A transcriptional regulator interacts with the transcription factor PIF1 to modulate hypocotyl cell growth in response to light.
Abstract
Seedling deetiolation, a critical process in early plant development, is regulated by an intricate transcriptional network. Here, we identified VQ MOTIF-CONTAINING PROTEIN29 (VQ29) as a novel regulator of the photomorphogenic response in Arabidopsis (Arabidopsis thaliana). We showed that 29 of the 34 VQ proteins present in Arabidopsis exhibit transcriptional activity in plant cells and that mutations in the VQ motif affect the transcriptional activity of VQ29. We then functionally characterized VQ29 and showed that the hypocotyl growth of plants overexpressing VQ29 is hyposensitive to far-red and low-intensity white light, whereas a vq29 loss-of-function mutant exhibits decreased hypocotyl elongation under a low intensity of far-red or white light. Consistent with this, VQ29 expression is repressed by light in a phytochrome-dependent manner. Intriguingly, our yeast (Saccharomyces cerevisiae) two-hybrid, bimolecular fluorescence complementation, and coimmunoprecipitation assays showed that VQ29 physically interacts with PHYTOCHROME-INTERACTING FACTOR1 (PIF1). We then showed that VQ29 and PIF1 directly bind to the promoter of a cell elongation-related gene, XYLOGLUCAN ENDOTRANSGLYCOSYLASE7, and coactivate its expression. Furthermore, the vq29 pif1 double mutant has shorter hypocotyls than either of the corresponding single mutants. Therefore, our study reveals that VQ29 is a negative transcriptional regulator of light-mediated inhibition of hypocotyl elongation that likely promotes the transcriptional activity of PIF1 during early seedling development.
Light is an important environmental signal that affects plant growth and development throughout its life cycle, directing processes such as seed germination, seedling deetiolation, phototropism, circadian rhythms, shade avoidance, and flowering timing. Dark-grown seedlings, which adopt a developmental program known as etiolation or skotomorphogenesis, exhibit elongated hypocotyls and closed cotyledons with apical hooks. Upon light irradiation, seedlings undergo deetiolation or photomorphogenesis, which slows hypocotyl growth and causes cotyledons to expand and chloroplasts and chlorophylls to develop (Von Arnim and Deng, 1996).
Intensive research has revealed the main signaling pathway governing photomorphogenesis (Chen et al., 2004; Lau and Deng, 2010; Arsovski et al., 2012). To initiate the light responses, plants rely on a set of photoreceptors, including the red/far-red light-absorbing phytochromes (phys) and the blue/UV-A light-absorbing cryptochromes (crys). Activation of photoreceptors transmits signals to key downstream negative factors, such as CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and members of the PHYTOCHROME INTERACTING FACTOR (PIF) protein family. COP1 is a RING-type E3 ubiquitin ligase that targets photomorphogenesis-promoting factors, such as ELONGATED HYPOCOTYL5 (HY5) and LONG HYPOCOTYL IN FAR-RED1 (HFR1), for 26S proteasome-mediated degradation, which desensitizes the light pathway initiated by both phys and crys (Wei and Deng, 1996; Lau and Deng, 2012). PIFs encode a group of basic helix-loop-helix (bHLH) transcription factors (TFs) that are phosphorylated and degraded in a phy-dependent manner in light. PIF proteins (including PIF1, PIF3, PIF4, and PIF5) play redundant roles in directly regulating gene expression and repressing photomorphogenic responses (Leivar et al., 2008; Shin et al., 2009; Leivar and Quail, 2010).
Genetic and molecular studies in Arabidopsis (Arabidopsis thaliana) have identified another series of signaling components that are involved in either negatively or positively regulating the light pathway (Jiao et al., 2007). Notably, many of the identified effectors are nucleus-localized TFs, such as FAR-RED ELONGATED HYPOCOTYLS3 and FAR-RED IMPAIRED RESPONSE1 (transposase-derived TFs), bZIP16 (bZIP TFs), HFR1 and MYC2 (bHLH TFs), LONG AFTER FAR-RED LIGHT1 (a MYB TF), and SALT TOLERANCE HOMOLOG2 and LIGHT-REGULATED ZINC FINGER1 (B-box-containing TFs; Hudson et al., 1999; Ballesteros et al., 2001; Wang and Deng, 2002; Yadav et al., 2005; Datta et al., 2007; Chang et al., 2011; Hsieh et al., 2012). Moreover, genes encoding F-box proteins (such as EMPFINDLICHER IM DUNKELROTEN LICHT1 and ATTENUATED FAR-RED RESPONSE), kinases (e.g. NUCLEOSIDE-DIPHOSPHATE KINASE2), and phosphatases (such as PROTEIN PHOSPHATASE7) were also isolated and shown to mediate light signaling (Choi et al., 1999; Dieterle et al., 2001; Harmon and Kay, 2003; Møller et al., 2003). However, the regulatory mechanisms underlying the actions of these factors are not well understood. In addition, we recently showed that a chromatin-remodeling factor, ENHANCED PHOTOMORPHOGENESIS1/PICKLE, interacts with HY5 to fine-tune light signaling by modulating tri-methylation of lysine27 on histone H3 levels in cell elongation-related genes (Jing et al., 2013). Even though a wide range of light signaling intermediates have been identified and extensively studied, a complete picture of the light signaling pathway has yet to emerge.
A group of genes encoding proteins containing a unique and conserved FxxxVQxxTG motif (termed the VQ motif) was recently identified (Xie et al., 2010; Cheng et al., 2012). These proteins include 34 members in Arabidopsis, and each is designated as VQ MOTIF-CONTAINING PROTEIN (VQ). The functions of a few members of this family has been characterized to date. For example, SIGMA FACTOR BINDING PROTEIN1 (SIB1/VQ23), its close homolog SIB2 (VQ16), and MITOGEN-ACTIVATED PROTEIN KINASE4 SUBSTRATE1 (VQ21) are required for the plant defense response (Andreasson et al., 2005; Narusaka et al., 2008; Xie et al., 2010; Lai et al., 2011). In addition, HAIKU1 (VQ14) is a regulator of endosperm growth and seed size (Wang et al., 2010), while CALMODULIN-BINDING PROTEIN25 (VQ15) and VQ9 function as negative effectors of osmotic and salinity stress tolerance, respectively (Perruc et al., 2004; Hu et al., 2013). It is anticipated that this family responds to various environmental signals and plays diverse roles in plant defense, growth, and development. However, the functions and regulatory mechanisms of most VQ family members remain unknown.
In this study, we demonstrate that the VQ family of proteins largely possesses transcriptional activities. Furthermore, we characterize the regulation and function of VQ29 in detail. We show that VQ29 expression is down-regulated by light. Overexpression of VQ29 results in hyposensitivity of hypocotyl growth to far-red light and low-light conditions, whereas the vq29 loss-of-function mutant exhibits decreased hypocotyl elongation under low-intensity far-red and white light during seedling deetiolation. We also demonstrate that VQ29 physically interacts with PIF1 and that these proteins cooperatively activate the expression of downstream genes. Our study identifies a novel factor in photomorphogenesis and provides insight into the roles of VQ family proteins in regulating diverse plant growth and developmental processes.
RESULTS
Analysis of VQ Genes from Arabidopsis, Rice, and Moss
Previous studies documented that the VQ gene family is found only in plants, and it was systematically studied in Arabidopsis (Xie et al., 2010; Cheng et al., 2012). To gain insight into the evolution of this family, we searched the GenBank database for sequences of the VQ genes from rice (Oryza sativa) and moss (Physcomitrella patens), which represent monocot and lower plants, respectively. Whereas the model dicot plant Arabidopsis has 34 VQ members, rice and moss have 39 and 25, respectively. Interestingly, Arabidopsis and rice VQ proteins are relatively small, with about 85% and 92% of the proteins containing fewer than 300 amino acid residues, respectively. In contrast, 68% of the moss VQ proteins are longer than 300 amino acids (Supplemental Fig. S1A). We further found that most VQ genes in higher plants (30 in Arabidopsis and 37 in rice) do not contain an intron. However, in moss, only seven VQ genes do not have an intron, whereas five VQs have one intron and 13 genes possess two or more introns (Supplemental Fig. S1B). These results suggest that VQ genes tend to be intronless and encode relatively small proteins in higher plants.
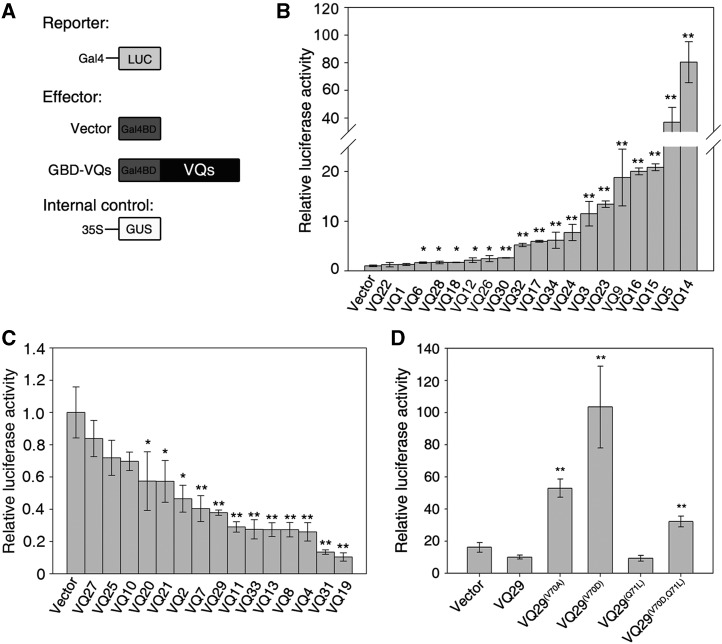
VQ Proteins Exhibit Transcriptional Activity
To investigate whether the VQ proteins are involved in transcriptional regulation, we attempted to isolate the open reading frame (ORF) of each VQ gene in Arabidopsis Columbia (Col) plants using reverse transcription (RT)-PCR (for VQ2) or PCR (for other intronless VQ genes). The PCR primers were designed according to the available complementary DNA (cDNA) sequence information or the predicted sequences. The ORFs of all VQ genes were amplified and cloned into the pEASY vector and verified by sequencing. We then subcloned the VQ genes in frame with the GAL4 DNA-binding domain (GBD) and under the control of the cauliflower mosaic virus (CaMV) 35S promoter in the pSAT-GAL4DB vector (Jing et al., 2013). The construct was cotransformed with a luciferase reporter gene (LUC), driven by the 35S minimal promoter and fused in frame to a GAL4-binding sequence, into Arabidopsis mesophyll protoplasts (Fig. 1A). We found that GBD fusion proteins with VQ14, VQ5, VQ15, VQ16, VQ9, VQ23, VQ3, VQ24, VQ34, VQ17, VQ32, or VQ30 drastically activated LUC reporter expression compared with GBD alone. The VQ26, VQ12, VQ18, VQ28, and VQ6 fusions promoted LUC expression to a lesser extent (Fig. 1B). However, GBD fusions with VQ19, VQ31, VQ4, VQ8, VQ13, VQ33, VQ11, VQ29, VQ7, VQ2, VQ21, or VQ20 remarkably repressed the expression of the LUC reporter (Fig. 1C). We also observed that GBD fusions with VQ22, VQ1, VQ27, VQ25, or VQ10 did not affect the transcription of LUC (Fig. 1, B and C). These data indicate that most members of the VQ family possess either transcriptional activation or repression activity in this heterologous gene reporter system. In this study, we focused on VQ29 (At4g37710) because it is involved in the seedling deetiolation response (see below).
Figure 1.
VQ proteins possess transcriptional activity. A, Diagrams of constructs used in the transient expression assay. B and C, Relative LUC reporter activity by various VQ effectors. D, Relative LUC reporter activity by VQ29 and its point mutants. For B to D, the effectors, LUC reporter, and GUS internal control were cotransformed into Arabidopsis protoplasts. Data denote means ± sd of three biological replicates. Asterisks in B and C indicate significant differences from the empty vector at P < 0.05 (single asterisk) or P < 0.01 (double asterisks) using Student’s t test. Asterisks in D indicate significant differences from the wild-type VQ29 at P < 0.01 (double asterisks) using Student’s t test.
Mutation in the VQ Motif Affects Transcriptional Activity
Since VQ family members contain only the VQ motif and possess activation or repression activity, we asked whether the VQ motif is required for transcriptional activity. We then altered conserved amino acids in the VQ motif of VQ29, where the very hydrophobic residue Val (V) was changed into the less hydrophobic residue Ala (A) or the hydrophilic residue Asp (D) and the hydrophilic residue Gln (Q) was mutagenized into the hydrophobic residue Leu (L). As shown in Figure 1D, single mutation of VQ29(V70A) or VQ29(V70D) abolished the repressive activity of VQ29 and greatly activated LUC reporter gene expression, whereas mutation in VQ29(Q71L) did not affect the activity. Furthermore, double mutations in VQ29(V70D,Q71L) led to a significant induction of LUC to a lesser extent than VQ29(V70D) (Fig. 1D). It should be noted that two point mutations (V70D and Q71L), as tested, did not affect the levels of the VQ29 protein (Supplemental Fig. S2). Taken together, these results suggest that the VQ motif is likely involved in mediating the transcriptional activity of VQ proteins.
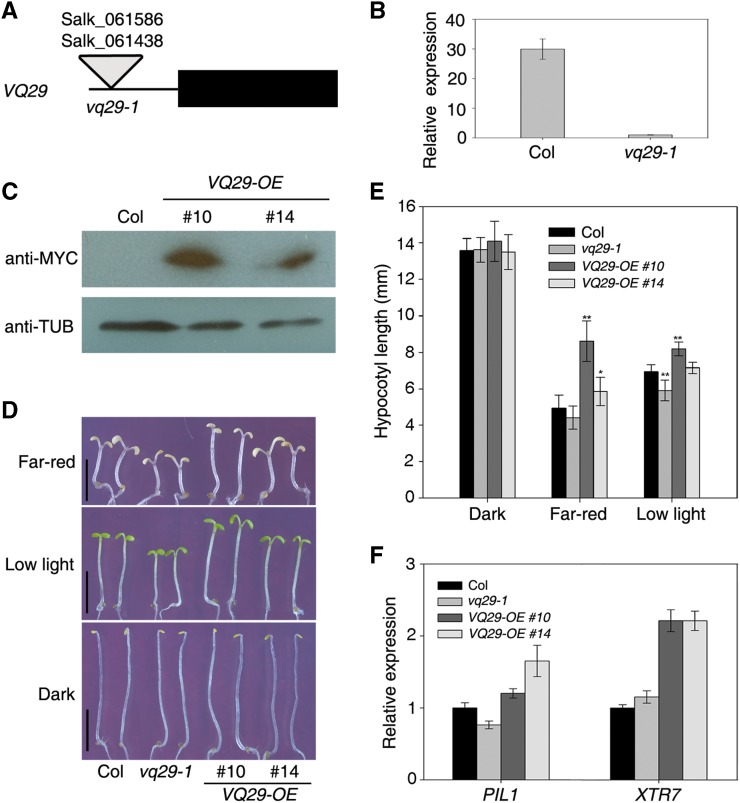
Overexpression of VQ29 Reduces the Hypocotyl Growth Response under Far-Red and Low-Intensity White Light Conditions
To elucidate the biological function of VQ29, we obtained two transfer DNA (T-DNA) insertion lines of VQ29, Salk_061586 and Salk_061438. PCR genotyping and sequencing studies revealed that the T-DNA is inserted in the promoter region 136 bp upstream of the ATG start codon of VQ29 in both mutants. These mutants thus represent the same allele (Fig. 2A). They had been backcrossed five times with the Col wild type, and thus the potential background mutations were largely eliminated. Quantitative RT-PCR analysis showed that the VQ29 transcripts were barely detectable in the mutant (hereafter referred to as vq29-1), suggesting that it is a null allele (Fig. 2B). We also generated transgenic plants overexpressing VQ29, fused with a MYC tag and driven by the CaMV 35S promoter (Pro-35S:Myc-VQ29, VQ29-OE). Over 40 transgenic lines were obtained, two of which (lines 10 and 14) were further studied in the following experiments. Immunoblot analysis using the MYC antibody showed that both lines accumulated the Myc-VQ29 fusion protein, with line 10 having a higher level of expression than line 14 (Fig. 2C).
Figure 2.
Phenotypic analysis of the VQ29 loss-of-function mutant and overexpression plants. A, Diagram of VQ29 gene structure and the position of the T-DNA insertion. The black rectangle is the exon of VQ29, and the triangle represents the T-DNA insertion. B, Detection of VQ29 transcript level in the wild type and the vq29-1 mutant by real-time RT-PCR. Seedlings were grown in white light conditions (10 µmol m−2 s−1) for 5 d. Data represent means ± sd of biological triplicates. C, Immunoblot analysis of two Pro-35S:Myc-VQ29 overexpression transgenic lines (VQ29-OE, lines 10 and 14) and the wild type using anti-MYC antibody. Immunoblotting against the tubulin antibody (anti-TUB) served as a loading control. Seedlings were grown in low white light (10 µmol m−2 s−1) for 5 d before harvesting. D, Photomorphogenic responses under far-red light (12 µmol m−2 s−1), low white light (10 µmol m−2 s−1), or darkness. Seedlings were grown in various conditions for 5 d. Bars = 5 mm. E, Quantification of hypocotyl length of the seedlings shown in D. Data are means ± sd of at least 20 seedlings. Asterisks indicate significant differences from the wild type at P < 0.05 (single asterisk) or P < 0.01 (double asterisks) using Student’s t test. F, Expression of PIL1 and XTR7 by quantitative RT-PCR. Four-day-old dark-grown seedlings were transferred to far-red light for 12 h. Error bars indicate the sd of biological triplicates. [See online article for color version of this figure.]
We then tested the hypocotyl growth response of the vq29-1 mutant and VQ29 overexpression lines under various light conditions. The hypocotyl length of vq29-1 was slightly but significantly shorter than that of the wild type under low-intensity white light (less than 20 µmol m−2 s−1) or low-intensity far-red light (less than 12 µmol m−2 s−1) but was indistinguishable from that of the wild type in red or blue light conditions with multiple fluence rates tested (Fig. 2, D and E; Supplemental Fig. S3). However, the hypocotyls of VQ29-OE transgenic seedlings were significantly longer than those of wild-type plants under far-red light conditions, with line 10 exhibiting the stronger phenotype, consistent with its higher VQ29 level (Fig. 2, C–E; Supplemental Fig. S3). The hypocotyls of overexpression line 10 were also longer than those of the wild type under low white light conditions but not in darkness or in red or blue light conditions (Fig. 2E; Supplemental Fig. S3). It has been documented that Suc promotes seedling growth (Stewart et al., 2011). Even without Suc supplement in the Murashige and Skoog medium, vq29-1 exhibited reduced hypocotyl elongation under low white light and different intensities of far-red light, and the hypocotyl length of VQ29-OE plants (line 10) was longer than that of the wild type under these conditions (Supplemental Fig. S4). Moreover, the expression of two light-responsive genes involved in cell elongation, PHYTOCHROME INTERACTING FACTOR3-LIKE1 (PIL1) and XYLOGLUCAN ENDOTRANSGLYCOSYLASE7 (XTR7), was increased in the VQ29-OE plants under the far-red light condition (Fig. 2F). Taken together, these results demonstrate that VQ29 is a negative regulator of seedling deetiolation under far-red and low white light conditions.
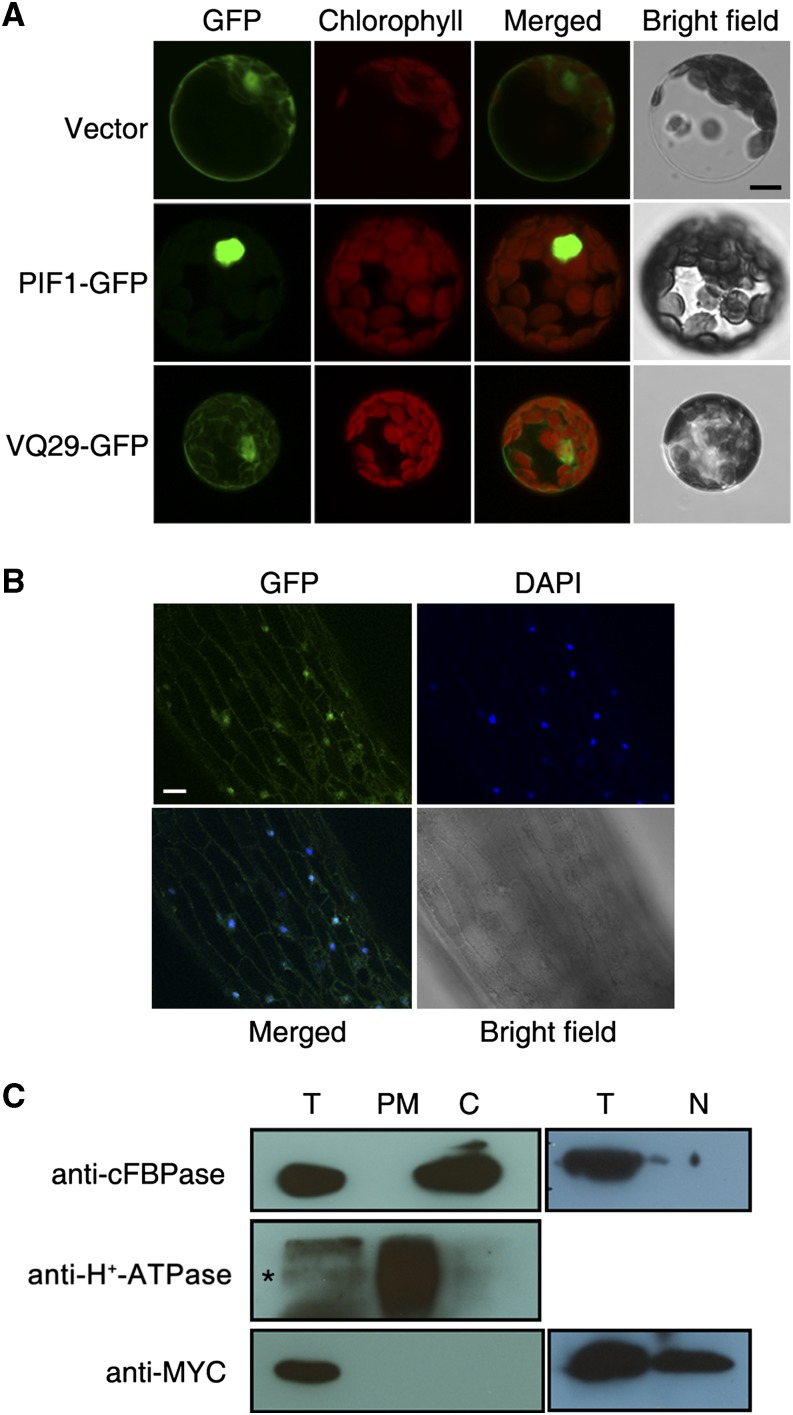
Subcellular Localization of VQ29
To determine the subcellular localization of VQ29, we first fused VQ29 with GFP and transiently expressed this construct in Arabidopsis protoplasts. The VQ29-GFP fusion protein was detected with multiple bands using the GFP antibody, likely due to partial degradation of the fusion protein in vitro (Supplemental Fig. S2). Confocal microscopy revealed that the VQ29-GFP fusion was likely localized to the nucleus, cytoplasm, and plasma membrane (Fig. 3A). Furthermore, we generated stable transgenic plants expressing a translational fusion of VQ29 with GFP under the control of the CaMV 35S promoter (Pro-35S:VQ29-GFP). GFP fluorescence was distributed mainly in the nucleus of hypocotyl cells from these transgenic plants (Fig. 3B). To substantiate the localization of VQ29, we isolated protein fractions from the nucleus, cytoplasm, and plasma membrane of Pro-35S:Myc-VQ29 (line 10) transgenic plants. As shown in Figure 3C, Myc-VQ29 fusion protein was only detected in cell fractions isolated from the nucleus but not from plasma membrane or cytosol. Its localization in the cytoplasm and plasma membrane in the protoplasts in Figure 3A might be caused by the overexpression of VQ29-GFP in the protoplasts.
Figure 3.
Localization of VQ29. A, The VQ29-GFP and PIF1-GFP constructs and empty vector controls were transformed into Arabidopsis protoplasts. Chlorophyll autofluorescence is shown in red. Representative images are shown. Bar = 5 µm. B, GFP fluorescence in the hypocotyl of a Pro-35S:VQ29-GFP transgenic seedling coincides with 4′,6-diamino-phenylindole (DAPI) staining (which marks the nucleus). Seedlings were grown in darkness for 4 d. Bar = 50 µm. C, Immunoblot assay of VQ29. The Pro-35S:Myc-VQ29 (line 10) transgenic plants were grown in 16-h-light/8-h-dark conditions for 3 weeks. Immunoblotting with antibodies against cytoplasm-localized Fru-1,6-bisphosphatase (cFBPase) and plasma membrane-targeted H+-ATPase (Iwata et al., 2008) served as positive controls. The asterisk indicates the specific band. T, Total protein; PM, plasma membrane; C, cytoplasm; N, nucleus.
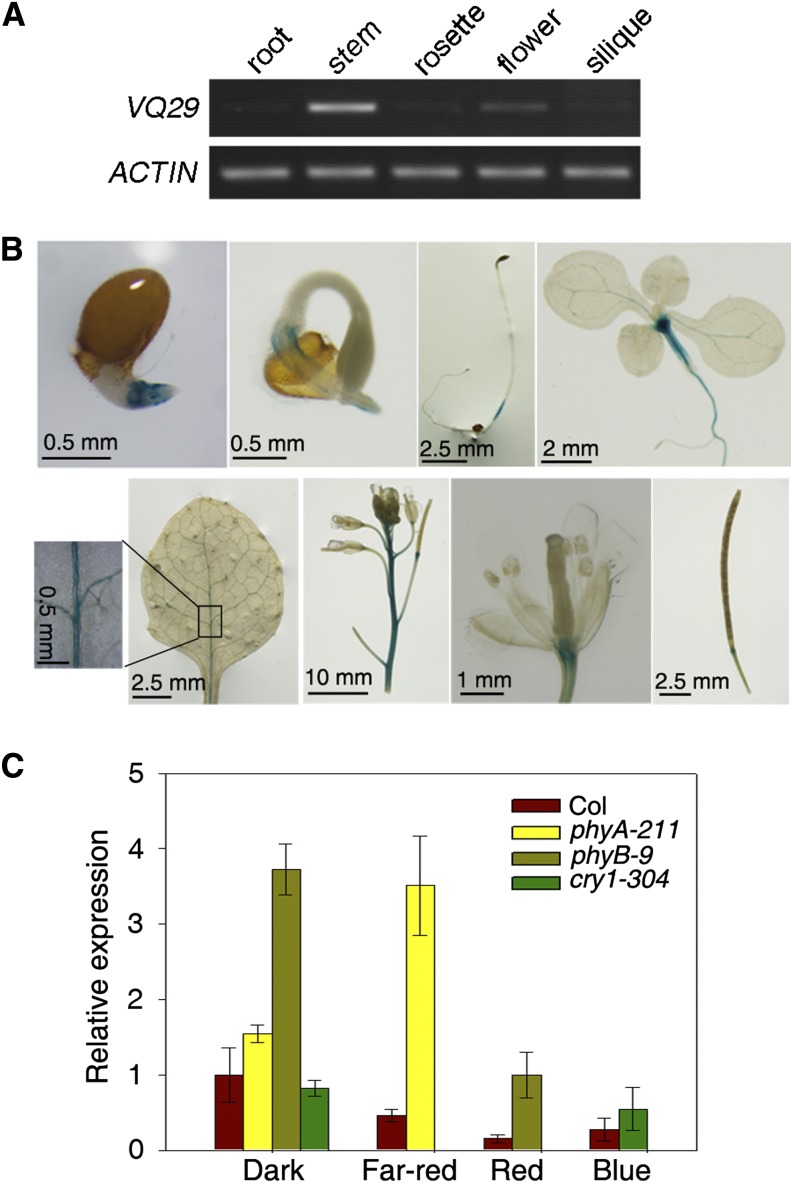
Expression Pattern of VQ29
To examine VQ29 expression in different tissues, we analyzed its transcript levels using RT-PCR. Relatively high levels of VQ29 transcript were observed in the stem, whereas expression was low in the root, rosette leaf, flower, and silique (Fig. 4A). To further visualize the expression pattern of VQ29, we generated transgenic plants expressing the GUS reporter gene under the control of the VQ29 promoter sequence (2.0 kb upstream of the ATG translational start codon). Histochemical staining showed that the GUS reporter gene was expressed in the radicle, hypocotyl, stem, leaf vein, flower, and silique base, indicating that VQ29 may function in these tissues (Fig. 4B).
Figure 4.
Expression pattern of VQ29. A, RT-PCR of VQ29 in various tissues. Amplified actin served as a loading control. B, GUS staining in various tissues of ProVQ29:GUS transgenic plants. C, Quantitative RT-PCR analysis of VQ29 in the photoreceptor mutants under various light conditions and darkness. Seedlings were grown in the indicated conditions for 5 d. Error bars indicate the sd of three biological triplicates.
We then asked whether VQ29 is regulated by light. phyA, phyB, and cry1 photoreceptor mutants, along with the Col wild type, were grown in far-red, red, and blue light conditions, respectively, for 5 d. As shown in Figure 4C, quantitative RT-PCR assays showed that VQ29 expression was repressed by various light treatments. Furthermore, the VQ29 transcript levels were up-regulated in the phyA-211 and phyB-9 mutants under far-red and red light conditions compared with the wild type, respectively. No pronounced difference in VQ29 expression was found between the cry1 mutant and the wild type. Surprisingly, in the dark, VQ29 expression was increased in the phyB mutant but was not affected by phyA and cry1 mutations, suggesting that phyB plays a role in regulating VQ29 in the etiolated seedlings (Fig. 4C). These observations suggest that light inhibits VQ29 expression in a phy-dependent manner, consistent with its role in regulating photomorphogenesis under far-red and low-intensity light conditions. However, an immunoblot assay using the MYC antibody and the VQ29-OE plants showed that the protein level of VQ29 was not regulated by light (Supplemental Fig. S5).
VQ29 Physically Interacts with PIF1
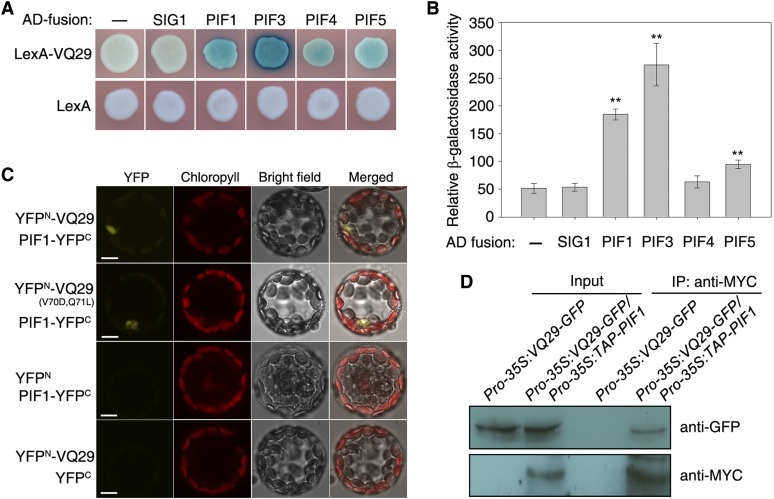
Previous studies reported that VQ proteins could interact with various WRKY TFs (Wang et al., 2010; Lai et al., 2011; Hu et al., 2013). The findings that VQ29 possesses transcriptional activity and is involved in regulating photomorphogenesis prompted us to hypothesize that VQ29 activity might depend on its interaction with other TF(s). Since PIF proteins, including PIF1, PIF3, PIF4, and PIF5, are bHLH-type TFs that play a critical role in repressing photomorphogenesis (Leivar et al., 2008; Shin et al., 2009), we tested their possible interaction with VQ29 in a yeast (Saccharomyces cerevisiae) two-hybrid system. VQ29 was fused with the LexA DNA-binding domain (LexA-VQ29), and various PIF proteins were ligated with the activation domain of B42 (AD-PIFs). Coexpression of LexA-VQ29 with AD-PIF1 or AD-PIF3 caused strong activation of the β-galactosidase gene reporter (Fig. 5, A and B), indicating that VQ29 interacts with PIF1 and PIF3 in yeast cells. However, only mild interaction was detected between VQ29 and PIF5. As negative controls, AD-SIG1 (for plastid-located SIGMA FACTOR1) or AD alone failed to interact with LexA-VQ29 (Fig. 5, A and B). In this study, the relationship between VQ29 and PIF1 was further analyzed.
Figure 5.
VQ29 physically interacts with PIF1. A, Yeast two-hybrid analysis of the interaction between LexA-VQ29 (fused with the LexA DNA-binding domain) and various proteins tagged with the B42 activation domain (AD). − means empty AD vector. B, Relative β-galactosidase activity of interactions between LexA-VQ29 and the AD-tagged proteins shown in A. Error bars indicate the sd of six individual yeast colonies. Asterisks indicate significant differences from the combination of LexA-VQ29 and AD at P < 0.01 using Student’s t test. C, BiFC assay showing the interaction between PIF1-YFPC and YFPN-VQ29 or YFPN-VQ29(V70D,Q71L). Chlorophyll autofluorescence is shown in red. Bars = 5 µm. D, Co-IP assay between VQ29 and PIF1. Pro-35S:VQ29-GFP and Pro-35S:VQ29-GFP/Pro-35S:TAP-PIF1 seedlings were grown in darkness for 5 d. After precipitation with anti-MYC antibody (TAP-PIF1 contains the MYC tag), proteins were immunoblotted with anti-MYC or anti-GFP antibody. IP, Immunoprecipitation.
To substantiate the interaction between VQ29 and PIF1 in plant cells, we performed a bimolecular fluorescence complementation (BiFC) assay in which we transiently coexpressed the N terminus of yellow fluorescent protein fused to VQ29 (YFPN-VQ29) and PIF1 fused to the C terminus of YFP (PIF1-YFPC) in Arabidopsis protoplasts (Walter et al., 2004). Coexpression of YFPN-VQ29 and PIF1-YFPC reconstituted a functional YFP in the nucleus, whereas coexpression with either control vector failed to generate YFP fluorescence (Fig. 5C). The in vivo interaction between VQ29 and PIF1 was further confirmed by coimmunoprecipitation (Co-IP) assay. Therefore, we generated double transgenic plants Pro-35S:VQ29-GFP/Pro-35S:TAP-PIF1 by crossing their single transgenic plants. Tandem affinity purification (TAP)-PIF1 (using MYC antibody) was able to pull down VQ29-GFP (detected by GFP antibody) in the double transgenic seedlings (Fig. 5D). In addition, two point mutations in the VQ motif of VQ29 (V70D and Q71L) did not affect its interaction with PIF1 (Fig. 5C). These data together indicate that VQ29 indeed interacts with PIF1 and that the VQ motif is likely not required for mediating the interaction.
VQ29 and PIF1 Coregulate Seedling Deetiolation
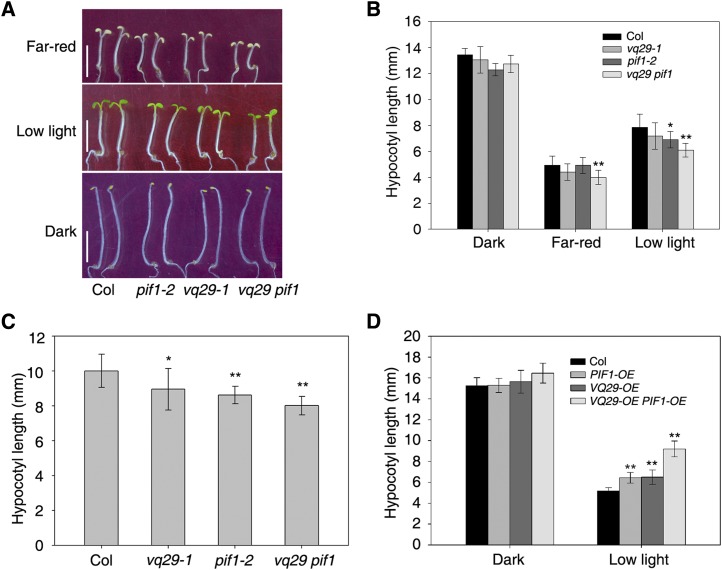
The physical interaction between VQ29 and PIF1 led us to investigate whether and how these proteins function together in the light signaling pathway in Arabidopsis. To this end, we generated the vq29 pif1 double mutant by crossing vq29-1 and pif1-2 mutants and VQ29-OE PIF1-OE double transgenic plants by crossing Pro-35S:Myc-VQ29 (line 10) and Pro-35S:TAP-PIF1. We examined the hypocotyl elongation phenotypes of the double homozygous lines, their single mutant or transgenic parent plants, and the wild type. The hypocotyl length of the vq29 pif1 double mutant was shorter than that of the single mutants and Col wild-type seedlings under both far-red and low light conditions (Fig. 6, A and B; Supplemental Fig. S3). Furthermore, the vq29 pif1 double mutant was pronouncedly shorter than the single mutants under far-red light/dark cycles or under far-red light conditions without Suc supplement in the medium (Fig. 6C; Supplemental Fig. S4B). By contrast, the VQ29-OE PIF1-OE double transgenic plants displayed much longer hypocotyls than their parent single overexpression lines and the wild-type plants under low light conditions (Fig. 6D). These data together indicate that VQ29 and PIF1 additively repress photomorphogenesis.
Figure 6.
Analysis of double mutants and overexpression plants of VQ29 and PIF1. A, Photomorphogenic phenotypes of the Col wild type and vq29, pif1, and vq29 pif1 mutants in darkness or under far-red light (12 µmol m−2 s−1) or low white light (10 µmol m−2 s−1) conditions. Bars = 5 mm. B, Hypocotyl length of the seedlings shown in A. Seedlings were grown in the indicated light conditions for 5 d. C, Hypocotyl length of seedlings grown under far-red light/dark photocycles. Three-day-old dark-grown seedlings were transferred to far-red light/dark conditions (12 h of far-red light/12 h of dark) at the beginning of the light treatment and grown for an additional 2 d. D, Hypocotyl length of Pro-35S:Myc-VQ29 (VQ29-OE, line 10) and Pro-35S:TAP-PIF1 (PIF1-OE) plants and their corresponding double transgenic lines under darkness or low light conditions (10 µmol m−2 s−1). Seedlings were grown in the indicated light conditions for 5 d. In B to D, data are means ± sd of at least 20 seedlings. Asterisks indicate significant differences from the wild type at P < 0.05 (single asterisk) or P < 0.01 (double asterisks) using Student’s t test. [See online article for color version of this figure.]
Since PIF1 is also involved in regulating phy-mediated seed germination and seedling growth during deetiolation (Huq et al., 2004; Oh et al., 2004; Leivar et al., 2008), we next tested whether mutation or overexpression of VQ29 leads to these responses. As reported previously, the pif1 mutant displayed a high germination rate after 5 min of far-red light treatment (Supplemental Fig. S6A). Seeds of the vq29 mutant and VQ29-OE did not show any germination difference compared with the wild type, and the germination efficiency of the vq29 pif1 double mutant was similar to that of pif1 (Supplemental Fig. S6A). In addition, mutation or overexpression of VQ29 did not show a seedling greening phenotype, and vq29-1 did not affect the defects of the pif1 mutant during deetiolation (Supplemental Fig. S6B). These observations indicate that VQ29 is not involved in seed germination and the seedling greening responses with PIF1 and that its responsiveness in hypocotyl elongation appears to be specific.
VQ29 Binds to the Promoters of PIL1 and XTR7 and Regulates Their Expression with PIF1
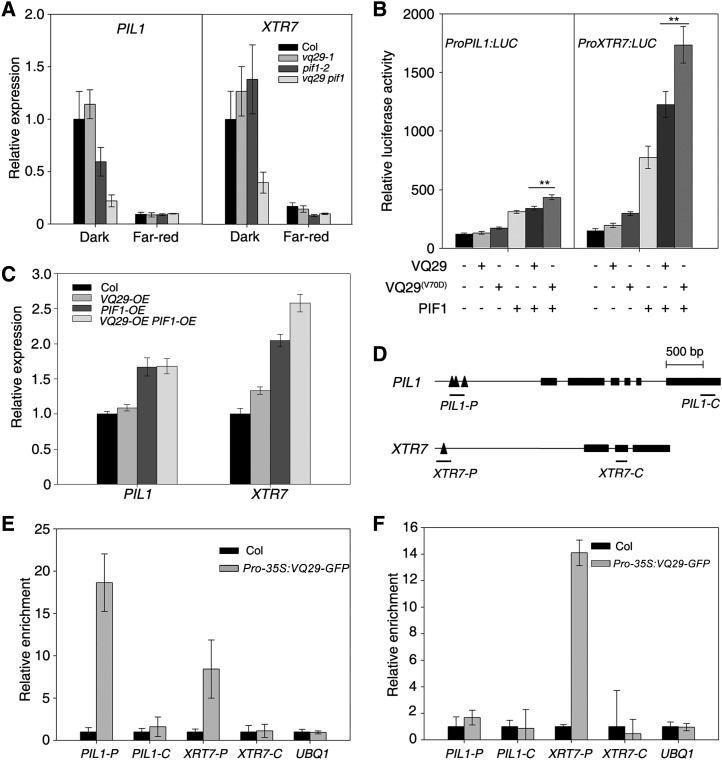
PIF1 also regulates the expression of light-responsive and cell elongation-related genes, such as PIL1 and XTR7 (Leivar et al., 2012). Quantitative RT-PCR analysis revealed that the expression of PIL1 and XTR7 was greatly down-regulated in darkness in the vq29 pif1 double mutant compared with its parent mutants and the wild type (Fig. 7A). In addition, the transcript levels of another two genes involved in cell elongation, EXTENSIN1 (EXT1) and EXT3, were decreased in vq29 and pif1 mutants and further reduced in the vq29 pif1 double mutant (Supplemental Fig. S7). We also tested the expression of EXPANSIN8 (EXP8) and EXP10, two genes that were repressed by PIF1 (Oh et al., 2009). Surprisingly, the EXP10 transcript level was increased in the vq29 pif1 double mutant compared with that in pif1 (Supplemental Fig. S7).
Figure 7.
VQ29 and PIF1 coregulate downstream gene expression. A, Relative expression of PIL1 and XTR7 by quantitative RT-PCR. Seedlings were grown in darkness for 4 d and then kept in darkness or transferred to far-red light (12 µmol m−2 s−1) for an additional 1 h. Error bars indicate the sd of triplicates. B, Relative LUC reporter activity in Arabidopsis protoplasts cotransformed with the effector constructs. The LUC gene was under the control of either the PIL1 or XTR7 promoter. Data represent means ± sd of triplicates. Asterisks indicate significant differences at P < 0.01 using Student’s t test. C, Relative expression of PIL1 and XTR7 in VQ29 and/or PIF1 overexpression plants by quantitative RT-PCR. Seedlings were grown in darkness for 5 d. Error bars indicate the sd of triplicates. D, Diagram of the genomic structures of PIL1 and XTR7. Rectangles denote exons, and triangles represent G-box motifs. Amplicons used in the ChIP assay are underlined. E and F, ChIP-quantitative PCR assay showing the relative enrichment of fragments corresponding to the promoters or coding regions of PIL1, XTR7, and the UBQ1 control. DNA was precipitated with anti-GFP antibody in Pro-35S:VQ29-GFP (E) or with anti-MYC antibody in Pro-35S:TAP-PIF1 (F) transgenic plants. Data represent means ± sd of triplicates.
Furthermore, vectors harboring the LUC reporter gene driven either by the PIL1 or XTR7 promoter were constructed and cotransformed with VQ29 and/or PIF1 into Arabidopsis protoplasts. As shown in Figure 7B, VQ29 itself did not significantly promote LUC expression under the control of either the PIL1 or XTR7 promoter, whereas PIF1 alone strongly activated LUC expression. Most remarkably, coexpression of PIF1 and VQ29 increased the expression levels of these LUC reporter genes. Interestingly, a mutated version of VQ29(V70D) further activated the levels of LUC expression when cotransformed with PIF1 (Fig. 7B), suggesting that the VQ domain of VQ29 indeed possesses transcription repression function. Consistent with these observations, the expression of PIL1 and XTR7 was increased in PIF1-OE transgenic plants, and XTR7’s level was further activated in VQ29-OE PIF1-OE double transgenic plants (Fig. 7C). Therefore, PIF1 and VQ29 coordinately regulate downstream gene expression, and VQ29 likely stimulates the activation activity of PIF1.
To assess whether the induction of PIL1 and XTR7 is directly affected by PIF1 and VQ29, we carried out chromatin immunoprecipitation (ChIP) followed by quantitative PCR assays. When precipitated with GFP antibodies, fragments of the PIL1 and XTR7 promoters containing the G-box, but not their coding regions, were greatly enriched in extracts from Pro-35S:VQ29-GFP transgenic seedlings but not from Col wild-type seedlings (Fig. 7, D and E). In addition, MYC antibody was able to pull down the promoter fragments of XTR7, but not of PIL1, in extracts from Pro-35S:TAP-PIF1 transgenic plants (Fig. 7F). These data indicate that VQ29 is associated with the promoter regions of PIL1 and XTR7 in plant cells.
PIF1 is rapidly degraded after light transition (Shen et al., 2005, 2008). To test whether VQ29 could affect the stability of PIF1, we performed an immunoblot assay using the MYC antibody and PIF1-OE and PIF1-OE VQ29-OE transgenic plants. Our result showed that overexpressing VQ29 did not affect the stability and degradation of PIF1 (Supplemental Fig. S8).
DISCUSSION
VQ29 Defines a Novel Repressor of Seedling Deetiolation
In this study, we present multiple lines of evidence that VQ29 is a novel transcription regulator that represses seedling deetiolation in Arabidopsis. First, hypocotyl elongation in plants overexpressing VQ29 exhibits reduced sensitivity specifically to far-red and low light conditions (Fig. 2). It should be noted that although no effect of the vq29-1 mutant in far-red light was observed in Figures 2E and 6B, a statistically significant short hypocotyl phenotype was observed in this mutant in low far-red light intensities (Supplemental Fig. S3) or in far-red light without Suc in multiple independent experiments (Supplemental Fig. S4). These opposite phenotypes of the loss-of-function mutant and overexpression plants of VQ29 suggest that its coding protein is involved in the phy signaling pathway. In agreement with this, VQ29 expression is down-regulated by light in a phy-dependent manner (Fig. 4C). Second, yeast two-hybrid, BiFC, and Co-IP assays showed that VQ29 is able to physically interact with PIF1, a key TF in the light signaling pathway, both in yeast and plant cells (Fig. 5). Third, the facts that the vq29 pif1 double mutant or VQ29-OE PIF1-OE double transgenic plants exhibited pronounced phenotypes compared with their corresponding single parent plants suggest that VQ29 and PIF1 have additive effects (Fig. 6; Supplemental Figs. S3 and S4). Fourth, VQ29 and PIF1 function together to activate the expression of four genes involved in promoting cell elongation. Furthermore, this regulation might be direct, as VQ29 is associated with the promoters of PIL1 and XTR7 in vivo (Fig. 7E). The less strong phenotype in the vq29 mutant could be alternatively caused by the functional redundancy of VQ29 with other related VQ genes. Future studies using double or higher order mutants with other VQs should further test this possibility.
PIF1 is stabilized in darkness and degraded by light (Shen et al., 2005, 2008). It directly binds to DNA via the G-box cis-element in the promoter of downstream genes and regulates their expression (Leivar and Quail, 2010). Consistently, a G-box motif is found in the promoter regions of both PIL1 and XTR7. Therefore, we propose that VQ29 transcript accumulates in darkness and that its coding protein acts as a transcription coregulator to promote the activity of PIF1 through physical interaction, leading to the activation of cell elongation-related genes (e.g. PIL1 and XTR7), thereby inhibiting seedling deetiolation (Fig. 8). In far-red light conditions, the levels of PIF1 protein and VQ29 transcript are reduced, resulting in the inhibition of cell elongation-related gene expression and, consequently, the promotion of photomorphogenesis. Alternatively, VQ29 might function as a TF that directly binds to the promoter of downstream genes independent of PIF1. Further study is warranted to examine this possibility.
Figure 8.
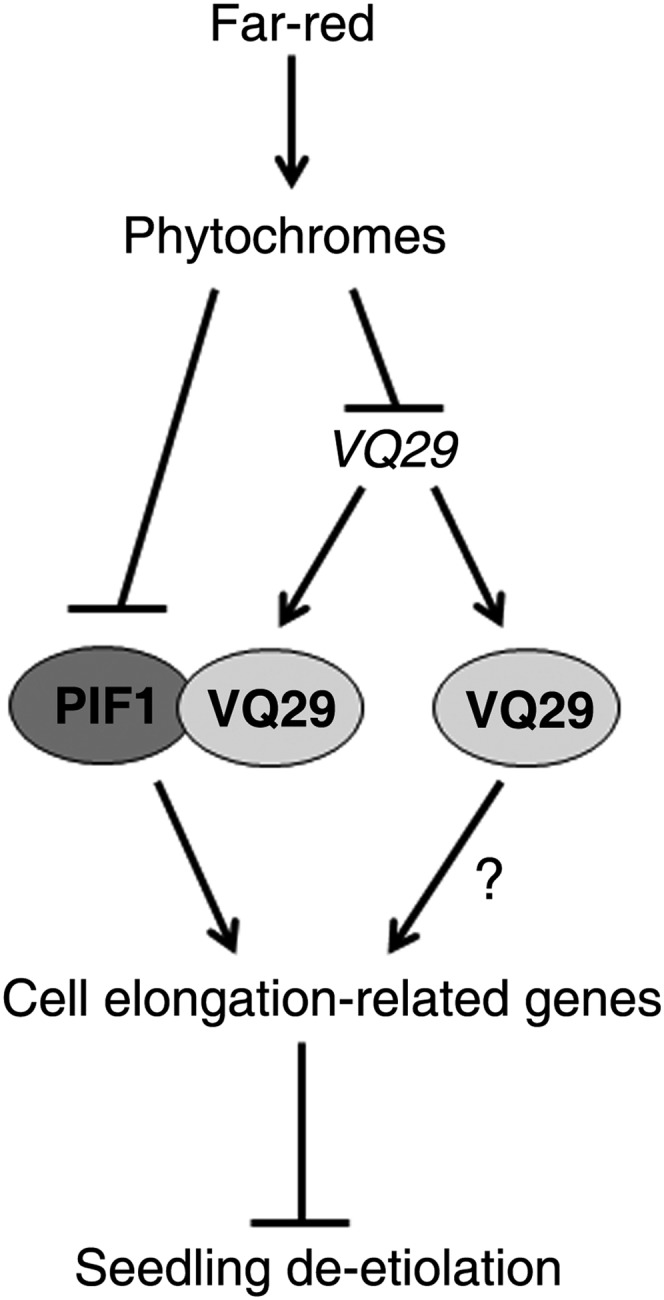
Proposed model of VQ29 in regulating seedling deetiolation. Far-red light, which is perceived by phys, represses PIF1 and VQ29 levels. VQ29 interacts with PIF1 and acts as a transcriptional coregulator to promote PIF1’s activation activity on cell elongation-related genes, leading to the suppression of seedling deetiolation. Meanwhile, VQ29 might regulate downstream gene expression independently of PIF1 as indicated by a question mark. Arrows denote positive effects, and bars indicate negative roles.
PIL1 (which encodes a PIF-regulated TF) and XTR7 (which encodes a xyloglucan endotransglycosylase-related protein) are early shade marker genes (Hornitschek et al., 2009) that are directly regulated by PIF1 and VQ29. VQ29 is required for regulating photomorphogenesis specifically in low white light and far-red light, which mimic the shade conditions. Hence, the involvement of VQ29 might fine-tune the responsiveness to shade-like environments by coordinating with PIF1. Consistently, PIF1 also plays a role, although moderate, in the shade avoidance response (Leivar et al., 2012). Our ChIP experiment showed that VQ29 is associated with the promoter region of XTR7. However, no difference in the expression of XTR7 in the vq29-1 mutant and the wild type was found, suggesting that some other factor(s) is required for regulating downstream gene expression with VQ29. In agreement with this notion, the XTR7 mRNA level was greatly reduced in the vq29 pif1 double mutant. In addition, it is also possible that some other VQ member(s) plays a role in regulating downstream gene expression with VQ29.
Accumulating studies suggest that members of the VQ family play diverse functions, such as regulating the plant defense response (Andreasson et al., 2005; Narusaka et al., 2008; Xie et al., 2010; Lai et al., 2011), abiotic stress resistance (Perruc et al., 2004; Hu et al., 2013), and seed development (Wang et al., 2010). Our study provides genetic and molecular evidence that VQ29 has a prominent role in plant growth and development in response to the light environment, thus extending the functionality of this plant-specific protein family.
VQ Proteins Are Involved in Transcriptional Regulation
VQs are small proteins with a short, unique VQ motif. Molecular studies indicate that VQ proteins likely interact with WRKY TFs and other proteins, such as kinases and plastid sigma factors (Morikawa et al., 2002; Andreasson et al., 2005; Wang et al., 2010; Lai et al., 2011; Cheng et al., 2012; Hu et al., 2013). WRKY and PIF are plant-specific TFs that play crucial biological roles (Leivar and Quail, 2010; Rushton et al., 2010). The interaction of VQ transcription regulators with other TFs appears to fine-tune the temporal and spatial expression patterns of specific targets during plant development or upon exposure to particular biotic and abiotic stresses. Our data showed that mutations in the VQ domain of VQ29 do not affect the interaction with PIF1 (Fig. 5C). However, a recent study revealed that the VQ motif of SIB1 is important for its interaction with WRKY33 (Lai et al., 2011). Therefore, there is variation in which domains of VQ proteins interact with other proteins.
VQ29 possesses transcriptional repression activity in a transient expression system. Our detailed analysis of the conserved V and Q residues in the VQ motif indicates that point mutations of VQ29 alter its transcriptional activity (Fig. 1D). Moreover, mutation in VQ29(V70D) further activates ProPIL1:LUC and ProXTR7:LUC reporter expression mediated by PIF1 (Fig. 7B). These observations demonstrate that the VQ domain of VQ29 indeed has a repressive function on gene expression. However, VQ29 helps PIF1 to promote PIF1-induced gene expression and to repress the expression of PIF1-inhibited genes during light-mediated early seedling growth. Hence, the activity of VQ29 and its effect on gene expression likely depend on specific interacting factors and the target genes in a given signaling process. Similarly, SIB1 simulates the DNA-binding and transcriptional activity of WRKY33 in the plant defense response (Lai et al., 2011). In contrast, recruitment of VQ9 leads to the inactivation of WRKY8, and these proteins act antagonistically to mediate the salt stress response (Hu et al., 2013).
Our transient expression assay in protoplasts suggests that the majority of VQ proteins exhibit either transcriptional activation or repression activities (Fig. 1). The fact that VQ29 localizes to the nucleus further supports its role in regulating transcription. Furthermore, almost all members of this family are thought to localize to the nucleus, based on subcellular proteomic database predictions (Cheng et al., 2012). The identification of more interacting proteins, particularly TFs in the nucleus, should further elucidate the functions and mechanisms of VQ proteins in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The vq29-1 (Salk_061586 and Salk_061438), pif1-2, phyA-211, phyB-9, and cry1-304 mutants and Pro-35S:TAP-PIF1 transgenic plants were derived from the Arabidopsis (Arabidopsis thaliana) Col ecotype (Reed et al., 1993, 1994; Mockler et al., 1999; Moon et al., 2008; Chen et al., 2013). vq29-1 was confirmed by PCR genotyping, and the T-DNA insertion site was verified by sequencing. Double mutant or transgenic plants were generated by genetic crossing, and homozygous lines were used in these experiments. Seedlings were grown on Murashige and Skoog medium containing 1% (w/v) Suc or without Suc as indicated in the text and Supplemental Figure S4. Far-red light (12 µmol m−2 s−1), red light (20 µmol m−2 s−1), and blue light (14 µmol m−2 s−1) were supplied by light-emitting diode light sources, and low white light (10 µmol m−2 s−1) was supplied by cool-white fluorescent lamps. For fluence rate analysis, the light intensities are indicated in the figures.
Determination of Hypocotyl Length, Seedling Greening, and Seed Germination Rate
For hypocotyl length measurement, seedlings of different genotypes were grown side by side on the same plate, and at least three independent plates were used in all experiments. Seedlings were then photographed, and their hypocotyl length was measured by using ImageJ software (http://rsb.info.nih.gov/ij). For seedling greening rate analysis, dark-grown seedlings were transferred to continuous white light for 2 d. Greening rate was determined by counting the percentage of dark-green cotyledons from 50 to 80 seedlings of each genotype. For seed germination assay, seeds of different genotypes were harvested on the same day from plants grown in identical conditions. Seed germination was observed with a microscope and determined based on the appearance of radicle protrusion.
Gene Expression Analysis
Plant total RNA was extracted using the RNAprep Pure Plant Kit (Tiangen), and first-strand cDNA was synthesized using reverse transcriptase (Invitrogen). Real-time PCR was carried out using the SYBR Premix ExTaq Kit (Takara) following the manufacturer’s instructions. The expression levels were normalized to the expression of the UBIQUITIN1 (UBQ1) gene. Primers are listed in Supplemental Table S1.
Plasmid Construction
To obtain ORFs of VQ2, PIF4, and PIF5, first-strand cDNA was reverse transcribed from total RNA extracted from Col wild-type seedlings using oligo(dT)18 primer and high-fidelity Pfu DNA polymerase (Invitrogen). The products were cloned into the pEASY vector (TransGen), resulting in pEASY-VQ2, pEASY-PIF4, and pEASY-PIF5, respectively. Due to the absence of introns in all VQ genes (except VQ2), the ORFs were amplified from genomic DNA using Pfu and cloned into the pEASY vector, resulting in pEASY-VQs. Mutations in VQ29 (V70A, V70D, Q71L, and V70D Q71L) were generated using the MutantBEST site-directed mutagenesis kit (Takara) according to the manufacturer’s instructions. Appropriate restriction enzyme sites were designed at the end of each primer (Supplemental Table S1). All amplified ORFs were validated by sequencing.
To generate constructs for the protoplast transient expression assay, the VQ1 fragment was released from pEASY-VQ1 digested with BamHI and XhoI and inserted into the BglII/SalI site of pSAT-GAL4DB (Jing et al., 2013), to yield pGAL4DB-VQ1. The ORFs of VQ4, VQ5, VQ6, VQ7, and VQ18 were released from the corresponding MfeI-XhoI-digested pEASY-VQ vectors and cloned into the EcoRI/SalI sites of pSAT-GAL4DB, giving rise to pGAL4DB-VQ4, pGAL4DB-VQ5, pGAL4DB-VQ6, pGAL4DB-VQ7, and pGAL4DB-VQ18, respectively. The ORFs of VQ24 and VQ30 were released from pEASY-VQ24 and pEASY-VQ30 digested with EcoRI and SalI and inserted into the EcoRI/SalI sites of pSAT-GAL4DB, generating pGAL4DB-VQ24 and pGAL4DB-VQ30, respectively. To obtain pGAL4DB-VQ34, the pEASY-VQ34 plasmid was cut with MfeI and SalI, and the VQ34 ORF was cloned into the EcoRI/SalI sites of pSAT-GAL4DB. The ORFs of VQ2, VQ3, VQ8, VQ9, VQ10, VQ11, VQ12, VQ13, VQ14, VQ15, VQ16, VQ17, VQ19, VQ20, VQ21, VQ22, VQ23, VQ25, VQ26, VQ27, VQ28, VQ29, VQ31, VQ32, and VQ33 were released from the corresponding EcoRI-XhoI-digested pEASY-VQ vectors and inserted into the EcoRI/SalI sites of pSAT-GAL4DB, resulting in the corresponding pGAL4DB-VQ constructs.
To construct vectors for the yeast (Saccharomyces cerevisiae) two-hybrid assay, the pEASY-VQ29 plasmid was cut with EcoRI and XhoI and the VQ29 fragment was ligated into EcoRI/XhoI-digested EG202 vector (Clontech), to give rise to pLexA-VQ29. The pAD-PIF1 and pAD-PIF3 plasmids were constructed in a previous study (Chen et al., 2013). The pEASY-PIF4 and pEASY-PIF5 plasmids were digested with EcoRI and SalI, and the PIF4 and PIF5 fragments were inserted into the EcoRI/XhoI sites of the JG4-5 vector (Clontech), resulting in pAD-PIF4 and pAD-PIF5, respectively.
To prepare constructs for the BiFC assay, the VQ29 fragment released from EcoRI/XhoI-digested pEASY-VQ29 was inserted into the pUC-SPYNE vector (Walter et al., 2004) and digested with EcoRI and XhoI to generate pYFPN-VQ29. pYFPC-PIF1 was described in a previous study (Chen et al., 2013).
To study the subcellular localization of VQ29, the fragment released from pEASY-VQ29 using EcoRI and XhoI was ligated into the EcoRI/SalI sites of the modified pdGN vector (Lee et al., 2005), to generate pGFP-VQ29.
To construct ProVQ29:GUS, a fragment spanning the region 2 kb upstream of the ATG start site of VQ29 was amplified by PCR and cloned into pEASY, resulting in pEASY-VQ29p. The pEASY-VQ29p plasmid was then digested with SalI and BamHI to release the VQ29 promoter fragment, which was then ligated into the pBI101 vector digested with the same enzymes, to generate ProVQ29:GUS.
To construct VQ29 overexpression vectors, the VQ29 coding sequence was released from pEASY-VQ29 using EcoRI and XhoI and then ligated into the EcoRI-SalI sites of modified pRI101 (Takara), in which three copies of MYC tag were inserted, resulting in Pro-35S:Myc-VQ29. The VQ29 ORF was amplified from pEASY-VQ29 by introducing an NcoI site at the N terminus and a SpeI site at the C terminus and cloned into pEASY to produce pEASY-VQ29G. The pEASY-VQ29G plasmid was then digested with NcoI and SpeI, and the VQ29 fragment was inserted into pCAMBIA1302 (www.cambia.org/daisy/cambia/585) cut with the same enzymes, to generate Pro-35S:VQ29-GFP.
To generate constructs for the transient expression of PIL1 and XTR7, the promoter sequences of both genes were amplified from Col genomic DNA and ligated into the pEASY vector, resulting in pEASY-PIL1p and pEASY-XTR7p, respectively. These plasmids were cut with HindIII and BamHI to release the PIL1 and XTR7 fragments, which were inserted into the HindIII/BamHI sites of the pUC-35sLUC vector (Chen et al., 2013), to produce ProPIL1:LUC and ProXTR7:LUC, respectively.
The binary constructs were electroporated into Agrobacterium tumefaciens strain GV3101 and then introduced into Col wild-type plants. Transgenic plants were selected on Murashige and Skoog plates in the presence of 50 mg L−1 kanamycin or hygromycin, and homozygous lines were used in this study.
LUC Transient Expression Assay
The LUC transient expression assay was carried out as described previously (Jing et al., 2013). LUC reporter activity was detected with a luminescence kit using the Luciferase Assay System (Promega), and the relative activity was expressed as the LUC-GUS ratios.
GUS Histochemical Assay
Seedlings of the ProVQ29:GUS transgenic line were harvested and incubated overnight in 0.1 m sodium phosphate buffer containing 50 mm K3Fe(CN)6, 50 mm K4Fe(CN)6, and 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide at 37°C. GUS expression was examined with a dissecting microscope, and images were captured by a digital camera (Olympus).
GFP Protein Localization Assay
For the transient assay, VQ29-GFP was transformed into Arabidopsis protoplasts, and the protoplasts were incubated under darkness for 16 h before observation. For GFP localization in stable transgenic plants, a homozygous line of Pro-35S:VQ29-GFP was used. The protoplasts and transgenic seedlings were mounted on a slide, and GFP fluorescence was visualized with a Leica TCS SP5 confocal microscope.
Yeast Two-Hybrid Assay
Yeast two-hybrid analysis was performed as described previously (Tang et al., 2012). Transformants were grown on synthetic dextrose/−Trp−uracil−His dropout plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside for color development. Relative β-galactosidase activity was quantified according to the method described in the Yeast Protocols Handbook (Clontech).
Co-IP and Immunoblotting
Seedlings were homogenized in extraction buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10 mm MgCl2, 0.1% (w/v) Tween 20, 1 mm phenylmethylsulfonyl fluoride, and 1× complete protease inhibitor cocktail (Roche). The extracts were centrifuged at 14,000g twice at 4°C for 10 min each, and protein concentration was determined using the Bradford assay (Bio-Rad). Protein fractionations from nucleus, plasma membrane, or cytoplasm were isolated according to the methods described (Larsson et al., 1994; Liu et al., 2001). Co-IP assay was carried out as described previously (Tang et al., 2012). Proteins were separated on 10% (w/v) SDS-PAGE gels and transferred onto polyvinylidene fluoride membranes. They were then blotted against anti-MYC (Abcam), anti-GFP (Sigma), anti-H+-ATPase (Agrisera), anti-cytoplasm-localized Fru-1,6-bisphosphatase (Agrisera), and anti-tubulin (Jing et al., 2013) antibodies. The protein bands were visualized using the standard enhanced chemiluminescence method.
BiFC
Plasmids containing N- and C-terminal YFP fusions were cotransformed into Arabidopsis protoplasts as described previously (Walter et al., 2004). The protoplasts were incubated under weak light for 12 to 16 h before observation. YFP fluorescence was captured with a confocal microscope (Leica).
ChIP
The Col wild-type and Pro-35S:TAP-PIF1 and Pro-35S:VQ29-GFP transgenic plants were used in the ChIP assay, which was carried out as described previously (Chen et al., 2013). The chromatin complexes were incubated with anti-MYC or anti-GFP polyclonal antibody. The precipitated DNA fragments were recovered and quantified by quantitative PCR with the primers shown in Supplemental Table S1.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: VQ29 (At4g37710), PIF1 (At2g20180), PIF3 (At1g09530), PIF4 (At2g43010), PIF5 (At3g59060), PIL1 (At2g46970), XTR7 (At4g14130), EXT1 (At1g76930), EXT3 (At1g21310), EXP8 (At2g40610), EXP10 (At1g26770), SIG1 (At1g08540), ACTIN (At3g18780), and UBQ1 (At3g52590). The accession numbers of all other VQ genes are listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of VQ genes in Arabidopsis, rice, and moss.
Supplemental Figure S2. VQ29 protein level in the wild type and various point mutants.
Supplemental Figure S3. Fluence rate response under different light conditions.
Supplemental Figure S4. Hypocotyl length of VQ29 mutant and overexpression plants grown in the medium without Suc.
Supplemental Figure S5. VQ29 protein level during the dark-to-light transition.
Supplemental Figure S6. Phenotypes in seedling greening and seed germination.
Supplemental Figure S7. Downstream gene expression in vq29 and/or pif1 mutants.
Supplemental Figure S8. The effect of VQ29 on PIF1 protein stability.
Supplemental Table S1. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing the vq29 mutant. We thank Dr. Enamul Huq (University of Texas, Austin) for providing the mutant and overexpression transgenic seeds of PIF1 and Dr. Tai Wang (Institute of Botany, Chinese Academy of Sciences) for providing plasma membrane- and cytosol-localized antibodies.
Glossary
- phy
phytochrome
- cry
cryptochrome
- bHLH
basic helix-loop-helix
- TF
transcription factor
- ORF
open reading frame
- Col
Columbia
- RT
reverse transcription
- cDNA
complementary DNA
- GBD
GAL4 DNA-binding domain
- CaMV
cauliflower mosaic virus
- T-DNA
transfer DNA
- BiFC
bimolecular fluorescence complementation
- ChIP
chromatin immunoprecipitation
- Co-IP
coimmunoprecipitation
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31170221 and 31325002) and the Ministry of Agriculture of China (grant no. 2011ZX08009–003 to R.L.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NHT, Zhu S, Qiu JL, Micheelsen P, Rocher A, Petersen M, et al. (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 24: 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Galstyan A, Guseman JM, Nemhauser JL. (2012) Photomorphogenesis. The Arabidopsis Book 10: e0147, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH. (2001) LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev 15: 2613–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Maloof JN, Wu SH. (2011) COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol 156: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R. (2013) Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zhou Y, Yang Y, Chi YJ, Zhou J, Chen JY, Wang F, Fan B, Shi K, Zhou YH, et al (2012) Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol 159: 810–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS. (1999) Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401: 610–613 [DOI] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M. (2007) SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Zhou YC, Schäfer E, Funk M, Kretsch T. (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon FG, Kay SA. (2003) The F box protein AFR is a positive regulator of phytochrome A-mediated light signaling. Curr Biol 13: 2091–2096 [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh WP, Hsieh HL, Wu SH. (2012) Arabidopsis bZIP16 transcription factor integrates light and hormone signaling pathways to regulate early seedling development. Plant Cell 24: 3997–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Chen L, Wang H, Zhang L, Wang F, Yu D. (2013) Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J 74: 730–745 [DOI] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. (1999) The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13: 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim CH, Apel K, Quail PH. (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N. (2008) Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jing Y, Zhang D, Wang X, Tang W, Wang W, Huai J, Xu G, Chen D, Li Y, Lin R. (2013) Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z. (2011) Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23: 3824–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C, Sommarin M, Widell S. (1994) Isolation of highly purified plant plasma membranes and separation of inside-out and right-side-out vesicles. Methods Enzymol 228: 451–469 [Google Scholar]
- Lau OS, Deng XW. (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW. (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Lee JY, Taoka K, Yoo BC, Ben-Nissan G, Kim DJ, Lucas WJ. (2005) Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17: 2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH. (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH. (2012) Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C. (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Møller SG, Kim YS, Kunkel T, Chua NH. (2003) PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15: 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Zhu L, Shen H, Huq E. (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA 105: 9433–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa K, Shiina T, Murakami S, Toyoshima Y. (2002) Novel nuclear-encoded proteins interacting with a plastid sigma factor, Sig1, in Arabidopsis thaliana. FEBS Lett 514: 300–304 [DOI] [PubMed] [Google Scholar]
- Narusaka M, Kawai K, Izawa N, Seki M, Shinozaki K, Seo S, Kobayashi M, Shiraishi T, Narusaka Y (2008) Gene coding for SigA-binding protein from Arabidopsis appears to be transcriptionally up-regulated by salicylic acid and NPR1-dependent mechanisms. J Gen Plant Pathol 74: 345–354 [Google Scholar]
- Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G. (2009) Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G. (2004) PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perruc E, Charpenteau M, Ramirez BC, Jauneau A, Galaud JP, Ranjeva R, Ranty B. (2004) A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J 38: 410–420 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. (1994) Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E. (2005) PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E. (2008) Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G. (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JL, Maloof JN, Nemhauser JL. (2011) PIF genes mediate the effect of sucrose on seedling growth dynamics. PLoS ONE 6: e19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Wang W, Chen D, Ji Q, Jing Y, Wang H, Lin R. (2012) Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Arnim A, Deng XW. (1996) Light control of seedling development. Annu Rev Plant Physiol Plant Mol Biol 47: 215–243 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang A, Garcia D, Zhang H, Feng K, Chaudhury A, Berger F, Peacock WJ, Dennis ES, Luo M. (2010) The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J 63: 670–679 [DOI] [PubMed] [Google Scholar]
- Wang H, Deng XW. (2002) Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J 21: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW. (1996) The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol 112: 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YD, Li W, Guo D, Dong J, Zhang Q, Fu Y, Ren D, Peng M, Xia Y. (2010) The Arabidopsis gene SIGMA FACTOR-BINDING PROTEIN 1 plays a role in the salicylate- and jasmonate-mediated defence responses. Plant Cell Environ 33: 828–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S. (2005) A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.