the GUS gene is driven by the promoter of the Oskn2 gene, which was described before (Postma-Haarsma et al., 2002). Calli at 2, 4, 6, 8, and 10 d post inoculation (dpi) with A. tumefaciens were stained with X-Gluc, and ProOskn2:GUS expression was quantified from stereomicroscope images using ImageJ. Agroinoculation with both OsGRF3 and OsGRF10 constructs resulted in a decrease of the Oskn2 promoter-driven GUS expression at 4, 6, 8, and 10 dpi compared with the expression at 2 dpi and compared with control calli that were treated with an A. tumefaciens strain transferring construct Pro-35S:GFP (Supplemental Fig. S10, A and B). These results demonstrate that when OsGRF3 and OsGRF10 are overexpressed in ProOskn2:GUS calli, Oskn2 promoter-driven GUS expression decreases, indicating that both GRF proteins act as repressors in this experimental set up.
GRF-KNOX Interactions Are Conserved in Barley and Arabidopsis
In a yeast one-hybrid approach, Osnato et al. (2010) cloned BGRF1, a member of the GRF family, from barley and one of several transcription factors interacting with a 305-bp sequence from intron IV of the barley KNOX class I gene Bkn3. A duplication of this sequence causes ectopic expression of Bkn3 that leads to the so-called Hooded phenotype (Müller et al., 1995). Here, we used rice protoplast assays to investigate the interaction of BGRF1 with the intron IV 305-bp element. It has been shown that expression of a GUS reporter construct consisting of the 305-bp element upstream of a minimal CaMV 35S promoter can be activated by transcription factor Barley Ethylene Insensitive-Like1 (BEIL1) (Osnato et al., 2010). We found that cotransformation of BGRF1 resulted in repression of BEIL1-activated expression of the reporter construct (Fig. 6). This result supports the ability of both BGRF1 and BEIL1 to bind in planta to the 305-bp element and is consistent with EMSA experiments that showed that BGRF1 binds distal to the 305-bp sequence, whereas BEIL1 binds in the center (Osnato et al., 2010). OsGRF10 could substitute for BGRF1 as a repressor of BEIL1-activated reporter gene expression (Fig. 6), indicating species-conserved binding specificity and repressor activity of the GRF proteins.
Figure 6.
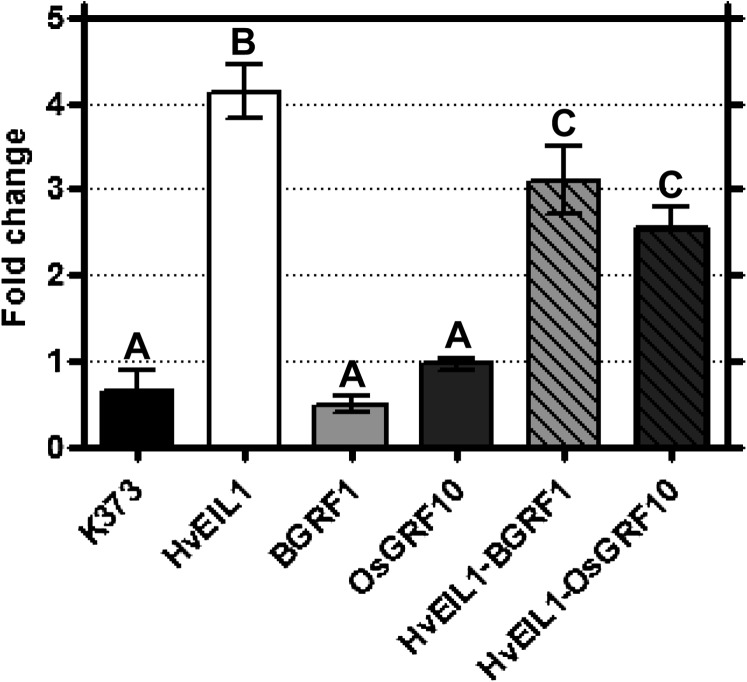
Interaction of BGRF1 and OsGRF10 with the 305-bp Bkn3 intron IV element in rice protoplasts. Effects of BGRF1 and OsGRF10 overexpression on down-regulation of BEIL1-activated GUS reporter gene expression were tested using reporter constructs K373 and K373+305 bp. Construct K373 harbors the GUS gene preceded by the –46 minimal promoter derived from ProCaMV 35S. Construct K373+305 bp contains the 305-bp enhancer sequence from intron IV of Bkn3 upstream of the minimal CaMV 35S promoter. GUS activities were normalized on FLUC activity, which served as a control for transformation efficiency. Bars in the histogram indicate GUS/FLUC activity ratio of effector with K373+305 bp with respect to the value obtained with the same effector and the control reporter K373. The graph reports mean values of six independent transformations of each construct combination, and error bars represent sd values. BEIL1 activates construct K373+305 bp with a factor 4.2, which is lowered to 3.1 and 2.6 in the presence of BGRF1 and OsGRF10, respectively. Six individual transformations per construct were used in each of the measurements, and the data were analyzed using ANOVA followed by Bonferroni corrections. Letters have been assigned to distinct groups based on 95% confidence intervals.
To obtain additional experimental support for the interaction between GRF proteins and KNOX genes, we used GUS reporter lines of Arabidopsis driven by promoter sequences of the KNOX class I genes BP/KNAT1 (Ori et al., 2000; ProBP:GUS) and KNAT2 (Dockx et al., 1995; ProKNAT2:GUS) and stably transformed these with overexpression constructs of OsGRF10 and AtGRF4, AtGRF5, and AtGRF6. ProKNAT2:GUS expression was concentrated in the SAM of seedlings and in part of the hypocotyl but absent from the petioles of leaves (Supplemental Fig. S11E). By contrast, seedlings overexpressing OsGRF10, AtGRF4, AtGRF5, or AtGRF6 showed developmental aberrations (Supplemental Fig. S11, L–O and Q–T), and the expression of ProKNAT2:GUS was reduced to a weak spot around the SAM (Supplemental Fig. S11, A–D). Thus, the same rice and Arabidopsis GRF proteins that showed conserved binding to a 34-bp subfragment of the Oskn2 promoter were able to down-regulate ProKNAT2:GUS expression. By contrast, ProBP:GUS expression was not affected by overexpression of any of these four GRF proteins (Supplemental Figure S11, F–J), indicating that the interaction with the KNAT2 promoter is specific. As discussed above, GRF binding activity to Oskn2 promoter fragments was associated with the presence of CTG or CAG repeats. We checked BP and KNAT2 promoter regions of 1 kb upstream of the ATG start codon and found 16 CTG and CAG repeats in the KNAT2 promoter, whereas the BP promoter, which was not repressed by GRFs, only contained three such sequences.
DISCUSSION
Here, we report on two factors of the GRF family, OsGRF3 and OsGRF10, interacting with the 5′ upstream sequence of the rice KNOX class I homeobox gene Oskn2. We found that these GRF proteins and homologs in barley and Arabidopsis act as transcriptional repressors on KNOX genes. KNOX expression is linked to SAM maintenance and organ formation and requires tight control by a network of transcription factors (Byrne et al., 2000; Semiarti et al., 2001; Lin et al., 2003; Uchida et al., 2010; Tsuda et al., 2011). Most regulators of KNOX genes were identified through analysis of mutants resembling plants with altered KNOX expression. In this study, yeast one-hybrid screening revealed a novel interaction between OsGRF3 and OsGRF10 and Oskn2. We mapped binding of these GRFs to three promoter fragments of the 5′ UTR region of Oskn2, suggesting that the interaction might prevent transcription. A similar regulatory mechanism was found for the murine Tumor Necrosis Factor-α gene, where mutation of a heat shock factor (HSF1) binding site in the 5′ UTR abolishes HSF1-mediated repression (Singh et al., 2002).
Little is known about the precise functions of GRFs. Previously, rice GRF genes were shown to be regulated by GA3 (van der Knaap et al., 2000; Choi et al., 2004). Several KNOX genes from rice, tobacco (Nicotiana tabacum), and Arabidopsis have been implicated in regulation of GA biosynthesis by repression of a Gibberellic Acid-20 oxidase (GA20ox) gene (Kusaba et al., 1998; Sakamoto et al., 2001; Hay et al., 2002). However, we examined expression of OsGA20ox1 and OsGA20ox2 in OsGRF3 overexpressors as well as RNAi plants but did not find a significant difference (data not shown) that could explain the phenotypes we observed. Mutant analysis is hampered by the fact that single mutants for GRF1, GRF2, and GRF3 in Arabidopsis do not show many morphological changes, which may be due to redundancy, as expression patterns of different GRF genes are overlapping (Kim et al., 2003a; Horiguchi et al., 2005). Double and triple grf mutants for GRF1, GRF2, and GRF3 in Arabidopsis showed smaller leaf sizes, which could be attributed to reduced cell proliferation (Kim and Kende, 2004; Kim and Lee, 2006). Addition of a grf4 mutation to the grf1/grf2/grf3 triple mutant resulted in spoon- and cup-shaped cotyledons, and a significant portion of the mutant population showed the phenotype of the Arabidopsis stm mutant lacking a SAM (Chuck et al., 1996; Kim and Lee, 2006). Single mutants of grf4 or grf5 had smaller leaves, and GRF5 overexpression increased leaf size, suggesting a role in controlling cell number in leaves as also observed for GRF1 and GRF2 overexpression (Horiguchi et al., 2005). Recently, a function for GRF7 from Arabidopsis was described in repression of abscisic acid and osmotic stress-responsive genes including the transcription factor Dehydration-Responsive Element Binding2A (DREB2A) protein (Kim et al., 2012). Horiguchi et al. (2005) showed that all nine Arabidopsis GRF promoter constructs are highly expressed in leaf primordia and have different expression patterns that partially overlap with each other. Formation of leaf primordia is dependent on down-regulation of KNOX genes in the SAM (Byrne et al., 2000; Semiarti et al., 2001; Lin et al., 2003). Therefore, it is possible that GRFs may contribute to the down-regulation of KNOX activity during leaf primordium development in Arabidopsis, together with other transcriptional repressors for which such function has previously been demonstrated (Byrne et al., 2000; Semiarti et al., 2001; Lin et al., 2003). In rice, it is more difficult to speculate about the possible function of GRFs in relation to KNOX activity, as expression of both OsGRF10 and Oskn2 was absent from the seedling SAM and leaf primordia. However, at the late stage of embryo development, ProOskn2:GUS expression was down-regulated in the SAM when ProOsGRF10:GUS was still highly expressed, suggesting the possibility that OsGRF10 might repress Oskn2 at this developmental stage. Other functional relationships between OsGRF10 and Oskn2 might also exist, as their expression patterns were partially overlapping in several other tissues, including the vascular system.
OsGRF3 and OsGRF10 overexpression resulted in taller, slender plants, with reduced tillering, increased internode number, extensive adventitious roots and ectopic shoots on the nodes, delayed flowering, and abnormalities in leaf epidermis development. Only OsGRF3 overexpressors were fertile, and their seedlings showed Oskn2 down-regulation. By contrast, RNAi silencing of GRF genes resulted in reduced plant height and tillering and in up-regulation of Oskn2 expression. The Oskn2 expression pattern in GRF overexpressors and RNAi plants is consistent with a repressor function for GRFs. The visible shortening of internodes in OsGRF RNAi-silencing plants is consistent with the proposed function of rice GRF genes in stem growth (van der Knaap et al., 2000). In addition to Oskn2, expression of Oskn1 and Oskn3 was also down-regulated in GRF overexpressors and up-regulated in RNAi plants. The effects on Oskn1 and Oskn3 expression might be indirect, because their respective promoters were not recognized by OsGRFs in yeast assays. Alternatively, it is possible that GRF binding sites might be located in the introns or further downstream of the open reading frame, as in the case of Bkn3 (Osnato et al., 2010). Such a direct interaction of GRFs with Oskn1 and Oskn3 is conceivable, because their respective expression patterns are overlapping in the SAM of developing embryos and in developing spikelets and leaf blades and sheaths (Sato et al., 1998; Postma-Haarsma et al., 1999, 2002; Sentoku et al., 1999). On the other hand, an indirect mechanism is also conceivable. Tsuda et al. (2011) showed, using ORYZA SATIVA HOMEOBOX1 (OSH1)/Oskn1 overexpressors and an osh1osh15 double mutant together with in vivo binding assays, that this gene positively regulates at least four other KNOX I genes, including OSH71/Oskn2 and OSH15/Oskn3. This regulation occurs very likely through binding to conserved TGAC sequences. Similar results were obtained with maize KNOTTED1, which was shown among other KNOX genes to positively regulate liguless4a (lg4a), which is orthologous to Oskn2 (Bolduc et al., 2012). It is plausible that Oskn2 has the same effect on expression of other KNOX class I genes in rice, because yeast assays have shown that Oskn2 recognizes the same conserved TGAC sequences (Postma-Haarsma et al., 2002).
Further evidence for a repressor function of members of the GRF family was obtained from transient expression experiments with the BGRF1 gene from barley and analysis with Arabidopsis GRF overexpressing lines in a ProKNAT2:GUS background. Similar as in rice, overexpression of GRFs in Arabidopsis did not result in typical KNOX overexpression phenotypes (Kuijt et al., 2004) but in plants showing severe defects in SAM and leaf development. This cannot be solely due to repression of KNAT2 because knat2 does not show any obvious phenotype despite the predominant expression of KNAT2 in the SAM (Byrne et al., 2002; Belles-Boix et al., 2006). Likely, the expression of other target genes involved in SAM development is also altered by GRF overexpression. Although we did not observe down-regulation of ProBP:GUS, we cannot exclude the possibility that the expression of the endogenous KNOX genes might be altered due to the presence of regulatory elements elsewhere in the gene as with barley Hooded (Müller et al., 1995; Osnato et al., 2010). In the rice OsGRF overexpressors, we did not observe similar SAM defects; however, the reduced tillering and adventitious root and shoot formation on the nodes are indications of aberrant meristematic activity. Further investigations will be required into the functional relationships between GRF and KNOX genes and biological processes involved. Furthermore, with the GRF7-DREB interaction confirmed (Kim et al., 2012), we also expect other genes than KNOX genes to be regulated by GRFs, and it will be of great interest to gain further insight into these regulatory networks.
MATERIALS AND METHODS
Yeast One-Hybrid Screenings and Domain Mappings
For yeast (Saccharomyces cerevisiae) one-hybrid screening, the amplified λACT-ZE6 cDNA library (constructed with cDNA derived from rice [Oryza sativa ssp. indica ‘IR58’] embryos, 6 d after pollination) described by Postma-Haarsma et al. (1999) was converted into a pACTII-ZE6 library by Cre-loxP-mediated subcloning in Escherichia coli BNN132 (Ouwerkerk and Meijer, 2001). All handlings with yeast were as described earlier (Meijer et al., 2000; Ouwerkerk and Meijer, 2001, 2011). Yeast strain Y187 (Clontech, genotype: MATα, ura3-52, his3-Δ200, ade2-101, trp1-901, leu2-3,112, met–, gal4Δ, gal80Δ, URA3::GAL1UAS-GAL1TATA-lacZ) derivatives with HIS3 reporter constructs were integrated via the pINT1 integrative vector system at the nonessential locus PDC6 as described earlier (Meijer et al., 1998) and were generated as follows. ProOskn2:HIS3 constructs were based on an earlier described promoter fragment in which the sequence context of the translation start codon was changed to an NcoI restriction site (Postma-Haarsma et al., 2002). Two promoter fragments, a 750-bp SacI-NcoI fragment (in GenBank accession no. AF323785) to construct ProOskn2S:HIS3 and a 1,349-bp SalI-NcoI (in GenBank accession no. AF323785) fragment to construct ProOskn2L:HIS3 were subcloned into pUC21 (GenBank accession no. AF223641). From the resulting pUC21 derivatives, NotI-NheI fragments were cloned in NotI/SpeI-cut pHIS3NX (GenBank accession no. AF272030), leading to promoter fragments of 662 and 1,264 bp fused to HIS3, respectively. Next, NotI-XbaI fragments (containing the ProOskn2:HIS3 fusions) were cloned into NotI/XbaI-cut pINT1 (GenBank accession no. AF289993). The resulting plasmids were linearized with NcoI and introduced in yeast to generate Y187::ProOskn2-662 and Y187::ProOskn2-1264. One-hybrid screenings were performed with selection on medium lacking Leu and His and containing 5 mm 3-AT. Library plasmids were isolated from positive yeast colonies as described earlier (Meijer et al., 2000; Ouwerkerk and Meijer, 2011) and transformed into E. coli DH5α for subsequent sequence analysis (Baseclear) and clonings.
A set of control yeast strains was used to study the binding specificity of the clones obtained from the yeast one-hybrid screenings. These control strains were Y187::ProOskn1-1130, Y187::ProOskn1-483, Y187::ProOskn3-1170, and the promoterless HIS3 reporter control strain YPO101 (Ouwerkerk and Meijer, 2001, 2011). The ProOskn1:HIS3 construct was derived from pVDH767 (kindly provided by Van der Have B.V.), which contains the Oskn1 promoter sequence as reported by Matsuoka et al. (1993; GenBank accession no. D16507; ATG start codon at 1,065 bp). From this plasmid, an AlwNI (blunt)-PstI (–521 to –38) fragment was cloned in EcoRV/PstI-cut pUC21 (GenBank accession no. AF223641) and subsequently subcloned as NotI-EcoRI fragment in pHIS3NX (GenBank accession no. AF272030). From the resulting plasmid, a ProOskn1:HIS3 fusion was excised as NotI-XbaI fragment and cloned in NotI/XbaI-cut pINT1 (GenBank accession no. AF289993). ProOskn3:HIS3 was based on an Oskn3 promoter fragment (in GenBank accession no. AF323786) isolated earlier (Postma-Haarsma et al., 2002). A 1,170-bp fragment (directly upstream of the ATG) was subcloned using NcoI-XhoI to the same sites in pIC-21H (GenBank accession no. AF294975). From the resulting plasmid, a NotI-EcoRI fragment was cloned into the same sites of pHIS3NX, and then the whole cassette comprising the Oskn3 promoter fused to the HIS3 gene was cloned into pINT1 as NotI-XbaI fragment.
The short Oskn2 promoter was dissected into three fragments, spanning from –753 to –508, –538 to –295, and –328 to –83 with respect to the translation start codon, respectively, using the PCR primer sets 2SN1N/2SN4S, 2SN2N/2SN5S, and 2SN3N/2SN6S (Supplemental Table S1) resulting in fragments of 246 (fragment A), 244 (fragment B), and 246 bp (fragment C), respectively. These were first cloned in pCR2.1-Topo (Invitrogen) for sequence analysis. Next, NotI-SpeI fragments were transferred to NotI/SpeI-cut pINT1-HIS3NB (GenBank accession no. AY061966). A fourth fragment (D) was obtained by annealing the primers Oskn2D-Fw and Oskn2D-Rev and cloning of the annealed 34-bp fragment D (spanning from –327 to –294) supplied with SpeI and NotI overhanging ends into pINT1-HIS3NB. From the resulting plasmids, NcoI-SacI or NcoI-AgeI fragments were introduced in yeast to generate Y187::ProOskn2-A, Y187::ProOskn2-B, Y187::ProOskn2-C, and Y187::ProOskn2-D.
pACTII constructs expressing Arabidopsis (Arabidopsis thaliana) GRF genes as translational fusion products with the Gal4p AD were made as follows. For cloning AtGRF4, AtGRF5, and AtGRF6, full-length cDNA clones were assembled from two parts. The N-terminal parts were PCR amplified using Phusion DNA polymerase (Finnzymes) on cDNA derived from Arabidopsis (ecotype Columbia) using primer sets AtGRF4-Fw/AtGRF4-Rev, AtGRF5-Fw/AtGRF5-Rev, and AtGRF6-Fw/AtGRF6-Rev (Supplemental Table S1) to introduce an NcoI restriction site with the ATG start codon embedded or, in case of the AtGRF5, an EcoRI restriction site just upstream of the starting ATG. The resulting PCR products were ligated into pTOPO2.1 vector (Invitrogen) for sequence analysis and digested with NcoI-PvuII (AtGRF4), EcoRI-XcmI (AtGRF5), and NcoI-NsiI (AtGRF6) for further cloning. The C-terminal parts of the AtGRF genes were excised as the PvuII-XhoI fragment from M25D3 (GenBank nos. BE522365, and BE522366, AtGRF4), the XcmI-XhoI fragment from M61B1 (GenBank accession no. BE525293, AtGRF5), and the NsiI-BamHI fragment from RAFL09-87-A03 (GenBank accession no. AY060586, AtGRF6). Finally, AtGRF N- and C-terminal subfragments were joined together in pACTII between NcoI and XhoI for AtGRF4, EcoRI-XhoI for AtGRF5, and NcoI-BamHI for AtGRF6, respectively.
For DNA-binding mapping assays in yeast, the WRC and QLQ domains from pACTII-OsGRF10 were subcloned into pACTIIa (Meijer et al., 1997) as follows. The fragment containing the WRC domain was amplified by PCR using primers WRC-Fw and WRC-Rev (Supplemental Table S1) and cloned into SK+ Bluescript II (Stratagene) by EcoRI and XhoI for sequencing and further cloning. From this plasmid, an XhoI-EcoRI fragment was transferred to SalI (partial)/EcoRI-cut pACTIIa resulting in pACTII-WRC. From pACTII-OsGRF10, an NcoI-NotI fragment (containing both QLQ and WRC domains) was cloned into NcoI/NotI-cut pUC28. From this plasmid, an NcoI-SalI fragment was cloned into pACTIIa and cut with NcoI-XhoI, resulting in pACTII-QLQ-WRC. A fragment containing only the QLQ domain was excised by NcoI-HaeIII from the pUC28 clone and was cloned into pACTIIa and digested with NcoI and SmaI, resulting in pACTII-QLQ.
Bioinformatics and Phylogenetic Analysis
BLAST searches of the rice genomic sequence were carried out with the BLAST programs running at National Center for Biotechnology Information (http://www.ncbi.nih.nlm.gov), TIGR (http://www.tigr.org/), and GRAMENE (http://www.gramene.org). To predict intron-exon boundaries of the rice and Arabidopsis genes, we used GENSCAN (http://genes.mit.edu/burgelab/) and the GENSCAN predictions of the Sanger Ensemble. The predicted sequences were adjusted manually based on comparisons with EST or cDNA sequences using Vector NTI10 (Invitrogen). Deduced amino acid sequences of GRF genes from Arabidopsis, barley (Hordeum vulgare), maize (Zea mays), and rice were used to construct a phylogenetic tree (procedure according to Harrison and Langdale, 2006). The whole protein sequences were aligned using ClustalW (Thompson et al., 1994) at the European Bioinformatics Institute server with default settings and then manually revised using BioEdit (Hall, 1999). The conserved QLQ and WRC domains of 160 amino acids (including caps) were used for phylogenetic study. Phylogenetic trees were obtained by maximum parsimony methods using PAUP, version 4.0b10 (Swofford, 2002). From 1,000 replications, three trees were obtained, and a strict consensus tree was calculated with PAUP, version 4.0b10 (Swofford, 2002). The bootstrap probability of each branch was estimated with 500 replications.
Binary Vector Constructs
All GRF overexpression constructs were made in binary vector pC1300intB-35SnosEX (GenBank accession no. AY560325; Kuijt et al., 2004). OsGRF3 and OsGRF10 overexpression constructs were made by subcloning EcoRI-XhoI fragments from pACTII-OsGRF3 and pACTII-OsGRF10, the original clones obtained in yeast one-hybrid screenings. AtGRF4 was overexpressed by cloning an EcoRI-XhoI fragment from clone M25D3. An AtGRF5 overexpression construct was made by transferring an EcoRI-XhoI fragment from pACTII-AtGRF5. AtGRF6 was overexpressed by transferring EST clone pda06900 (GenBank accession no. AY060586), firstly in pUC21 between EcoRI and KpnI and then as EcoRI-XhoI fragment.
To silence GRF genes in rice, an RNAi approach was taken. For this, a binary construct using Gateway technology was made. An attB-flanked PCR product, produced by the amplification of BGRF1 cDNA with attB-specific primers (AttB1 BGRF1Fw and AttB2 BGRF1Rev; Supplemental Table S1), was introduced in pDONR207 by BP recombination. Subsequently, the RNAi construct was obtained by attL/attR recombination using LR clonase enzyme (Invitrogen) between vector pBios738 (kindly provided by Biogemma) and available pDONR207 containing the BGRF1 open reading frame. This sequence was cloned in both orientations, with the tubulin intron (1 kb) in the middle allowing the formation of the hairpin that mediates RNAi. To get constitutive silencing, the expression of the transgene was driven by the strong promoter of the Cassava vein mosaic virus.
For expression pattern analysis, a 1,560-bp fragment of the 5′ regulatory sequence of OsGRF10 was amplified with the Expand High Fidelity PCR System (Roche) with primers ProOsGRF10-Fw and ProOsGRF10-Rev (Supplemental Table S1). With the ProOsGRF10-Rev primer, an NcoI restriction site is created around the ATG by changing 2 bp (GAATGG to CCATGG). PCR products were ligated into the pCR2.1-TOPO vector (Invitrogen) for sequence analysis. The correct clone of ProOsGRF10 was digested with SacI and NcoI and cloned into binary vector pCAMBIA-1391Z (GenBank accession no. AF234312) for translational fusion to the GUS (uidA) gene. Prior to plant transformation, binary vectors were transferred from E. coli strain DH5α to Agrobacterium tumefaciens strain LBA 4404 (for rice transformation) or LBA 1115 (for Arabidopsis transformation) via triparental mating. Ten independent plant lines were made and analyzed for GUS expression using X-Gluc stainings.
Plant Transformation and Growth Conditions
Transformation of japonica rice cultivar Taipei 309 was performed as previously described (Scarpella et al., 2000), except that A. tumefaciens strain LBA 4404 was used instead of LBA 1119. Plantlets were maintained in tissue culture on one-half-strength Murashige and Skoog medium with 10 g L–1 Suc until transfer to the greenhouse, where they were grown at 28°C under a 16-h photoperiod and 85% humidity. With the (shorter) cDNA isolated in the original yeast screens (OsGRF3s), seven independent rice overexpressor lines were analyzed, and with the full-length OsGRF3 cDNA, two independent lines were analyzed. Overexpressor lines with the shorter and longer version of OsGRF3 showed similar phenotypical alterations, although effects of the longer version were more severe. Ten independent overexpressor lines harboring the full-length OsGRF10 cDNA construct were analyzed.
In the case of the RNAi plants, a transformed A. tumefaciens (strain EHA105) was cocultivated with rice calli (cv Nipponbare, japonica rice), and plants were regenerated as in Sallaud et al. (2003). A primary screening based on the resistance to Geneticin was conducted, and positive regenerants were transferred to test tubes. A secondary RT-PCR screening was conducted using primers GRF222Fw and GRF500Rev designed on a conserved DNA binding domain sequence (WRC motif). Selected plants were transferred to soil pots and grown until maturity.
Arabidopsis ProBP:GUS (in Columbia background) and ProKNAT2:GUS (in C24 background) lines were transformed by the floral dip method (Clough and Bent, 1998). Arabidopsis seeds were surface sterilized using chlorine vapor, sown on one-half-strength Murashige and Skoog medium containing 20 mg L–1 hygromycin, 100 mg L–1 nystatin, and 100 mg L–1 timentin, and incubated in the dark at 4°C for 72 h before germination at 21°C under a 16-h photoperiod. Plants 3 to 4 weeks old were transferred to soil and grown at 21°C under a 16-h photoperiod and 60% relative humidity.
EMSA and Constructs for Expression of Recombinant GRF Proteins
OsGRF3 and OsGRF10 cDNA fragments were fused in frame with the GST sequence in expression vector pGEX-KG (Guan and Dixon, 1991) by subcloning EcoRI-XhoI fragments from the pACTII clones isolated in the library screening. pGEX-AtGRF4 and pGEX-AtGRF6 were made by subcloning NcoI-XhoI fragments from the respective pACTII plasmids between NcoI and XhoI of pGEX-KG. pGEX-AtGRF5 was made by cloning an EcoRI-XhoI fragment from pACTII-AtGRF5 between EcoRI and XhoI of pGEX-KG. To extract recombinant protein, 5 mL of overnight cultures of BL21(DE3)pLys cells (Novagen) carrying pGEX plasmids were used to inoculate 500 mL Luria-Bertani medium containing 200 µg mL–1 carbenicillin and 25 µg mL–1 chloramphenicol and grown at 37°C to an optical density at 600 nm of 0.5. Next, protein synthesis was induced by the addition of solid isopropylthio-β-galactoside to final concentration of 1 mm, and cultures were grown for 4 h at 29°C. The harvested cells were suspended in 20 mL phosphate-buffered saline (PBS) and frozen in liquid nitrogen. After thawing on ice, the bacteria were lysed by sonication (eight times, 10-s burst; 5-s pause between bursts) and centrifuged at 18,000 rpm for 30 min at 4°C, and the supernatant was filtered through a 0.45-µm membrane. Protein purification was performed using Poly-Prep Chromatography columns (Bio-Rad 731-1550) containing 0.5 mL settled Glutathione-Sepharose 4B beads (Amersham Biosciences). Columns were first washed two times with 10 mL PBS before bacterial extract was passed through. After binding, columns were washed with 10 mL PBS, and bound proteins were eluted in 2.5 mL (10 × 0.25 mL) glutathione elution buffer (100 mm glutathione, 500 mm Tris-HCl, pH 8.0). Eluted protein was concentrated using Microcone centrifugal filter devices (Millipore) according to manufacturer’s protocol, and the protein content was determined by the method of Bradford. All EMSA reactions contained 100 ng poly-(dI-dC)-poly-(dI-dC; Amersham Pharmacia) and 0.5 ng of 32P end-labeled probe (approximately 108 cpm µg–1) in nuclear extraction buffer (Green et al., 1987) supplemented with 75 mm KCl. Probes used in labelings originated from combinations of annealed oligonucleotides (primers Oskn2D-Fw and Oskn2D-Rev; Supplemental Table S1) or gel-purified PCR reactions using primer sets 2SN1/2SN4, 2SN2/2SN5, and 2SN3/2SN6 (Supplemental Table S1) resulting in fragments A, B, and C, respectively, similar to the fragments used in yeast strains Y187::pOskn2-A to Y187::pOskn2-C. DNA-binding reactions were loaded onto 4% (w/v) acrylamide/bisacrylamide (37.5:1) gels with the current switched on (10 V cm–1), and one-half-strength Tris-borate/EDTA was used as running buffer. The gels were dried on Whatman DE81 paper on top of Whatman 3MM paper and autoradiographed.
Histological and Morphological Observations
Arabidopsis and rice tissues were stained for GUS expression by incubation of the samples overnight at 37°C in 100 mm sodium phosphate buffer, pH 7.5–7.7, 10 mm sodium EDTA, 0.5 mm potassium ferricyanide, 0.1% Triton X-100, and 2 mm X-Gluc. The reaction was stopped by adding 70% ethanol for direct observations using a Leica MZ12 stereomicroscope equipped with a Sony 3CCD camera (DKC 5000). For more detailed analysis, 70-µm fresh vibratome sections (made with a Leica VT1000S vibrating microtome) were incubated in prechilled (–20°C) 90% acetone for 1 h at –20°C. The samples were then stained overnight under the same reaction conditions as above. Phenotypes of Arabidopsis plants in tissue culture were observed and photographed using a Leica MZ12 stereomicroscope equipped with a Sony 3CCD camera (DKC 5000), and greenhouse-grown Arabidopsis and rice plants were photographed with a Nikon Coolpix 5000 camera.
Transient Expression Analysis in Onion Cells
Particle bombardment assays were performed on the abaxial side of adaxial epidermal peels from onion (Allium cepa) bulb scales as described earlier (Kuijt et al., 2004).
Transient Expression Using A. tumefaciens-Mediated Transformation of Rice Calli
Transformation of calli derived from ProOskn2:GUS seeds (Postma-Haarsma et al., 2002) was as previously described by Scarpella et al. (2000) using A. tumefaciens strain LBA 4404 harboring OsGRF3 and OsGRF10 constructs driven by the CaMV 35S promoter. Two independent transgenic ProOskn2:GUS lines were used for the transformation experiments, and a ProCaMV 35S:GFP overexpression construct was used in parallel transformations as control. In brief, embryonic calli were induced on scutella by growing germinating seeds on callus induction medium (Rueb et al., 1994) and transformed using a suspension of 3 to 5 x109 A. tumefaciens cells mL–1. After immersion in the A. tumefaciens suspension in liquid cocultivation medium (Basal R2 medium supplemented with 10 g L–1 Glc, 2.5 mg L–1 2,4-Dichlorophenoxyacetic acid, 100 μm acetosyringone, pH 5.2), calli were transferred to plates with cocultivation medium for 3 d. Following cocultivation, selection of transgenic calli was performed on Basal R2 medium supplemented with 30 g L–1 Suc and 2.5 mg L–1 2,4-Dichlorophenoxyacetic acid together with 100 mg L–1 hygromycin, 100 mg L–1 cefotaxime, and 100 mg L–1 vancomycin to prevent overgrowth by A. tumefaciens. All binary overexpression constructs were based on hygromycin phosphotransferase-intron vectors, and the presence of the intron in the hygromycin phosphotransferase gene renders the A. tumefaciens strain sensitive toward hygromycin. Transformed embryonic ProOskn2:GUS calli grown on cocultivation medium or selection media were collected at several time points after inoculation with A. tumefaciens, stained for GUS expression with X-Gluc as indicated in Supplemental Figure S10, and photographed. The photos were opened in ImageJ and split into blue, red, and green channels. Next, the GUS density was measured as the negative of the average pixel value in the red channel over single calli.
Rice Protoplast Isolation and Transient Expression Assays with GUS and Firefly Luciferase (FLUC) Reporters
Preparation of rice protoplasts, transfection with effector/reporter gene constructs, and protein extraction for GUS and FLUC assay measurements were according to Chen et al. (2006) or to the manufacturer’s instructions (Promega). One hundred rice seeds (cv IR58, indica rice) were grown in soil (28°C, 85% humidity, and 12-h dark and 12-h light) in a 10-cm-diameter pot for about 12 to 14 d (3 d in the dark, and the last 10–11 d under light/dark). Plantlets were collected and kept wrapped in aluminum foil on ice. Using razor blades, stems and leaves were cut into approximately 0.5 mm pieces, which were collected in 50-mL centrifuge tubes with 25 mL Enzyme Solution (containing 1.5% cellulase and 0.3% Macerozyme; see Chen et al., (2006)). Further handlings to prepare protoplasts and transformation with effector and reporter gene constructs were also according to Chen et al. (2006). Usually, about 107 cells were obtained from 100 rice seedlings, and 200 μL (1.5 to 2.5 × 106 cells mL–1) of suspended protoplasts were used for transfection with 4 μg effector plasmid, 5 μg effector plasmid, and 3 μg FLUC plasmid.
Plasmid FLUC is the firefly luciferase gene driven by the CaMV 35S promoter and Ω leader (Osnato et al., 2010) and was added as a control for transformation efficiency. Activity of the effectors was derived as GUS activity normalized on FLUC activity. The effector construct with OsGRF10 is the same transcriptional fusion with the CaMV 35S promoter as in the binary vector construct used to make stable overexpression plants but was cloned in the high copy vector SK+ Bluescript II (Stratagene). The effector ProUbi:BEIL1, in which the effector gene is driven by the constitutive maize Ubiquitine promoter, and the reporter gene constructs K373+305 bp are described in Osnato et al. (2010). ProUbi:BGRF1 was cloned by coupling the BGRF1 cDNA to the maize Ubiquitine promoter and the Nos terminator in vector pCRII-TOPO (Invitrogen).
Proteins were extracted as follows. After overnight incubation, the protoplasts were resuspended by gently pipetting up and down and transferred to 2-mL Eppendorf tubes, kept on ice, and centrifuged at 16,000 rpm for 10 min at 4°C. Next, the supernatant was removed and 100 μL Cell Culture Lysis Reagent (Promega) buffer was added to the protoplast pellet. Cells were shared by pipetting up and down, vortexed, and kept on ice. Next, the tubes were centrifuged at 16,000 rpm for 10 min at 4°C, and the supernatant with the protein extracts was transferred to 1.5-mL tubes and kept on ice until the FLUC assays were ready and then frozen immediately with liquid nitrogen and stored at –80°C.
Detection of GUS activity was essentially according to Jefferson (1987). Twenty microliters cell lysate was incubated at 37°C with 180 μL 4-methylumbelliferyl-β-D-glucuronide for the fluorimetrical assay. Ten-microliter samples of the reaction mixes were taken at 3 and 6 h, added to 190 μL 0.2 m Na2CO3 to stop the reaction, and measured for 4-methyl-umbelliferon fluorescence against a 4-methyl-umbelliferon calibration series in a Cytofluor 2350 fluorimeter (Millipore).
For detection of FLUC activity, 10 μL cell lysate was transferred in duplicate to white plates for luminescence assays containing 90 μL Cell Culture Lysis Reagent, and, next, 100 μL of Bright-Glo Luciferase Assay buffer (Promega) was added. The reactions were immediately read during 5 s in a Perkin Elmer Victor 3 with 10-min intervals.
SEM
Tissue was fixed overnight at room temperature in 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 m sodium cacodylate buffer (pH 7.3). Fixed tissue was dehydrated in a graded acetone series and critical point dried with CO2. Specimens were coated with a thin layer of gold using a Polaron E5100 sputter coater and were examined with a JEOL 6400 SEM with a tungsten filament operating at an accelerating voltage of 5 kV.
RT-PCR
Total RNA extraction from rice materials was performed using Trizol according to the manufacturer’s instructions (Invitrogen). Genomic DNA contaminants were removed from RNA samples by incubating with DNA-free (Ambion) at 37°C for 30 min according to the manufacturer’s instructions. First-strand cDNA was synthesized starting from 1 μg of total RNA using primer B26 with SuperScript III reverse transcriptase (Invitrogen) as described by the manufacturer. RT-PCR reactions were performed on single-strand cDNA using primer sets Oskn2-K4-5/Oskn2-K4-6 for Oskn2, OsGRF3-Fw2/OsGRF3-Rev2 for OsGRF3, and OsGRF10-Fw3/OsGRF10-Rev4 for OsGRF10 (Supplemental Table S1). The concentrations of the cDNA samples were equalized using actin primers OsActin U/OsActin L (Monna et al., 2002). Aliquots of the amplified products were separated on 1× Tris-borate/EDTA and 1% agarose gels, blotted on Hybond N+ filters, and hybridized to Oskn2, OsGRF3, and OsGRF10 probes. qPCR for Oskn2 mRNA in OsGRF3-Overexpression seedlings was done by using the SYBR Green I Master Mix and a Light Cycler 480 (Roche) with primers Oskn2-qFw and Oskn2-qRev. Normalization of Oskn2 expression was done by using primers OsUbi-Fw and OsUbi-Rev to amplify the ubiquitin gene (Supplemental Table S1).
qPCR Experiments
T2 and T3 rice plants were grown under 12-h light/12-h dark at 28°C in a growth chamber, together with the wild type (cv Taipei 309 for OsGRF3 overexpressors and cv Nipponbare for RNAi GRF lines). RNA was extracted from pools of five plants by using TRIzol (Invitrogen) and treated with DNaseI RNase free (Ambion), and 1 µg was retrotranscribed with oligo(dT) and SuperScript III (Invitrogen). The expression levels of genes of interest were monitored by qPCR using SYBR Green I Master Mix and Light Cycler 480 (Roche). Primers used were GRF3Fw, GRF3Rev, GRF4Fw, GRF4Rev, GRF5Fw, GRF5Rev, Oskn1Fw, Oskn1Rev, Oskn2-1803Fw, Oskn2-1805Rev, Oskn3Fw, and Oskn3Rev. The OsUbi gene was used to normalize data. Experiments were performed with three biological replicates, of which each was performed in three technical replicates.
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers FJ546693 for OsGRF3 and FJ546694 for OsGRF10, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Yeast one-hybrid assays with OsGRF3 and OsGRF10 expression constructs and rice promoter KNOX constructs.
Supplemental Figure S2. Interaction of OsGRF3 and OsGRF10 proteins with Oskn2 promoter subfragments B, C, and D and DNA-binding properties of the QLQ and WRC domains.
Supplemental Figure S3. Interaction of OsGRF3 and Arabidopsis GRF proteins with the Oskn2 promoter and subfragments.
Supplemental Figure S4. Expression pattern analysis of OsGRF10 in rice with promoter-GUS fusion constructs and nuclear localization of OsGRF10.
Supplemental Figure S5. Node numbers, internode lengths, and total plant lengths of OsGRF3 and OsGRF10 overexpression plants.
Supplemental Figure S6. Unrooted phylogenetic tree showing the predicted relationship between GRF proteins from rice, maize, barley, and Arabidopsis.
Supplemental Figure S7. Chromosomal positions of the GRF genes on the 12 rice chromosomes.
Supplemental Figure S8. Alignments of the full-length GRF protein sequences from rice (OsGRF), maize (ZmGRF), barley (BGRF1), and Arabidopsis (AtGRF).
Supplemental Figure S9. Intron-exon organization of rice and Arabidopsis GRF genes.
Supplemental Figure S10. Effects of OsGRF3 and OsGRF10 overexpression on ProOskn2:GUS in rice calli.
Supplemental Figure S11. Effects of ectopic expression of rice and Arabidopsis GRF genes on ProKNAT2:GUS expression and SAM development in Arabidopsis.
Supplemental Table S1. Overview of all oligonucleotides used in this paper.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for clones for AtGRF4 (clone M25D3) and AtGRF5 (clone M61B1), RIKEN for AtGRF6 clone (pda06900), Dr. Sarah Hake (Plant Gene Expression Center) for the Arabidopsis line ProBP:GUS, Dr. Jan Traas (Institut National de la Recherche Agronomique) for line ProKNAT2:GUS, Evelyn Wiese, Nicolas André, and Serdar Arslan for technical assistance, and Elly Schrijnemakers for plant care.
Glossary
- SAM
shoot apical meristem
- cDNA
complementary DNA
- AD
activation domain
- 3-AT
3-aminotriazole
- GST
glutathione S-transferase
- EMSA
electrophoretic mobility shift assay
- UTR
untranslated region
- RT
reverse transcription
- CaMV
Cauliflower mosaic virus
- SEM
scanning electron microscopy
- qPCR
quantitative PCR
- T-DNA
transfer DNA
- RNAi
RNA interference
- dpi
d post inoculation
- PBS
phosphate-buffered saline
Footnotes
This work was supported by the European Union FP5 project CerealGene Tags (QLGT–CT–2001–01453 to S.J.H.K., P.B.F.O., R.G., and C.C.J.‘t.H.), the European Union FP5 project TF–STRESS (QLK3–CT–2000–00328 to A.H.M.), the European Union FP6 INCO–MPC2 project CEDROME (INCO–CT–2005–015468 to A.A. and P.B.F.O.), and the CSC (grant no. 2007103056 to J.S.). Part of the work was carried out on the Rice Functional Genomics (REFUGE) platform, Montpellier, France, which is funded by the Agropolis Fondation (to E.G. and D.M.).
References
- Aida M, Ishida T, Tasaka M. (1999) Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126: 1563–1570 [DOI] [PubMed] [Google Scholar]
- Belles-Boix E, Hamant O, Witiak SM, Morin H, Traas J, Pautot V. (2006) KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18: 1900–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N, Hake S. (2009) The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S. (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi L, Bureau M, Simon R. (2007) Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell 19: 1795–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Grünewald M, Hobe M, Simon R. (2002) Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol 129: 565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. (2002) ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129: 1957–1965 [DOI] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Choi D, Kim JH, Kende H. (2004) Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol 45: 897–904 [DOI] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S. (1996) KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8: 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dean G, Casson S, Lindsey K. (2004) KNAT6 gene of Arabidopsis is expressed in roots and is required for correct lateral root formation. Plant Mol Biol 54: 71–84 [DOI] [PubMed] [Google Scholar]
- Dockx J, Quaedvlieg N, Keultjes G, Kock P, Weisbeek P, Smeekens S. (1995) The homeobox gene ATK1 of Arabidopsis thaliana is expressed in the shoot apex of the seedling and in flowers and inflorescence stems of mature plants. Plant Mol Biol 28: 723–737 [DOI] [PubMed] [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. (1996) The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J 10: 967–979 [DOI] [PubMed] [Google Scholar]
- Freeling M, Hake S. (1985) Developmental genetics of mutants that specify knotted leaves in maize. Genetics 111: 617–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ, Kay SA, Chua NH. (1987) Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J 6: 2543–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem 192: 262–267 [DOI] [PubMed] [Google Scholar]
- Guo M, Thomas J, Collins G, Timmermans MC. (2008) Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CM, Kim GT, Kim BC, Jun JH, Soh MS, Ueno Y, Machida Y, Tsukaya H, Nam HG. (2003) The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130: 161–172 [DOI] [PubMed] [Google Scholar]
- Hake S. (1996) Shootmeristemless ties the knot. Trends Plant Sci 1: 75–76 [Google Scholar]
- Hake S, Char BR, Chuck G, Foster T, Long J, Jackson D. (1995) Homeobox genes in the functioning of plant meristems. Philos Trans R Soc Lond B Biol Sci 350: 45–51 [DOI] [PubMed] [Google Scholar]
- Hake S, Ori N. (2002) Plant morphogenesis and KNOX genes. Nat Genet 31: 121–122 [DOI] [PubMed] [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Harrison CJ, Langdale JA. (2006) A step by step guide to phylogeny reconstruction. Plant J 45: 561–572 [DOI] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. (2002) The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12: 1557–1565 [DOI] [PubMed] [Google Scholar]
- Hibara K, Takada S, Tasaka M. (2002) The Arabidopsis CUC1 gene induces the KNOX genes. Plant Cell Physiol 43: S137 [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Ito M, Sato Y, Matsuoka M. (2002) Involvement of homeobox genes in early body plan of monocot. Int Rev Cytol 218: 1–35 [DOI] [PubMed] [Google Scholar]
- Jefferson RA. (1987) Assaying chimaeric genes in plants: The GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Kim JH, Choi D, Kende H. (2003a) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J 36: 94–104 [DOI] [PubMed] [Google Scholar]
- Kim JH, Kende H. (2004) A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA 101: 13374–13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee BH. (2006) GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J Plant Biol 49: 463–468 [Google Scholar]
- Kim JS, Mizoi J, Kidokoro S, Maruyama K, Nakajima J, Nakashima K, Mitsuda N, Takiguchi Y, Ohme-Takagi M, Kondou Y, et al. (2012) Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 24: 3393–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Pham T, Hamidi A, McCormick S, Kuzoff RK, Sinha N. (2003b) Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX genes in generating compound leaves. Development 130: 4405–4415 [DOI] [PubMed] [Google Scholar]
- Kuijt SJH, Lamers GEM, Rueb S, Scarpella E, Ouwerkerk PBF, Spaink HP, Meijer AH. (2004) Different subcellular localization and trafficking properties of KNOX class 1 homeodomain proteins from rice. Plant Mol Biol 55: 781–796 [DOI] [PubMed] [Google Scholar]
- Kumar R, Kushalappa K, Godt D, Pidkowich MS, Pastorelli S, Hepworth SR, Haughn GW. (2007) The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell 19: 2719–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran MK, Bowman JL, Sundaresan V. (2002) YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14: 2761–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba S, Fukumoto M, Honda C, Yamaguchi I, Sakamoto T, Kano-Murakami Y. (1998) Decreased GA1 content caused by the overexpression of OSH1 is accompanied by suppression of GA 20-oxidase gene expression. Plant Physiol 117: 1179–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS. (2003) The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15: 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Ichikawa H, Saito A, Tada Y, Fujimura T, Kano-Murakami Y. (1993) Expression of a rice homeobox gene causes altered morphology of transgenic plants. Plant Cell 5: 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AH, Scarpella E, van Dijk EL, Qin L, Taal AJ, Rueb S, Harrington SE, McCouch SR, Schilperoort RA, Hoge JHC. (1997) Transcriptional repression by Oshox1, a novel homeo domain leucine zipper protein from rice. Plant J 11: 263–276 [DOI] [PubMed] [Google Scholar]
- Meijer AH, Ouwerkerk PBF, Hoge JHC. (1998) Vectors for transcription factor cloning and target site identification by means of genetic selection in yeast. Yeast 14: 1407–1415 [DOI] [PubMed] [Google Scholar]
- Meijer AH, Schouten J, Ouwerkerk PBF, Hoge JHC (2000) Yeast as versatile tool in transcription factor research. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands pp 1–28 [Google Scholar]
- Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y. (2002) Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res 9: 11–17 [DOI] [PubMed] [Google Scholar]
- Müller KJ, Romano N, Gerstner O, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W. (1995) The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature 374: 727–730 [DOI] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. (2000) Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Osnato M, Stile MR, Wang Y, Meynard D, Curiale S, Guiderdoni E, Liu Y, Horner DS, Ouwerkerk PBF, Pozzi C, et al. (2010) Cross talk between the KNOX and ethylene pathways is mediated by intron-binding transcription factors in barley. Plant Physiol 154: 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwerkerk PBF, Meijer AH (2001) Yeast one-hybrid screening for DNA-protein interactions. In FM Ausubel, R Brent, RE Kingston, DD Moore, JG Seidman, JA Smith, K Struhl, eds, Current Protocols in Molecular Biology. John Wiley and Sons, Inc., New York, pp 1–22. [DOI] [PubMed] [Google Scholar]
- Ouwerkerk PBF, Meijer AH. (2011) Yeast one-hybrid screens for detection of transcription factor DNA interactions. Methods Mol Biol 678: 211–227 [DOI] [PubMed] [Google Scholar]
- Postma-Haarsma AD, Rueb S, Scarpella E, den Besten W, Hoge JHC, Meijer AH. (2002) Developmental regulation and downstream effects of the knox class homeobox genes Oskn2 and Oskn3 from rice. Plant Mol Biol 48: 423–441 [DOI] [PubMed] [Google Scholar]
- Postma-Haarsma AD, Verwoert IIGS, Stronk OP, Koster J, Lamers GEM, Hoge JHC, Meijer AH. (1999) Characterization of the KNOX class homeobox genes Oskn2 and Oskn3 identified in a collection of cDNA libraries covering the early stages of rice embryogenesis. Plant Mol Biol 39: 257–271 [DOI] [PubMed] [Google Scholar]
- Reiser L, Sánchez-Baracaldo P, Hake S. (2000) Knots in the family tree: evolutionary relationships and functions of knox homeobox genes. Plant Mol Biol 42: 151–166 [PubMed] [Google Scholar]
- Rosin FM, Hart JK, Horner HT, Davies PJ, Hannapel DJ. (2003) Overexpression of a knotted-like homeobox gene of potato alters vegetative development by decreasing gibberellin accumulation. Plant Physiol 132: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueb S, Leneman RA, Schilperoort RA, Hensgens LAM. (1994) Efficient plant regeneration through somatic embryogenesis from callus induced on mature rice embryos (Oryza sativa L.). Plant Cell Tissue Organ Cult 36: 259–264 [Google Scholar]
- Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M. (2001) KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C, Meynard D, van Boxtel J, Gay C, Bès M, Brizard JP, Larmande P, Ortega D, Raynal M, Portefaix M, et al. (2003) Highly efficient production and characterization of T-DNA plants for rice ( Oryza sativa L.) functional genomics. Theor Appl Genet 106: 1396–1408 [DOI] [PubMed] [Google Scholar]
- Sato Y, Sentoku N, Nagato Y, Matsuoka M. (1998) Isolation and characterization of a rice homebox gene, OSH15. Plant Mol Biol 38: 983–998 [DOI] [PubMed] [Google Scholar]
- Scarpella E, Rueb S, Boot KJ, Hoge JHC, Meijer AH. (2000) A role for the rice homeobox gene Oshox1 in provascular cell fate commitment. Development 127: 3655–3669 [DOI] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. (2001) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771–1783 [DOI] [PubMed] [Google Scholar]
- Sentoku N, Sato Y, Kurata N, Ito Y, Kitano H, Matsuoka M. (1999) Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11: 1651–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh IS, He JR, Calderwood S, Hasday JD. (2002) A high affinity HSF-1 binding site in the 5′-untranslated region of the murine tumor necrosis factor-α gene is a transcriptional repressor. J Biol Chem 277: 4981–4988 [DOI] [PubMed] [Google Scholar]
- Swofford DL (2002) PAUP* Phylogenetic Analysis Using Parsimony (and Other Methods). Sinauer Associates, Sunderland, MA. [Google Scholar]
- Takada S, Hibara K, Ishida T, Tasaka M. (2001) The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128: 1127–1135 [DOI] [PubMed] [Google Scholar]
- Tanaka-Ueguchi M, Itoh H, Oyama N, Koshioka M, Matsuoka M. (1998) Over-expression of a tobacco homeobox gene, NTH15, decreases the expression of a gibberellin biosynthetic gene encoding GA 20-oxidase. Plant J 15: 391–400 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MC, Hudson A, Becraft PW, Nelson T. (1999) ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284: 151–153 [DOI] [PubMed] [Google Scholar]
- Tsiantis M, Schneeberger R, Golz JF, Freeling M, Langdale JA. (1999) The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284: 154–156 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Ito Y, Sato Y, Kurata N. (2011) Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23: 4368–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Kimura S, Koenig D, Sinha N. (2010) Coordination of leaf development via regulation of KNOX1 genes. J Plant Res 123: 7–14 [DOI] [PubMed] [Google Scholar]
- van der Knaap E, Kim JH, Kende H. (2000) A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol 122: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, Martienssen R, Selvaraj G, Datla R. (2002) The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc Natl Acad Sci USA 99: 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites R, Selvadurai HR, Oliver IR, Hudson A. (1998) The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93: 779–789 [DOI] [PubMed] [Google Scholar]
- Zhang DF, Li B, Jia GQ, Zhang TF, Dai JR, Li JS, Wang SC. (2008) Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in Maize (Zea mays, L.). Plant Sci 175: 809–817 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.