A tomato transcriptional repressor regulates development and salinity response in Arabidopsis and tomato plants.
Abstract
The zinc finger superfamily includes transcription factors that regulate multiple aspects of plant development and were recently shown to regulate abiotic stress tolerance. Cultivated tomato (Solanum lycopersicum Zinc Finger2 [SIZF2]) is a cysteine-2/histidine-2-type zinc finger transcription factor bearing an ERF-associated amphiphilic repression domain and binding to the ACGTCAGTG sequence containing two AGT core motifs. SlZF2 is ubiquitously expressed during plant development, and is rapidly induced by sodium chloride, drought, and potassium chloride treatments. Its ectopic expression in Arabidopsis (Arabidopsis thaliana) and tomato impaired development and influenced leaf and flower shape, while causing a general stress visible by anthocyanin and malonyldialdehyde accumulation. SlZF2 enhanced salt sensitivity in Arabidopsis, whereas SlZF2 delayed senescence and improved tomato salt tolerance, particularly by maintaining photosynthesis and increasing polyamine biosynthesis, in salt-treated hydroponic cultures (125 mm sodium chloride, 20 d). SlZF2 may be involved in abscisic acid (ABA) biosynthesis/signaling, because SlZF2 is rapidly induced by ABA treatment and 35S::SlZF2 tomatoes accumulate more ABA than wild-type plants. Transcriptome analysis of 35S::SlZF2 revealed that SlZF2 both increased and reduced expression of a comparable number of genes involved in various physiological processes such as photosynthesis, polyamine biosynthesis, and hormone (notably ABA) biosynthesis/signaling. Involvement of these different metabolic pathways in salt stress tolerance is discussed.
Osmotic stresses such as salinity or drought strongly impair plant development and growth, thus compromising yield and causing important economic losses for essential crops such as cultivated tomato (Solanum lycopersicum). Upon stress perception, plants rapidly respond by modulating expression of key genes encoding transcription factors. Transcription factors subsequently target a plethora of structural genes in order to orchestrate biochemical and physiological modifications critical for tolerance to stress and plant growth adaptation. Widespread zinc finger transcription factors regulate several plant developmental and cellular processes, including RNA binding, flowering time, plant defense, and osmotic stress response, among others (Putterill et al., 1995; Kim et al., 2004; Kielbowicz-Matuk, 2012). This large family encompasses 176 members in Arabidopsis (Arabidopsis thaliana), 189 members in rice (Oryza sativa; Englbrecht et al., 2004; Agarwal et al., 2007; Ciftci-Yilmaz and Mittler, 2008), and at least 87 in papaya (Carica papaya; Jiang and Pan, 2012). Cys-2/His-2-type (C2H2) zinc finger transcription factors are an abundant type of zinc finger proteins in plants. They bear the CX2-4CX3FX5LX2HX3-5H signature of about 30 amino acids, including two pairs of Cys and His (C and H, respectively), repeated one to four (and more rarely five) times within a protein. This short motif defines two β sheets and a single α helix, which when folded, permit the C and H residues to bind tetrahedrally to a zinc ion (for review, see Pabo et al., 2001). The α helices of both fingers allow specific binding to an AGT core motif (Kubo et al., 1998; for review, see Pabo et al., 2001).
Typically, Arabidopsis C2H2-type zinc fingers involved in stress response belong to the C1-2i subclass harboring two zinc finger domains, including at least 20 members in Arabidopsis, and are classified as active repressors (Englbrecht et al., 2004; for review, see Ciftci-Yilmaz and Mittler, 2008; Kielbowicz-Matuk, 2012). These repressors are thought to limit stress-activated gene expression under optimal growth conditions, and to moderate subsequent adjustments to stress to avoid cellular burst (for review, see Kazan, 2006). In addition, repressors may inhibit plant growth to limit photosynthesis and energy necessary for plant development, and thus efficiently redirect energy for stress accommodation (Seki et al., 2002). Stress-regulated zinc fingers classically carry the ERF-associated amphiphilic repression (EAR) motif (L/F)DLN(L/F)xP in their C-terminal end (Ohta et al., 2001), and represent 15.4% of the identified C2H2 zinc finger proteins in papaya for instance (Jiang and Pan, 2012). However, the EAR motif is directly involved in plant salinity tolerance and is not associated with growth suppression, because deletion of this motif renders Arabidopsis transgenic plants more sensitive to salinity (Ciftci-Yilmaz et al., 2007).
In Arabidopsis, key C2H2 zinc finger candidates sharing the EAR motif involved in salinity and additional osmotic stress responses have been characterized, such as ZAT7, salt tolerance zinc finger (STZ)/ZAT10, ZAT12, and ARABIDOPSIS ZINC FINGER PROTEINS1 (AZF1) and AZF2. Overexpression of these zinc finger repressors increases tolerance to the array of abiotic stresses by which they are induced. ZAT12 responds to wounding and biotic and environmental stresses (e.g. cold, heat, salinity, drought, hydrogen peroxide [H2O2], UV light), in addition to auxin and abscisic acid (ABA) treatments (Davletova et al., 2005; Mittler et al., 2006). ZAT12 loss-of-function plants are more sensitive to salinity and sorbitol osmotic stresses, whereas ZAT12 overexpressors are more tolerant to the same stresses. ZAT12 is involved in several signaling pathways, probably in response to H2O2 accumulation in cells, and regulates expression of several genes implicated in light and oxidative stress responses (Davletova et al., 2005). STZ/ZAT10 is strongly induced by salinity, drought, and cold, in addition to ABA and ethephon (Sakamoto et al., 2004). Like ZAT12, ZAT10 also contributes to the oxidative stress response (Mittler et al., 2006). Interestingly, both ZAT10 overexpressors and loss-of-function lines showed enhanced osmotic stresses tolerance. A dual function of ZAT10 for both reactive oxygen species (ROS) response activation, and thus stress tolerance, in parallel to repression of a different network of defense mechanisms, has been proposed (Mittler et al., 2006). AZF1 and AZF2 are induced by salt, drought, ABA, and ethephon (Sakamoto et al., 2000, 2004). In contrast with results reported for ZAT10 or ZAT12, overexpression of AZF1 and AZF2 under control of the RD29A stress-responsive promoter hampered plant growth and viability. These transgenic plants showed enhanced salt sensitivity by suppressing expression of many osmotic stress-regulated genes, as well as those inhibited by ABA treatment (Kodaira et al., 2011). In general, only few zinc finger proteins regulating osmotic stresses have been characterized in other plant species, specifically those holding the EAR motif. For instance, the chrysanthemum (Chrysanthemum grandiflorum) C2H2 ZINC FINGER PROTEIN1 (CgZFP1) is up-regulated by salinity and drought, and its heterologous expression in Arabidopsis enhanced plant tolerance to both stresses (Gao et al., 2012). The potato (Solanum tuberosum) ZINC FINGER PROTEIN1 (StZFP1) is induced by drought and salinity, and its ectopic expression in tobacco (Nicotiana tabacum) enhances tolerance to salinity (Tian et al., 2010).
In wild or cultivated tomatoes, no candidates with the EAR motif have yet been characterized, and only a single zinc finger involved in abiotic stress regulation has been identified thus far. Solanum lycopersicum COLD ZINC FINGER PROTEIN1 (SICZFP1) is rapidly induced by cold, salinity, and drought treatments (Zhang et al., 2011). Under standard growth conditions, Arabidopsis and rice plants constitutively overexpressing SlCZFP1 showed no phenotypic differences compared with wild-type plants. However, the transgenic plants showed a specific enhanced tolerance to chilling and freezing, by altering expression of some cold-regulated genes (Zhang et al., 2011).
Our study reports the identification and functional characterization of the first tomato repressor-type zinc finger, SlZF2. SlZF2 encodes a ubiquitously expressed transcription factor, harboring an EAR motif in its C-terminal end. SlZF2 is transiently induced by osmotic stresses, such as sodium chloride (NaCl) or drought, and seems to be involved in ABA biosynthesis/signaling. Ectopic expression of SlZF2 in Arabidopsis and tomato impaired seedling development, and led to a general state of stress in plants, as revealed by increased malonyldialdehyde and polyamine (PA) levels. In Arabidopsis, SlZF2 increased plant sensitivity to salinity but inserting the same construct in tomato generally improved tolerance to salinity, mainly by maintaining photosynthesis. Transcriptomic analysis of SlZF2 transgenic lines revealed that a similar number of genes were up- or down-regulated, indicating that SlZF2 is putatively involved in the regulation of several biosynthesis pathways (PAs, carotenoids, alkaloids, etc) and developmental processes (photosynthesis, nucleic acid processing, etc).
RESULTS
SlZF2 Encodes a Ubiquitously Expressed Tomato Zinc Finger Transcription Factor
A suppression subtractive hybridization coupled to a microarray approach involving both salt-tolerant LA2711 and salt-sensitive ZS292 cultivated tomato genotypes subjected to short-term salt stress in roots was performed (up to 6 h after treatment with 150 mm NaCl; Ouyang et al., 2007). This approach allowed identification of transcription factors putatively involved in salt tolerance. Among up-regulated genes, a zinc finger transcription factor (initial accession DY523398) encoding gene was selected for further functional characterization, and was named SlZF2 (accession no. HQ738351). Access to the tomato genome database (http://www.solgenomics.net; Tomato Genome Consortium, 2012) allowed the design of primers amplifying a 1,086-bp coding sequence, using tomato salt-stressed root cDNA (2 h, 150 mm NaCl) as a template. Identical cloning of SlZF2 genomic DNA sequence indicated that this gene is intronless. A 933-bp open reading frame was identified, and predicted to encode a 310-amino acid protein of 32.9 kD.
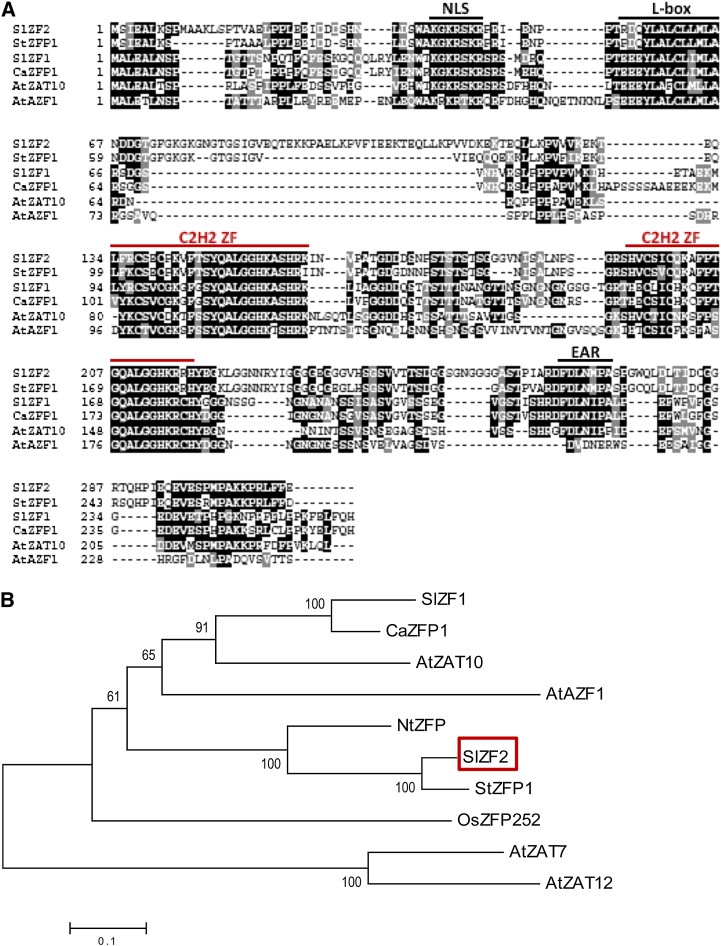
SlZF2 harbors two dispersed C2H2 zinc finger domains of 26 and 23 amino acids, respectively, located between residues 134 to 159 and 195 to 217, thus separated from one another by 35 residues (Fig. 1A). α-Helices of both zinc fingers bear the plant-specific QALGGH motif, which is essential for DNA binding (Takatsuji and Matsumoto, 1996; Kubo et al., 1998). SlZF2 displays additional features regarding the flanking residues of the QALGGH motif, which allow its classification into the group G1 of zinc finger proteins (Gourcilleau et al., 2011). These additional signatures include a K conserved residue in the first finger QALGGHK (amino acid 154) and K/Y residues in the second finger QALGGHKRRHY (amino acids 214 and 218), increasing DNA recognition specificity. Likewise, SlZF2 exhibits a putative nuclear localization site in its N-terminal extremity (KGKRSKR) known as B box, in addition to a hydrophobic Leu-rich box (Fig. 1A) putatively involved in protein-protein dimerization or protein conformation stability (Sakamoto et al., 2000). Alignments of SlZF2 with some orthologs belonging to diverse species, including potato StZFP1, tomato SlZF1, pepper (Capsicum annuum) ZINC FINGER PROTEIN1 (CaZFP1), and Arabidopsis AtZAT10 and AtAZF1 indicate a high conservation of these motifs. In addition, SlZF2 encompasses a FDLNMPA motif (amino acids 266 to 272) in its C-terminal end, carrying the DLN core sequence of the EAR (Fig. 1A). Finally, a Gly-rich domain (GX)6GXXG, specific to SlZF2, has been observed next to the L box (amino acids 72 to 85), and a second Gly-rich domain is also present between the second zinc finger motif and the EAR (Fig. 1A).
Figure 1.
Identification of SlZF2. A, Amino acid sequence comparison of SlZF2 and StZFP1, SlZF1, CaZFP1, AtZAT10, and AtAZF1. Identical residues are shown in black and conserved residues are in dark gray. The C2H2 domains, as well as the nuclear localization site (B box), L box, and EAR motif, are labeled. B, Phylogenetic relationships between SlZF2 (ADZ15317) and zinc finger proteins from different species: tomato SlZF1 (ACG50000), Arabidopsis ZAT10 (Q96289), AZF1 (BAA85108), ZAT7 (Q42453), ZAT12 (Q42410), tobacco NtZFP (AAC06243), potato StZFP1 (ABK78777), pepper CaZFP1 (AAQ10954), and rice OsZFP252 (AAO46041).
A phylogenetic analysis including SlZF2 and nine additional orthologs belonging to different species was constructed by means of the neighbor-joining method using full-length amino acid sequences (Fig. 1B). This analysis revealed that SlZF2 clusters with StZFP1 (76% identity), which is induced by salt, dehydration, and ABA, and increases salt tolerance when expressed in Arabidopsis (Tian et al., 2010), and with NtZFT1 (62% identity), which is induced by osmotic stress such as mannitol and is involved in spermine signaling pathway in tobacco (Mitsuya et al., 2007). Apart from the Solanaceae family, SlZF2 also clusters with other transcription factors such as OsZFP252, which enhances salinity and drought tolerance in rice (Xu et al., 2008). Together, these data strongly suggest that SlZF2 could also be involved in osmotic stresses signaling in tomato.
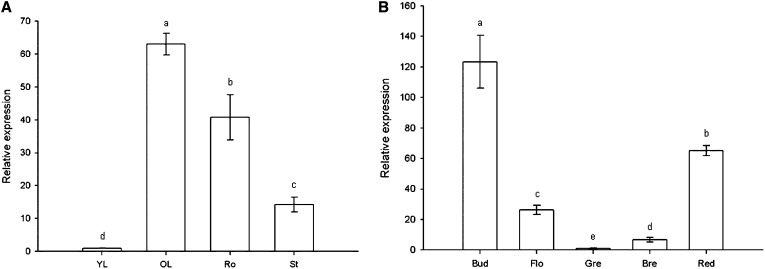
During plant development, SlZF2 is highly expressed both in vegetative (aerial and subterranean) and reproductive organs. Indeed, real-time reverse transcription PCR analysis of the SlZF2 spatiotemporal expression profile in vegetative tissues showed the highest transcript accumulation in old tomato leaves, followed by the roots and the stem, respectively, whereas SlZF2 was barely detected in young leaves (Fig. 2A). In the reproductive organs, SlZF2 transcript abundance was the highest in buds, and then declined during flower maturation and early stages of fruit development (green and breaker stages), before increasing again during fruit ripening (red stage; Fig. 2B).
Figure 2.
qRT-PCR analysis of the SlZF2 spatiotemporal expression profile during tomato plant development in vegetative organs (A) and reproductive organs (B). Actin and GAPDH were used as internal controls. Data represent means and sd of three replicates. Bre, Breaker stage; Flo, flower; Gre, green stage; OL, old leaf; Red, red stage of tomato development; Ro, root; St, stem; YL, young leaf. Letters indicate values of SlZF2 expression that differ significantly between tomato organs according to the Student-Newman-Keuls test at P < 0.05.
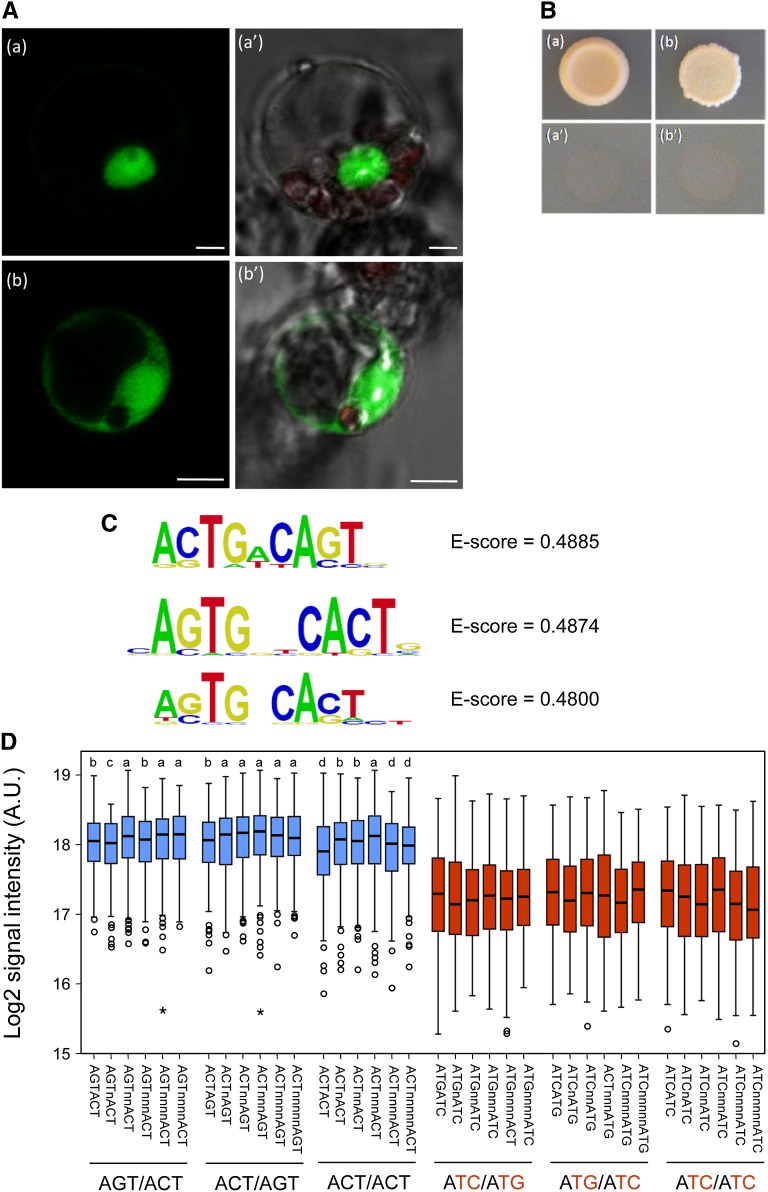
To confirm its putative function as a transcription factor, three SlZF2 transcriptional properties were assessed. First, two putative nuclear localization sites of four amino residues (positions 212/213 and 302) in addition to the B box were identified (PSORT, http://psort.ims.u-tokyo.ac.jp/; Nakai and Horton, 1999), with a probability of 0.7 for SlZF2 to be a nuclear protein. To verify SlZF2 subcellular targeting, the whole protein coding sequence was fused in its N-terminal part to the yellow fluorescent protein (YFP) under control of the Cauliflower mosaic virus (CaMV) 35S promoter. The construct was used to transiently transform tomato protoplasts (Fig. 3A). In accordance with the predicted function of a transcription factor, the YFP-SlZF2 fusion protein was exclusively detected in the nucleus (Fig. 3, Aa and Aa′), in contrast with the free YFP protein, which was distributed in both the cytosol and the nucleus (Fig. 3, Ab and Ab′).
Figure 3.
SlZF2 transcriptional properties. A, Transient expression of YFP-SlZF2 fusion protein in tomato leaf protoplasts: YFP-SlZF2 (Aa and Aa′), YFP control fluorescence (Ab and Ab′), and bright field/chlorophyll/YFP fluorescence respectively. B, SlZF2 transactivation ability. The SlZF2 coding region was fused to GAL4 DNA binding domain (DBD) into the pGBKT7 vector carrying the nutritional marker TRYPTOPHAN1 as the reporter gene. Yeasts were transformed with pGBKT7 empty vector (Ba and Ba′) or with the GAL4-DBD-SlZF2 construct (Bb and Bb′). Both constructs were transformed into the yeast strain Y8930 harboring the ß-galactosidase L, ADE2, and HIS3 reporter genes. Yeasts were separately grown on synthetic dropout medium lacking either Trp (Ba and Bb), or A and His (Ba′ and Bb′). C, Position weight matrix representation of the three top-scoring 8 mers obtained in a seed-and-wobble algorithm. D, Box plot representation of signal intensities of the probes containing the elements indicated. Different combinations of the AGT modules in bipartite motifs are shown in blue, whereas their corresponding mutant versions are shown in red. Letters above the boxes represent different groups of statistical significance, relative to the motif ACTnnnAGT with highest median intensity, as follows: a, P > 0.05 (no significant differences); b, P < 0.01; c, P < 0.001; and d, P < 0.0001 (Wilcoxon exact test). All of the mutant derivatives grouped together at the same significance group (P < 2.2e-16). The number of probes containing the elements indicated ranges from 243 to 303.
Next, the putative ability of SlZF2 to activate downstream target gene expression was investigated in yeast (Saccharomyces cerevisiae). SlZF2 cDNA was fused in frame to the yeast GALACTOSE4 (GAL4) DNA binding domain coding sequence and the construct used to transform the Y8930 yeast strain carrying the ADENINE2 (ADE2) and HISTIDINE3 (HIS3) reporter genes under the GAL4 promoter, used as nutritional markers for A and His auxotrophy, respectively. SlZF2 was not able to activate ADE2 and HIS3 transcription (Fig. 3B), indicating either that SlZF2 has no transcriptional activation properties, or that it could be a transcriptional repressor as supported by the presence of the EAR repression motif in its C-terminal end.
Finally, SlZF2 DNA binding capability was assessed in vitro using a protein binding microarray assay (Godoy et al., 2011). The SlZF2 coding sequence was fused in frame to the maltose-binding protein (MBP), and fusion protein expressed into Escherichia coli, and were then hybridized to the Protein-Binding-Microarray11 (PBM11; Godoy et al., 2011). The top-scoring motif corresponded to the sequence ACTGACAGT, which contains two AGT core motifs (Fig. 3C). Close inspection of top-scoring motifs revealed that SlZF2 binds with high affinity to several DNA sequences containing bipartite AGT core motifs, with variable spacing between the two modules (Fig. 3C). DNA binding affinity of SlZF2 to DNA sequences containing different combinations of AGT modules, spaced between 0 and 5 nucleotides, was evaluated (Fig. 3D). We measured signal intensities of the probes containing either the elements under study or, as a control, of probes containing mutant bipartite elements which core AGT motifs were changed to ATG. As expected, the signal intensity corresponding to bipartite sequences was significantly higher than their corresponding mutant versions (P < 2.2e-16, Wilcoxon exact test; Fig. 3D). When we focused on bipartite elements, we obtained the higher median signal intensity for the motif ACTnnnAGT, which is consistent with the primary motif obtained for SlZNF2. However, no significant differences between this element and some others containing different combinations of the AGT module were obtained (Fig. 3D). These results suggest that SlZF2 binds with high affinity to a broad range of DNA sequences containing different combinations of two AGT core modules, spaced at 1 to 5 nucleotides. In spite of this, a slight preference for ACT/AGT motifs, particularly spaced by 3 nucleotides, was observed (Fig. 3D).
SlZF2 Is Induced by Salinity and Additional Osmotic Stresses
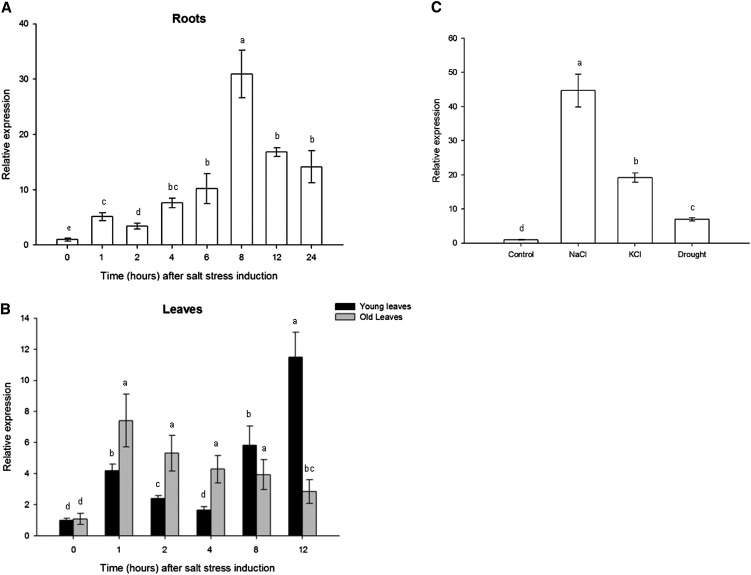
Analysis of SlZF2 expression patterns under a short duration of salt (150 mm NaCl) stress was conducted by real-time reverse transcription-PCR, separately on roots and leaves (young and old) of tomato plants. In roots, SlZF2 transcripts gradually increased up to 6 h after onset of treatment (approximately 10 times more transcripts than control roots), before reaching their maximum 8 h after stress began (approximately 32 times more transcripts than control roots; Fig. 4A). SlZF2 expression then started to decrease until 24 h after stress, with SlZF2 transcript levels exceeding those in control roots. In young leaves, SlZF2 transcripts were four times more abundant than untreated leaves as early as 1 h after salt stress application, before decreasing in the following hours. Eight hours after salt stress onset, SlZF2 transcripts started to increase again, before peaking (approximately 11 times more transcripts) 12 h after salinity was applied (Fig. 4B). In old leaves, the SlZF2 expression profile is different. Indeed, SlZF2 was rapidly induced by salt, with approximately seven times more transcripts 1 h after commencement of stress than in untreated leaves. SlZF2 expression then steadily declined (Fig. 4B).
Figure 4.
A and B, qRT-PCR analysis of SlZF2 expression pattern in response to salinity (150 mm NaCl) in roots (A) or leaves (B; young and old leaves for 50-d-old plants). C, Expression of SlZF2 in roots of young tomato plants (3 weeks old) submitted to 150 mm NaCl, 150 mm KCl, or drought. Actin and GAPDH were used as internal controls. Data represent means and sd of three replicates. Letters indicate values of SlZF2 expression that differ significantly between treatments according to the Student-Newman-Keuls test at P < 0.05.
To analyze SlZF2 response to additional stresses, potassium chloride (KCl; 150 mM) and air-drying treatments were performed on tomato roots (3 weeks old) for 3 h, along with salinity treatment (150 mm NaCl; Fig. 4C). Quantitative PCR analysis of SlZF2 expression indicated that its transcripts were the most elevated after NaCl treatment (44.7 times more transcripts than control roots), then by KCl (19.2 times more transcripts than control roots), and finally by drought (6.9 times more transcripts than control roots).
Overexpression of SlZF2 in Arabidopsis Impairs Development and Increases Salt Sensitivity
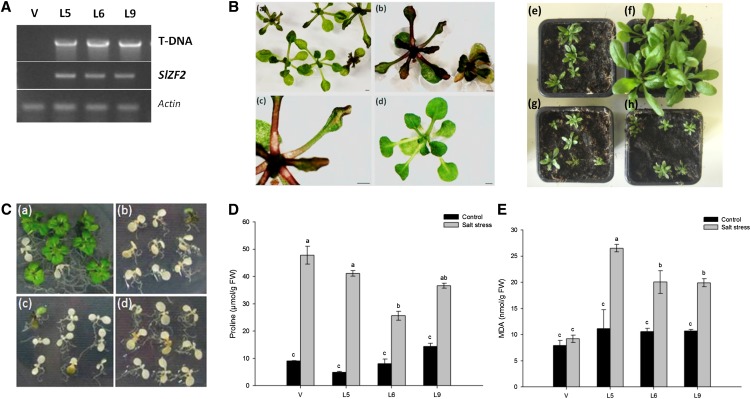
To examine its function in planta, SlZF2 was constitutively expressed in Arabidopsis under control of the CaMV 35S promoter, and analysis conducted on three F3 homozygous independent lines (L5, L6, and L9) and compared with those of a control line (V) transformed with an empty binary vector (Fig. 5A). Under normal growth conditions either on one-half-strength Murashige and Skoog (MS) medium or in soil, transgenic seedlings were strongly impaired in their development compared with control plants, either in their shoot or in their very limited root system, with leaves curling at their extremity (Fig. 5B). In addition, SlZF2-transformed plants visibly accumulated more anthocyanin pigments than controls. To estimate salt sensitivity of the three Arabidopsis 35S::SlZF2 lines, seeds were geminated on salt-enriched (80 mm NaCl) one-half-strength MS medium; 5 d after germination, seeds were transferred to 150 mm NaCl medium. The survival rate was scored 1 week after transfer compared with that of V plants. With a null rate of survival, 35S::SlZF2 Arabidopsis plants appeared much more sensitive to salinity than controls (Fig. 5C).
Figure 5.
Overexpression of SlZF2 enhances salinity sensitivity in Arabidopsis. A, Transfer DNA (T-DNA) insertion in the Arabidopsis genome and expression of SlZF2 in L5, L6, and L9 transgenic lines and a control line transformed with empty vector (V) were analyzed by PCR. B, Phenotype of transgenic Arabidopsis plants grown on one-half-strength MS medium and then transferred to soil, and ectopically expressing SlZF2 (Ba, Bc, Be, Bg, and Bh) compared with control plants (Bd and Bf). C, Phenotype of Arabidopsis V (Ca), L5 (Cb), L6 (Cc), and L9 (Cd) lines 5 d after transfer on 125 mM NaCl medium. D, Pro accumulation in control (V) and SlZF2 transgenic lines grown for 5 d on 80 mm NaCl. E, MDA accumulation in control (V) and SlZF2 transgenic lines grown 5 d on 80 mm NaCl. Data represent means and se of three replicates. Letters indicate values that differ significantly between SlZF2-transgenic Arabidopsis and control V lines according to the Student-Newman-Keuls test at P < 0.05.
Effects of salt stress on plant oxidative status were evaluated by measurements of some osmoprotective and oxidative molecule content (Pro and malonyldialdehyde [MDA], respectively). Analyses were conducted on Arabidopsis seedlings grown for 5 d on 80 mm NaCl. Under control growth conditions, the L5, L6, L9, and V lines showed Pro concentrations ranging between 4.9 and 14.4 µmol g−1 fresh weight (FW; Fig. 5D). In the presence of salt, Pro content was less abundant in SlZF2-Arabidopsis than in control plants, with line L6 significantly (P < 0.05) accumulating less Pro (25.6 µmol g−1 FW) than lines L5 (41.2 µmol g−1 FW), L9 (36.5 µmol g−1 FW), or V (47.8 µmol g−1 FW).
On one-half-strength MS medium, SlZF2-transgenic Arabidopsis and the V line accumulated approximately 10 nmol g−1 FW MDA, although the former lines showed, on average, higher MDA content (Fig. 5E). Five days after growth on 80 mm NaCl, lines L5, L6, and L9 accumulated >20 nmol g−1 FW MDA, whereas the V line displayed only about one-half of this amount. Altogether, these results underline that under salt stress, Arabidopsis seedlings ectopically expressing SlZF2 accumulated fewer osmoprotectants such as Pro, and showed more severe oxidative stress than control plants.
SlZF2 Regulates Multiple Developmental Processes
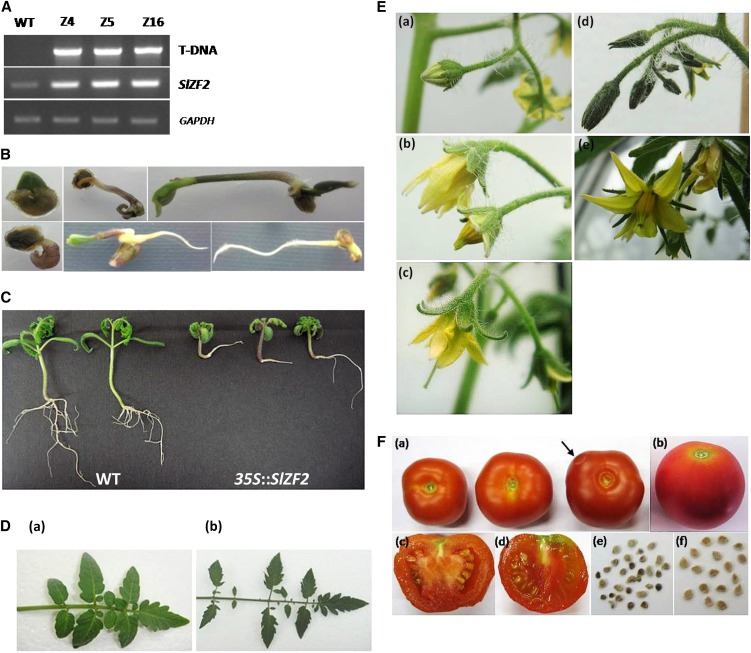
To further characterize SlZF2 in vivo, we generated transgenic tomato plants harboring the CaMV 35S::SlZF2 construct, and performed various analyses on F2 homozygous transgenic lines (Z4, Z5, and Z16) showing different levels of SlZF2 expression compared with wild-type plants (Fig. 6A). Transgenic plants displayed multiple phenotypes under optimal growth conditions. Seedlings displayed numerous germination defects, affecting either the root or the shoot system (Fig. 6B), associated with strong anthocyanin accumulation, as described above for SlZF2-Arabidopsis plants. Developing seedlings, especially from F1 generation, were dwarfs compared with control plants (Fig. 6C).
Figure 6.
Effects of SlZF2 ectopic expression in tomato. A, Transfer DNA (T-DNA) insertion in tomato genome and expression of SlZF2 in Z4, Z5, and Z16 transgenic lines and wild-type (WT) plants were analyzed by PCR. B, Defaults of germination of SlZF2 transgenic seeds on MS medium supplemented with 100 mg/L kanamycin. C, Dwarfism of 35S::SlZF2 tomato seedlings grown in vitro compared with wild-type tomatoes. D, Fully expanded leaf of transgenic (Da) and wild-type (Db) plants. E, Flowers of SlZF2-transgenic (Ea, Eb, and Ec) or wild-type (Ed and Ee) plants. F, Details of 35S::SlZF2 and wild-type tomato fruits (Fa and Fb, respectively) and according to a longitudinal section (Fc and Fd, respectively), and seeds of transgenic (Fe) and wild-type (Ff) tomatoes.
Ectopic expression of SlZF2 affected leaf shape, because transgenic leaves presented reduced interfoliole spacing and showed less pronounced and serrated borders and extremities (Fig. 6Da), with roundish angles compared with wild-type plants (Fig. 6Db). Transgenic flowers likewise displayed a more conical shape with sepals shorter than petals (Fig. 6, Ea and Eb), whereas wild-type flowers were more elongated with sepals totally covering the petals (Fig. 6, Ed and Ee). In addition, pistils of transgenic flowers could punctually top the stamens (Fig. 6Ec), which has not been observed for wild-type flowers. Finally, SlZF2 seems to also be involved in fruit development (Fig. 6F). SlZF2-transgenic tomato fruits were generally smaller and less round than wild-type tomatoes (Fig. 6, Fa and Fb). Likewise, the transgenic fruits presented an early senescence phenotype visible by their softened state at harvest compared with wild-type fruits. This advanced senescence state was visualized by a longitudinal section of SlZF2 tomato fruit in which at the same stage of harvest, transgenic fruits showed a larger central placenta, reduced pulp surrounding the seeds (larger cavity endocarp/mesocarp), and smaller/dark coat seeds (Fig. 6, Fc and Fe) compared with wild-type fruits (Fig. 6, Fd and Ff). Together, these results show the pleiotropic effects of SlZF2 on tomato development.
Overexpression of SlZF2 in Tomato Raised Its Tolerance to Salinity
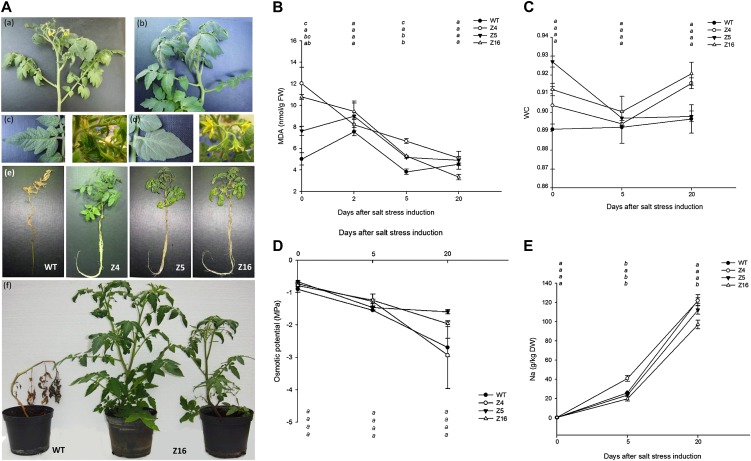
Because SlZF2 was differentially expressed between salt-tolerant and salt-sensitive tomato cultivars (Ouyang et al., 2007), the effects of SlZF2 constitutive expression on plant salinity response were investigated in hydroponics (125 mm NaCl) for 20 d by measuring plant oxidative and water status, in addition to photosynthesis. Transgenic plants exhibited delayed salt-induced senescence compared with wild-type plants (Fig. 7A). Visible wilting in wild-type plants started 10 d after stress initiation (Fig. 7Aa), followed by leaf chlorosis, then flower necrosis/abortion (Fig. 7Ac), and ultimately death (Fig. 7, Ae and Af). In transgenic plants, salt-induced senescence was delayed (Fig. 7Ab) because leaves remained green and healthy and flowers developed normally (Fig. 7Ad). When transferred to soil, transgenics quickly recovered and could continue normal development (Fig. 7Af).
Figure 7.
Characterization of hydroponic cultures of SlZF2 transgenic and wild-type tomatoes exposed to salinity (125 mm NaCl) during 3 weeks. A, Phenotype of wild-type (WT; Aa) and transgenic (Ab) tomatoes after 10 d. Accelerated senescence of the wild type (Ac and Ae) compared with transgenic tomatoes (Ad and Ae) is revealed by leaf chlorosis and flower abortion. Transfer of transgenic and wild-type tomatoes from hydroponics to soil results in death of wild-type plants (Af). B, MDA accumulation in transgenic (Z4, Z5, and Z16) and wild-type tomato lines exposed to salinity. C, Water content of transgenic and wild-type leaf 3. D, Osmotic potential of transgenic and wild-type leaf 5. E, Sodium accumulation in leaf 3 of Z4, Z5, Z16, and wild-type plants. Data represent means and se of at least two replicates. Letters indicate values that significantly differ between SlZF2 transgenic tomatoes and the wild type according to the Student-Newman-Keuls test at P < 0.05.
Levels of membrane lipid peroxidation were investigated before and after salinization in tomato seedlings. Under control conditions, all transgenic lines significantly (P < 0.05) accumulated up to 2.4 times more MDA than wild-type plants (Fig. 7B). However, after salinization, MDA content kept decreasing for Z4 and Z16, whereas it increased for Z5 and the wild type. At d 20, MDA content was very comparable for Z4, Z5, and the wild type. Together, these results indicated that although transgenic plants initially showed higher oxidative stress than the wild-type plants, salt stress induced less MDA accumulation during the stress period.
Water content represents an important physiological index because salt stress classically decreases it, and its value was determined on leaf 4. Under control conditions, all transgenic lines tend to show higher average water content than wild-type plants, and this tendency was observed again 20 d after stress initiation for the Z4 and Z16 lines (Fig. 7C). Similar results were observed with leaf 5 osmotic potential. Indeed, leaf osmotic potential was comparable between Z4 (−0.72 MPa), Z5 (−0.66 MPa), and Z16 (−0.78 MPa) transgenic and wild-type (−0.89 MPa) plants under control conditions (Fig. 7D). After salt stress, leaf osmotic potential decreased for all transgenic and wild-type plants, with Z5 and Z16 showing higher values than the wild type on average; this tendency was again observed on d 20.
Finally, analysis of Na+ content was conducted on leaf 3 of Z4, Z5, Z16, and wild-type plants (Fig. 7E). Before stress initiation, wild-type and transgenic tomatoes accumulated <0.5 g kg−1 dry weight (DW) sodium, and these values increased after stress onset. On d 20, Z4, Z5, and Z16 showed sodium content (121.18, 112.17, and 97.11 g kg−1 DW, respectively) that was similar (Z4 and Z5) or less (Z16) than that of the wild type (122.23 g kg−1), and very comparable results were observed in independent experiments and in different organs such as tomato roots (detailed data not shown).
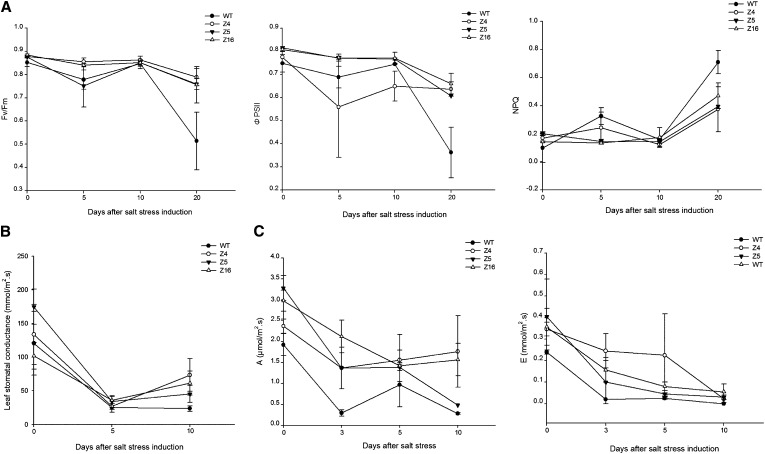
Transgenic plants displayed a higher photosynthetic rate than the wild-type plants during long-term exposure to salt (Fig. 8). Under control conditions and during the first 10 d of salt application, no difference of maximum photochemical efficiency of PSII in the dark-adapted state (Fv/Fm) was significant between wild-type and transgenic plants (ranging between 0.85 and 0.89). Three weeks after stress initiation, the Fv/Fm ratio decreased in wild-type plants, reaching a value of 0.514 (Fig. 8A). A similar pattern was observed for the quantum yield of PSII (ΦPSII). Indeed, ΦPSII declined over the stress period, particularly for Z4 and wild-type plants, and 3 weeks of salinization induced a sharp decrease in ΦPSII value for wild-type plants (P < 0.05). Finally, evolution of the nonphotochemical quenching (NPQ) parameter was also considered. Salt stress classically increases NPQ, which represents thermal dissipation, and thus energy loss for photosynthesis (Maxwell and Johnson, 2000). NPQ remained constant for Z5 and Z16 during the first 10 d of salinization, whereas it increased for Z4 and even more for the wild type as early as 5 d after stress (P < 0.05), before decreasing by d 10. Three weeks after stress began, the NPQ value augmented for all transgenic lines and to a greater extent for wild-type plants.
Figure 8.
Photosynthesis parameters of SlZF2 transgenic and wild-type (WT) tomatoes exposed to salt (125 mm NaCl) for 3 weeks. A, Fv/Fm, ΦPSII, and NPQ of transgenic and wild-type leaf 3. B and C, Leaf stomatal conductance (gs; B) and net assimilation of CO2 (A) and net transpiration rate (E) of transgenic and wild-type tomato lines (C). Data represent means and se of at least two replicates. Letters indicate values that significantly differ between SlZF2 transgenic tomatoes and the wild type according to the Student-Newman-Keuls test at P < 0.05.
Gas exchanges were examined, including leaf stomatal conductance (gs), the net assimilation of CO2 (A), and the transpiration rate (E; Fig. 8B) for 10 d after salt stress initiation, because no values could have been recorded later than d 10 because of modification of the leaf structure leading to higher organ fragility (mainly for the wild type and Z5).
Before salt stress onset, leaf stomatal conductance of transgenic lines Z4 and Z5 was on average 40% higher than that of wild-type tomatoes. Five days after establishment of the salt stress, gs values started to decrease up to five times for Z4, Z5, Z16, and wild-type plants. By day 10, gs values remained constant and low for wild-type plants, whereas gs values increased for all transgenic lines and could reach up to 2.6 times that of wild-type plants.
To measure A and E evolution during salinization stress, tomatoes were grown in individual pots and watered every 3 d with saltwater (150 mm NaCl; Fig. 8C). Under control conditions, Z4, Z5, and Z16 presented a higher A value (3.28, 2.845, and 2.385 µmol /m2⋅s respectively) than that of wild-type plants (1.925 µmol/m2⋅s). The A value sharply decreased for all plants as early as 3 d after exposure to salt, before increasing for Z4 and the wild type, declining for Z16, and remaining constant for Z5. By the end of the experiment, Z4, Z5, and Z16 displayed on average higher assimilation of CO2 values than that of the wild type (P < 0.05). Finally, transpiration was also measured during salinization. SlZF2-transgenic and wild-type plants showed similar reductions in E values after treatment. Under control conditions, Z4, Z5, and Z16 showed a higher E value (0.405, 0.605, and 0.425 mmol/m2⋅s respectively) than wild-type plants (0.24 mmol/m2⋅s). All transgenic lines maintained higher E values until d 10. Taken together, these results indicate that transgenic lines tend to retain, under salt stress, higher photosynthesis efficiency compared with wild-type plants.
SlZF2 Regulates PA Biosynthesis in Tomato
Free PAs were extracted from tomato seedlings grown in hydroponics, under control conditions and 5 d after initiation of salt stress (125 mm NaCl; Fig. 9). PAs such as spermine, spermidine, and putrescine (spermine and spermidine precursor) are small aliphatic polycationic compounds that are ubiquitously synthesized during plant development, and also known as antistress and antisenescence molecules allowing plants to face abiotic stresses (Gill and Tuteja, 2010).
Figure 9.
Free PA (spermine, spermidine, and putrescine) content in tomato seedlings grown in hydroponics under control conditions or 5 d of salt (125 mm NaCl) stress. Data represent means and se of six replicates. Letters indicate values that significantly differ between SlZF2 transgenic tomatoes and wild-type (WT) lines according to the Student-Newman-Keuls test at P < 0.05.
On average, transgenic plants tend to accumulate more spermine (up to 1.16 times), spermidine (up to 1.8 times), and putrescine (up to 1.38 times) under control conditions than wild-type plants, although these values were only significant (P < 0.05) for spermidine (Z4, Z5, and Z16 lines) and putrescine. Salt stress increased spermine accumulation in all tomato lines, whereas it decreased the content of spermidine and putrescine. In all cases, transgenic lines tended to accumulate more PAs than the wild type after 5 d of salt stress, particularly for spermidine (Z5 and Z16 lines). Together, these results indicate that SlZF2 modulates PA biosynthesis in tomato and that PA content is on average higher in transgenic lines than in wild-type plants under salt stress.
SlZF2 Mediates ABA Signaling
In silico analysis of the SlZF2 putative proximal promoter (1092 bp upstream from cloned 5′ untranslated region) was conducted using the Plant cis-Acting Regulatory DNA Elements database (http://www.dna.affrc.go.jp/PLACE/index.html; Higo et al., 1999). Assessment of the SlZF2 promoter first revealed the occurrence of scattered cis-elements present in the promoter of genes involved in light/phytochrome induction, as well as the circadian cycle. Further analysis pointed out the presence of ABA-responsive element and drought-responsive element sequences, with some additional consensus sequences bound by MYB, basic Helix-Loop-Helix (bHLH), and Leu-zipper transcription factors involved in drought and cold stress responses in an ABA-dependent manner. Likewise, the gibberellic acid (GA)-responsive element, along with supplementary cis-elements present in GA down-regulated genes and W boxes recognized by WRKY transcription factors repressing GA signaling pathway, was also discovered in the SlZF2 promoter.
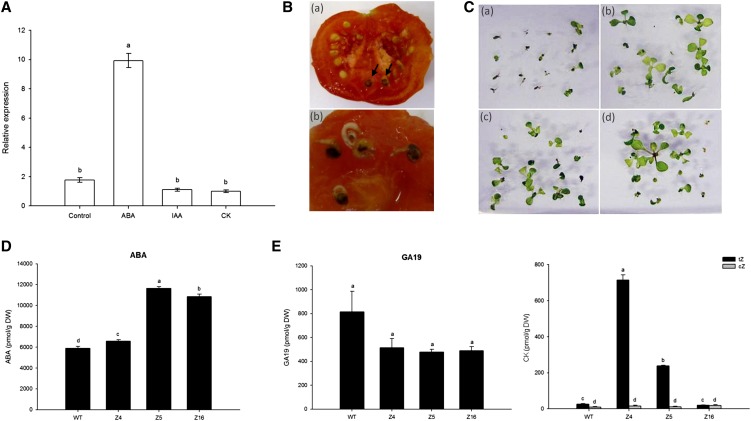
Expression of SlZF2 in response to different phytohormonal treatments was investigated in tomato roots by quantitative reverse transcription PCR (qRT-PCR; Fig. 10A). Neither cytokinin (CK) nor indole-3-acetic acid (IAA) could significantly (P < 0.05) affect SlZF2 expression. By contrast, ABA treatment caused a 5-fold induction of SlZF2 transcript accumulation. SlZF2-plants seemed ABA insensitive, because ripened fruits from 35S::SlZF2 line Z4 tomato plants occasionally exhibited germinating seeds (Fig. 10B). Likewise, transgenic Arabidopsis plants overexpressing SlZF2 showed better germination and early seedling development compared with the V control line when grown in vitro on 0.2 µM ABA (Fig. 10C).
Figure 10.
SlZF2 hormonal regulation. A, qRT-PCR analysis of SlZF2 expression in response to different hormonal treatments. EF1α and GAPDH were used as internal controls for ABA, CK, and IAA treatments. Data represent means and sds of three replicates. B, Viviparous phenotype of SlZF2 seeds in Z4 transgenic fruits. Germinating seeds developing in transgenic tomato fruits at harvest (Ba). Details of emerging radicals and hypocotyls (Bb). C, Insensitivity of Arabidopsis L5 (Cb), L6 (Cc), and L9 (Cd) transgenic seeds to ABA compared with a control line (Ca). Plants were grown on one-half-strength MS medium supplemented with 0.2 μM ABA. D and E, Phytohormone content in Z4, Z5, and Z16 compared with wild-type tomato seedlings grown in vitro on regular MS medium. Data represent means and se of two readings of the same extract. Each extract concerns at least four plants, and the experiment was repeated twice. Different letters indicate significant differences between treatments or tomato lines according to the Student-Newman-Keuls test at P < 0.05. cZ, cis-Zeatin; tZ, trans-Zeatin; WT, Wild type.
Total hormonal content was subsequently analyzed in wild-type and transgenic tomato seedlings (Fig. 10, D and E). Under nonstress growth conditions, all SlZF2-transgenic lines accumulated significantly more ABA (6,580 pmol/g DW, 11,639 pmol/g DW, and 10,842 pmol/g DW for Z4, Z5, and Z16, respectively) than control plants (5,900 pmol/g DW). Likewise, levels of trans-zeatin and cis-zeatin (CKs) were also significantly higher in transgenic tomatoes (Fig. 10E), whereas Z4 and Z5 had greater ethylene precursor 1-aminocyclopropane-1-carboxylic acid accumulation than control plants (data not shown). By contrast, SlZF2-transgenic lines showed reduced content of gibberellins GA19 (Fig. 10E) and GA4 (data not shown).
Transgenic and wild-type plants displayed similar accumulation of IAA and its conjugated forms (IAA-Asp and IAA-Glu) and oxo-IAA compared with the wild type (data not shown). Altogether, these results indicate that overexpression of SlZF2 in tomato affected multiple hormonal biosynthetic and metabolic pathways, mainly ABA accumulation.
Transcriptome Analysis of SlZF2-Tomato Z4
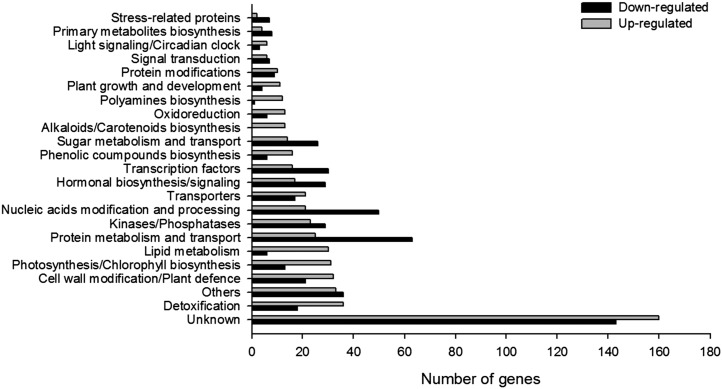
Genes differentially expressed between the Z4 transgenic line and wild-type tomatoes were determined by a transcriptomic analysis. The Z4 transgenic line was selected because germinating seeds were specifically observed in its fruits. A technical repetition was conducted and allowed identification of 1,084 differentially expressed genes (log-fold 2.5, P ≤ 0.05), with 532 down-regulated genes and 552 up-regulated genes. Functional classification of these genes indicated that ectopic expression of SlZF2 affects many physiological and biochemical pathways (Fig. 11).
Figure 11.
Microarray analysis of differentially regulated genes between wild-type and Z4 transgenic lines: number and categories of genes up- and down-regulated in tomato Z4 transgenic line compared with the wild type.
Among the differentially expressed genes, 29 encoding transcription factors were down-regulated, whereas 16 of them were up-regulated (Table I). The down-regulated transcription factors chiefly included CONSTANS-like zinc fingers and GIGANTEA (GI), which regulate circadian rhythm (Mizoguchi et al., 2005), in addition to the Phytochrome Interacting Factor1 bHLH, which is also involved in phytochrome-mediated light response. Likewise, repressed transcription factors included salt tolerance/stress-related zinc fingers (A20/AN1 group). Up-regulated transcription factors encoding genes comprised the same category of zinc finger proteins, in addition to four MYB transcription factors. Together, these results indicate that constitutive expression of SlZF2 affected different types of downstream regulators (Table I).
Table I. Transcription factors encoding genes differently up-regulated or down-regulated (P ≤ 0.05) between SlZF2-transgenic tomato line Z4 and wild-type tomatoes.
| Accession | Transcription Factor Family | Log2 Fold |
|---|---|---|
| Up-regulated genes | ||
| GH623042 | Putative zinc finger protein At1g68190 | 9.3 |
| AJ784616 | Zinc finger A20 and AN1 domain-containing stress-associated protein 7-like | 6.9 |
| EU636698 | CONSTANS-like protein | 5.9 |
| AK323090 | Zinc Finger CONSTANS-LIKE16-like | 4.1 |
| AK325209 | CONSTANS 1 | 3.4 |
| AK247503 | MYB | 14.8 |
| AK323291 | MYB-R1 | 14.4 |
| AW442364 | MYB48 | 10.2 |
| AK319692 | MYB | 8.9 |
| BI203387 | Transcription factor ORG2-like (bHLH) | 7.8 |
| DB714081 | NAC domain protein | 7.1 |
| AK322575 | SlWRKY3 | 5 |
| Down-regulated genes | ||
| DB690655 | Zinc finger protein CONSTANS-LIKE16-like | 17 |
| DB697244 | Zinc finger protein CONSTANS-LIKE9-like | 11.8 |
| AW624869 | Zinc finger CONSTANS-LIKE 9-like | 10.6 |
| ES896246 | Probable salt tolerance-like protein At1g78600-like | 5.1 |
| AK326301 | Probable salt tolerance-like protein At1g75540-like | 4.8 |
| AK320956 | Zinc finger A20 and AN1 domain-containing stress-associated protein8-like isoform1 | 4.5 |
| BT014337 | Zinc finger A20 and AN1 domain-containing stress-associated protein8-like isoform1 | 3.3 |
| TA43977_4081 | GI-like | 29.5 |
| DB718245 | GI-like | 17.9 |
| TA39072_4081 | GI-like | 8.7 |
| BM413010 | GI-like | 8.6 |
| BM409095 | GI-like | 7.5 |
| AW737518 | GI-like | 6.8 |
| AK247135 | PIF1 (bHLH) | 8.3 |
| TA38942_4081 | bHLH143-like | 3.7 |
| BI935893 | bHLH49-like | 3.7 |
| AK319263 | GRAS7 | 4.5 |
Induced genes involved in secondary metabolite biosynthesis encompassed 12 genes encoding PA biosynthesis enzymes such as Arg decarboxylase, S-adenosyl-Met decarboxylase, and spermidine synthase1 (Table II). Among induced genes, 13 encoded for enzymes regulating carotenoid/alkaloid biosynthesis (Table II) and 16 genes coded for phenolic compounds and biosynthesis enzymes. Likewise, it is also worth noting that 36 induced genes encoded enzymes of detoxification such as glutathione-S-transferase (AK327211), peroxidase (BI923252), and ATP-binding cassette (ABC) transporters (AW033521), whereas 16 repressed genes coded for the same category of enzymes, mainly catalases (DY523553).
Table II. Up-regulated genes putatively involved secondary metabolites biosynthesis differently (P ≤ 0.05) expressed between SlZF2-line Z4 and wild-type tomatoes.
| Accession | Description | Log2 Fold |
|---|---|---|
| Polyamine biosynthesis | ||
| AK319876 | Arg decarboxylase | 5.8 |
| AW216422 | Arg decarboxylase | 5.6 |
| A_96_P082314 | S-adenosyl-Met decarboxylase2 | 11.2 |
| EF550528 | S-adenosyl-Met decarboxylase | 3.3 |
| BT012829 | Hop-interacting protein THI107 | 2.9 |
| AK324059 | Spermidine synthase SPM1 | 2.8 |
| Carotenoid/alkaloid biosynthesis | ||
| TA40730_4081 | Carotenoid cleavage dioxygenase4 | 8.8 |
| DQ335097 | Phytoene synthase | 6.6 |
| AK327384 | Zeaxanthin epoxidase | 5.6 |
| AK319553 | Lycopene beta cyclase | 4.4 |
| AK321659 | Hyoscyamine 6-dioxygenase | 9.6 |
| AK319704 | Salutaridinol 7-O-acetyltransferase-like | 6.2 |
| AK323868 | Secologanin synthase-like isoform2 | 5.5 |
| Phenolic compound biosynthesis | ||
| AK320988 | Phe ammonia-lyase-like | 5.6 |
| AK326062 | Anthocyanin 5-aromatic acyltransferase-like | 5 |
| AK328438 | 4-coumarate-CoA ligase1-like | 4.9 |
| AK322767 | Anthocyanin 3′-O-β-glucosyltransferase | 3.6 |
| AW738087 | Shikimate O-hydroxycinnamoyltransferase-like | 3.2 |
A high proportion of genes induced in 35S::SlZF2 tomatoes included photosynthesis/chlorophyll biosynthesis-related genes (31 genes induced versus 11 repressed) such as those coding for a Rubisco activase (BG129244), chlorophyll a-b-binding proteins (AK319645), PSI (BG128097) and PSII (AK325903) proteins, or chlorophyllide a oxygenase (BE461802). Because part of our current work consisted of deciphering the phytohormonal pathways putatively regulated by SlZF2 in parallel to identification of the hormonal compounds contrastingly accumulated between transgenic and wild-type lines (Fig. 10), differentially expressed genes related to hormone biosynthesis/signaling present on the chip were also selected (Table III). The 17 induced genes were involved in ethylene, auxin, and gibberellin signaling pathways, whereas the 24 repressed genes were chiefly related to auxin signaling. Finally, repressed genes include some coding for histone deacetylase (TA51311_4081) and Lys demethylase (AK321177), which are responsible for chromatin silencing, as well as the Argonaute protein, which is part of the RNA-induced silencing complex (BT013764).
Table III. Differentially regulated genes putatively involved hormones biosynthesis/signaling differently expressed between SlZF2-line Z4 and wild-type tomatoes (P ≤ 0.05).
| Accession | Description | Log2 Fold |
|---|---|---|
| Up-regulated genes | ||
| Y14809 | β-carotene hydroxylase | 20.6 |
| AK324589 | 2-oxoglutarate-dependent dioxygenase | 18.2 |
| AK247698 | Zeatin O-glucosyltransferase-like | 11.6 |
| AK324766 | Gibberellin 20 oxidase 1-like | 9.1 |
| AW217769 | S-adenosyl-l-Met: salicylic acid carboxyl methyltransferase | 7.8 |
| AK320908 | Auxin-induced protein IAA4 | 4 |
| X04792 | 1-aminocyclopropane-1-carboxylate oxidase1 | 2.9 |
| Down-regulated | ||
| AW041529 | Auxin-binding protein | 5.9 |
| AK325743 | Auxin:hydrogen symporter | 4.6 |
| GQ144956 | EIN3-binding F-box protein2 | 4.4 |
| AF118844 | Ethylene receptor homolog (ETR5) | 2.9 |
| AF258808 | Aldehyde oxidase | 5.3 |
| AY584532 | DWARF1/DIMINUTO | 5 |
| EF501822 | 1-aminocyclopropane-1-carboxylate oxidase | 4.2 |
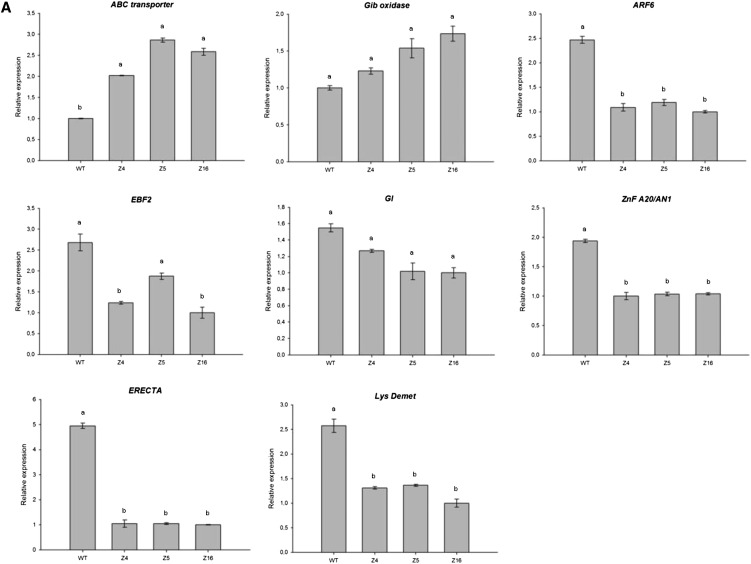
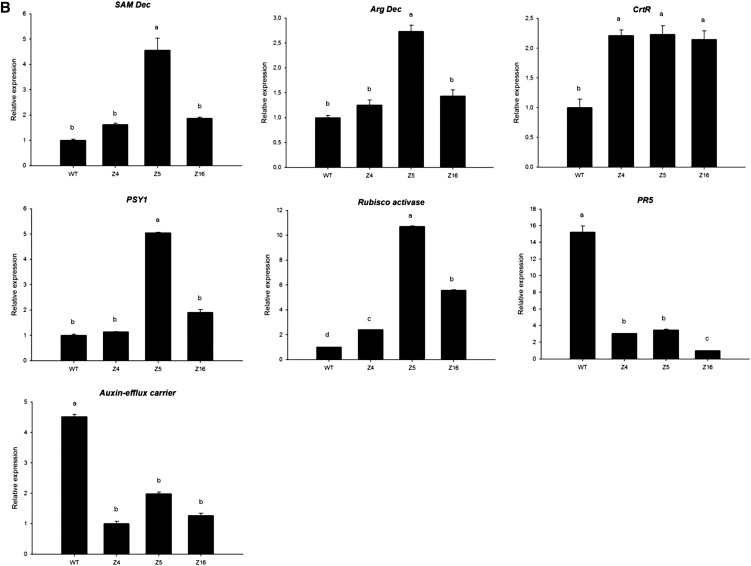
To validate the microarray data, 15 genes involved in the different biosynthesis pathways examined in this study were selected for further qRT-PCR analysis and accumulation of the corresponding transcripts measured in wild-type tomato seedlings and compared with the three Z4, Z5, and Z16 transgenic lines (Fig. 12) under control growth conditions (Fig. 12A) or under salinity (48 h, 125 mm NaCl; Fig. 12B). This group encompassed genes coding for the following: transcription factors GI (DB718245), zinc finger A20/AN1 (AK320956), and EIN3-binding F-box protein2 (GQ144956); auxin response factor6 (BE436460), auxin-efflux carrier protein (CK715455), and gibberellin20 oxidase (AK324766) for hormonal signaling; β-carotene hydroxylase (CrtR, Y14809) and phytoene synthase1 (PSY1; BT012712) for carotenoid biosynthesis; and Arg decarboxylase (Arg Dec; AW649598) and S-adenosyl-Met decarboxylase (SAM Dec; AK319251) for PA biosynthesis. Additional genes related to various physiological/metabolic processes were tested, including Rubisco activase (BT012795), ABC transporter (AW033521), LRR receptor-like Ser/Thr-protein kinase ERECTA-like (ERECTA; DB682292), Lys demethylase (AK321177), and pathogenesis-related protein5 (PR5; AK322591; the full list of primers used is given in Supplemental Table S1). Under control conditions (Fig. 12A), qRT-PCR analysis could confirm microarray data for the selected genes. Under salt stress (Fig. 12B), it is interesting to note that SAM Dec, Arg Dec, CrtR, PSY1, and Rubisco activase are more often expressed in transgenic lines than in the wild type, whereas PR5 and Auxin-efflux carrier are repressed in the same conditions, as was already the case under control conditions (data not shown).
Figure 12.
qRT-PCR analysis of SlZF2 putative target genes expression in transgenic tomato lines Z4, Z5, and Z16 compared with wild-type (WT) tomatoes, under control conditions (A) or after salt stress (B; 125 mm NaCl, 48 h). Actin and GAPDH were used as internal controls. Data represent means and sds of three replicates.
DISCUSSION
Environmental stresses strongly affect expression of both activators and repressors of transcription. However, roles of repressors such as zinc fingers are still under investigation, because it is unknown whether tolerance is only attributed to growth (and thus energy needs) reduction, or is really a result of a specific targeting of key genes. Herein we decipher, for the first time in tomato, the regulation of different physiological and biochemical pathways modulated by the repressor SlZF2 during salinization.
SlZF2 Impacts Multiple Aspects of Plant Development
SlZF2 belongs to the C1-2i subgroup encompassing stress-regulated zinc finger proteins induced by salinity and supplementary abiotic stresses, in addition to ROS. In Arabidopsis, the STZ/AZF repressors and related proteins include at least 18 members, and can be further divided into G1 (ZAT10) or G2 (ZAT7 and ZAT12) subgroups according to a conserved motif in the N-terminal end (Sakamoto et al., 2004; Gourcilleau et al., 2011). In cultivated or wild tomato, such zinc finger type proteins have not been characterized prior to this report. Nevertheless, another category of zinc finger proteins induced by environmental stresses and/or ABA treatment have been identified in tomato and belong to the Stress Associated Protein family, which includes 13 members containing A20/AN1 zinc finger domains (Solanke et al., 2009). SlZF2 is ubiquitously expressed in tomato (except in young leaves), as illustrated by its spatiotemporal profile of expression, and an unrestricted pattern of expression has also been observed for AZF2 or ZAT10, for instance (Sakamoto et al., 2004).
Heterologous expression of SlZF2 in Arabidopsis and tomato inhibited plant development and led to obvious dwarfism, but also affected leaf, flower, and fruit shape, and seedling germination. Growth inhibition, directly linked to the transgene expression level, was also reported for Arabidopsis plants constitutively expressing ZAT10 (Sakamoto et al., 2004), ZAT7 (Ciftci-Yilmaz et al., 2007), AZF1, and AZF2 (Kodaira et al., 2011). However, this phenomenon is not specific to the STZ/AZF group, because growth and leaf shape defects, in addition to altered flowering time, have also been reported for CCCH-type zinc finger proteins (Lin et al., 2011) and for a single C2H2 zinc finger (Dinkins et al., 2002). Whether growth suppression is a result of a direct activation or repression of specific target genes by the above-mentioned zinc fingers constitutes the key point of all of these studies. In tomato, overexpression of SlZF2 induced and repressed a similar number of genes belonging to various cellular, physiological, or biochemical processes, from photosynthesis to secondary metabolites, strengthening the idea that SlZF2 is directly regulating these events. Growth suppression is associated in transgenic plants with elevated levels of ABA and reduced content of GA, among others, along with repression of several auxin-binding proteins. Inhibition of protein and carbohydrate metabolism-related genes, together with the hormonal imbalance, could also limit growth.
Growth repression was first attributed to the DLN box. However, the rice ZFP179 and the chrysanthemum CgZFP1do also present a DLN box in their C-terminal end and function as transcriptional activators in yeast (Sun et al., 2010; Gao et al., 2012). Likewise, overexpression of ZAT7 devoid of its C-terminal (ZAT7Δ) end also inhibited growth in Arabidopsis (Ciftci-Yilmaz et al., 2007). The DLN box was then suggested to enhance salt stress tolerance. Indeed, ZAT7Δ plants showed salt sensitivity (Ciftci-Yilmaz et al., 2007). Nevertheless, the C2H2 rice zinc finger ZFP182, for instance, is related to Arabidopsis ZAT12, but does not present a DLN box, and ZFP182 overexpression in tobacco enhanced salt tolerance in transgenic plants (Huang et al., 2007). In addition, both ZAT10 overexpressors and loss-of-function mutants display enhanced tolerance to drought and salinity stresses, emphasizing a possible dual function of ZAT10 (Mittler et al., 2006). These contrasting data emphasize the existence of complex networks of interactions between zinc finger proteins and different partners to regulate both plant development and osmotic stress tolerance.
These networks seem to be somehow conserved between species. Indeed, ZAT7 interacted with AtWRKY70 in the yeast two-hybrid system, and AtWRKY70 transcripts are more abundant in ZAT7 overexpressors than in control plants (Ciftci-Yilmaz et al., 2007). Similarly, in SlZF2 overexpressors, SlWRKY3, a close ortholog to AtWRKY70, is also induced, suggesting comparable mechanisms of ZAT7/SlZF2 downstream targeting. Besides the zinc finger domain itself and the DLN box that could be a platform for protein-protein interaction, SlZF2 also presents two Gly-rich domains, which seem to be only common, although with less importance in size/number, to the transcription factors from the Solanaceae family (StZFP1 and NtZFP). Gly-rich proteins (GRPs) are classified into several subgroups, including class IV, which includes a RNA binding motif together with a C2HC zinc finger motif, and class V, which is a new class of GRPs presenting mixed patterns of Gly repeats (for review, see Mangeon et al., 2010). No RNA binding domain was identified in SlZF2. However, it is worth noting that class IV GRPs are involved in osmotic stress responses by modulating H2O2 accumulation, do also respond to ABA, and can also interfere with stomata aperture in an ABA-independent pathway (Mangeon et al., 2010). Investigating Gly-rich domain functions as putative interaction or RNA stabilization domains would greatly help to decipher SlZF2 mode of action and interacting partners.
SlZF2 Regulates ABA Biosynthesis/Signaling
SlZF2 expression is rapidly induced by ABA. In the Arabidopsis AZF/STZ zinc fingers group, ABA up-regulates the expression of no less than AZF1, AZF2, Zat10, and Zat12 (Sakamoto et al., 2004; Davletova et al., 2005; Kodaira et al., 2011). ABA increases during drought and salinity stresses both in leaves and roots (Albacete et al., 2008; Ghanem et al., 2008; Huang et al., 2008; Lovelli et al., 2012) and promotes stomatal closure (Mishra et al., 2006). Under optimal growth conditions, ABA induces seed dormancy and represses germination and seedling establishment (Koornneef et al., 1989; for review, see Finkelstein et al., 2002). AZF2 expression is reduced in Arabidopsis ABA-deficient1 (aba1) and ABA-insensitive1 (abi1) mutants (deficient in ABA biosynthesis/signaling respectively) during dehydration, demonstrating that AZF2 expression is partly ABA dependent (Sakamoto et al., 2004). By contrast, ZAT10 expression is comparable under salinity in aba1 and abi1 mutants, indicating that ZAT10 must be induced in an ABA-independent pathway during salt stress and underlying complexity of hormonal signaling/crosstalk after stress establishment, according to the type of stress (Sakamoto et al., 2004).
Higher accumulation of ABA in SlZF2-transgenic tomato lines was expected to induce stomatal closure, in contrast with our observations (Fig. 8, B and C). Besides ABA signaling, additional complex patterns of hormonal cross regulation can be at the origin of the Z4 plant phenotype. Indeed, although control of stomata aperture is chiefly attributed to ABA, other hormones participate in regulating this physiological process because auxins promote stomatal opening (Dodd, 2003), whereas ethylene may induce stomata closure (Desikan et al., 2006) or inhibit ABA-induced stomatal closure (Tanaka et al., 2005). Microarray data from Z4 transgenic lines indicated that 38% of down-regulated genes involved in hormone signaling are related to auxin, whereas 24% are linked to ethylene signaling. In general, all transgenic lines displayed increased levels of ABA and reduced levels of GA19 compared with the wild type, but contents of the other hormones tested (jasmonates, salicylic acid, CK, and auxin) greatly vary depending on the transgenic lines (Fig. 10E; detailed data not shown), making it difficult to attribute stomata opening/closure to hormonal imbalance only. Studies conducted on Arabidopsis emphasized the involvement of AZF/STZ transcription factors in hormonal signaling. Indeed, AZF1 and AZF2 directly bind to and down-regulate expression of several auxin-responsive genes (Kodaira et al., 2011). Arabidopsis seedlings overexpressing AtTZF1 and strongly impaired in their development can be rescued by external applications of GA3, but levels of ABA and GA4 were comparable in wild-type plants, overexpressors, and loss-of-function mutants (Lin et al., 2011).
In the Z4 transgenic line, several genes related to carotenoid biosynthesis were up-regulated (Fig. 11), in parallel to ABA overaccumulation. However, no typical ABA marker genes such as PYRABACTIN RESISTANCE1 (PYR1)/PYRABACTIN RESISTANCE1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR), DEHYDRATION-RESPONSIVE ELEMENT-BINDING and MYB2/MYC2 transcription factors, desiccation-responsive rd29A gene, ABI type2C phosphatase, or heat shock proteins were differentially regulated compared with wild-type plants in our microarray data (Fig. 11; Wang et al., 2011). Our results suggest that ABA signaling could thus be affected in Z4-transgenic plants. Indeed, contradictory phenotypes were observed between ABA overaccumulation and release of seed dormancy (Fig. 10B) or insensitivity to ABA (Fig. 10C). Microarray data revealed that several kinases encoding genes were repressed in the Z4 transgenic line. Phosphorylation cascade through action of specific kinases and PP2C phosphatases such as ABI1/2 are indicative of ABA signaling (Nakashima and Yamaguchi-Shinozaki, 2013). Among down-regulated kinases encoding genes in the Z4 transgenic line, some candidates such as SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE1 (SnRK1; γ-subunit) and the LRR receptor-like Ser/Thr-protein kinase ERECTA seemed important. The SnRK1 complex consists of the SUCROSE NONFERMENTING1 kinase (α) and some regulatory subunits (γ), among others, and mainly regulates plant metabolism (especially in seeds) in response to nutritional and environmental conditions (Kulik et al., 2011). In Medicago truncatula seeds, genes encoding γ-subunits of the SnRK1 complex were differently regulated by desiccation, polyethylene glycol, and ABA (Buitink et al., 2004). Likewise, ABA and GA, in addition to drought, differently regulate expression of Lycopersicon esculentum SUCROSE NONFERMENTING4, encoding a γ-subunit of SnRK1 complex in tomato seeds and during early germination steps, as well as in tomato leaves (Bradford et al., 2003). In Arabidopsis, ERECTA genes regulate plant growth and inflorescence architecture, because erecta loss-of-function mutants show reduced growth and lateral organ size as a result of reduced cell proliferation, in addition to abnormal flower development (Shpak et al., 2004). Overexpression of the poplar PdERECTA in Arabidopsis increased plant biomass and improved water use efficiency and the photosynthetic rate, whereas it decreased the transpiration rate by diminishing the stomata number (Xing et al., 2011). Together, these studies strongly suggest that modification of kinase expression after SlZF2 constitutive expression could affect both plant development and abiotic stress response.
Occasional vivipary was also observed in Z4 transgenic fruits. Viviparous mutants are affected in ABA response or biosynthesis (Finkelstein et al., 2002). The maize seeds exhibiting the red embryonic axis (rea) phenotype, for instance, are deficient for ABA metabolism. They accumulate anthocyanins in the embryonic axis in the dormant seed, and occasionally vivipary leading to precocious germination, but rea mutants accumulate levels of ABA that are comparable with the wild type (Sturaro et al., 1996). However, regulation of seed maturation and dormancy cannot be solely attributed to ABA. Indeed, antagonistic effects of ABA and GA regulate seed maturation, dormancy, and germination, but additional hormones can also either promote (ethylene) or inhibit (jasmonate) germination (Linkies and Leubner-Metzger, 2012); in SlZF2 transgenic tomatoes, at least GA and 1-aminocyclopropane-1-carboxylic acid (ethylene precursor) tend to differentially accumulate compared with the wild type (Fig. 10; data not shown). Moreover, GI was down-regulated in the Z4 tomato line. In Arabidopsis, circadian clock genes such as GI regulate seed dormancy and germination through light signaling but also through control of ABA and GA metabolism genes in seeds, and response to environmental conditions, and ABI3 expression is reduced in gi-11 mutants (Penfield and Hall, 2009). Together, these data provide some possible insights to elucidate the vivipary phenotype.
In summary, SlZF2 can affect several aspects of plant development and stress adaptation through ABA biosynthesis/signaling.
SlZF2 Modulates Salt Stress Response
Phylogenetic analysis of SlZF2 indicated that its closest related proteins are involved in salt stress tolerance. Salinity inhibits plant growth (biomass, leaf number, and area) and increases root/shoot ratio in tomato, whereas it reduces photosynthesis and transpiration (Lovelli et al., 2012). Salinity first causes osmotic and ionic stresses, followed by oxidative damage. Proteomic analysis indicated that majority of the salt stress-induced proteins are involved in photosynthesis and energy metabolism, antioxidant scavenging mechanisms, ion exclusion and/or compartmentalization, and compatible solute biosynthesis (Zhang et al., 2012). No specific gene directly involved in salinity or other osmotic stresses was differently expressed in SlZF2 overexpressors under control conditions, although overexpression of SlZF2 altered expression of different types of kinases, photosynthesis-related genes, detoxification mechanisms, or diverse secondary metabolite biosynthesis. Comparable results were described for Arabidopsis ZAT10 overexpressors, in which no transcripts of genes related to salinity, drought, or cold stresses were differently accumulated under control conditions of growth or even under stress, although overexpressors displayed enhanced tolerance (Mittler et al., 2006).
Ectopic expression of SlZF2 induced a general state of stress, in both Arabidopsis and tomato, visible by MDA and anthocyanin accumulation in transgenic seedlings. Increased MDA production as a result of SlZF2 ectopic expression accompanied induction of detoxification enzymes such as peroxidases, glutathione S-transferases, and ABC transporters. Expression of ROS scavenging genes such as Arabidopsis thaliana MANGANESE SUPEROXIDE DISMUTASE, Arabidopsis thaliana PEROXIDASE, and Arabidopsis thaliana ASCORBATE PEROXIDASE1 is also significantly higher in Arabidopsis lines overexpressing CgZFP1, both under control conditions and under salinity constraints (Gao et al., 2012), and similar results have been described for Arabidopsis ZAT10 overexpressors (Mittler et al., 2006). The ROS scavenging system was thus already active in SlZF2 transgenic plants before salt stress onset (Fig. 11) and could alleviate salt stress damage in transgenic plants when it occurred. Indeed, ROS cause irreversible cellular damage at high concentrations. At low levels, ROS act as secondary messengers relaying hormonal adjustments after abiotic and biotic stresses, and are involved in stomata closure (e.g., ABA induces ROS synthesis; Yan et al., 2007; Kar, 2011). Wild tomato (Solanum pennelli) contained about twice as much chloroplastic H2O2 and MDA as cultivated tomato under control growth conditions, and these contents decreased for wild tomato, whereas they increased for cultivated tomato during salt stress (Mittova et al., 2002). These results are comparable with those reported for SlZF2-transgenic lines, suggesting possible similar mechanisms of salt stress tolerance.
Likewise, retention of photosynthesis is critical for plant survival. In Z4 transgenic lines and under control conditions (Fig. 11) as well as under salt constraints (Fig. 12B), photosynthesis-related genes involved in light harvesting, electron transfer, CO2 assimilation, and additional mechanisms such as photosystem protein, light harvesting complex I/II binding proteins, Rubisco activase, and chlorophyll a/b-binding proteins are up-regulated and are presented as potential candidates in salt stress tolerance, as suggested by Walia et al. (2007) in barley (Hordeum vulgare). Most of the photosynthesis-related genes are induced under salt stress in different species such as Arabidopsis, wheat (Triticum durum), rice, or grapevine (Vitus vinifera; for review, see Zhang et al., 2012). Different strategies can nevertheless be adopted by plants to modulate their growth under stress conditions. In grapevine, VvZFPL enhanced tolerance to drought and salinity in Arabidopsis transgenic plants and down-regulated expression of photosynthesis-related genes, probably to reduce growth under adverse conditions (Kobayashi et al., 2012). In SlZF2 overexpressors, higher expression of photosynthesis-related genes under both control and salt conditions seems to be the strategy adopted to overcome stress effects.
Secondary metabolites such as flavonoids, carotenoids, and PAs constitute an important source of antioxidants to protect the photosynthetic machinery from photo-oxidative damages, and their synthesis increases after abiotic stress (Chen et al., 2011). At least seven genes coding for enzymes of the carotenoid pathway were up-regulated in Z4 transgenic lines compared with the wild type, both under control and salinity conditions. In tomato, salinity induced up to three times the lycopene accumulation in fruits, whereas it affected anthocyanin accumulation differently depending on the cultivar (Borghesi et al., 2011). Overexpression of a Salicornia europea β-lycopene cyclase encoding gene in Arabidopsis and tobacco enhanced salt tolerance of transgenic plants by retention of higher carotenoid levels, photosynthetic activity, and reduction of H2O2 accumulation (Chen et al., 2011), underlining the importance of carotenoids in salt stress tolerance.
Among SlZF2 protein closest orthologs, the tobacco zinc finger NtZFT1 regulates the spermine signaling pathway (Mitsuya et al., 2007). Transcripts corresponding to key genes of PA pathways such as Arg decarboxylase and SAM decarboxylase were more abundant in SlZF2-transgenic lines 48 h after salinity onset than in control plants (Fig. 12B), and are consistent with the PA content in transgenic lines (Fig. 9). PAs accumulate in response to various environmental stresses such as drought or salinity in tomato, and plant treatment with PAs improves plant stress tolerance (Hu et al., 2012). PAs are important signaling molecules involved in activation of the antioxidative mechanisms of defense, retention of plant photosynthesis efficiency, alleviation of growth inhibition, ion homeostasis, and modulation of expression of different genes related to osmotic stresses tolerance (Kasinathan and Wingler, 2004; Kasukabe et al., 2004; Hazarika and Rajam, 2011). Arabidopsis Arg decarboxylase1 loss-of-function mutants accumulated fewer PAs and were more sensitive to salinity than wild-type plants (Kasinathan and Wingler, 2004). In cultivated tomato, overexpression of the human SAM decarboxylase encoding gene triggered PA accumulation and improved plant tolerance to fungal pathogens, salinity, and drought, among others (Hazarika and Rajam, 2011). Hu et al. (2012) showed that salt-tolerant cultivated tomato cultivars displayed higher SAM decarboxylase activity in roots under salinity. Together, these data strongly suggest that PA accumulation in SlZF2 transgenic lines is an important mechanism of stress tolerance. In addition, PAs can also remarkably influence expression of carotenoid biosynthesis-related genes and accumulation of carotenoids in tomato fruits, because overexpression of a spermidine synthase in tomato led to enhancement of lycopene content in ripe fruits (Neily et al., 2011). Together, these data illustrated the importance of PAs directly in tolerance to salinity, but also in modulating additional biosynthesis pathways.
CONCLUSION
This study characterized the first tomato EAR zinc finger type repressor, and provided a comprehensive overview of SlZF2 involvement both in plant development and adaptation to salinity. SlZF2 regulates expression of several classes of genes involved in secondary metabolism that are crucial for plant adaptation to environmental conditions, as well as photosynthesis and hormone biosynthesis/signaling-related genes. Deciphering roles of this repressor in a Solanaceae species shed considerable light on fundamental understanding of plant complex network regulation during stress adaptation. SlZF2 can thus be considered as a molecular marker for stress tolerance in tomato, and can be included in genetic engineering processes of selection/generation of tolerant cultivars.
MATERIALS AND METHODS
Plant Material and Osmotic Stress/Hormonal Treatment Assays
Wild-type (untransformed) cultivated tomato (Solanum lycopersicum, cv Ailsa Craig) seeds were germinated for a few days on Whatman 3MM paper soaked with sterile water until radicule emergence, and were then shifted to trays filled with perlite-vermiculite (1:3 v/v) mixture. Four-week-old seedlings intended for hydroponics were transferred in 52-L tanks containing aerated one-half-strength Hoagland nutrient solution as described by Ghanem et al. (2008), and were allowed to develop for an additional 10 d before applying stress. For the stress assay, tanks were refilled with one-half-strength Hoagland solution whenever needed. The nutrient solution contained the following chemicals (in g/L; concentrated 200×): 8 NH4NO3, 82.6 Ca(NO3)4H2O, 40.7 KNO3, 27.4 KH2PO4, 24.6 MgSO47H2O, 0.053 MnSO45H2O, 0.14 H3BO3, 0.015 CuSO45H2O, 0.008 (NH4)Mo7O244H2O, 0.06 ZnSO47H2O, and 1.87 FeEDDHA. Tomato plants were grown in a growth chamber at 24°C/22°C under a 16-h-d/8-h-night photoperiod. Arabidopsis (Arabidopsis thaliana Heynh cv Columbia) plants were grown in individual pots filled with commercial soil at 20°C/19°C under a 16-h-d/8-h-night photoperiod.
For plant transformation as well as for plant in vitro culture, tomato seeds were surface sterilized for 15 min (2.5% [v/v] potassium hypochlorite and 0.02% [v/v] Triton X-100) and washed five times with sterile water. Transgenic tomato seeds were germinated on MS medium containing 100 mg L−1 kanamycin, at 24°C under a 16-h-d/8-h-night regime. Arabidopsis seeds were surface sterilized as described above, and underwent 48 h of vernalization at 4°C. Seeds were subsequently plated on one-half-strength MS medium (with 1.5% [w/v] Suc), supplemented with 35 mg L−1 kanamycin for transgenic plant selection.
For tomato salt stress (125 mm or 150 mm NaCl) treatment, salt was directly added to the culture tank. To simultaneously conduct additional NaCl (150 mM), KCl (150 mM), and drought (air-drying) stresses as well as hormonal assays, 3-week-old plants were grown in smaller tanks with a capacity of 4.5 L. CK (zeatin; mixed isomers including approximately 80% [w/w] trans-zeatin; Sigma Z0164), ABA (Sigma A7383), and auxin (IAA; Sigma I2886) were used at final concentrations of 10 µM. At least three plants were sampled at each time.
RNA Extraction and qRT-PCR Analysis
For RNA extraction, tissues were either sampled from plants grown in the greenhouse (from August to November 2010) to determine SlZF2 expression profile during tomato plant development, or from hydroponics in case of osmotic stresses and hormonal treatments. Tomato total RNA was extracted, treated with RNase-free DNase I (Promega), and purified as described by Hichri et al. (2010). Total RNA was quantified on a NanoDrop spectrophotometer (ND-1000; Isogen Life Science), and nucleic acid purity was evaluated by the absorbance ratio (A260/A280), in parallel to verification by agarose gel electrophoresis. Two µg of DNAse-treated RNA was used as a template for reverse transcription (RevertAid H Minus First Strand cDNA Synthesis Kit; Fermentas) using oligo d(T)18 following the supplier’s protocol. Transcript levels of the different genes were measured by qRT-PCR using SYBR Green on an iCycler iQ (Bio-Rad). PCR reactions were performed in triplicate using 0.2 µM of each primer, 5 µL of SYBR Green mix (Bio-Rad), and 300 ng of DNAse-treated cDNA in a final volume of 10 µL. Negative controls were included in each run. PCR conditions were as follows: initial denaturation at 95°C for 90 s followed by 40 cycles of 95°C for 30 s and 60°C for 1 min. Amplification was followed by melting curve analysis to check the specificity of each reaction. The forward primer 5′-AGTGGAAAGTCCAATGCCTG-3′ and the reverse primer 5′-ATAATATCATCACATGTAAAACCA-3′ were used to specifically amplify a 112-bp sequence including a partial sequence of the SlZF2 3′ untranslated region.
Data were normalized according to the S. lycopersicum GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE and S. lycopersicum ACTIN or S. lycopersicum ELONGATION FACTOR1α housekeeping gene expression. GAPDHF (5′-GGTGCCAAGAAGGTTGTGAT-3′) and GAPDHR (5′-TTTTCTGGGTGGCAGTCAT-3′) primers generated a 217-bp PCR product for GAPDH, whereas ActinF (5′-ATGGTGGGTATGGGTCAAAA-3′) and ActinR (5′-GAGGACAGGATGCTCCTCAG-3′) allowed the formation of a 183-bp PCR product, and EF1αF (5′-TACTGGTGGTTTTGAAGCTG-3′) and EF1αR (5′-AACTTCCTTCACGATTTCATCATA-3′) amplified a 166-bp DNA fragment. Normalized expression of SlZF2 was calculated using Gene Expression Analysis for iCycle iQ Real-Time PCR Detection System software (Bio-Rad) with a method derived from the algorithms outlined by Vandesompele et al. (2002).
Cloning, Transient, and Stable Plant Transformation
SlZF2 cDNA was amplified by PCR with the Pfu DNA polymerase (Promega) in a final reaction volume of 50 µL following the manufacturer’s instructions, including 300 ng of cDNA synthesized from RNA isolated from salt-stressed (150 mm NaCl, 2 h of stress) tomato roots grown in vitro. SlZF2 1086-bp cDNA was amplified with primers 5′-TATCACACACTTCTCTTTCAGT-3′ and 5′-ATAATATCATCACATGTAAAACCA-3′, and the PCR product was subcloned into a pGEM-T-easy vector (Promega) before DNA sequencing (Macrogen) with universal T7 and SP6 primers. Subsequently, a SlZF2 933-bp open reading frame was cloned into the pDONR221 entry vector (Gateway Technology; Invitrogen) by a BP recombination reaction prior to sequencing, and then transferred to the pK7WG2D binary vector (Karimi et al., 2002) by a LR recombination reaction (Gateway Technology). The construct was introduced into the Agrobacterium tumefaciens strain GV3101 for Arabidopsis transformation and in the LBA44A4 strain for tomato transformation. Competent cells were prepared as described by Jyothishwaran et al. (2007).
For subcellular localization, a SlZF2 open reading frame was introduced in frame into the pYFP-attR vector (Subramanian et al., 2006), which allowed in planta transitory expression of the protein via N-terminal fusion to the YFP. Isolation, purification, and polyethylene glycol-mediated transformation of tomato protoplasts starting from young leaves of in vitro grown plants were performed as described by Hichri et al. (2010). YFP alone (positive control), YFP-SlZF2 fusion protein, and chlorophyll fluorescence were visualized using a Zeiss 710 confocal microscope (Carl Zeiss). YFP was excited at 514 nm and detected between 530 and 570 nm, whereas chlorophyll was excited at 643 nm and detected at 772 nm. Images were analyzed using Zen Software (Zeiss).
Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998) and analysis was conducted on self-fertilized F3 homozygous (single-copy insertion) transgenic lines. Tomato stable transformation was adapted from Ellul et al. (2003).
Tomato genomic DNA extraction was conducted as described by Fulton et al. (1995). DNA pellets were resuspended in 30 µL of sterile water and 2 μL of the resuspension was used in a 20-μL PCR reaction volume.
Constructs and Yeast Transformation
To verify any putative transcriptional activation capability of SlZF2, its cDNA was amplified by PCR using the Pfu DNA polymerase (Promega) with oligonucleotides introducing the restriction sites EcoRI (5′-GAATTC-3′) for the forward primer and BamHI (5′-GGATCC-3′) for the reverse primer. After enzymatic digestion, the PCR fragment was cloned into the pGBKT7 vector (BD Biosciences) as a fusion in frame to the GAL4 DNA binding domain coding region under control of the ALCOHOL DEHYDROGENASE1 promoter. The pGBKT7 vector carries the TRYPTOPHAN1 nutritional marker for yeast selection and ADE2 and HIS3 as reporter genes. The yeast strain Y8930 was transformed by the lithium acetate method as described by Gietz et al. (1992).
Protein Purification and Protein Binding Microarray
SlZF2 cDNA was amplified by PCR with the Pfu DNA polymerase (Promega) with primers introducing the EcoRI (5′-GAATTC-3′) restriction site in the 5′ region and the XbaI (5′-TCTAGA-3′) restriction site in the 3′ region. After enzymatic digestion, SlZF2 was cloned into the pMAL-c2 expression vector (New England Biolabs), allowing in-frame fusion of SlZF2 with the MBP coding sequence. The construct was introduced into the Escherichia coli BL2.1 strain after sequencing (Macrogen). MBP-SlZF2 protein production was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside, for 5 h at 37°C.
For protein binding array experiments, a 25-mL E. coli culture pellet was resuspended in 1 mL 1× binding buffer [(2.5 mm 2-amino-2-(hydroxymethyl)-1,3-propanediol (Tris)-HCl pH 8, 15 mm KCl, 1 mm MgCl2, 25 μm EDTA, 2.5% (v/v) glycerol, 50 mg/mL bovine serum albumin], sonicated (2 × 30 s) and centrifuged twice to obtain cleared extracts of soluble proteins as described by Godoy et al. (2011); 150 μL of these extracts supplemented with 2% (w/v) dry milk were directly used for DNA binding reactions. Incubation of protein extracts and antibodies, washing, scanning, quantification, and analysis were performed as described by Godoy et al. (2011). The PBM version used in this study contained all possible 10-mer combinations compacted in a 4 × 44k Agilent microarray, as described by Berger et al. (2006).
Malonyldialdehyde, Pro, Polyamine, Ion and Hormone Extraction, and Quantification
MDA was extracted from whole tomato plants (root and shoot) by the thiobarbituric acid reaction as described by Heath and Packer (1968). Extraction was conducted as detailed by Quinet et al. (2012). Pro was extracted and quantified as described by Carillo et al. (2008), adapted from Gibon et al. (2000). Fifty to 100 mg of plant fresh material was extracted with 1 mL of 1% (w/v) solution of ninhydrin in 60% (v/v) acetic acid. Samples were heated at 95°C for 20 min, and chilled before measuring A520.
For free PA extraction from tomato plants, approximately 250 mg of ground tissues was extracted with 500 µL of cold 4% (v/v) perchloric acid containing 5 mg L−1 1,7-diaminoheptane (as internal standard) by vortexing vigorously, and incubated 1 h at 4°C. The homogenate was centrifuged at 13,000g at 4°C for 20 min. The pellet was then reextracted with 500 µL 4% (v/v) perchloric acid and centrifuged again as described above. The two supernatants were pooled and stored at −20°C until used for free PA determination. For fluorescence detection, PAs were derivatized by dansylation according to Lefèvre et al. (2001) with some modifications. Two hundred-µL aliquots of the previous supernatant were added to 200 µL of saturated sodium carbonate and 500 µL of dansyl chloride in acetone (20 mg mL−1), and vortexed for 10 s. The mixture was incubated at 60°C for 1 h in the dark. To remove the excess dansyl chloride, 250 µL of Pro (150 mg mL−1) was subsequently added; after 1 h of incubation in the dark, acetone was evaporated under nitrogen. The PAs were then extracted with 1 mL ethyl acetate with vigorous vortexing for 60 s. The mixture was incubated for 15 min at 4°C in order to separate organic and aqueous phases. The upper organic phase was collected and dried under nitrogen. The dried extract was dissolved in 1 mL of methanol, filtered through a 0.45-µm microfilter (Chromafil PES-45/15; Macherey-Nagel), and 5 µL of sample was injected on a Nucleodur C18 Pyramid column (125 × 4.6 mm internal diameter; 5-μm particle size; Macherey-Nagel) maintained at 40°C.
HPLC coupled with a fluorescence detector was performed on a Shimadzu HPLC system, equipped with a solvent delivery unit LC-20AT, an SIL-HTc autosampler, and a RF-20A fluorescence detector (Shimadzu) with the excitation wavelength at 340 nm and the emission wavelength at 510 nm. The flow of the mobile phase was 1.0 mL⋅min−1. The mobile phase consisted of water (eluent A) and acetonitrile (eluent B). The gradient program was as follows: 40% B to 91% B (20 min), 91% B to 100% B (2 min), 100% B (8 min), 100% B to 40% B (1 min), and column equilibration at 40% B during 4 min. Total run time was 35 min.
Free PAs were quantified using six-point calibration curves with custom-made external standard solutions and internal standard (1,7-diaminoheptane), ranging from 3.125 to 100 µM. Every 10 injections, a check standard solution was used to confirm the calibration of the system. Internal standards indicated the recovery of the extraction and derivatization during the evaluation of PA content.
For mineral extraction (conducted on the third fully expanded leaf from the bottom of the plant, leaf 3), leaves were allowed to dry at least for 3 d at 70°C, and 50 to 100 mg DW of plant samples was digested in 4 mL of 67% (v/v) nitric acid at 80°C. After evaporation to dryness, samples were treated as detailed by Quinet et al. (2012). Sodium and potassium ion concentrations were determined using an atomic absorption spectrometer (ICE 3300; Thermo Scientific). All measurements were performed in triplicate.
For phytohormone quantification, fresh tomato leaves were extracted by methanol:formic acid:water (15:1:4, v/v/v; 5 mL/g FW). After centrifugation (20,000g; 15 min), the supernatant was purified using the dual-mode solid-phase method according to Dobrev and Kamínek (2002). Hormonal analysis and quantification was performed by HPLC (Ultimate 3000; Dionex) coupled to hybrid triple quadrupole/linear ion trap mass spectrometer (3200 Q TRAP; Applied Biosystems) using a multilevel calibration graph with [2H]-labeled internal standards as described by Dobrev and Vankova (2012) and Djilianov et al. (2013).
Physiological and Photosynthesis-Related Parameters
Leaf gas exchange was measured on leaf 3 (third leaf from the bottom of the plant, fully expanded) with an infrared gas analyzer (LCA4 8.7; ADC) using a PLC Parkinson leaf cuvette on intact leaves for 1 min (20 records/min), with an air flow of 300 mL min−1. Air taken from the greenhouse was introduced to a chamber (6.25 cm2 maximum leaf area) containing the leaf. The net CO2 assimilation rate (A) and instantaneous transpiration rate (E) were estimated on leaf 3, between 10 am and 12 am The stomatal conductance (gs) was measured using an AP4 system (Delta-T Devices) on leaf 3 between 2 pm and 4 pm.
To determine leaf osmotic potential on leaf 4, leaves were cut into small pieces and immediately frozen into a perforated Eppendorf tube. After two thaw-freeze cycles, sap was collected after 15 min of centrifugation (15,000g) at 4°C. Leaf osmotic potential was measured with a vapor pressure osmometer (model 5500; Wescor).
Chlorophyll fluorescence was measured on leaf 3 with a Fluorescence Monitoring System II (Hansatech Instruments) on three plants per sampling point. Part of the leaf was maintained in the dark for 30 min and was then illuminated with a first pulse of saturating light (18,000 μmol m−2 s−1) before 1 min of constant light intensity (1,200 μmol m−2 s−1). A second saturating pulse was then sent again. The Fv/Fm ratio, which represents the maximum quantum yield of open PSII, was calculated as (Fm − F0)/Fm, and NPQ and ΦPSII were calculated as (Fm − F′m)/F′m and (F′m − Fs)/F′m, respectively (Maxwell and Johnson, 2000).
Microarray Analysis
A commercial Affymetrix tomato microarray representing 43,803 tomato probes was used. Transcripts represented on the array have been selected from RefSeq, UniGene, TIGR Plant Transcript Assemblies, and TIGR Gene Indices databases. Total RNA was extracted from at least four tomato seedlings (4 weeks old) grown in vitro on MS medium. RNA purity and yield were first assessed as described above, and RNA (500 ng) quality was again examined with the Agilent 6000 RNA Nano kit by the Agilent Bioanalyzer (Agilent Technologies). Ten µg of total RNA was used for each fluorescence labeling reaction (Invitrogen). RNA fluorescence labeling, hybridization, data extraction, and gene expression analysis were conducted as described by Quinet et al. (2012). Statistical selection of differentially expressed genes between wild-type and Z4 transgenic tomato lines was based on a minimal 2-fold ratio, together with a P value ≤0.05 for the t test, for each technical repetition. Annotation of genes was completed by BLAST searches in the National Center for Biotechnology Information database.
Statistical Analysis
Statistical analyses were conducted with SAS software for Windows (version 9.1; SAS Institute). Shapiro-Wilk and Bartlett/Levene tests were performed to check normality and variance equality of the data, respectively. An ANOVA using the mean discrimination was performed on all data set using the Student-Newman-Keuls test at the 5% level. Depending on the considered data, one-way or two-way ANOVA with a cross fixed factor (treatment × genotype) was considered. Percentage values were arc-sine transformed before analysis. Data presented for Arabidopsis and tomato evaluation of tolerance to salt stress are from representative experiments among three conducted (additional data related to other experiments are presented in Supplemental Fig. S1).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers SlZF2 (ADZ15317), SlZF1 (ACG50000), NtZFP (AAC06243), StZFP1 (ABK78777), CaZFP1 (AAQ10954), ZAT10 (Q96289), AZF1 (BAA85108), ZAT7 (Q42453), ZAT12 (Q42410), and OsZFP252 (AAO46041).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotype, physiological behavior, and hormonal content of wild-type and transgenic tomato lines.
Supplemental Table S1. List of primers used to validate the microarray data by qRT-PCR.
Supplementary Material
Acknowledgments
We Brigitte Vanpée, Jacques Bar, and Thomas Dagbert for technical assistance, André Clippe for the microarray analysis, and Abdelmounaim Errachid for help with confocal microscope.
Glossary
- ABA
abscisic acid
- H2O2
hydrogen peroxide
- ROS
reactive oxygen species
- NaCl
sodium chloride
- PA
polyamine
- YFP
yellow fluorescent protein
- CaMV
Cauliflower mosaic virus
- KCl
potassium chloride
- MS
Murashige and Skoog
- MDA
malonyldialdehyde
- FW
fresh weight
- DW
dry weight
- Fv/Fm
maximum photochemical efficiency of PSII in the dark-adapted state
- ΦPSII
quantum yield of PSII
- NPQ
nonphotochemical quenching
- GA
gibberellic acid
- qRT-PCR
quantitative reverse transcription-PCR
- CK
cytokinin
- IAA
indole-3-acetic acid
- ABC
ATP-binding cassette
- GRP
Gly-rich protein
- H2O2
hydrogen peroxide
Footnotes
This work was supported by the Fonds National pour la Recherche Scientifique (convention no. 1–5117–11), the Czech Science Foundation (P506/11/0774, to E.Z., P.I.D., and V.M), the Wallonie-Bruxelles International program (Rhéa 2011/35047), the Université Catholique de Louvain (postdoctoral fellowship to I.H.), and the Spanish Ministry for Science and Innovation (grant nos. BIO2010–21739, CSD2007–00057, and EUI2008–03666 to R.S.) for work in the Solano Laboratory.
The online version of this article contains Web-only data.
References
- Agarwal P, Arora R, Ray S, Singh AK, Singh VP, Takatsuji H, Kapoor S, Tyagi AK. (2007) Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol Biol 65: 467–485 [DOI] [PubMed] [Google Scholar]
- Albacete A, Ghanem ME, Martínez-Andújar C, Acosta M, Sánchez-Bravo J, Martínez V, Lutts S, Dodd IC, Pérez-Alfocea F. (2008) Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J Exp Bot 59: 4119–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, III, Bulyk ML. (2006) Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol 24: 1429–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghesi E, González-Miret ML, Escudero-Gilete ML, Malorgio F, Heredia FJ, Meléndez-Martínez AJ. (2011) Effects of salinity stress on carotenoids, anthocyanins, and color of diverse tomato genotypes. J Agric Food Chem 59: 11676–11682 [DOI] [PubMed] [Google Scholar]
- Bradford KJ, Downie AB, Gee OH, Alvarado V, Yang H, Dahal P. (2003) Abscisic acid and gibberellin differentially regulate expression of genes of the SNF1-related kinase complex in tomato seeds. Plant Physiol 132: 1560–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitink J, Thomas M, Gissot L, Leprince O. (2004) Starvation, osmotic stress and desiccation tolerance lead to expression of different genes of the regulatory β and γ subunits of the SnRK1 complex in germinating seeds of Medicago truncatula. Plant Cell Environ 27: 55–67 [Google Scholar]
- Carillo P, Mastrolonardo G, Nacca F, Parisi D, Verlotta A, Fuggi A. (2008) Nitrogen metabolism in durum wheat under salinity: accumulation of proline and glycine betaine. Funct Plant Biol 35: 412–426 [DOI] [PubMed] [Google Scholar]
- Chen X, Han H, Jiang P, Nie L, Bao H, Fan P, Lv S, Feng J, Li Y. (2011) Transformation of beta-lycopene cyclase genes from Salicornia europaea and Arabidopsis conferred salt tolerance in Arabidopsis and tobacco. Plant Cell Physiol 52: 909–921 [DOI] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Mittler R. (2008) The zinc finger network of plants. Cell Mol Life Sci 65: 1150–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, Warren D, Cushman J, Connolly EL, Mittler R. (2007) The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem 282: 9260–9268 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Last K, Harrett-Williams R, Tagliavia C, Harter K, Hooley R, Hancock JT, Neill SJ. (2006) Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J 47: 907–916 [DOI] [PubMed] [Google Scholar]
- Dinkins R, Pflipsen C, Thompson A, Collins GB. (2002) Ectopic expression of an Arabidopsis single zinc finger gene in tobacco results in dwarf plants. Plant Cell Physiol 43: 743–750 [DOI] [PubMed] [Google Scholar]
- Djilianov DL, Dobrev PI, Moyankova DP, Vankova R, Georgieva DT, Gajdošová S, Motyka V. (2013) Dynamics of endogenous phytohormones during desiccation and recovery of the resurrection plant species Haberlea rhodopensis. J Plant Growth Regul 32: 564–574 [Google Scholar]
- Dobrev PI, Kamínek M. (2002) Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J Chromatogr A 950: 21–29 [DOI] [PubMed] [Google Scholar]
- Dobrev PI, Vankova R. (2012) Quantification of abscisic acid, cytokinin, and auxin content in salt-stressed plant tissues. In Shabala S, Cuin TA, eds, Plant Salt Tolerance: Methods and Protocols, Methods in Molecular Biology, Vol 913, Springer Science + Business Media, New York, pp 251–261 [DOI] [PubMed] [Google Scholar]
- Dodd IC. (2003) Hormonal interactions and stomatal responses. J Plant Growth Regul 22: 32–46 [Google Scholar]
- Ellul P, Garcia-Sogo B, Pineda B, Ríos G, Roig LA, Moreno V. (2003) The ploidy level of transgenic plants in Agrobacterium-mediated transformation of tomato cotyledons ( Lycopersicon esculentum Mill.) is genotype and procedure dependent [corrected]. Theor Appl Genet 106: 231–238 [DOI] [PubMed] [Google Scholar]
- Englbrecht CC, Schoof H, Böhm S. (2004) Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl): S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton TM, Chunwongse J, Tanksley SD. (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13: 207–209 [Google Scholar]
- Gao H, Song A, Zhu X, Chen F, Jiang J, Chen Y, Sun Y, Shan H, Gu C, Li P, et al. (2012) The heterologous expression in Arabidopsis of a chrysanthemum Cys2/His2 zinc finger protein gene confers salinity and drought tolerance. Planta 235: 979–993 [DOI] [PubMed] [Google Scholar]
- Ghanem ME, Albacete A, Martínez-Andújar C, Acosta M, Romero-Aranda R, Dodd IC, Lutts S, Pérez-Alfocea F. (2008) Hormonal changes during salinity-induced leaf senescence in tomato (Solanum lycopersicum L.). J Exp Bot 59: 3039–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Sulpice R, Larher F. (2000) Proline accumulation in canola leaf discs subjected to osmotic stress is related to the loss of chlorophylls and to the decrease of mitochondrial activity. Physiol Plant 110: 469–476 [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. (2010) Polyamines and abiotic stress tolerance in plants. Plant Signal Behav 5: 26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy M, Franco-Zorrilla JM, Pérez-Pérez J, Oliveros JC, Lorenzo O, Solano R. (2011) Improved protein-binding microarrays for the identification of DNA-binding specificities of transcription factors. Plant J 66: 700–711 [DOI] [PubMed] [Google Scholar]
- Gourcilleau D, Lenne C, Armenise C, Moulia B, Julien JL, Bronner G, Leblanc-Fournier N. (2011) Phylogenetic study of plant Q-type C2H2 zinc finger proteins and expression analysis of poplar genes in response to osmotic, cold and mechanical stresses. DNA Res 18: 77–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazarika P, Rajam MV. (2011) Biotic and abiotic stress tolerance in transgenic tomatoes by constitutive expression of S-adenosylmethionine decarboxylase gene. Physiol Mol Biol Plants 17: 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RL, Packer L. (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125: 189–198 [DOI] [PubMed] [Google Scholar]
- Hichri I, Heppel SC, Pillet J, Léon C, Czemmel S, Delrot S, Lauvergeat V, Bogs J. (2010) The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol Plant 3: 509–523 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Zhang Y, Shi Y, Zhang Z, Zou Z, Zhang H, Zhao J. (2012) Effect of exogenous spermidine on polyamine content and metabolism in tomato exposed to salinity-alkalinity mixed stress. Plant Physiol Biochem 57: 200–209 [DOI] [PubMed] [Google Scholar]
- Huang D, Wu W, Abrams SR, Cutler AJ. (2008) The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J Exp Bot 59: 2991–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Yang X, Wang MM, Tang HJ, Ding LY, Shen Y, Zhang HS. (2007) A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochim Biophys Acta 1769: 220–227 [DOI] [PubMed] [Google Scholar]
- Jiang L, Pan LJ. (2012) Identification and expression of C2H2 transcription factor genes in Carica papaya under abiotic and biotic stresses. Mol Biol Rep 39: 7105–7115 [DOI] [PubMed] [Google Scholar]
- Jyothishwaran G, Kotresha D, Selvaraj T, Srideshikan SM, Rajvanshi PK, Jayabaskaran C. (2007) A modified freeze–thaw method for efficient transformation of Agrobacterium tumefaciens. Curr Sci 93: 770–772 [Google Scholar]
- Kar RK. (2011) Plant responses to water stress: role of reactive oxygen species. Plant Signal Behav 6: 1741–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kasinathan V, Wingler A. (2004) Effect of reduced arginine decarboxylase activity on salt tolerance and on polyamine formation during salt stress in Arabidopsis thaliana. Physiol Plant 121: 101–107 [DOI] [PubMed] [Google Scholar]
- Kasukabe Y, He L, Nada K, Misawa S, Ihara I, Tachibana S. (2004) Overexpression of spermidine synthase enhances tolerance to multiple environmental stresses and up-regulates the expression of various stress-regulated genes in transgenic Arabidopsis thaliana. Plant Cell Physiol 45: 712–722 [DOI] [PubMed] [Google Scholar]
- Kazan K. (2006) Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci 11: 109–112 [DOI] [PubMed] [Google Scholar]
- Kiełbowicz-Matuk A. (2012) Involvement of plant C(2)H(2)-type zinc finger transcription factors in stress responses. Plant Sci 185-186: 78–85 [DOI] [PubMed] [Google Scholar]
- Kim SH, Hong JK, Lee SC, Sohn KH, Jung HW, Hwang BK. (2004) CAZFP1, Cys2/His2-type zinc-finger transcription factor gene functions as a pathogen-induced early-defense gene in Capsicum annuum. Plant Mol Biol 55: 883–904 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Horiuchi H, Fujita K, Takuhara Y, Suzuki S. (2012) Characterization of grape C-repeat-binding factor 2 and B-box-type zinc finger protein in transgenic Arabidopsis plants under stress conditions. Mol Biol Rep 39: 7933–7939 [DOI] [PubMed] [Google Scholar]
- Kodaira KS, Qin F, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. (2011) Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol 157: 742–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Hilhorst HW, Karssen CM. (1989) In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol 90: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Ki, Sakamoto A, Kobayashi A, Rybka Z, Kanno Y, Nakagawa H, Takatsuji H. (1998) Cys2/His2 zinc-finger protein family of petunia: evolution and general mechanism of target-sequence recognition. Nucleic Acids Res 26: 608–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulik A, Wawer I, Krzywińska E, Bucholc M, Dobrowolska G. (2011) SnRK2 protein kinases—key regulators of plant response to abiotic stresses. OMICS 15: 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre I, Gratia E, Lutts S. (2001) Discrimination between the ionic and osmotic components of salt stress in relation to free polyamine level in rice (Oryza sativa). Plant Sci 161: 943–952 [Google Scholar]
- Lin PC, Pomeranz MC, Jikumaru Y, Kang SG, Hah C, Fujioka S, Kamiya Y, Jang JC. (2011) The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J 65: 253–268 [DOI] [PubMed] [Google Scholar]
- Linkies A, Leubner-Metzger G. (2012) Beyond gibberellins and abscisic acid: how ethylene and jasmonates control seed germination. Plant Cell Rep 31: 253–270 [DOI] [PubMed] [Google Scholar]
- Lovelli S, Scopa A, Perniola M, Di Tommaso T, Sofo A. (2012) Abscisic acid root and leaf concentration in relation to biomass partitioning in salinized tomato plants. J Plant Physiol 169: 226–233 [DOI] [PubMed] [Google Scholar]
- Mangeon A, Junqueira RM, Sachetto-Martins G. (2010) Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal Behav 5: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. (2000) Chlorophyll fluorescence—a practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Mishra G, Zhang W, Deng F, Zhao J, Wang X. (2006) A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312: 264–266 [DOI] [PubMed] [Google Scholar]
- Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK. (2006) Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett 580: 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittova V, Tal M, Volokita M, Guy M. (2002) Salt stress induces up-regulation of an efficient chloroplast antioxidant system in the salt-tolerant wild tomato species Lycopersicon pennellii but not in the cultivated species. Physiol Plant 115: 393–400 [DOI] [PubMed] [Google Scholar]
- Mitsuya Y, Takahashi Y, Uehara Y, Berberich T, Miyazaki A, Takahashi H, Kusano T. (2007) Identification of a novel Cys2/His2-type zinc-finger protein as a component of a spermine-signaling pathway in tobacco. J Plant Physiol 164: 785–793 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, et al. (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17: 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34–36 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K. (2013) ABA signaling in stress-response and seed development. Plant Cell Rep 32: 959–970 [DOI] [PubMed] [Google Scholar]
- Neily MH, Matsukura C, Maucourt M, Bernillon S, Deborde C, Moing A, Yin YG, Saito T, Mori K, Asamizu E, et al. (2011) Enhanced polyamine accumulation alters carotenoid metabolism at the transcriptional level in tomato fruit over-expressing spermidine synthase. J Plant Physiol 168: 242–252 [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang B, Yang T, Li H, Zhang L, Zhang Y, Zhang J, Fei Z, Ye Z. (2007) Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. J Exp Bot 58: 507–520 [DOI] [PubMed] [Google Scholar]
- Pabo CO, Peisach E, Grant RA. (2001) Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem 70: 313–340 [DOI] [PubMed] [Google Scholar]
- Penfield S, Hall A. (2009) A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 21: 1722–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Quinet M, Vromman D, Clippe A, Bertin P, Lequeux H, Dufey I, Lutts S, Lefèvre I. (2012) Combined transcriptomic and physiological approaches reveal strong differences between short- and long-term response of rice (Oryza sativa) to iron toxicity. Plant Cell Environ 35: 1837–1859 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Araki T, Meshi T, Iwabuchi M. (2000) Expression of a subset of the Arabidopsis Cys(2)/His(2)-type zinc-finger protein gene family under water stress. Gene 248: 23–32 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 136: 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU. (2004) Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131: 1491–1501 [DOI] [PubMed] [Google Scholar]
- Solanke AU, Sharma MK, Tyagi AK, Sharma AK. (2009) Characterization and phylogenetic analysis of environmental stress-responsive SAP gene family encoding A20/AN1 zinc finger proteins in tomato. Mol Genet Genomics 282: 153–164 [DOI] [PubMed] [Google Scholar]
- Sturaro M, Vernieri P, Castiglioni P, Binelli G, Gavazzi G. (1996) The rea (red embryonic axis) phenotype describes a new mutation affecting the response of maize embryos to abscisic acid and osmotic stress. J Exp Bot 47: 755–762 [Google Scholar]
- Subramanian C, Woo J, Cai X, Xu X, Servick S, Johnson CH, Nebenführ A, von Arnim AG. (2006) A suite of tools and application notes for in vivo protein interaction assays using bioluminescence resonance energy transfer (BRET). Plant J 48: 138–152 [DOI] [PubMed] [Google Scholar]
- Sun SJ, Guo SQ, Yang X, Bao YM, Tang HJ, Sun H, Huang J, Zhang HS. (2010) Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J Exp Bot 61: 2807–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji H, Matsumoto T. (1996) Target-sequence recognition by separate-type Cys2/His2 zinc finger proteins in plants. J Biol Chem 271: 23368–23373 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Sano T, Tamaoki M, Nakajima N, Kondo N, Hasezawa S. (2005) Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol 138: 2337–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian ZD, Zhang Y, Liu J, Xie CH. (2010) Novel potato C2H2-type zinc finger protein gene, StZFP1, which responds to biotic and abiotic stress, plays a role in salt tolerance. Plant Biol (Stuttg) 12: 689–697 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: Research0034.1–Research0034.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Close TJ. (2007) Large-scale expression profiling and physiological characterization of jasmonic acid-mediated adaptation of barley to salinity stress. Plant Cell Environ 30: 410–421 [DOI] [PubMed] [Google Scholar]
- Wang RS, Pandey S, Li S, Gookin TE, Zhao Z, Albert R, Assmann SM. (2011) Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HT, Guo P, Xia XL, Yin WL. (2011) PdERECTA, a leucine-rich repeat receptor-like kinase of poplar, confers enhanced water use efficiency in Arabidopsis. Planta 234: 229–241 [DOI] [PubMed] [Google Scholar]
- Xu DQ, Huang J, Guo SQ, Yang X, Bao YM, Tang HJ, Zhang HS. (2008) Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.). FEBS Lett 582: 1037–1043 [DOI] [PubMed] [Google Scholar]
- Yan J, Tsuichihara N, Etoh T, Iwai S. (2007) Reactive oxygen species and nitric oxide are involved in ABA inhibition of stomatal opening. Plant Cell Environ 30: 1320–1325 [DOI] [PubMed] [Google Scholar]
- Zhang H, Han B, Wang T, Chen S, Li H, Zhang Y, Dai S. (2012) Mechanisms of plant salt response: insights from proteomics. J Proteome Res 11: 49–67 [DOI] [PubMed] [Google Scholar]
- Zhang X, Guo XP, Lei CL, Chen Z, Lin Q, Wang J, Wu FQ, Wang J, Wan J. (2011) Overexpression of SlCZFP1, a novel TFIIIA-type zinc finger protein from tomato, confers enhanced cold tolerance in transgenic Arabidopsis and rice. Plant Mol Biol Rep 29: 185–196 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.