Rice heat-shock proteins modulate long-term acquired thermotolerance, which can be decoupled from basal thermotolerance in different rice cultivars.
Abstract
Heat stress is an important factor that has a negative impact on rice (Oryza sativa) production. To alleviate this problem, it is necessary to extensively understand the genetic basis of heat tolerance and adaptability to heat stress in rice. Here, we report the molecular mechanism underlying heat acclimation memory that confers long-term acquired thermotolerance (LAT) in this monocot plant. Our results showed that a positive feedback loop formed by two heat-inducible genes, HEAT SHOCK PROTEIN101 (HSP101) and HEAT STRESS-ASSOCIATED 32-KD PROTEIN (HSA32), at the posttranscriptional level prolongs the effect of heat acclimation in rice seedlings. The interplay between HSP101 and HSA32 also affects basal thermotolerance of rice seeds. These findings are similar to those reported for the dicot plant Arabidopsis (Arabidopsis thaliana), suggesting a conserved function in plant heat stress response. Comparison between two rice cultivars, japonica Nipponbare and indica N22 showed opposite performance in basal thermotolerance and LAT assays. ‘N22’ seedlings have a higher basal thermotolerance level than cv Nipponbare and vice versa at the LAT level, indicating that these two types of thermotolerance can be decoupled. The HSP101 and HSA32 protein levels were substantially higher in cv Nipponbare than in cv N22 after a long recovery following heat acclimation treatment, at least partly explaining the difference in the LAT phenotype. Our results point out the complexity of thermotolerance diversity in rice cultivars, which may need to be taken into consideration when breeding for heat tolerance for different climate scenarios.
Rice (Oryza sativa) is an important staple food crop that feeds a major portion of the world population. High temperatures have been shown to negatively affect rice yield and quality (Peng et al., 2004; Lin et al., 2005; Ishimaru et al., 2009; Welch et al., 2010). Climate change may aggravate this problem in the future due to a higher probability of higher growing season temperatures (Battisti and Naylor, 2009). To alleviate this threat, it is necessary to improve the adaptability of rice genetically to cope with the warming trends. However, the genetic basis for heat tolerance or adaptability to heat stress in crop plants, including rice, is poorly understood. Association between the adaptability of crop plants to high temperature stress and heat acclimation has been reported (Chen et al., 1982).
Heat acclimation is a physiological response manifested in the acquired thermotolerance of organisms or cells to noxious high temperature following exposure to a moderate heat stress condition. In general, acclimation under moderate heat stress induces the expression of a large number of genes that encode proteins with diverse biological functions, a process known as heat shock or heat stress response (Lindquist, 1986). The effect of heat acclimation is transient in nature, as the level of acquired thermotolerance gradually decreases to the preacclimation state in a few days (Kregel, 2002; Charng et al., 2006). In mammalian cells, decay of acquired thermotolerance was shown to be associated with the decay of heat-induced heat shock protein70 (HSP70; Landry et al., 1982).
Reverse genetic studies conducted on HEAT STRESS-ASSOCIATED 32-KD PROTEIN (HSA32) showed that it retards the decay of acquired thermotolerance in Arabidopsis (Arabidopsis thaliana; Charng et al., 2006). HSA32 is a heat-induced protein mainly present in land plants (Liu et al., 2006). It is encoded by a single-copy gene in many higher plant species, including Arabidopsis, tomato (Solanum lycopersicum), and rice (Liu et al., 2006). In the HSA32 knockout (KO) or knockdown mutants in Arabidopsis, acquired thermotolerance was induced to wild-type level after a short recovery of 2 h following acclimation treatment at 37°C but was compromised significantly after a long recovery for 48 h. Based on this phenotype, acquired thermotolerance retained after a long recovery period was named long-term acquired thermotolerance (LAT), as opposed to the short-term acquired thermotolerance (SAT) that is attained after a short recovery (Yeh et al., 2012). Recently, it was shown that HSA32 regulates the degradation rate of HSP101, a cytosolic caseinolytic peptidase B (ClpB)/HSP100 family protein in plants (Wu et al., 2013). Cytosolic ClpB/HSP100 proteins are molecular chaperones that promote the renaturation of protein aggregates (Mogk et al., 2008) and are required for the development of acquired thermotolerance (Sanchez and Lindquist, 1990; Hong and Vierling, 2000; Queitsch et al., 2000; Nieto-Sotelo et al., 2002). In the absence of HSA32, heat-induced HSP101 is degraded faster. Interestingly, the level of HSA32 protein itself is positively regulated by HSP101, suggesting that HSA32 and HSP101 form a positive feedback loop to prolong the memory of heat acclimation (Wu et al., 2013).
In cultivated rice, heat-induced HSP101 was shown to decay faster in the seedlings of the indica rice varieties than in that of the japonica varieties (Agarwal et al., 2003). Because the indica and japonica cultivars are tropical and temperate rice varieties, respectively, the differential response of HSP101 suggests an ecophysiological role of modulating the decay of the molecular chaperone in adaptation to different climate zones. How decay of HSP101 is modulated in rice is not clear. Previously, expression of rice HSA32 was shown to be highly induced by heat treatment (Charng et al., 2006). However, the biological function of HSA32 in rice has not been shown. Based on the working model found in Arabidopsis, we hypothesized that interplay between HSP101 and HSA32 also exists in rice and that this interplay affects LAT.
In this study, we used rice retrotransposon Tos17 insertion lines of HSA32 and RNA interference (RNAi) lines of HSP101 to test the above hypothesis. Our results showed that disruption of HSA32 led to faster decay of HSP101 and a defect in LAT in rice, and suppression of HSP101 by RNAi-mediated gene silencing compromised the expression of HSA32 posttranscriptionally. Comparison between a japonica rice cultivar, Nipponbare, and an indica cultivar, N22, showed opposite performance in basal thermotolerance and LAT assays. ‘N22’ has a higher basal thermotolerance level than cv Nipponbare, and cv Nipponbare has a higher LAT level than cv N22, indicating that basal thermotolerance is dissociable from LAT. The higher LAT level in cv Nipponbare was associated with a slower decay of HSPs, including HSP101, than in cv N22. These results shed light on the thermotolerance diversity in rice cultivars, which may need to be taken into consideration in breeding heat tolerance trait for different climate scenarios.
RESULTS
Expression Pattern of HSA32 and HSP101 in Rice Tissues
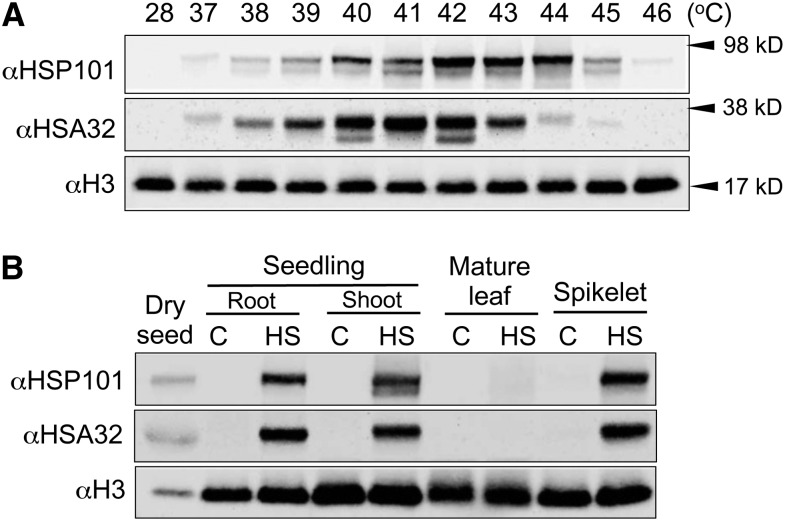
To investigate the function of HSA32 and its relationship with HSP101 in rice, we first analyzed the expression patterns of the proteins in different rice tissues under heat-stressed and normal conditions with immunoblotting. To this end, the japonica rice cultivar Nipponbare was used because HSA32 KO mutants were found in this genetic background. The antibodies against rice HSA32 and HSP101 were raised as described in “Materials and Methods.” The antibodies specifically recognized the heat-inducible proteins corresponding to the sizes of HSA32 and HSP101 that were missing in the HSA32 KO and HSP101 knockdown lines, respectively (refer to results below). We then determined the optimal temperature for inducing the proteins by treating the rice seedlings at various temperatures from 37°C to 46°C for 2 h. Figure 1A shows that the responses of HSA32 and HSP101 to elevated temperature were different. HSA32 was induced to its highest level at 41°C, while the optimal temperature for inducing HSP101 was 42°C to 43°C. For convenience, we subsequently used 42°C for inducing both proteins. Figure 1B shows that HSA32 and HSP101 in seedling shoots and roots and spikelets were barely detectable under normal conditions but were induced to high level by heat treatment (Fig. 1B); however, such drastic induction by heat was not obvious in the mature leaf. Moreover, these proteins could be detected in nonheated mature seeds (Fig. 1B).
Figure 1.
Rice HSP101 and HSA32 protein levels induced under varied high temperatures and in different tissues. A, Immunoblot analysis of HSP101 and HSA32 proteins in 4-d-old seedlings exposed to varied high temperatures. The seedlings were subjected to heat treatment at 37°C to 46°C for 2 h and recovered at 28°C for 2 h before being used for protein extraction. A sample kept at 28°C was used as a nonheated control. B, HSP101 and HSA32 proteins in different rice tissues detected by immunoblotting. Total proteins were extracted from dry seeds, shoots, and roots of 4-d-old seedlings, the first leaf below the flag leaf (labeled as the mature leaf), and spikelets collected 3 to 4 d after anthesis. The samples were treated at 28°C (C) or 42°C (HS) for 2 h and recovered at 28°C for 2 h before being used for protein extraction. In each lane, 60 μg of total protein was loaded. The staining of histone H3 was used as a loading control in all panels. The tests were repeated at least once, and similar results were obtained.
Isolation and Characterization of Tos17 Insertion Mutants of Rice HSA32
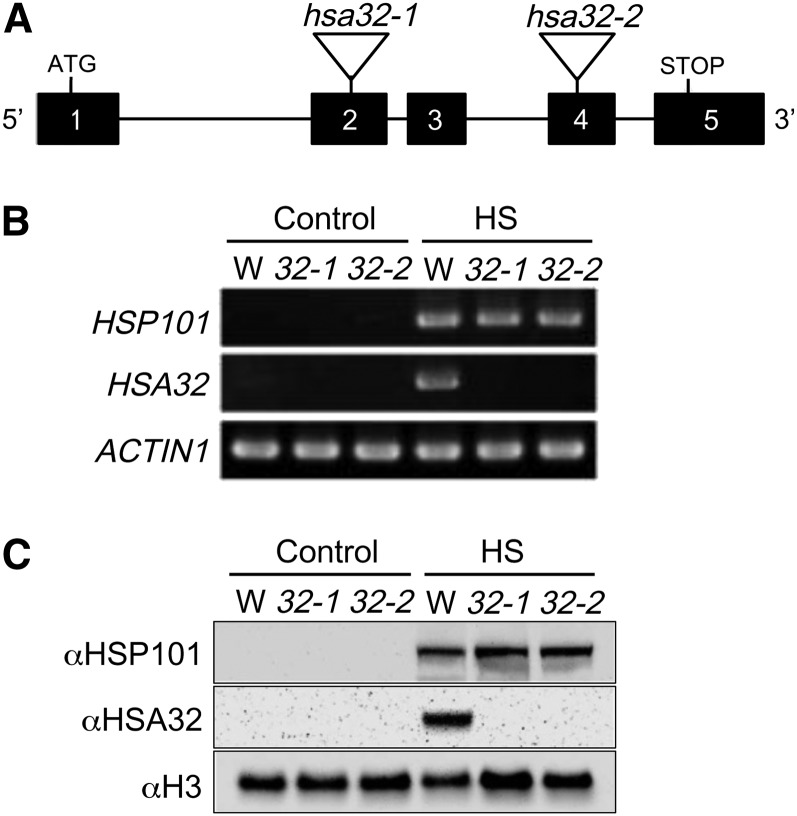
To elucidate the function of HSA32 in rice, we obtained two rice transposon Tos17 insertion mutants of HSA32 in cv Nipponbare background, one with the transposon inserted in the exon 2 (hsa32-1) and another in exon 4 (hsa32-2; Fig. 2A). The locations of Tos17 insertions were confirmed by genotyping, and homozygous insertion lines were isolated for further studies. Reverse transcription (RT)-PCR analysis showed that in wild-type seedlings, HSA32 was expressed at very low level under control conditions (28°C) but was substantially induced under heat stress at 42°C for 90 min (Fig. 2B). In Tos17 homozygous insertion mutants, HSA32 transcripts were not detectable after heat treatment, but HSP101, as a positive control, was expressed to a wild-type level (Fig. 2B). Similarly, immunoblotting analysis showed that the heat-induced HSA32 was not detectable in the mutants, while the level of HSP101 was not affected (Fig. 2C). These data indicate that the two independent Tos17 insertion events disrupt the integrity of rice HSA32 and generate two allelic KO mutants. Under normal conditions, no obvious phenotypic differences between the mutant and wild-type plants were observed in terms of germination and growth rate, time to flowering, and seed setting, suggesting that HSA32 is not essential for normal growth and development. However, we could not exclude the possibility that HSA32 has a minor effect at a certain growth stage, which requires rigorous examination.
Figure 2.
Characterization of homozygous Tos17 insertion mutants of rice HSA32. A, Schematic representation of the gene structure of rice HSA32 and locations of Tos17 in two allelic mutants, hsa32-1 and hsa32-2. The numbered black boxes and horizontal lines represent exons and introns, respectively. The locations of start and stop codons are shown. Tos17 insertion sites are marked by triangles. B, RT-PCR analysis of the transcript levels of HSA32 in 4-d-old wild-type (W), hsa32-1 (32-1), and hsa32-2 (32-2) seedlings treated at 28°C (Control) or 42°C (HS) for 90 min. The RT-PCR products of HSP101 and ACTIN1 are shown as heat treatment and loading controls, respectively. Twenty-five cycles of PCR were performed. C, Immunoblot analysis of HSA32 protein in the crude extracts prepared from seedling samples described in B. In each lane, 60 μg of total protein was loaded. HSP101 and histone H3 are shown as heat treatment and loading controls, respectively.
HSP101 and Basal Thermotolerance Levels of HSA32 KO Mutants Are Lower Than Those of the Wild Type in Rice Seeds
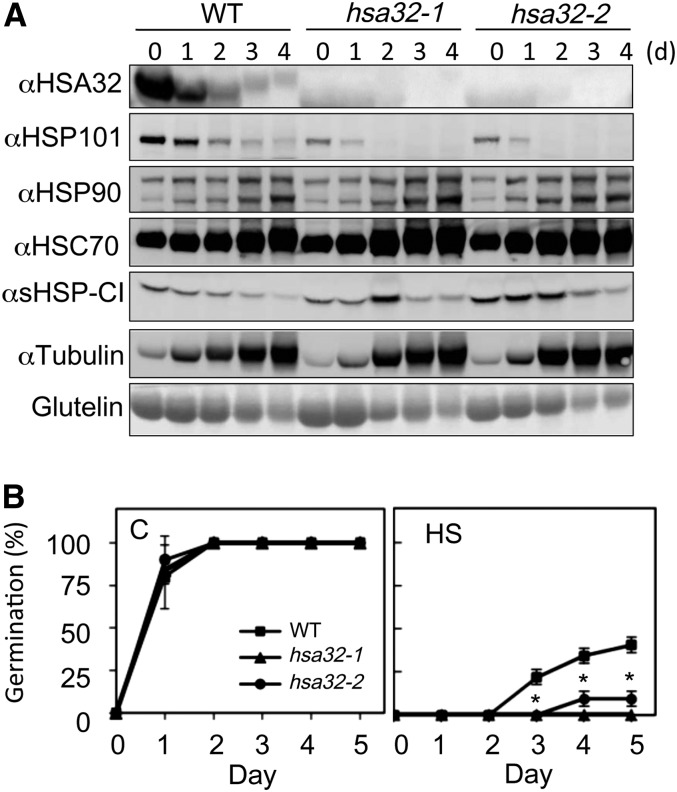
Figure 1B shows that HSA32 is present in mature seeds. To determine the effect of the HSA32 null mutations, we first examined the levels of HSPs and thermotolerance in rice seeds of the wild type and KO mutants. The levels of HSA32, HSP101, HSP90, the eukaryotic 70-kD heat shock cognate HSC70, and class I small HSPs (sHSP-CI) during seed imbibition were analyzed by immunoblot analysis. The results confirmed that HSA32 protein was not detected in the mutant seeds (Fig. 3A). The patterns of other HSPs in the mutants did not substantially differ from those in the wild type except for HSP101. HSP101 level in the wild-type seeds was highest before imbibition and then gradually degraded during imbibition up to 4 d (Fig. 3A). In both the hsa32-1 and hsa32-2 seeds, HSP101 level was substantially lower than in the wild-type seeds. We then measured the seed thermotolerance by treating the imbibed seeds of the wild type and HSA32 mutants at 58°C for 30 min. Without heat treatment, the wild-type and mutant seeds showed no difference in germination rate, while the heat treatment retarded seed germination more severely in the mutants than in the wild type (Fig. 3B). These results suggest that HSA32 is required for maintaining the levels of HSP101 and basal thermotolerance in rice seeds.
Figure 3.
Effect of disrupting rice HSA32 on the profile of HSPs in germinating seeds and thermotolerance of imbibed seeds. A, Immunoblot analysis of the indicated HSPs in the crude extracts prepared from imbibed seeds of the wild type (WT) and the hsa32 mutants. The seeds were imbibed for 0 to 5 d at 28°C. In each lane, 60 μg of total protein was loaded. Tubulin and storage protein glutelin stained with naphthol blue black are shown as loading controls. Two biological repeats were performed, and similar results were obtained. B, Germination rates of the wild-type and hsa32 mutant seeds were assessed after treatment at 28°C (C) or 58°C (HS) for 30 min. Results are presented as mean values of three biological replicates ± sd (n = 48). *P < 0.01 (versus the wild type, Student’s t test).
Heat-Induced HSP101 Decays Faster in HSA32 KO Mutants Than in the Wild Type during Poststress Recovery in Rice Seedlings
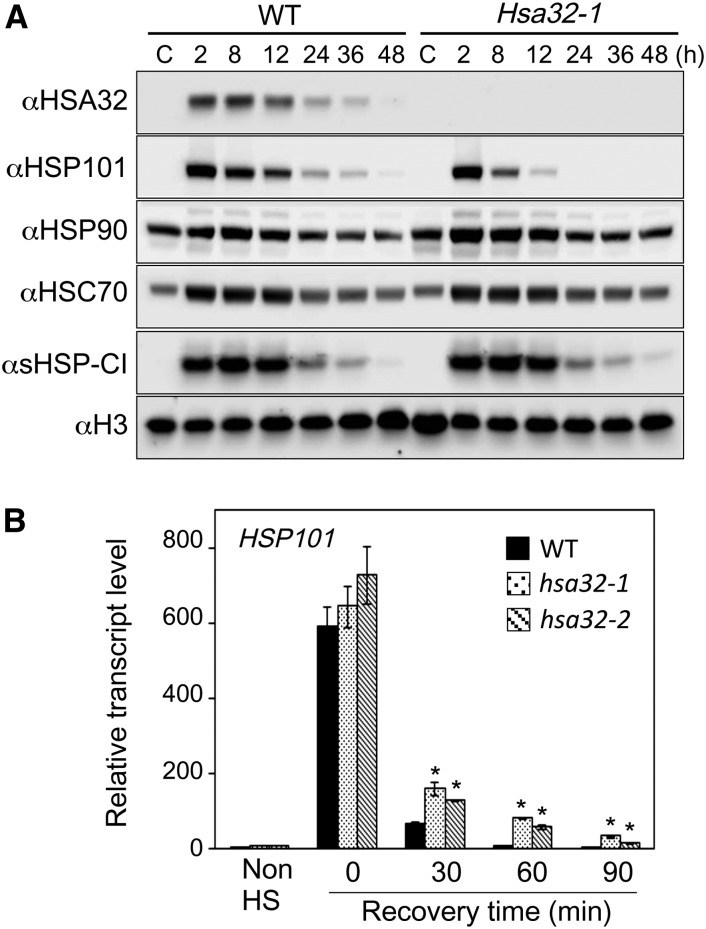
In Arabidopsis, HSA32 was shown to retard the degradation of heat-induced HSP101 (Wu et al., 2013). To see whether rice HSA32 has similar function, HSP101 level was examined in HSA32 KO mutant and the wild-type seedlings during recovery after heat treatment. Figure 4A shows that HSP101 was induced to similarly high levels in the wild type and hsa32-1 after heat treatment at 42°C for 2 h and recovery at 28°C for 2 h. Gradual decline in HSP101 level after longer recovery was observed in both lines, but the protein declined faster in hsa32-1. HSP101 was barely detectable after recovery for 24 h in the mutant, while it was still detectable in the wild type after 48 h of recovery (Fig. 4A). Other HSPs, such as HSP90, HSC70/HSP70, and sHSP-CI, were not decayed at substantially different rates in the wild type and the mutant, suggesting the effect of HSA32 on HSP101 is relatively specific. A similar result was observed in another mutant line, hsa32-2 (Supplemental Fig. S1). We examined the transcript level of HSP101 in the seedlings during recovery after heat treatment using quantitative RT-PCR. Unlike the slow decay in protein level, HSP101 transcripts were reduced to the prestress level after recovery for 60 min in the wild type (Fig. 4B). In the HSA32 KO lines, the levels of HSP101 transcripts were slightly higher than that in the wild type, suggesting that the faster decay of HSP101 in the HSA32 KO mutants was not due to a lower transcript level of HSP101.
Figure 4.
Faster decay of heat-induced HSP101 protein, not its transcripts, in the absence of HSA32. A, Immunoblot analysis of the indicated HSPs in 4-d-old wild-type and hsa32-1 mutant seedlings, from which the crude extracts were prepared after recovery for 2 to 48 h following heat treatment at 42°C for 2 h. The nonheated sample was included as a control (C). In each lane, 60 μg of total protein was loaded. Histone H3 is shown as a loading control. B, Quantitative RT-PCR analysis of the transcript levels of HSP101 in wild-type (WT), hsa32-1, and hsa32-2 seedlings before (Non HS) or after heat treatment at 42°C for 2 h followed by recovery at 28°C for 0 to 90 min. Relative transcript levels of HSP101 were obtained by normalization to the nonheated wild-type sample, which is set as one. Results are presented as mean values of three technical replicates ± sd. *P < 0.01 (versus the wild type, Student’s t test). The tests were repeated at least once, and similar results were obtained.
HSA32 KO Mutants Show a Defect in LAT But Not in SAT
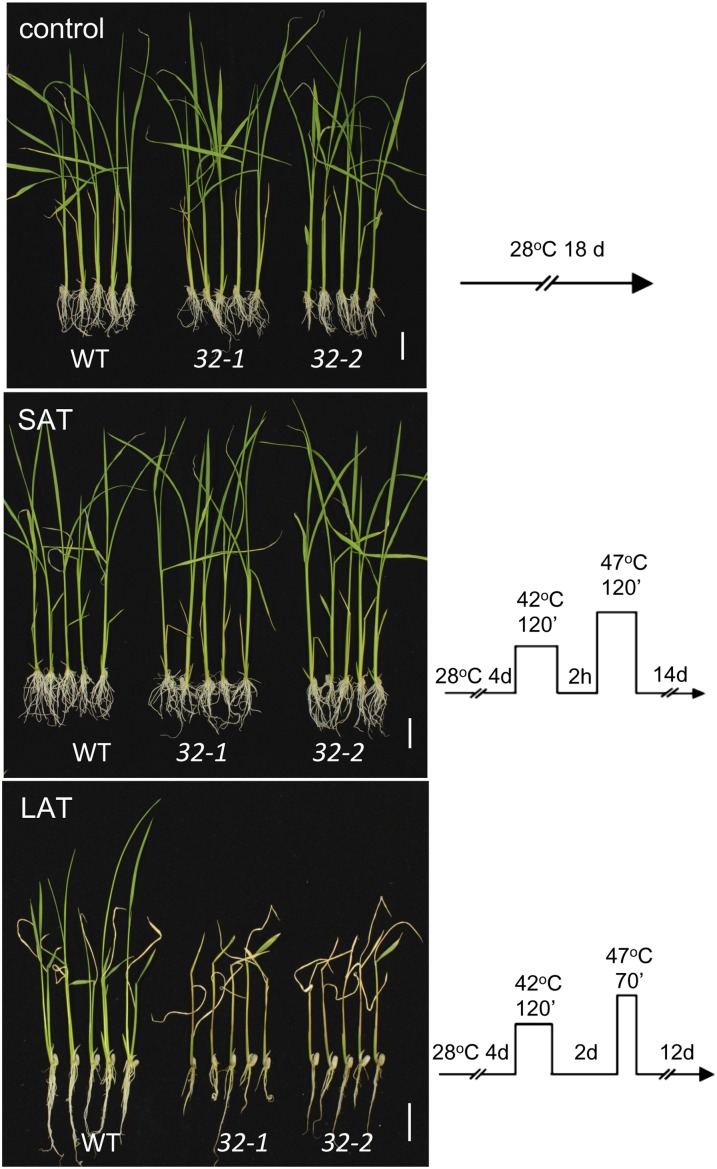
To investigate whether disruption of rice HSA32 leads to a defect in acquired thermotolerance, SAT and LAT assays were performed on 4-d-old seedlings of the wild type and hsa32-1 and hsa32-2 mutants. Our results showed that there was no significant difference between the wild-type and mutant seedlings in morphology (Fig. 5) or fresh weight (Supplemental Fig. S2) under nonstressed conditions or after SAT assay treatment. However, after the LAT assay treatment, the HSA32 KO mutant plants all died, whereas the wild-type plants were alive and continued to grow by forming new tillers (Fig. 5). This result suggests that HSA32 is required for LAT but not for SAT in rice seedlings.
Figure 5.
Substantial defect in LAT but not in SAT observed in HSA32 KO mutants. SAT and LAT assays of wild-type (WT), hsa32-1 (32-1), and hsa32-2 (32-2) seedlings were performed under the corresponding heat stress regimes shown schematically on the right. The control group contains seedlings of the same age without heat treatments. The representative plants were photographed after recovery for the indicated time. White bars represent 2 cm in each picture. Three biological repeats were performed, and similar results were obtained.
HSP101 Positively Regulates the Level of HSA32 in Rice Seeds and Heat-Treated Seedlings
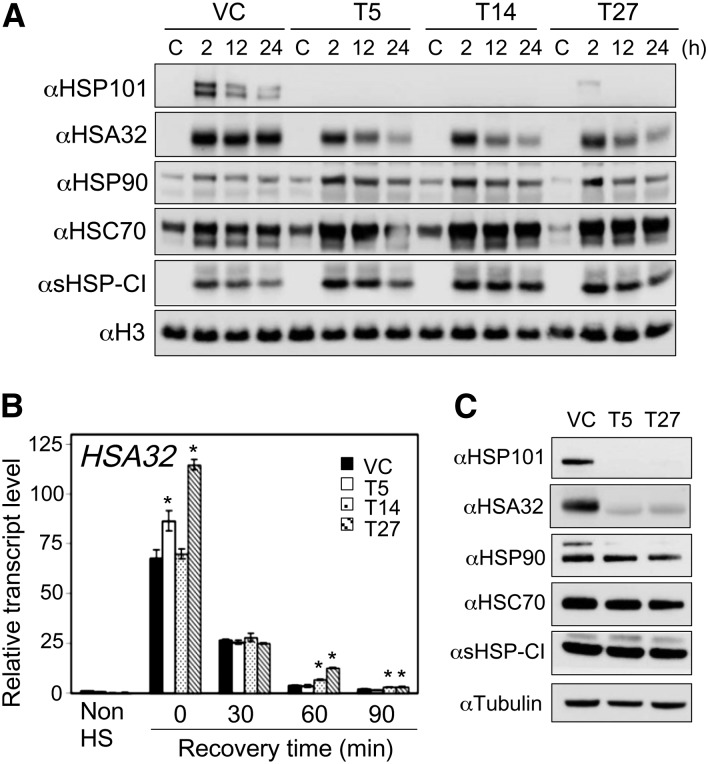
To investigate whether HSP101 positively affects the protein level of HSA32 in rice after heat treatment, as is the case in Arabidopsis, we generated HSP101 knockdown lines by introducing an RNAi construct into the japonica rice cultivar Tainung 67, which is readily transformable for functional genomic studies (Hsing et al., 2007). From 28 putative RNAi lines, three independent lines were selected for further analysis after confirmation by PCR genotyping and Southern-blot hybridization (Supplemental Fig. S3). Quantitative RT-PCR analysis of these transgenic lines at the T2 generation showed a drastic reduction in the heat-induced expression of HSP101 at the transcript level compared with the control line containing the empty vector (Supplemental Fig. S4). We then examined the protein levels of HSP101 and other HSPs in the seedlings after 2, 12, or 24 h recovery following heat treatment at 42°C for 2 h. The results of immunoblotting analysis showed that the levels of heat-induced HSP101 in all three RNAi lines were drastically reduced (Fig. 6A), which is consistent with the transcript levels. Interestingly, the levels of HSA32 in all three RNAi lines were substantially lower than that of the control line during recovery. The difference was more obvious after a longer recovery time. Other HSPs tested were not reduced in the RNAi lines compared with the control. On the contrary, the levels of HSP90, HSC70, and sHSP-CI were higher in the RNAi lines than in the control line. Quantitative RT-PCR analysis further showed that the reduction in HSA32 protein level was not due to a reduced HSA32 transcript level in these lines (Fig. 6B). In the seeds, we also observed association of lower level of HSA32 with suppression of HSP101 in the RNAi lines (Fig. 6C).
Figure 6.
Effect of suppressing the expression of rice HSP101 on HSA32 protein levels in heated seedlings and imbibed seeds of RNAi lines. VC indicates transgenic line with an empty vector only; T5, T14, and T27 are independent transgenic lines with the RNAi construct targeting HSP101. A, Immunoblot analysis of the indicated HSPs in 4-d-old seedlings, from which the crude extracts were prepared after recovery for 2 to 24 h following heat treatment at 42°C for 2 h. The nonheated sample was used as control (C). In each lane, 60 μg of total protein was loaded. Histone H3 is shown as a loading control. B, Quantitative RT-PCR analysis of the transcript levels of HSA32 in 4-d-old seedlings before (Non HS) or after heat treatment at 42°C for 2 h followed by recovery at 28°C for 0 to 90 min. Relative transcript levels of HSA32 were obtained by normalization to the nonheated VC sample, which is set as one. Results are presented as mean values of three technical replicates ± sd. *P < 0.01 (versus VC, Student’s t test). C, Immunoblot analysis of the indicated HSPs in imbibed seeds of transgenic lines. Tubulin is shown as a loading control. The tests were repeated at least once, and similar results were obtained.
N22, an indica Rice Cultivar, Has Higher Basal Thermotolerance But Lower LAT Than the japonica Rice Cultivar Nipponbare
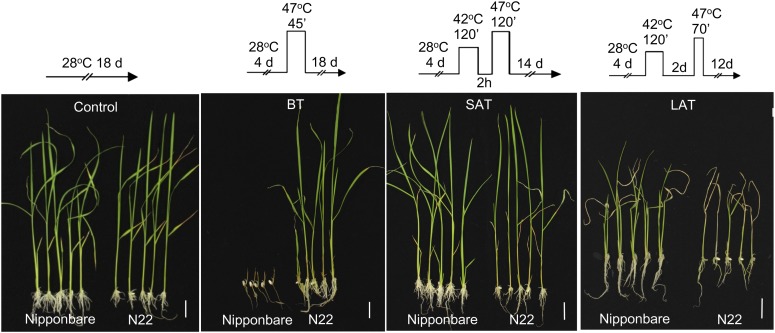
So far, our data show that there is a close relationship between the decay rate of HSP101, which is modulated by HSA32, and the level of LAT in rice. Because the decay of heat-induced HSP101 has been shown to be faster in indica than in japonica rice cultivars (Agarwal et al., 2003), it was of interest to know whether japonica rice could have a higher LAT level than indica rice. To address this question, two rice cultivars, N22 and Nipponbare, were compared directly. ‘N22’ has been shown to be highly heat tolerant during anthesis (Jagadish et al., 2010b). However, its heat tolerance level has not been determined at the seedling stage. Figure 7 shows that the seedlings of cv N22 tolerated the heat treatment well at 47°C for 45 min, a heat stress regime for basal thermotolerance, but the cv Nipponbare seedlings stopped growing and died. However, when subjected to a LAT stress regime, the cv Nipponbare seedlings survived and resumed growth by forming new tillers, whereas the cv N22 seedlings died (Fig. 7). There was no substantial difference in these two rice varieties after subjection to the SAT assay conditions. These results suggest that cv N22 has a higher basal thermotolerance level than cv Nipponbare, while the latter has a higher LAT level than the former.
Figure 7.
Thermotolerance phenotypes of japonica cultivar Nipponbare and indica cultivar N22. Basal thermotolerance, SAT, and LAT assays of cvs N22 and Nipponbare seedlings were performed under the corresponding heat stress regimes shown schematically at the top of each section. The representative plants were photographed after recovery for the indicated time. White bars represent 2 cm in each picture. Three biological repeats were performed, and similar results were obtained.
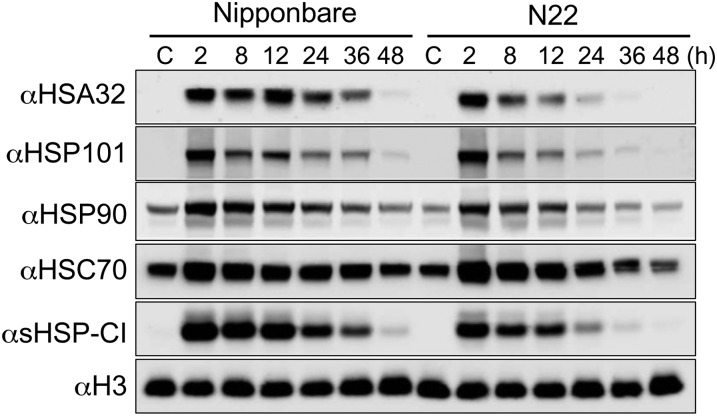
To explain the difference in LAT phenotype, immunoblotting analysis of HSP101 level was conducted in these two cultivars during the postheat stress period. Figure 8 shows that heat treatment induced the production of HSP101 to a similar level after 2 h of recovery in cv N22 and Nipponbare, and HSP101 was decayed faster in the indica varieties, which is consistent with a previous report (Agarwal et al., 2003). Although the lower LAT level in cv N22 seemed to be associated with faster decay of HSP101, we noticed that other HSPs also decayed faster in cv N22 than in cv Nipponbare. Immunoblotting results showed that the heat-induced expression of HSA32 and sHSP-CI exhibited a shorter duration in cv N22 than in cv Nipponbare (Fig. 8). Moreover, the amounts of HSC70 and HSP90 were slightly lower in cv N22 than in cv Nipponbare after a long recovery.
Figure 8.
Decay of HSPs in seedlings of japonica cultivar Nipponbare and indica cultivar N22 during recovery after heat treatment. Immunoblot analysis of the indicated HSPs in 4-d-old cv Nipponbare and N22 seedlings, from which the crude extracts were prepared after recovery for 2 to 48 h following heat treatment at 42°C for 2 h. The nonheated sample was used as a control (C). In each lane, 60 μg of total protein was loaded. Histone H3 is shown as a loading control. The test was repeated at least once, and similar results were obtained.
DISCUSSION
Rice is an important staple food crop whose productivity is threatened by high growing season temperature (Welch et al., 2010). Some studies of the heat tolerance phenotype in rice at the molecular level have been conducted, but progress is limited (Lin et al., 2005; Jagadish et al., 2010a, 2010b). Heat tolerance is a complex trait that likely involves multiple mechanisms and various genetic components. Diverse thermotolerance mechanisms could have evolved in rice to cope with various heat stress situations, as was reported recently for Arabidopsis (for review, see Yeh et al., 2012). Exploring the thermotolerance diversity in rice plants may provide useful insights for the improvement of heat-tolerant traits to cope with the projected warming scenarios in the future (IPCC, 2007).
In this study, we elucidated the function of HSA32 in rice by characterizing the biochemical and physiological responses to heat stress conditions in HSA32 KO mutants. Our data show that HSA32 plays an important role in LAT in heat-acclimated rice seedlings and is not essential for growth and development under normal conditions. These findings correlate well with those reported for Arabidopsis (Charng et al., 2006). We showed that HSA32 retarded the decay of heat-induced HSP101 in rice during the poststress period (Fig. 4A). This is likely due to an increased degradation rate rather than reduced synthesis of HSP101 in the absence of HSA32, as is the case in Arabidopsis (Wu et al., 2013). Degradation of HSP101 is slightly inhibited by the autophagic proteolysis inhibitor E-64d in Arabidopsis (Wu et al., 2013). However, it is not clear whether and how HSA32 modulates the degradation of HSP101 via the autophagy-mediated pathway. On the other hand, suppression of HSP101 expression reduced the level of HSA32, but not other HSPs examined, in heat-acclimated rice seedlings (Fig. 6A), which is also similar to the function of HSP101 in Arabidopsis (Wu et al., 2013). The suppression of rice HSP101 actually led to enhanced expression of other HSPs, such as HSP90 (Fig. 6A). A similar phenomenon was previously observed in Arabidopsis HSP101 mutants (Wu et al., 2013), and the cause is not clear. We speculate that reduced HSP101 chaperone activity is complemented in part by up-regulation of other chaperones via a feedback control. Taken together, these results suggest that there is posttranscriptional interplay between HSA32 and HSP101 in heat-treated rice seedlings and that the interplay prolongs the effect of heat acclimation. This notion agrees with the findings in Arabidopsis and suggests that the mechanism of LAT modulation involving HSA32 and HSP101 evolved earlier than the divergence of monocots and eudicots. Actually, a HSA32 homolog can be found in primitive land plants, such as the moss Physcomitrella patens (Liu et al., 2006). It is likely that this LAT modulating mechanism evolved during the terrestrial colonization of plants. LAT is probably important for plants to adapt to temperature fluctuation in the air. However, so far, the LAT phenotype has only been seen in plants subjected to heat stress treatment in the laboratory. The physiological importance of LAT for plants in the natural environment remains to be shown.
The interplay between HSA32 and HSP101 also takes place developmentally. We showed that in rice seeds, HSA32 is required for maintaining a high level of HSP101 (Fig. 3) and that HSP101 positively affects the level of HSA32 (Fig. 6C). A similar phenomenon was also found in Arabidopsis seeds (S.-h. Wu, Y.-t. Juan, and Y.-y. Charng, unpublished data), indicating that this developmental process is conserved. The absence of HSA32 or HSP101 has no effect on germination of rice seeds under nonstress conditions but is detrimental under severe heat stress, such as a brief exposure to 58°C for 30 min, which is likely to happen on the soil surface in the field (Peacock et al., 1993). Therefore, we suggest that the interplay between HSA32 and HSP101 enables plant seeds to cope with adverse high temperature in the natural environment.
Despite the interplay between HSA32 and HSP101 being conserved in rice and Arabidopsis, a difference in the accumulation patterns of the proteins was observed. Accumulation of HSA32 in Arabidopsis exhibits a dramatic delay after heat treatment. The maximal level of HSA32 was observed after 16 h recovery following heat treatment at 37°C for 1 h (Wu et al., 2013). However, we did not observe an obvious delay in the accumulation of rice HSA32. Of note, the transcripts of HSA32 were degraded much faster in rice than in Arabidopsis during the recovery period. In rice, HSA32 transcripts were reduced to the prestress level in heated samples after 1 h of recovery (Fig. 6B), while in Arabidopsis, the transcript level of HSA32 remained unchanged after 16 h of recovery (Wu et al., 2013). Moreover, Arabidopsis HSP101 seems to be more stable than the rice protein in heat-treated seedlings. The half-life of HSP101 in Arabidopsis (Columbia ecotype) is about 48 h (Wu et al., 2013), while rice HSP101 decayed more than 50% after 24 h of recovery in three different cultivars tested (Figs. 4A, 6A, and 8). Currently, the cause of this variation is unknown.
Establishing the LAT assay for rice is valuable for exploring the thermotolerance diversity in this important crop. In this study, we showed that the indica rice cultivar N22 displayed a substantially higher basal thermotolerance level than the japonica rice cultivar Nipponbare; by contrast, the latter has a higher LAT level than the former (Fig. 7). These results indicate that different types of thermotolerance can be decoupled. A rice cultivar tolerant to one type of heat stress is not necessarily tolerant to other types of heat stress. Hence, the diverse thermotolerance response in rice should be treated separately. Of course, we could not exclude the possibility that a trade-off between basal thermotolerance and LAT exists for local adaptation (Anderson et al., 2013). It remains to be seen whether such a trade-off is common in other rice cultivars.
The LAT level seemed to correlate well with the protein level of HSP101 after a longer recovery in the two different rice varieties tested (Fig. 8). ‘Nipponbare’ has a slower decay of HSP101 than cv N22, which is consistent with a previous report of HSP101 being decayed faster in indica rice (Agarwal et al., 2003). However, in addition to HSP101, we also noticed a general faster decay of other HSPs in cv N22 as well (Fig. 8), which may also lead to a lower LAT level in cv N22. Thus, HSP101 level may not be the sole factor in determining LAT in rice. The shorter duration of HSA32 may explain the faster decay of HSP101 in cv N22, owing to the interplay of these proteins. The cause of the differential decay rates in HSA32 and HSP101 in the two rice cultivars is not yet clear. There are minor differences in the protein sequences of HSA32 and HSP101 in the two cultivars, but the effect of this difference is unknown. Further studies on the structure-function relationship of these proteins may shed light on the underlying LAT mechanism involving them. It will also be of interest to examine the LAT response in different Arabidopsis accessions (Weigel and Mott, 2009; Cao et al., 2011), which should be instrumental in identifying genetic variations contributing to this thermotolerance type.
MATERIALS AND METHODS
Plant Materials
The wild-type rice (Oryza sativa) and two Tos17 insertion lines of HSA32 in japonica cultivar Nipponbare background were obtained from the National Institute of Agrobiological Sciences, Japan (Hirochika, 2001). The detailed information about the mutant lines hsa32-1 (NG7371) and hsa32-2 (ND1779) can be accessed at the Rice Tos17 Insertion Mutant Database (http://tos.nias.affrc.go.jp/). The Tos17 insertions were confirmed by PCR amplification with specific primers (Supplemental Table S1). The indica rice cultivar N22 was kindly provided by the International Rice Research Institute. The japonica rice cultivar Tainung 67 for producing RNAi transgenic lines was originally obtained from the Taiwan Agricultural Research Institute. Rice seeds of all cultivars used in this work were propagated under a natural light source in the greenhouse facility of the Biotechnology Center in Southern Taiwan, Academia Sinica. Rice seeds were stored at 4°C until use. To produce rice seedlings for heat stress treatment, dehulled rice seeds were sterilized with 4% (v/v) sodium hypochloride for 15 min, washed extensively with Milli-Q water, and imbibed in the dark for 2 d at 28°C in a square petri dish (12 × 12 cm) with 70 mL of 0.5× Murashige and Skoog (MS) solid medium containing 0.8% (w/v) agar. The germinating seeds were then exposed to 12-h/12-h photoperiod and a light intensity of 180 µmol m–2 s–1 at 28°C in a growth chamber. The petri dishes were positioned vertically during seed imbibition and growth of seedlings. To produce flowering plants, rice seeds were imbibed in Milli-Q water in the dark for 3 d. The germinating seeds were transplanted to potted soil in a greenhouse with 12-h/12-h photoperiod, temperature set at 30°C during the day and 25°C at night, and 60% humidity. Mature leaves and spikelets were collected 3 d after anthesis.
RNAi-Mediated Gene Silencing of HSP101
To generate an RNAi construct to suppress the expression of HSP101, a 258-bp fragment from the 5′ untranslated region of HSP101 was amplified by PCR with specific primers (Supplemental Table S1) and cloned into pCR8/TOPO (Invitrogen) to yield an entry vector. The HSP101 fragment was subcloned into the binary vector pANDA, kindly provided by Dr. Ko Shimamoto (Nara Institute of Science and Technology, Japan), as previously described (Miki and Shimamoto, 2004). The resulting construct was introduced into ‘Tainung 67’ embryogenic calli via Agrobacterium tumefaciens-mediated transformation according to Sallaud et al. (2003). Plants were regenerated from transformed calli by selection of hygromycin resistance. Homozygous transgenic lines with a single transfer DNA insertion event were obtained through segregation analysis of hygromycin-resistant test at T2 generation. Three independent RNAi lines, T5, T14, and T27, were chosen for further analysis. ‘Tainung 67’ plants transformed with pANDA were produced in the same way as the RNAi lines and used as control plants. The presence of transgenes in the transgenic lines was confirmed by PCR amplification with specific primers (Supplemental Table S1) and by Southern-blot analysis according to a standard procedure (Sambrook et al., 1989). Digoxigenin-labeled DNA for probing the hygromycin phosphotransferase (HPT) gene was generated for Southern hybridization by using PCR DIG Labeling Mix (Roche) with HPT-specific primers (Supplemental Table S1) and pANDA as a template.
Heat Treatment and Thermotolerance Assays
Heat treatment of rice seedlings and detached tissues was performed in the dark at the indicated temperatures for the indicated lengths of time in a water bath. Thermotolerance assays on rice seedlings were modified from those previously established for Arabidopsis (Arabidopsis thaliana; Charng et al., 2006). In brief, 12 seedlings grown on the 0.5× MS medium plates were subjected to treatments under the indicated heat stress regimes. After assay treatments, the seedlings were transferred to 50-mL Falcon tubes (four seedlings per tube) containing 20 mL Milli-Q water for assessment after recovery under normal growth conditions in a growth chamber. The fresh weight of the seedlings was measured, and their morphology was photographed. For basal thermotolerance assay of rice seeds, sterilized seeds were imbibed in the dark on 0.5× MS medium plates at 28°C for 1.5 h and then subjected to heat treatment at 58°C for 30 min in a water bath. Germination rates of the heat-treated seeds were counted daily during recovery at 28°C.
RNA Isolation and RT-PCR Analysis
Total RNA was isolated from 4-d-old seedlings as previously described (Charng et al., 2006). Three seedlings were pooled for RNA extraction for each treatment. Semiquantitative RT-PCR analysis of HSA32, HSP101, and ACTIN1 was performed as previously described with minor modifications (Wang et al., 2001). PCR reactions were performed for 25 cycles in a volume of 10 μL using complementary DNA reverse transcribed from 10 ng of total RNA. The PCR product was analyzed by agarose gel electrophoresis and staining with ethidium bromide. Quantitative RT-PCR was performed as described in Liu et al. (2011) with minor modifications. The expression level of the target genes was normalized to that of 18S ribosomal RNA (rRNA) by subtracting the cycle threshold value of 18S rRNA from the cycle threshold values of the target genes. The sequences of the primers for RT-PCR analysis are listed in Supplemental Table S1.
Immunoblotting Analysis
Protein extraction, quantification, and immunoblotting were performed as previously described (Charng et al., 2006). Three seedlings were pooled for protein extraction for each treatment. Rabbit antiserum against rice HSP101 was produced by employing a synthetic peptide (LKKLPSQSPPPDSVPAS) as antigen. To generate antibody against rice HSA32, the full-length recombinant HSA32 protein fused to glutathione S-transferase at the N terminus was produced by using pET (pET42a) and Escherichia coli BL21 (DE3, Novagen) system. The recombinant protein was purified from the insoluble fraction of cell lysate by solubilizing with SDS-PAGE sample buffer and resolving on SDS-PAGE. The identity of the gel-purified protein was confirmed by liquid chromatography-electrospray ionization-tandem mass spectrometry performed by the Core Facilities for Proteomics Research at Academia Sinica, Taiwan. The purified recombinant protein was then used as antigen to immunize mice. Peptide synthesis, immunization of animals and serum collection were performed by LTK Biotechnology, Taoyuan, Taiwan. Antibodies against HSP90 and sHSP-Cl and tubulin were as described in Charng et al. (2006) and Chi et al. (2009). The antibodies against HSC70/HSP70 and histone H3 were purchased from Stressgen (SPA-807) and Abcam (ab1791), respectively.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HSA32 (Os06g46900.1), HSP101 (Os05g0519700), ACTIN1 (Os03g0718100), and 18S rRNA (Os03g0786600).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Faster decay of heat-induced HSP101 protein in the absence of HSA32.
Supplemental Figure S2. Analysis of fresh weight change after SAT assay.
Supplemental Figure S3. PCR genotyping and Southern blot analysis of RNAi lines.
Supplemental Figure S4. Quantitative RT-PCR analysis of HSP101 transcripts in RNAi lines.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Yao-Pin Lin, Hsiang-Chin Liu, Chia-an Huang, and Hsiu-Yin Ho for technical assistance, Miranda Loney for English editing, the DNA Sequencing Core Facility of the Institute of Biomedical Sciences at Academia Sinica for providing DNA sequencing services, and three anonymous reviewers for helpful comments on a previous draft of the paper.
Glossary
- LAT
long-term acquired thermotolerance
- SAT
short-term acquired thermotolerance
- RNAi
RNA interference
- KO
knockout
- RT
reverse transcription
- MS
Murashige and Skoog
- rRNA
ribosomal RNA
Footnotes
This work was supported by the Sustainability Science Research Projects at Academia Sinica (grant no. AS–102–SS–A03).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Agarwal M, Sahi C, Katiyar-Agarwal S, Agarwal S, Young T, Gallie DR, Sharma VM, Ganesan K, Grover A. (2003) Molecular characterization of rice hsp101: complementation of yeast hsp104 mutation by disaggregation of protein granules and differential expression in indica and japonica rice types. Plant Mol Biol 51: 543–553 [DOI] [PubMed] [Google Scholar]
- Anderson JT, Lee CR, Rushworth CA, Colautti RI, Mitchell-Olds T. (2013) Genetic trade-offs and conditional neutrality contribute to local adaptation. Mol Ecol 22: 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battisti DS, Naylor RL. (2009) Historical warnings of future food insecurity with unprecedented seasonal heat. Science 323: 240–244 [DOI] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Hsu FC, Ko SS. (2006) Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 140: 1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Shen ZY, Li PH. (1982) Adaptability of crop plants to high temperatures stress. Crop Sci 22: 719–725 [Google Scholar]
- Chi WT, Fung RWM, Liu HC, Hsu CC, Charng YY. (2009) Temperature-induced lipocalin is required for basal and acquired thermotolerance in Arabidopsis. Plant Cell Environ 32: 917–927 [DOI] [PubMed] [Google Scholar]
- Hirochika H. (2001) Contribution of the Tos17 retrotransposon to rice functional genomics. Curr Opin Plant Biol 4: 118–122 [DOI] [PubMed] [Google Scholar]
- Hong SW, Vierling E. (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing YI, Chern CG, Fan MJ, Lu PC, Chen KT, Lo SF, Sun PK, Ho SL, Lee KW, Wang YC, et al. (2007) A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol Biol 63: 351–364 [DOI] [PubMed] [Google Scholar]
- Intergovernmental Panel on Climate Change (2007) Climate Change 2007: The Physical Science Basis. Cambridge University Press, Cambridge, UK [Google Scholar]
- Ishimaru T, Horigane AK, Ida M, Iwasawa N, San-Oh YA, Nakazono M, Nishizawa NK, Masumura T, Kondo M, Yoshida M. (2009) Formation of grain chalkiness and changes in water distribution in developing rice caryopses grown under high-temperature stress. J Cereal Sci 50: 166–174 [Google Scholar]
- Jagadish SVK, Cairns J, Lafitte R, Wheeler TR, Price AH, Craufurd PQ. (2010a) Genetic analysis of heat tolerance at anthesis in rice. Crop Sci 50: 1633–1641 [Google Scholar]
- Jagadish SVK, Muthurajan R, Oane R, Wheeler TR, Heuer S, Bennett J, Craufurd PQ. (2010b) Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). J Exp Bot 61: 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kregel KC. (2002) Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol (1985) 92: 2177–2186 [DOI] [PubMed] [Google Scholar]
- Landry J, Bernier D, Chrétien P, Nicole LM, Tanguay RM, Marceau N. (1982) Synthesis and degradation of heat shock proteins during development and decay of thermotolerance. Cancer Res 42: 2457–2461 [PubMed] [Google Scholar]
- Lin SK, Chang MC, Tsai YG, Lur HS. (2005) Proteomic analysis of the expression of proteins related to rice quality during caryopsis development and the effect of high temperature on expression. Proteomics 5: 2140–2156 [DOI] [PubMed] [Google Scholar]
- Lindquist S. (1986) The heat-shock response. Annu Rev Biochem 55: 1151–1191 [DOI] [PubMed] [Google Scholar]
- Liu HC, Liao HT, Charng YY. (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34: 738–751 [DOI] [PubMed] [Google Scholar]
- Liu NY, Ko SS, Yeh KC, Charng YY. (2006) Isolation and characterization of tomato Hsa32 encoding a novel heat-shock protein. Plant Sci 170: 976–985 [Google Scholar]
- Miki D, Shimamoto K. (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Mogk A, Haslberger T, Tessarz P, Bukau B. (2008) Common and specific mechanisms of AAA+ proteins involved in protein quality control. Biochem Soc Trans 36: 120–125 [DOI] [PubMed] [Google Scholar]
- Nieto-Sotelo J, Martínez LM, Ponce G, Cassab GI, Alagón A, Meeley RB, Ribaut JM, Yang R. (2002) Maize HSP101 plays important roles in both induced and basal thermotolerance and primary root growth. Plant Cell 14: 1621–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock J, Soman P, Jayachandran R, Rani A, Howarth C, Thomas A. (1993) Effects of high soil surface temperature on seedling survival in pearl millet. Exp Agric 29: 215 [Google Scholar]
- Peng S, Huang J, Sheehy JE, Laza RC, Visperas RM, Zhong X, Centeno GS, Khush GS, Cassman KG. (2004) Rice yields decline with higher night temperature from global warming. Proc Natl Acad Sci USA 101: 9971–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S. (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C, Meynard D, van Boxtel J, Gay C, Bès M, Brizard JP, Larmande P, Ortega D, Raynal M, Portefaix M, et al. (2003) Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor Appl Genet 106: 1396–1408 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sanchez Y, Lindquist SL. (1990) HSP104 required for induced thermotolerance. Science 248: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Wang NN, Yang SF, Charng YY. (2001) Differential expression of 1-aminocyclopropane-1-carboxylate synthase genes during orchid flower senescence induced by the protein phosphatase inhibitor okadaic acid. Plant Physiol 126: 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Mott R. (2009) The 1001 genomes project for Arabidopsis thaliana. Genome Biol 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JR, Vincent JR, Auffhammer M, Moya PF, Dobermann A, Dawe D. (2010) Rice yields in tropical/subtropical Asia exhibit large but opposing sensitivities to minimum and maximum temperatures. Proc Natl Acad Sci USA 107: 14562–14567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TY, Juan YT, Hsu YH, Wu SH, Liao HT, Fung RW, Charng YY. (2013) Interplay between heat shock proteins HSP101 and HSA32 prolongs heat acclimation memory posttranscriptionally in Arabidopsis. Plant Physiol 161: 2075–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CH, Kaplinsky NJ, Hu C, Charng YY. (2012) Some like it hot, some like it warm: phenotyping to explore thermotolerance diversity. Plant Sci 195: 10–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.