Genome structural variations in Arabidopsis-related extremophile modify the transcriptome, endowing competence to survive multiple-ion salt stress in its natural habitat.
Abstract
Schrenkiella parvula (formerly Thellungiella parvula), a close relative of Arabidopsis (Arabidopsis thaliana) and Brassica crop species, thrives on the shores of Lake Tuz, Turkey, where soils accumulate high concentrations of multiple-ion salts. Despite the stark differences in adaptations to extreme salt stresses, the genomes of S. parvula and Arabidopsis show extensive synteny. S. parvula completes its life cycle in the presence of Na+, K+, Mg2+, Li+, and borate at soil concentrations lethal to Arabidopsis. Genome structural variations, including tandem duplications and translocations of genes, interrupt the colinearity observed throughout the S. parvula and Arabidopsis genomes. Structural variations distinguish homologous gene pairs characterized by divergent promoter sequences and basal-level expression strengths. Comparative RNA sequencing reveals the enrichment of ion-transport functions among genes with higher expression in S. parvula, while pathogen defense-related genes show higher expression in Arabidopsis. Key stress-related ion transporter genes in S. parvula showed increased copy number, higher transcript dosage, and evidence for subfunctionalization. This extremophyte offers a framework to identify the requisite adjustments of genomic architecture and expression control for a set of genes found in most plants in a way to support distinct niche adaptation and lifestyles.
The sum of adaptations of an organism forecasts the range of environments a given species can survive. Schrenkiella parvula (formerly Thellungiella parvula and Eutrema parvulum in the Brassicaceae), native to the shores of Lake Tuz, is adapted to an extremely saline habitat (Orsini et al., 2010). Lake Tuz, in central Anatolia, Turkey, is one of the largest hypersaline lakes in the world. The lake’s water composite ion content varies seasonally to often exceed Na+ concentrations greater than 6 times that of seawater. Its water also shows extremely high concentrations of other ions, including K+, Li+, Mg2+, and borate (Helvaci et al., 2004). Not only the lake water, but also soils around the shore where vegetation is found, contain extreme levels of cations as well as sulfates and borates (Nilhan et al., 2008). Combined with high ion concentrations, extreme aridity for most of the year generates a unique habitat with a highly specialized flora. The Lake Tuz ecosystem is considered a UNESCO world heritage site partly because it represents a refuge for wild relatives of crop species adapted to multiple abiotic stresses. These species can serve as natural repositories of genetic resources for future crop improvement (http://whc.unesco.org/en/tentativelists/5824/). S. parvula is a close relative of Brassica crop species (Cheng et al., 2013; Haudry et al., 2013) and thus has the potential to provide unique genetic resources for crop improvement.
The recently sequenced genome of S. parvula (Dassanayake et al., 2011a) provides a powerful resource to study the genomic basis underlying adaptations to multiple abiotic stresses (Dittami and Tonon, 2012; Oh et al., 2012). A comparison with the genome of Arabidopsis (Arabidopsis thaliana) strengthens the validity of conclusions that become possible (Oh et al., 2010; Dassanayake et al., 2011b). The evolution into diverse lifestyles between Arabidopsis and S. parvula is estimated to have taken place within the last 12 million years, after the most recent whole-genome duplication in the crucifer lineage (Oh et al., 2010; Haudry et al., 2013). As the gene-rich regions of the S. parvula genome show genome-wide macrosynteny and extensive colinearity with Arabidopsis, these enable informative cross-species comparisons.
Gene copy number variation (CNV), observed within closely related species, has now been recognized as a major underlying principle in adaptation to new or changing environments (Hanada et al., 2008; Bratlie et al., 2010; Kondrashov, 2012). For example, the regulation of flowering time, crucial for reproductive success in a narrow seasonal window, has been associated with copy numbers of FLOWERING LOCUS T1 (FT1) genes in barley (Hordeum vulgare; Nitcher et al., 2013). Increased copy numbers of related genes or gene clusters have conferred adaptive traits such as an enhanced defense against pathogens (Keeling et al., 2008; Cook et al., 2012) and metal ion tolerance (Hanikenne et al., 2008; Craciun et al., 2012; Maron et al., 2013) in various plant species. Additional copies of genes involved in wax and abscisic acid biosynthesis pathways have been suggested to support increased drought endurance of an Arabidopsis-relative extremophyte, Eutrema salsugineum (formerly Thellungiella salsuginea), when compared with Arabidopsis (Wu et al., 2012).
Variations in genome structure and gene copy number, in turn, affect the transcriptomes via modifications of regulatory sequences (Harewood et al., 2012) and, often, by subfunctionalization of duplicated genes (Duarte et al., 2006; Freeling, 2009). Comparative transcriptomics have demonstrated that different basal-level expression strengths of homologous genes support niche adaptation. This has been highlighted for metal hyperaccumulators such as Arabidopsis halleri (Becher et al., 2004), Alyssum lesbiacum (Ingle et al., 2005), and Thlaspi caerulescens (van de Mortel et al., 2006) among the Brassicaceae, when compared with their respective less-adapted species. Modules of coexpressed genes with different expression strengths in the salt-adapted species E. salsugineum were identified compared with Arabidopsis, even in the absence of salt (Gong et al., 2005). Indeed, variations in basal or constitutive expression strengths of stress-related genes have been recently recognized as a recurring theme in the adaptation to changing environments in studies on various species encompassing all kingdoms of life (Juenger et al., 2010; Geisel, 2011; Barshis et al., 2013; Bedulina et al., 2013). However, studies are scarce that link variations in genome structures to transcriptome profiles and phenotypes showing adaptations to distinct habitats and lifestyles.
In this study, we identify the intrinsic capacity of S. parvula to tolerate multiple-ion stress, enabled by its genome structure and transcriptome variations, which bring about ecological niche adaptations, compared with Arabidopsis. We demonstrate the capacity in S. parvula to grow and complete its life cycle in the presence of high concentrations of a number of ions all lethal to Arabidopsis. Using RNA sequencing (RNA-seq), we compared transcriptomes of root and aerial tissues of S. parvula and Arabidopsis and identified genes and pathways with significantly different basal expression strengths. Many stress-related ion transporter genes in S. parvula showed increased copy numbers and basal expression strengths compared with Arabidopsis, with evidence of subfunctionalization. These results provide a blueprint for mechanisms of plant ion stress tolerance as well as potential genetic resources for crop improvement in closely related species.
RESULTS
S. parvula Completes Its Life Cycle in the Presence of High Na+, K+, Li+, Mg2+, and Borate at Concentrations Lethal to Arabidopsis
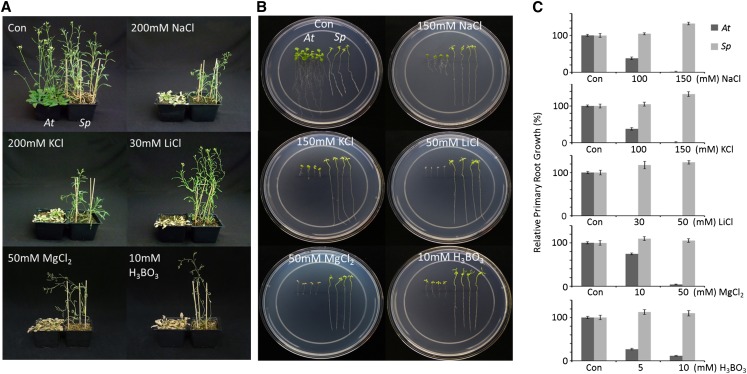
The competence of S. parvula to survive high concentrations of various ions was compared with that of Arabidopsis. Pairs of 4-week-old plants from each species were supplied with 200 mm NaCl, 200 mm KCl, 30 mm LiCl, 50 mm MgCl2, or 10 mm H3BO3 for 2 weeks. While all Arabidopsis plants tested (n = 12, repeated three times) turned yellow and eventually died within 2 weeks, all S. parvula plants continued to grow, completed their life cycle, and produced seeds (Fig. 1A). Root growth of S. parvula seedlings was maintained or even enhanced by the addition of ions at these concentrations. This is contrasted by the inhibition of primary root growth of Arabidopsis seedlings at the same concentrations of ions (Fig. 1, B and C).
Figure 1.
Treatment of S. parvula and Arabidopsis with high concentrations of various salts. A, Four-week-old Arabidopsis (At) and S. parvula (Sp) plants were treated with the indicated concentrations of salts for 2 weeks. Shown are representative examples from three biological replicates (12 plants per species in each replicate) with the same result. B and C, Two-week-old seedlings of Arabidopsis and S. parvula, grown on modified Murashige and Skoog medium as described in “Materials and Methods,” were transferred to the same medium supplemented with the indicated concentrations of salts. Photographs were taken after 10 d of incubation (B), during which root growth of the seedlings was recorded (C). Error bars indicate sd of four measurements. [See online article for color version of this figure.]
E. salsugineum, another Arabidopsis-related extremophyte, exhibited root growth comparable to S. parvula in the presence of NaCl but showed sensitivity to LiCl and high concentrations of KCl (Supplemental Fig. S1). Replacing MgCl2 with MgSO4 produced the same results, indicating that the root growth phenotypes (Fig. 1, B and C) were caused by cations rather than Cl− (Supplemental Fig. S2A). Unlike alkali cations, treatment with heavy metal ions including Ni2+, Zn2+, and Fe2+ inhibited S. parvula root growth more severely compared with Arabidopsis at micromolar concentrations (Supplemental Fig. S2B).
Ion contents of leaf tissues were measured in S. parvula and Arabidopsis treated with NaCl, KCl, LiCl, and MgCl2 from plants grown as in Figure 1A. When treated with lower concentrations that are not stressful to either species, S. parvula accumulated slightly higher amounts of Na+ and K+ and a lower amount of Li+ than Arabidopsis. However, the differences were not statistically significant. Arabidopsis accumulated significantly higher amounts of Mg2+ in the leaves, compared with S. parvula (Supplemental Fig. S3).
Identification of Differently Expressed Homologous Gene Pairs between S. parvula and Arabidopsis
Using RNA-seq, we compared the basal-level expression strengths from nonstressed plants between homologous gene pairs in S. parvula and Arabidopsis. Supplemental Figure S4 summarizes the RNA-seq data-processing pipeline and statistical analysis. RNA samples were prepared from the entire root and shoots of 4-week-old plants. Illumina reads of 100 nucleotides with a total average yield of 51 million reads per sample were generated for three biological replicates. RNA-seq Unified Mapper was used for aligning the reads to the genome and gene models of S. parvula and Arabidopsis. On average, 93.4% and 96.0% of all reads were uniquely mapped, covering 38.4% and 40.0% of the genome sequences of S. parvula and Arabidopsis, respectively. Less than 5% of reads were mapped to multiple gene models. The number of uniquely mapped reads to a gene model becomes a weak proxy for the true expression counts for certain gene models that carry repetitive sequences. Therefore, in our downstream analysis, we excluded gene models with a ratio of nonuniquely mapped reads to those uniquely mapped higher than 20%.
We defined homologous gene pairs between S. parvula and Arabidopsis based on sequence identity using OrthoMCL and reciprocal BLAST searches as described in “Materials and Methods.” For each homologous gene pair, expression strengths of S. parvula and Arabidopsis homologs were compared using DESeq. Among 20,005 homologous gene pairs tested, 3,918 showed significantly different expression between homologs from the two species, either in root or shoot tissues, with a false discovery rate (FDR) smaller than 5%, based on the three biological repeats. These were considered as differently expressed gene pairs (DEGPs).
Genome Structural Variations and Repetitive Sequences Are Overrepresented among DEGPs
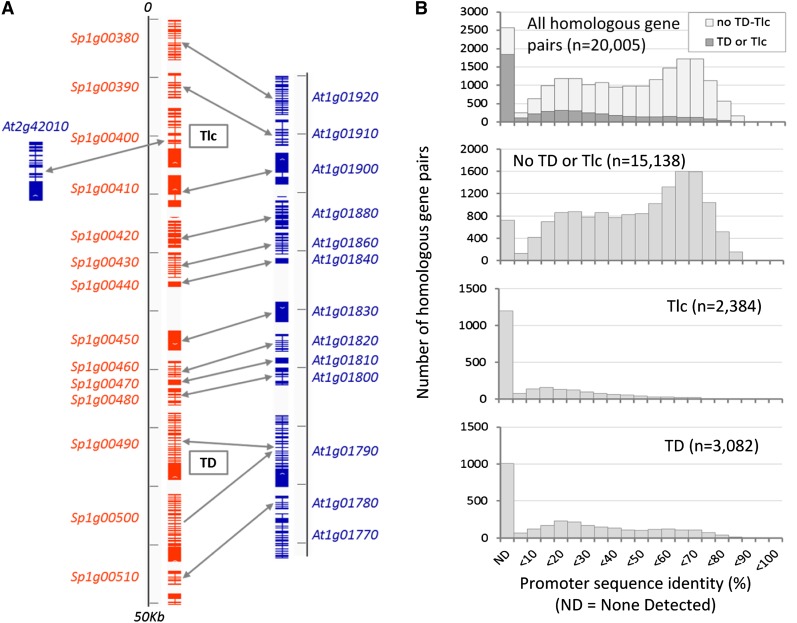
We identified differences between S. parvula and Arabidopsis genome structures that possibly contribute to the differences in expression strengths of homologous genes. While the two genomes show extensive genome-wide colinearity, genomic structural variations including tandem duplication (TD) and translocation (Tlc) events disrupt local synteny between the two species, as exemplified in Figure 2A. We tested whether these structural variations affected the promoter sequences of homologous gene pairs. We used LASTZ to align the promoter sequences of each Arabidopsis and S. parvula homologous gene pair and calculated promoter sequence similarity (see “Materials and Methods”). Homologous gene pairs that underwent TD or Tlc showed clearly lower promoter sequence similarity compared with homologous gene pairs with no TD or Tlc (Fig. 2B). Out of 20,005 S. parvula-Arabidopsis homologous gene pairs, we could not detect any sequence similarity between promoters in 2,574 gene pairs, of which 1,848 (71.8%) were associated with TD or Tlc events (Fig. 2B, top). We tested whether genes affected by TD and Tlc were overrepresented among DEGPs. Additionally, we included insertion of a transposable element or repetitive sequences within 1 kb of the open reading frame (ORF) in the test (Table I). Only 19.6% of the total S. parvula-Arabidopsis homologous gene pairs were identified as DEGPs. However, among S. parvula-Arabidopsis homologous gene pairs associated with TD and Tlc, the proportions of DEGPs were significantly higher, at 37.4% and 37.2%, respectively. This shows a significant enrichment (P < 10−4, χ2 test) of DEGPs among homologous genes involved in structural variations. Containing a transposable element or a repeat longer than 100 nucleotides, within 1 kb of the ORF in either of the two genomes, increased the chance of the homologous gene pair being a DEGP (P = 0.0036, χ2 test). In contrast, homologous gene pairs located next to a TD or Tlc event did not show significant enrichment among DEGPs (Table I).
Figure 2.
Genome structural variation influences promoter sequence divergence between homologous genes. A, A 50-kb S. parvula genome segment was aligned with its colinear Arabidopsis genome segment, showing examples of synteny and structural variations involving TD and Tlc of homologous genes. Exons of S. parvula and Arabidopsis gene models are shown as red and blue boxes, respectively. Arrows connect homologous gene pairs between S. parvula and Arabidopsis. B, Histograms showing the distribution of promoter sequence similarity between S. parvula and Arabidopsis genes in homologous gene pairs with and without TD and Tlc events, respectively. Sequence similarity of the upstream 1-kb sequences of S. parvula and Arabidopsis genes was calculated for each homologous gene pair, using LASTZ as described in “Materials and Methods.” ND denotes no detectable sequence similarity. [See online article for color version of this figure.]
Table I. Enrichment DEGPs among homologous gene pairs showing genome structural variations between S. parvula and Arabidopsis.
| Categories | Total | DEGPsa | DEGPs per Total | P b |
|---|---|---|---|---|
| % | ||||
| All S. parvula-Arabidopsis homologous gene pairs | 20,005 | 3,918 | 19.6 | Background |
| TD c | 3,082 | 1,153 | 37.4 | <10−4 |
| Tlc c | 2,384 | 886 | 37.2 | <10−4 |
| TD or Tlcc | 4,866 | 1,776 | 36.5 | <10−4 |
| Neighboring TD or Tlc to the 5′ sidec | 4,531 | 907 | 20.0 | 0.51 |
| Neighboring TD or Tlc to the 3′ sidec | 4,535 | 896 | 19.8 | 0.99 |
| Near a repetitive sequenced | 3,363 | 732 | 21.8 | 0.0048 |
| TD, Tlc, or near a repetitive sequencec | 7,154 | 2,113 | 29.5 | <10−4 |
DEGPs are based on RNA-seq results (FDR < 0.05). bTwo-tailed χ2 test (Yates correction) for enrichment of DEGPs in each category, with all S. parvula-Arabidopsis homologous gene pairs as the background. cEither the S. parvula or Arabidopsis homolog. dEither the S. parvula or Arabidopsis homolog contains a transposable element or repetitive sequence larger than 100 nucleotides, within 1 kb of the ORF.
Gene Ontology Enrichment Analysis in DEGP Groups
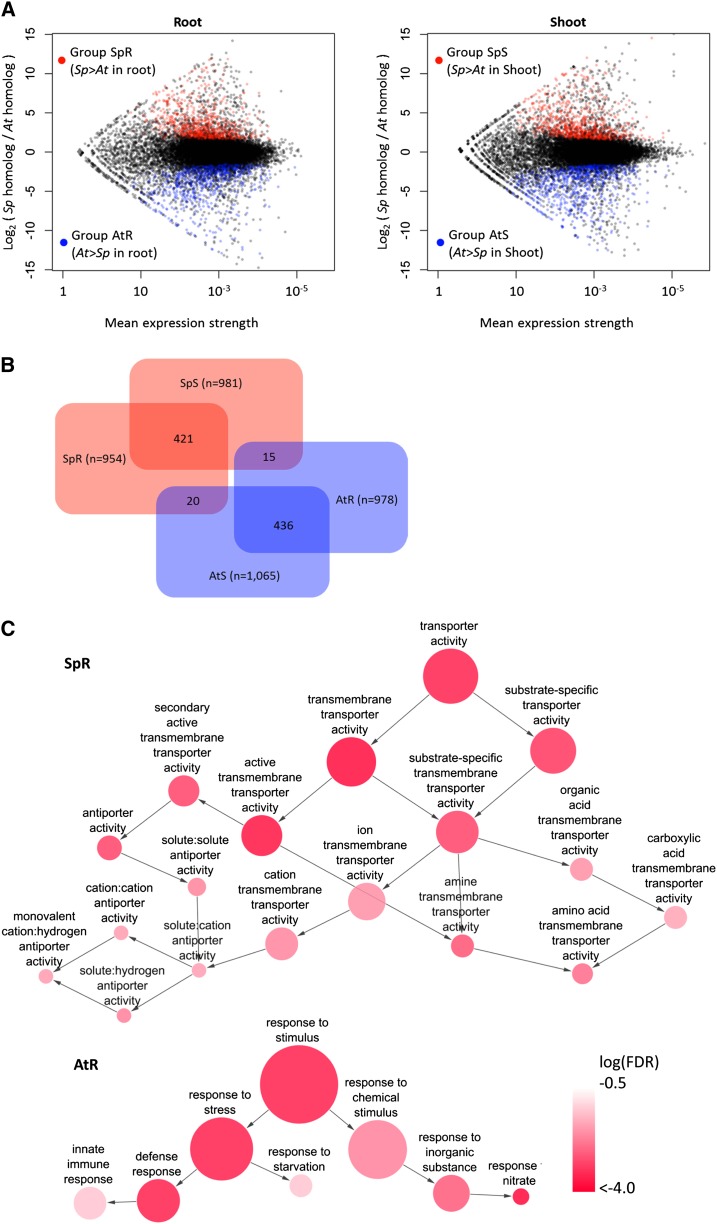
We identified four groups within DEGPs based on differential expression in root and shoot tissues (Fig. 3, A and B) and identified Gene Ontology (GO) terms enriched in each DEGP group, as described in “Materials and Methods.” Figure 3A presents DEGPs included in each group in M-A (fold change versus average expression level) plots (Yang et al., 2002) representing the results of RNA-seq analyses comparing root and shoot samples of S. parvula and Arabidopsis. Groups SpR and SpS, which included 954 and 981 members, had DEGPs with higher expression strength of the S. parvula homolog than the Arabidopsis homolog, in root and shoot tissues, respectively. Similarly, groups AtR and AtS included 978 and 1,065 DEGPs with higher expression strengths of the Arabidopsis homolog than the S. parvula homolog, in the tissues compared (Fig. 3A; Supplemental Data S1). Numbers of homologous gene pairs that belong to more than one DEGP group are given as a Venn diagram in Figure 3B. DEGP groups SpS and SpR shared 421 gene pairs, while DEGP groups AtS and AtR had 436 gene pairs in common. However, gene pairs shared between SpS and AtR or between SpR and AtS were rare at 15 and 20 gene pairs, respectively (Fig. 3B).
Figure 3.
DEGPs between S. parvula (Sp) and Arabidopsis (At). A, M-A plots showing means (x axis) and log2 ratios (y axis) of expression strengths of S. parvula and Arabidopsis homologous gene models, based on normalized uniquely mapped RNA-seq read counts. DEGPs were identified based on FDR < 0.05 (Benjamini-Hochberg multiple testing adjustment) using DESeq (Anders and Huber, 2010). Groups SpR, AtR, SpS, and AtS include DEGPs showing stronger expression in S. parvula and Arabidopsis, in root and shoot tissues, respectively, identified with consideration for duplicated genes, as described in “Materials and Methods.” The complete DESeq result is listed in Supplemental Data S1. B, Venn diagram presenting overlaps among the four DEGP groups. C, Examples of networks representing GO terms enriched in DEGP groups SpR and AtR. Enriched GO terms were identified using PlantGSEA and visualized with Cytoscape, as described in “Materials and Methods.” GO terms are connected based on their ancestor-child relationships. Colors of circles indicate log FDR (Fisher’s test with Yekutieli correction) of enrichment. The size of circles represents the size of GO terms in the background The Arabidopsis Information Resource 10 annotation. The complete networks and lists of enriched GO terms are summarized in Supplemental Tables S1 and S2. [See online article for color version of this figure.]
GO terms enriched in the four SpR, SpS, AtR, and AtS DEGP groups were further analyzed (see “Materials and Methods”). To describe the large numbers of GO terms, we built networks of enriched GO terms based on parent-child relationships in the GO classification. Figure 3C shows representative networks of GO terms enriched in DEGP groups SpR and AtR. A summary of all networks of GO terms enriched in each DEGP group is presented in Supplemental Tables S1 and S2. Compared in Supplemental Table S1 are networks of GO terms enriched in DEGP groups SpR and AtR. Similarly, networks of GO terms enriched in DEGP groups SpS and AtS are shown in Supplemental Table S2. The full list of genes within each GO term is given as Supplemental Data S2. A few patterns emerged.
(1) Enrichment exists for the different child GO terms “transporter activity” and “transport” processes in the DEGP group SpR (higher in S. parvula roots) compared with DEGP groups with higher expression in Arabidopsis (AtR and AtS). SpR was enriched for “monovalent cation transporter activity” (Supplemental Table S1, SpR-1F), while AtR and AtS were enriched for GO terms related to “nitrate transport” and “metal ion transport” processes (Supplemental Tables S1 [AtR-1P] and S2 [AtS-3P]).
(2) Defense-related GO terms were overrepresented in both AtR (Supplemental Table S1, AtR-2P) and AtS (Supplemental Table S2, AtS-1P). However, no observable symptoms of disease were found for either species in any of the plant batches grown for the three biological repeats (data not shown). PATHOGENESIS-RELATED genes and other marker genes related to disease symptoms did not show significantly different basal-level expression between the two species. Rather, the observed enrichment of defense-related GO terms is based on higher basal-level expression of specific gene families in Arabidopsis. This was represented by enrichment of TOLL/INTERLEUKIN1 RECEPTOR-LIKE NUCLEOTIDE BINDING SITE LEUCINE-RICH REPEAT (TIR-NBS-LRR) gene families. The GO term “ADP binding,” enriched in the DEGP groups AtR (Supplemental Table S1, AtR-6F) and AtS (Supplemental Table S2, AtS-5F), almost exclusively consisted of genes of the TIR-NBS-LRR family (Supplemental Data S2).
(3) Different GO subgroups “catalytic activity” and “secondary metabolic process” were enriched in SpR, compared with AtR. The child GO term catalytic activity enriched in SpR included “esterase activity” and “acyltransferase activity,” while AtR was enriched for “glycosyltransferase activity.” SpR and AtR also showed enrichment of different child GO terms “oxidoreductase activities” (Supplemental Table S1, SpR-3F and AtR-3F). Notably, “heme binding” was overrepresented only in the Arabidopsis DEGP groups AtR (Supplemental Table S1, AtR-7F) and AtS (Supplemental Table S2, AtS-8F). More than half of genes included in this GO term encode cytochrome P450 family proteins (Supplemental Data S2). In addition, the GO term “ligand-gated channel,” which almost exclusively consisted of glutamate receptors, was overrepresented in the group AtS (Supplemental Table S2, AtS-9F; Supplemental Data S2).
Ion Transporter Family Genes in S. parvula and Arabidopsis
Supplemental Table S3 lists Arabidopsis loci that are associated with salt stress or other ion toxicity-related phenotypes, and their homologs in S. parvula show different basal-level expression or gene copy number. The majority of the stress-related genes found in a DEGP group encode for ion transporters. The few exceptions are ABA INSENSITIVE5, Δ1-PYRROLINE-5-CARBOXYLATE SYNTHASE1, REPRESSOR OF SILENCING1, and CALCINEURIN B-LIKE10 (CBL10), whose S. parvula homologs show either increased copy numbers or basal-level expression (Supplemental Table S3).
To gain detailed insights on the basis of the multiple-ion tolerance of S. parvula, we focused on the ion transporter family, comparing S. parvula and Arabidopsis. We identified 381 and 372 gene models in S. parvula and Arabidopsis, respectively, that encoded ion transporters from 27 subfamilies, as described by Maathuis et al. (2003). The unequal pairing between S. parvula and Arabidopsis gene models was due to different duplication events present in each species. Supplemental Table S4 summarizes the ion transporter subfamilies in the two species. The complete list of homologous gene pairs encoding ion transporters is given in Supplemental Data S3. Out of the 27 ion transporter subfamilies, S. parvula had nine and Arabidopsis had seven gene families higher in gene copy number as the result of duplications (Supplemental Table S4). The homologous gene pairs encoding ion transporters that were included in at least one of the four DEGP groups were 18.3%, or 66 out of 360 cases (Supplemental Tables S4 and S5). Gene families encoding aquaporins, K+ channels, Na+-H+ antiporters, cation-H+ antiporters, and sulfate transporters contained higher number of DEGPs in groups SpR and SpS. In contrast, nitrate transporters, metal transporters, and P-type pump gene families showed more DEGPs in groups AtR and AtS (Supplemental Table S4).
We could detect increased copy numbers due to duplications leading to different outcomes at the transcriptome level. In some cases, increased copy numbers resulted in overall higher expression strengths, with expression observed for all duplicates, as exemplified by the K+ EFFLUX ANITIPORTER1 (KEA1), K+ UPTAKE PERMEASE9 (KUP9), and ARABIDOPSIS VACUOLAR H+-PYROPHOSPHATASE1 (AVP1) homologs of S. parvula (Supplemental Table S5). In other duplication events, such as those involving HIGH-AFFINITY K+ TRANSPORTER (HKT1) and Na+/H+ EXCHANGER8 (NHX8), subfunctionalization was observed, where duplicates exhibit compartmentalized expression between root and shoot tissues (Supplemental Table S5).
Genomic Structural Variation and Expression Strengths of Stress-Related Ion Transporters
TD and Tlc events among ion transporter genes support the S. parvula multiple-ion tolerance phenotypes. We characterized these events and their effects on gene expression by comparing colinear genomic regions (Figs. 4 and 5).
Figure 4.
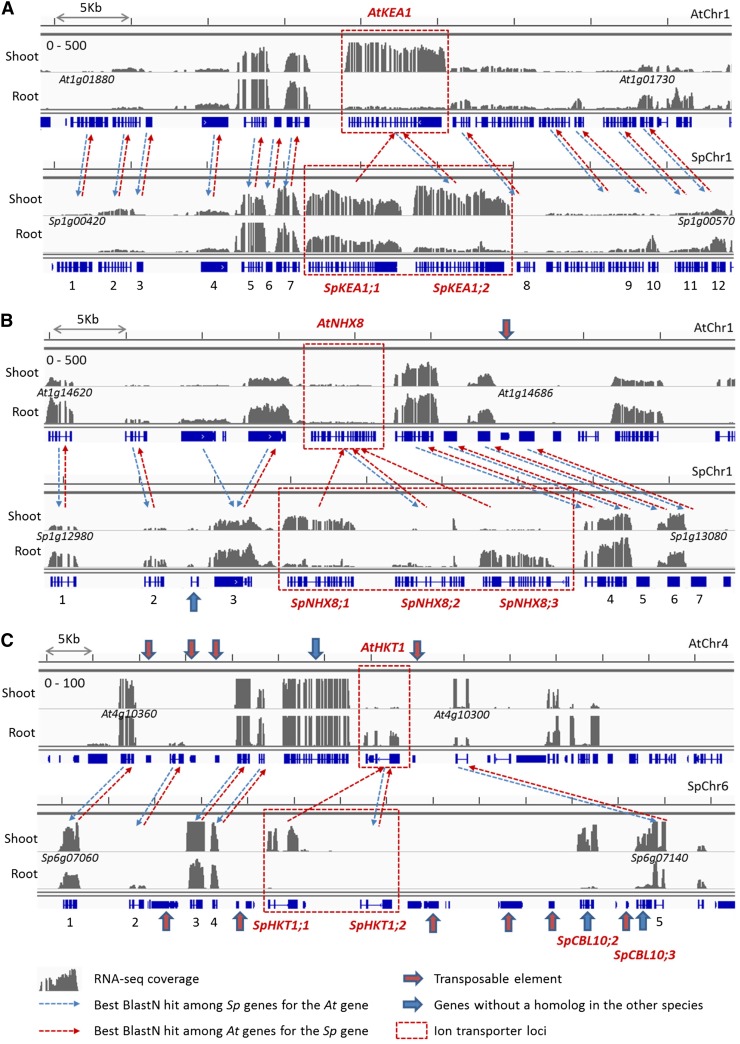
TD of ion transporter genes in S. parvula. RNA-seq read coverage landscapes and homologous gene pairs are shown for colinear genomic regions of Arabidopsis (top) and S. parvula (bottom) around AtKEA1 (At1g01790) and SpKEA1;1 (Sp1g00490)/SpKEA1;2 (Sp1g00500; A), AtNHX8(At1g14660) and SpNHX8;1 (Sp1g13020)/SpNHX8;2 (Sp1g13030)/SpNHX8;2 (Sp1g13040; B), and AtHKT1 (At4g10310) and SpHKT1;1 (Sp6g07110)/SpHKT1;2 (Sp6g07120; C) loci. Gray histograms indicate normalized genome coverage by RNA-seq reads for shoot and root samples. Gene and putative transposon models are presented in blue below each histogram. For each gene in S. parvula and Arabidopsis, dashed arrows connect the homologous gene with the best BLASTN hit scores among all gene models in the genome of the other species. Ion transporter genes tandemly duplicated in S. parvula are marked as red dashed boxes. Putative transposons and gene models unique to either species are indicated by red and blue arrows, respectively. Detailed information on numbered homologous gene pairs, including quantified expression values, is presented as Supplemental Table S6. [See online article for color version of this figure.]
Figure 5.
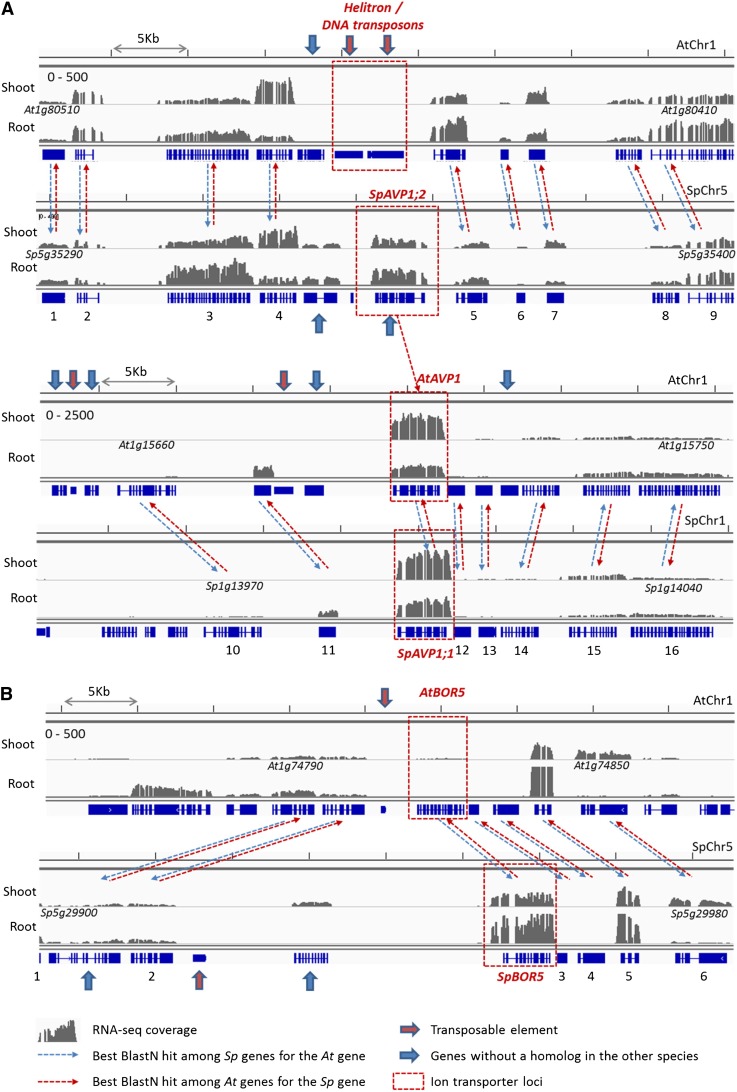
Comparison of Arabidopsis and S. parvula genome segments near AVP1 and BOR5 loci. Colinear genomic regions including AtAVP1 (At1g05690) and SpAVP1;1 (Sp1g13990)/SpAVP1;2 (Sp5g35350; A) and AtBOR5 (At1g74810) and SpBOR5 (Sp5g29940; B) loci are compared, exemplifying Tlc events that affect ion transporter gene copy numbers and gene expression. Arrows, boxes, and histograms were used in the same manner as in Figure 4. Detailed information on numbered homologous gene pairs is presented as Supplemental Table S7. [See online article for color version of this figure.]
The K+ and Li+ transporters KEA1, NHX8, and HKT1 exemplify TD events that resulted in different expression strengths in the two species (Fig. 4; Supplemental Table S6). The genomic regions surrounding KEA1 loci showed extensive colinearity and very similar RNA-seq coverage between S. parvula and Arabidopsis homologous gene pairs. SpKEA1;2/Sp1g00500, which showed higher similarity with AtKEA1 among duplicates, had similar expression strengths with AtKEA1. The other duplicate, SpKEA1;1/Sp1g00490, was expressed significantly higher in root tissue compared with AtKEA1 (Fig. 4A; Supplemental Table S6). While AtNHX8 showed substantially weaker expression compared with adjacent genes, TD in S. parvula resulted in NHX8 copies with significantly higher expression strengths both in S. parvula shoots (SpNHX8;1; Fig. 4B; Supplemental Table S6) and roots (SpNHX8;3; Fig. 4B; Supplemental Table S6). The neighboring colinear genes presented similar expression strengths between Arabidopsis and S. parvula, except for an Arabidopsis-specific TD of genes of unknown function located next to AtNHX8 (Fig. 4B; Supplemental Table S6). Genomic regions near HKT1 are characterized by extensive structural variations between S. parvula and Arabidopsis (Fig. 4C). In S. parvula, putative transposons (Fig. 4C, red arrows) and two tandemly duplicated copies of CBL10 homologs (Fig. 4C, blue arrows) were inserted near the tandemly duplicated SpHKT1;1 and SpHKT1;2. While AtHKT1 showed a stronger expression in the roots compared with shoots, the expression of both SpHKT1;1 and SpHKT1;2 was significantly lower in roots. SpHKT1;1 was expressed specifically in the shoots (Fig. 4C; Supplemental Table S6).
AVP1 and BORON TRANSPORTER5 (BOR5) represent Tlc events that were detected together with differential gene expression between S. parvula and Arabidopsis (Fig. 5; Supplemental Table S7). AtAVP1, encoding a vacuolar proton transporter, is homologous to two nontandem putative AVP1 genes in S. parvula (Fig. 5A). Genomic regions surrounding SpAVP1;1/Sp1g13990 and AtAVP1/At1g15690 showed extensive colinearity, with no significant differences between colinear homologous gene pairs. Both SpAVP1;1 and AtAVP1 are strongly expressed, with a mean normalized RNA-seq read count higher than those of 99.5% of the entire S. parvula and Arabidopsis genes, respectively. AtAVP1 showed higher sequence similarity with its colinear homolog, SpAVP1;1, with which it also showed similar expression (Supplemental Table S7). Similarly, the genomic region around SpAVP1;2/Sp5g35350 in S. parvula chromosome 5 showed similar expression strengths with the colinear region in Arabidopsis chromosome 1, with one exception. The genomic locus in Arabidopsis chromosome 1 corresponding to SpAVP1;2 was replaced by two Helitron family transposon sequences (Fig. 5A, red arrows). SpAVP1;2 showed substantial expression both in root and shoot, although weaker than that of SpAVP1;1 and AtAVP1 (Fig. 5A; Supplemental Table S7).
In the case of the BOR5 loci encoding a putative boron transporter family protein, the immediate 5′ upstream region including the promoter of SpBOR5 was found with an insertion of about 15 kb (Fig. 5B). The sequence insertion in the S. parvula genome harbors a transposon of the Mutator family and an expressed gene (Fig. 5B) showing partial sequence similarity with an isopropyl-malate dehydrogenase gene (Supplemental Table S7). While the adjacent colinear genes showed similar RNA-seq expression strength and patterns, SpBOR5 was expressed significantly higher than AtBOR5. This is one of the largest fold differences observed for all homologous gene pairs between the two species (Fig. 5B; Supplemental Table S7).
DISCUSSION
S. parvula, Adapted to Multiple-Ion Stresses
Mirroring its hypersaline natural habitat, S. parvula completes its life cycle in the presence of a variety of ions at levels that Arabidopsis cannot tolerate (Fig. 1). In fact, the extreme concentrations of sodium, potassium, lithium, magnesium, and boron ions that this plant encounters are high enough to prevent the growth of most plants. High concentrations of sodium ions in the soil limit water uptake and inhibit potassium uptake (Munns and Tester, 2008; Kronzucker and Britto, 2011). Lithium ions can be toxic at even low millimolar concentrations by directly inhibiting the functions of various enzymes (Quiroz et al., 2004). While potassium functions as an essential nutrient for plant growth, excessive amounts of K+ can retard growth by causing energy-consuming futile cycles of ion transport and impairing the acquisition of other nutrients (Wang et al., 1996; Britto and Kronzucker, 2006; ten Hoopen et al., 2010). Finally, borates can cause leaf chlorosis at a concentration that does not constitute an osmotic stress (Miwa and Fujiwara, 2010). S. parvula is prepared to tolerate high concentrations of these ions combined to survive in its natural habitat (Helvaci et al., 2004; Nilhan et al., 2008).
Studies on the salt-tolerant species S. parvula and E. salsugineum (previously T. parvula and T. salsuginea) have revealed higher basal-level expression of a couple of stress-related ion transporters even in the absence of salt stress, compared with their salt-sensitive relative Arabidopsis (Dassanayake et al., 2011b; Wu et al., 2012). Perturbing the basal-level expression of these ion transporters in E. salsugineum resulted in salt sensitivity of the naturally halophytic species (Oh et al., 2009; Ali et al., 2012). The increased basal-level expression of genes and pathways that are known to be stress related in functional studies using the model plant Arabidopsis suggests an adaptation strategy of modifying transcriptome profiles in species evolved to ecological niches distinct from the model plant (Juenger et al., 2010; Dassanayake et al., 2011b; Geisel, 2011). By comparing the transcriptomes of S. parvula and Arabidopsis grown simultaneously at similar developmental stages under stress-neutral experimental conditions, we identified homologous genes and pathways whose basal-level expression show significant differences between the two species. Utilizing gene models derived from the S. parvula and Arabidopsis genomes (Arabidopsis Genome Initiative, 2000; Dassanayake et al., 2011a) and RNA-seq data, our approach provides greater resolution, especially in characterizing the expression divergence of duplicated genes that were unable to be resolved in previous studies relying on microarrays or partial transcriptome sequencing. Moreover, the genomic basis and evolutionary mechanisms that resulted in the observed diversification of gene expression could be explored by focusing on differences in genome architecture and sequence complexity between the two species.
The Transcriptomes of S. parvula and Arabidopsis Suggest Distinct Niche Adaptation
Our analyses began with the careful identification of homologous gene pairs based on sequence similarity (Supplemental Fig. S4). OrthoMCL was used to cluster S. parvula and Arabidopsis gene models into homologous gene groups, and reciprocal BLASTN was used to identify best matching gene pairs within the homologous gene groups, as detailed in “Materials and Methods.” We include RNA-seq analyses on root and shoot tissues, since adaptation to ionic stress in plants has been suggested to require distinct responses from both belowground and aboveground tissues (Maathuis et al., 2003; Munns and Tester, 2008). Normalized expression levels were compared between homologous genes in S. parvula and Arabidopsis to identify DEGPs. With consideration for duplicated genes (see “Materials and Methods”), four groups, SpR, AtR, SpS, and AtS, were identified within DEGPs (Fig. 3, A and B), signifying homologous gene pairs with higher expression in either of species in either root and shoot tissues. We identified networks of GO terms enriched in each of the four DEGP groups, SpR, AtR, SpS, and AtS (Supplemental Tables S1 and S2; Supplemental Data S2), as detailed in “Materials and Methods.”
The GO enrichment analysis suggested niche adaptation through the modification of basal-level expression of transcriptomes not only for S. parvula but also for Arabidopsis. The enrichment of ion transporters appears to support the multiple-ion tolerance of S. parvula, as discussed below. It is not obvious why the basal-level expression of homologous genes encoding defense-related proteins, including TIR-NBS-LRR receptor family genes, should be higher in Arabidopsis compared with S. parvula. Arabidopsis colonized the Eurasian continent after the last glacial period from the Iberian peninsula to central Asia, including areas of northern Africa (Shimizu and Purugganan, 2005). The Arabidopsis ecotype Columbia-0 (Col-0) used in this study is derived from a natural population in Limburg, Germany (http://www.lehleseeds.com/). While the evolutionary history of S. parvula at the tribal level has yet to be established, the species distribution appears to be confined to Turkey and parts of central Asia (Al-Shehbaz, 2012). One possibility is that the habitat of arid salt flats where S. parvula is found (Orsini et al., 2010) shows relatively low pathogen density in comparison with Arabidopsis habitats in more temperate and moist regions. Therefore, S. parvula may not have had a strong evolutionary pressure to develop or sustain resistance genes for biotic stresses compared with Arabidopsis. It is also possible that S. parvula’s adaptation to an extreme environment had evolutionary tradeoffs that did not favor equal adaptation to biotic stress, in contrast to the resource partitioning favoring defense against pathogens over abiotic stress in Arabidopsis (Tian et al., 2003). The diversity of plant-microbe interactions has been suggested as the driving force behind the diversity in plant secondary metabolites (Bednarek and Osbourn, 2009) and the evolution of many lineage-specific metabolic pathway modules (Field and Osbourn, 2008; Kliebenstein and Osbourn, 2012). The enrichment of GO terms including gene families encoding P450s and glycosyltransferases in DEGP groups AtR and AtS further suggests an emphasis on metabolic diversification related to defense in Arabidopsis.
Expression Strength of Ion Transporter Genes Geared for Multiple-Ion Tolerance in S. parvula
Table II summarizes candidate ion transporter genes showing variations in copy number and basal-level expression strength between S. parvula and Arabidopsis. The increased basal-level expression of many of the ion transporters in S. parvula that had a functionally characterized, well-defined homolog in Arabidopsis supports the plant’s multiple-ion tolerance phenotypes. Increased copy numbers via duplication and subfunctionalization provide further diversification of gene functions. The observed correlation between phenotype and the nature and expression of ion transporters provides grounds for testable hypotheses to further our understanding of the functions and evolution of ion-transporter gene families.
Table II. Candidate ion transporter genes responsible for the multiple-ion tolerance of S. parvula and their Arabidopsis homologs.
| Ion Toxicity | Ion Transporter Family | Annotation | S. parvula Homolog | Arabidopsis Homolog | Root |
Shoot |
Structural Variationb | DEGP Group | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean S. parvulaa | Mean Arabidopsisa | FDR | Mean S. parvula | Mean Arabidopsisa | FDR | ||||||||
| % | % | ||||||||||||
| Li+ | Na+-H+ antiporter | NHX8 | Sp1g13020 | At1g14660 | 1,110 | 506 | 17.8 | 5,124 | 660 | 0 | TD | SpR, SpS | An et al. (2007) |
| Sp1g13030c | 1,247 | 51.4 | 438 | 85.8 | |||||||||
| Sp1g13040 | 3,733 | 0 | 484 | 74.2 | |||||||||
| Li+ and Na+ | Na+-H+ antiporter | SOS1 | Sp2g13410 | At2g01980 | 8,356 | 1,828 | 0.3 | 11,039 | 1,628 | 0 | SpR, SpS | Oh et al. (2009) | |
| Na+ | Pyrophosphatase | AVP1 | Sp1g13990c | At1g15690 | 30,195 | 21,806 | 73.9 | 59,813 | 43,620 | 72.8 | Tlc | Gao et al. (2006) | |
| Sp5g35350 | 6,264 | 1.7 | 3,510 | 0 | |||||||||
| Na+ and K+ | Na+-K+ transporter | HKT1 | Sp6g07110 | At4g10310 | 16 | 1,627 | 0 | 1,078 | 92 | 0.5 | TD | AtR, SpS | Ali et al. (2012) |
| Sp6g07120c | 14 | 0 | 3 | 0.4 | |||||||||
| K+ | K+ efflux system | KEA1 | Sp1g00490 | At1g01790 | 6,867 | 965 | 0 | 10,601 | 12,926 | 90.2 | TD | SpR | Chanroj et al. (2012) |
| Sp1g00500 | 2,212 | 21.7 | 13,075 | 99.2 | |||||||||
| K+ transporter | KUP9 | Sp7g12130 | At4g19960 | 1,901 | 941 | 57.6 | 4,830 | 3,181 | 84.9 | Tlc | SpR | Turner et al. (2010) | |
| Sp7g18400c | 4,540 | 0.8 | 2,636 | 85.9 | |||||||||
| Ammonium transporter | AMT1;5 | Sp3g22070 | At3g24290 | 8,802 | 47 | 4.8 | 42 | 2 | 31.2 | SpR | Yuan et al. (2007) | ||
| Aquaporins | TIP2;3 | Sp2g12160 | At5g47450 | 28,456 | 8,171 | 4 | 157 | 181 | 97.8 | SpR | Loqué et al. (2005) | ||
| Mg2+ | Mg2+ transporter | MGT7 | Sp6g33680 | At5g09690 | 674 | 833 | 87.2 | 15 | 295 | 0 | AtS | Gebert et al. (2009) | |
| Borate | Anion exchanger | BOR5 | Sp5g29940 | At1g74810 | 10,582 | 26 | 0 | 3,841 | 794 | 0.3 | Tlc d | SpR, SpS | Takano et al. (2008) |
| Aquaporins | NIP5;1 | Sp6g07050 | At4g10380 | 4,743 | 2,917 | 53.5 | 6,459 | 985 | 0 | SpS | Kato et al. (2009) | ||
| Aquaporins | NIP6;1 | Sp5g30030 | At1g80760 | 937 | 271 | 8.9 | 49 | 756 | 0 | TD | Tanaka et al. (2008) | ||
| Sp5g35000c | 778 | 57.4 | 3,718 | 28 | |||||||||
Mean normalized expression values from three biological repeats. Significantly different expression values (FDR < 5%) between species are in boldface. bStructural variation including TD and Tlc. cS. parvula duplicate copy showing the highest similarity with the Arabidopsis homolog. dBOR5 was not translocated; instead, it was located next to a Tlc event in the S. parvula genome.
The lithium tolerance of S. parvula (Fig. 1; Supplemental Fig. S1) is supported by increased copy numbers of NHX8 (Fig. 4B). TD at the NHX8 loci is unique to S. parvula and not observed in close relatives (Supplemental Fig. S5) or any other species analyzed so far. In Arabidopsis, loss of the single-copy AtNHX8 increased sensitivity, while overexpression improved tolerance, specifically to lithium (An et al., 2007). The three tandem duplicates of SpNHX8 demonstrate subfunctionalization of their expression pattern. SpNHX8;1 and SpNHX8;3 are significantly higher in basal-level expression compared with AtNHX8 in the shoot and root tissues, respectively (Fig. 4B; Supplemental Table S6). The expression strength of SpNHX8;2 was lower than that of the other copies but was induced by treatment with LiCl in the roots (Supplemental Fig. S6).
Many studies have associated high AVP1 and SALT OVERLY SENSITIVE1 (SOS1) expression levels with increased salt tolerance (Park et al., 2005; Gao et al., 2006; Oh et al., 2009). AVP1 exist as two copies in S. parvula, with their combined expression higher than AtAVP1 (Table II). The duplication of AVP1 is conserved in E. salsugineum at the same genomic locus as in S. parvula (Supplemental Fig. S7). Both the Arabidopsis and Arabidopsis lyrata genomes contain a single AVP1, while the locus colinear to SpAVP1;2 has been replaced by transposable elements or deleted (Fig. 5A; Supplemental Fig. S7). Salt stress did not significantly alter the expression of SpAVP1 (Supplemental Fig. S8). In yet another example, compared with Arabidopsis, the higher expression of SOS1 is shared by both S. parvula and E. salsugineum. While no obvious structural variation was observed near the SOS1 loci in Arabidopsis, S. parvula, and E. salsugineum, the conservation of promoter sequences and short simple repeats upstream to SOS1 genes unite S. parvula and E. salsugineum but exclude Arabidopsis in sharing this pattern (Oh et al., 2010; Dassanayake et al., 2011b). AVP1 and SOS1 loci in the genomes of the halophytes S. parvula and E. salsugineum, previously grouped together as genus Thellungiella (Amtmann, 2009), illustrate genomic architecture and regulatory sequences shared between the two halophytes but not the glycophyte Arabidopsis.
In S. parvula, HKT1, encoding an Na+/K+ transporter, exists as tandemly duplicated genes, with evidence for subfunctionalization. TD of HKT1 was also observed in E. salsugineum (Wu et al., 2012); however, the pattern of subfunctionalization of each duplicate’s expression distinguishes the two halophyte species. SpHKT1;2, shows strong shoot-specific expression, while both SpHKT1 duplicates are expressed at significantly lower levels in roots compared with AtHKT1 (Fig. 4C; Supplemental Table S6). In contrast, the HKT1 homologs in E. salsugineum show higher expression in both roots and shoots relative to AtHKT1 expression (Wu et al., 2012). This differential pattern of subfunctionalization may explain another adaptation for the unique environment of the S. parvula habitat, where both [Na+] and [K+] are extremely high in the soil (Helvaci et al., 2004), compared with the soil high in [Na+] but without high [K+] where E. salsugineum is found (Orsini et al., 2010). In further support, continued root growth in high [K+] media was observed only in S. parvula but not in E. salsugineum (Supplemental Fig. S1).
Studies on model plants have focused on deficiencies of macronutrients such as potassium and magnesium (Maathuis, 2009; Wang and Wu, 2013). On the other hand, plant responses to excessive [K+] or [Mg2+] have not received much attention. Homologs encoding two potassium transporters, KEA1 and KUP9, are duplicated in S. parvula and show higher basal-level expression in roots, compared with their Arabidopsis homologs (Fig. 4A; Supplemental Table S5). KEA1 shares sequence similarity with a bacterial K+/H+ antiporter, but precise functions of the family members are still unknown (Chanroj et al., 2012). KUP9, encoding a potassium transporter of unknown function, was considered a candidate polymorphism responsible for the adaptation of A. lyrata to serpentine soil (Turner et al., 2010). Elucidation of the intracellular and tissue localization and functions of these S. parvula-specific duplicates will provide clues about S. parvula’s adaptation capacity as well as on the functions of these two cryptic ion transporter genes. Contrastingly, homologs of Na+/H+ EXCHANGER1 (NHX1) and NHX2 and ARABIDOPSIS K+ TRANSPORTER1, whose functions have been studied in Arabidopsis in relation with K+ ion transport and sequestration (Xu et al., 2006; Barragán et al., 2012), do not show differences in copy number or basal-level expression in S. parvula. Excessive potassium in the soil also inhibits ammonium uptake (Wang et al., 1996; ten Hoopen et al., 2010). S. parvula homologs of AMMONIUM TRANSPORTER1;5 (AMT1;5), a high-affinity ammonium uptake transporter (Yuan et al., 2007), and TONOPLAST INTRINSIC PROTEIN2;3 (TIP2;3), a vacuolar ammonium channel gene (Loqué et al., 2005), exhibited higher basal-level expression in S. parvula roots (Table II). This suggests an adaptation in S. parvula for increased ammonium uptake in the soil with high [K+]. Among Mg2+ transporters, a homolog of Mg2+ TRANSPORTER7 (MGT7), critical in Arabidopsis for growth under low Mg2+ (Gebert et al., 2009), is expressed significantly lower in the shoot of S. parvula compared with Arabidopsis (Table II). The significance of this difference in explaining the observed survival of S. parvula under high Mg2+ (Fig. 1A) requires further assessments through functional studies.
Finally, SpBOR5, a member of a boron transporter family (Takano et al., 2008), shows approximately 400-fold higher expression in roots relative to the expression of the Arabidopsis homolog (Fig. 5B; Supplemental Table S7). This dramatic increase of the expression level of the BOR5 homolog in S. parvula was confirmed by quantitative real-time PCR (Supplemental Fig. S9). A Tlc of a 15-kb DNA fragment immediately 5′ of the BOR5 locus exists in S. parvula. This insertion, altering the promoter sequence of SpBOR5, coincides with one of the largest increments of basal-level expression strength among all S. parvula genes compared with their Arabidopsis homologs. This result leads to another testable hypothesis, that the increased expression of BOR5 may be related to S. parvula’s ability to survive high concentrations of borates, as illustrated in Figure 1. Interestingly, both S. parvula and E. salsugineum share the Tlc event in the 5′ upstream region of BOR5 (data not shown), suggesting a genomic feature conserved among the two closely related species that share tolerance to boron toxicity (Lamdan et al., 2012). The tolerance to boron toxicity is further supported by increased copy numbers and basal-level expression of aquaporin genes related to boron transport, such as NOD26-LIKE INTRINSIC PROTEIN5;1 (NIP5;1) and NIP6;1 (Tanaka et al., 2008; Kato et al., 2009), in S. parvula (Table II).
Genome Structure Shapes the Transcriptome
With a focus on homologous gene pairs at otherwise colinear chromosome regions, we observed structural variations breaking synteny (colinearity) for up to 20% of homologous gene pairs. S. parvula and Arabidopsis diverged after the most recent whole-genome duplication event in the crucifer lineage (Oh et al., 2010; Haudry et al., 2013). Except for the rearrangement of 24 conserved ancestral karyotype blocks into different numbers of chromosomes (Mandáková and Lysak, 2008; Dassanayake et al., 2011a), the most conspicuous mode of structural variation between the two species are TD and Tlc of individual genes, as exemplified in Figure 2A. Structural variation can lead to CNV of genes, which, in turn, affects gene dosage and transcription strength (Stranger et al., 2007; Schlattl et al., 2011; Massouras et al., 2012; Haraksingh and Snyder, 2013). In plants, many examples exist for the copy number of a gene or genome segment associated with phenotype changes (Cook et al., 2012; Li et al., 2012; Maron et al., 2013; Nitcher et al., 2013), and genome-wide CNVs appear to predict phenotypes to some extent (Wu et al., 2012; Muñoz-Amatriaín et al., 2013). The S. parvula genome is characterized by a higher copy number of genes associated with transporter activity (Dassanayake et al., 2011a). A number of abiotic stress-related qualitative trait loci show increased copy numbers in S. parvula (Supplemental Table S3), when compared with Arabidopsis.
Structural variations that do not cause CNVs (i.e. “copy-neutral” structural variants) can affect gene expression as well, either by changing regulatory elements or creating positional effects (Harewood et al., 2010, 2012; Haraksingh and Snyder, 2013). Homologous genes involved in structural variations between the genomes of S. parvula and Arabidopsis contained more divergent promoters (Fig. 2B) and were enriched with DEGPs (Table I). These alterations in genome structure, including TD, Tlc, and insertion of transposable elements and repetitive sequences, shaped or contributed to transcriptome differences between the two species.
We have characterized in detail the consequences of TD and Tlc events in the divergence of ion transporter gene expression (Figs. 4 and 5). During evolution, duplicated genes may have been retained when the increased transcript dosage proved beneficial for survival or adaptability or through the diversification of duplicates via subfunctionalization or neofunctionalization (Freeling, 2009; Innan and Kondrashov, 2010; Jiang et al., 2013). In S. parvula, examples abound that suggest the subfunctionalization of tandemly duplicated ion transporters and structural variations that resulted in higher transcript dosages. Based on functional studies in Arabidopsis, homologs of these ion transporters and their divergence in S. parvula may explain how multiple-ion tolerance has been acquired to support survival in an environment characterized by extreme levels of several typically detrimental ions.
CONCLUSION
S. parvula represents a plant that epitomizes adaptations for growth under multiple high ion levels coincidental in varying concentrations that are toxic to most plants. A complex phenotype such as tolerance to multiple salts cannot be explained by one or even a few genes. Intuitively, several pathways, or networks, are likely requiring changes. Given the relatively small divergence time between Arabidopsis and S. parvula, estimated at around 12 million years ago, macrosyntenic regions are evident throughout the two genomes, but structural variations, including tandem gene duplications, gene Tlc, and transposable element insertions, interrupt colinearity. The genomic structural variations, especially TD and Tlc of genes associated with Li+, Na+, K+, and borate management, distinguish the S. parvula genome from that of Arabidopsis. These genomic rearrangements are mirrored by and converge into transcriptome adaptations via differential gene expression and subfunctionalization that, in turn, can support the phenotypic adaptations for multiple-ion tolerance observed for S. parvula. Both Arabidopsis and S. parvula accumulated similar numbers of duplications and other structural variations. However, the changes retained in each genome followed different trajectories leading to distinct ecological niches. The genomic structural variations and the associated transcriptome reorganization in this extremophyte represent a model to study niche adaptation in the evolution of stress tolerance. Improvements can now be envisioned aimed at augmenting single-ion or multiple-ion salt stress tolerances in related Brassica species crops that share the majority of functionally characterized homologs with S. parvula.
MATERIALS AND METHODS
Plant Growth and Stress Treatment
Schrenkiella parvula and Arabidopsis (Arabidopsis thaliana Col-0) plants were grown on root wash mix soil (Plant Care Facility, University of Illinois at Urbana-Champaign) in a growth chamber with a 14-h-day/10-h-night cycle, 130 µmol m−2 s−1 light intensity, and 22°C to 24°C temperature. Plants were watered every other day, with a supplement of one-twentieth-strength Hoagland solution once every 6 d. For stress treatments, salts were added to irrigation as indicated. At least 12 4-week-old plants for each species were tested. Root growth assays were performed as described before (Oh et al., 2009), using 4-d-old S. parvula and Arabidopsis (Col-0) seedlings.
Identification of S. parvula-Arabidopsis Homologous Gene Pairs
Genome annotations version 2.0 (http://thellungiella.org/data/; National Center for Biotechnology Information BioProject PRJNA63667) and version 10 (http://www.arabidopsis.org/) were used for S. parvula and Arabidopsis, respectively. When multiple spliced forms exist in Arabidopsis, the longest version was considered. Homologous gene pairs were identified based on sequence similarity. OrthoMCL (Li et al., 2003) grouped genes showing deduced amino acid sequence alignment by all-to-all BLASTP (e < 10−5) for more than 70% of the entire gene length. To account for lineage-specific gene duplications in both species, homologous gene pairs were searched reciprocally between the two species. Each gene in a species was paired with a gene from the other species that showed the highest BLASTN hit score only if the pair was in the same gene group identified by OrthoMCL and their gene lengths were not different by more than 20%. Among a total of 27,207 Arabidopsis nontransposon, chromosomal, and putative protein-coding gene models, 19,783 were paired with an S. parvula homolog. Similarly, 19,292 out of 26,920 S. parvula gene models were paired with an Arabidopsis homolog. The two reciprocal searches were merged, and redundant pairs were removed to generate 20,005 S. parvula-Arabidopsis homologous gene pairs.
Identification of DEGPs
For RNA-seq experiments, root and shoot tissues were pooled from 10 4-week-old plants (Supplemental Fig. S4). Plants grown in different batches were sampled for three biological repeats. Total RNAs extracted with the RNeasy Plant kit (Qiagen) were processed using TruSeq RNA Sample Prep Kits version 2 (Illumina) and sequenced by the HiSeq2000 system (Illumina). The resulting 100-nucleotide, single-end reads were aligned to genome and gene model sequences of S. parvula and Arabidopsis, respectively, using RNA-seq Unified Mapper version 2.0.2 (Grant et al., 2011) with default parameters. S. parvula-Arabidopsis homologous gene pairs with significant differences between normalized RNA-seq read numbers uniquely aligned to the S. parvula and Arabidopsis gene model in the pair were identified as DEGPs using the DESeq package (Anders and Huber, 2010) with an FDR cutoff set to 0.05. A small number of DEGPs where the decision was affected by nonunique mapping were removed from downstream analyses. Bedgraph files generated by RNA-seq Unified Mapper were used for visualizing RNA-seq coverage in genomic regions with Integrated Genomics Viewer version 2.3 (Thorvaldsdóttir et al., 2013).
Analyses of Structural Variations and Promoter Sequences of Homologous Genes
TD indicates an event where two or more genes in the same homologous gene group identified by OrthoMCL are adjacent to, or separated by, one nonhomologous gene from each other. A Tlc event is defined as homologous gene pairs that are displaced to either a different chromosome or, if on the same chromosome, more than 20 gene loci away from the original colinear location. For transposable elements and repetitive sequences, we used annotations generated by the REPET package (Flutre et al., 2011) and RepeatMasker (http://www.repeatmasker.org/) for S. parvula and Arabidopsis genomes, respectively. To identify the promoter sequence similarity of a homologous gene pair, we used LASTZ (Kiełbasa et al., 2011) to align the 1-kb upstream sequences of the S. parvula and Arabidopsis homologs for each homologous gene pair. The percentage of identical nucleotides was counted for all alignments detected with the seed pattern “111101110010111” optimized for comparison of noncoding sequences.
Visualization of GO Terms Enriched in DEGP Groups
DEGPs identified by DESeq were further grouped into four DEGP groups, SpR, SpS, AtR, and AtS (Fig. 3A), based on different expression in root and shoot samples. When a single gene in one species was paired with multiple genes in the other species due to gene duplication, the single gene was included in the respective DEGP group only if its expression value was significantly higher than the highest expression value among the duplicated homologs in the other species. PlantGSEA was used to detect GO terms and gene families enriched in each of the four DEGP groups, using Fisher’s test with Yekutieli correction (FDR cutoff of 0.05), with The Arabidopsis Information Resource 10 Arabidopsis annotation as the background (Yi et al., 2013). Networks based on a parent-children relationship of GO terms significantly enriched in each of the four DEGP groups were constructed and visualized with the BiNGO plugin in Cytoscape (Maere et al., 2005).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Root growth comparison between Arabidopsis, S. parvula, and Eutrema salsugineum.
Supplemental Figure S2. Comparison of root growth of Arabidopsis and S. parvula under different concentrations of salts and metal ions.
Supplemental Figure S3. Comparison of ion contents between S. parvula and Arabidopsis.
Supplemental Figure S4. Flowchart depicting the process of homologous gene pair identification, RNA-seq, and downstream bioinformatics analyses.
Supplemental Figure S5. Comparison of genomic regions near NHX8 loci of Arabidopsis lyrata, S. parvula, and E. salsugineum, using CoGE GEvo.
Supplemental Figure S6. Quantitative RT-PCR analysis of transcript abundance of NHX8 homologs in S. parvula.
Supplemental Figure S7. Comparison of colinear genomic regions near AVP1;2 loci, of A. lyrata, S. parvula, and E. salsugineum.
Supplemental Figure S8. Quantitative RT-PCR analysis of transcript abundance of AVP1 homologs in S. parvula.
Supplemental Figure S9. Quantitative RT-PCR comparing relative transcript abundances of BOR5 homologs between S. parvula and Arabidopsis.
Supplemental Table S1. Summary of networks of GO terms overrepresented in DEGP groups SpR and AtR.
Supplemental Table S2. Summary of networks of GO terms overrepresented in DEGP groups SpS and AtS.
Supplemental Table S3. List of salt or ion stress-related Arabidopsis qualitative trait loci that are either included in DEGPs or showing CNVs in S. parvula.
Supplemental Table S4. Overview of ion transporter gene families in S. parvula and Arabidopsis.
Supplemental Table S5. List of DEGPs encoding ion transporters and channels.
Supplemental Table S6. Detailed information of homologous gene pairs shown in Figure 4.
Supplemental Table S7. Detailed information of homologous gene pairs shown in Figure 5.
Supplemental Data S1. RNA-seq results comparing expression strengths of homologs between S. parvula and Arabidopsis, including a list of S. parvula-Arabidopsis homologous gene pairs, genes involved in genomic structural variations, and promoter sequence similarity between S. parvula and Arabidposis homologs.
Supplemental Data S2. Network of GO terms enriched in DEGP groups.
Supplemental Data S3. Homologous gene pairs encoding putative ion transporter family proteins in S. parvula and Arabidposis.
Supplementary Material
Acknowledgments
We thank Dr. John M. Cheeseman (University of Illinois at Urbana-Champaign) for his advice on plant growth conditions and critical comments on the manuscript and Jeff Haas (University of Illinois at Urbana-Champaign) for help with server maintenance and program installation.
Glossary
- CNV
copy number variation
- RNA-seq
RNA sequencing
- FDR
false discovery rate
- DEGP
differently expressed gene pair
- TD
tandem duplication
- Tlc
translocation
- ORF
open reading frame
- GO
Gene Ontology
- Col-0
Columbia-0
Footnotes
This work was supported by the National Research Foundation of Korea (grant no. 2013R1A2A1A01005170) and the Next-Generation BioGreen21 Program (grant no. PJ009495), Rural Development Administration, Republic of Korea.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ali Z, Park HC, Ali A, Oh DH, Aman R, Kropornicka A, Hong H, Choi W, Chung WS, Kim WY, et al. (2012) TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiol 158: 1463–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shehbaz I. (2012) A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon 61: 931–954 [Google Scholar]
- Amtmann A. (2009) Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol Plant 2: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An R, Chen QJ, Chai MF, Lu PL, Su Z, Qin ZX, Chen J, Wang XC. (2007) AtNHX8, a member of the monovalent cation:proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li/H antiporter. Plant J 49: 718–728 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Barragán V, Leidi EO, Andrés Z, Rubio L, De Luca A, Fernández JA, Cubero B, Pardo JM. (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24: 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barshis DJ, Ladner JT, Oliver TA, Seneca FO, Traylor-Knowles N, Palumbi SR. (2013) Genomic basis for coral resilience to climate change. Proc Natl Acad Sci USA 110: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher M, Talke IN, Krall L, Krämer U. (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37: 251–268 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Osbourn A. (2009) Plant-microbe interactions: chemical diversity in plant defense. Science 324: 746–748 [DOI] [PubMed] [Google Scholar]
- Bedulina DS, Evgen’ev MB, Timofeyev MA, Protopopova MV, Garbuz DG, Pavlichenko VV, Luckenbach T, Shatilina ZM, Axenov-Gribanov DV, Gurkov AN, et al (2013) Expression patterns and organization of the hsp70 genes correlate with thermotolerance in two congener endemic amphipod species (Eulimnogammarus cyaneus and E. verrucosus) from Lake Baikal. Mol Ecol 22: 1416–1430 [DOI] [PubMed] [Google Scholar]
- Bratlie MS, Johansen J, Sherman BT, Huang W, Lempicki RA, Drabløs F. (2010) Gene duplications in prokaryotes can be associated with environmental adaptation. BMC Genomics 11: 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britto DT, Kronzucker HJ. (2006) Futile cycling at the plasma membrane: a hallmark of low-affinity nutrient transport. Trends Plant Sci 11: 529–534 [DOI] [PubMed] [Google Scholar]
- Chanroj S, Wang G, Venema K, Zhang MW, Delwiche CF, Sze H. (2012) Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Front Plant Sci 3: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Mandáková T, Wu J, Xie Q, Lysak MA, Wang X. (2013) Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25: 1541–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE, et al. (2012) Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338: 1206–1209 [DOI] [PubMed] [Google Scholar]
- Craciun AR, Meyer CL, Chen J, Roosens N, De Groodt R, Hilson P, Verbruggen N. (2012) Variation in HMA4 gene copy number and expression among Noccaea caerulescens populations presenting different levels of Cd tolerance and accumulation. J Exp Bot 63: 4179–4189 [DOI] [PubMed] [Google Scholar]
- Dassanayake M, Oh DH, Haas JS, Hernandez A, Hong H, Ali S, Yun DJ, Bressan RA, Zhu JK, Bohnert HJ, et al (2011a) The genome of the extremophile crucifer Thellungiella parvula. Nat Genet 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake M, Oh DH, Hong H, Bohnert HJ, Cheeseman JM. (2011b) Transcription strength and halophytic lifestyle. Trends Plant Sci 16: 1–3 [DOI] [PubMed] [Google Scholar]
- Dittami SM, Tonon T. (2012) Genomes of extremophile crucifers: new platforms for comparative genomics and beyond. Genome Biol 13: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, Ma H, Altman N, dePamphilis CW. (2006) Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol 23: 469–478 [DOI] [PubMed] [Google Scholar]
- Field B, Osbourn AE. (2008) Metabolic diversification: independent assembly of operon-like gene clusters in different plants. Science 320: 543–547 [DOI] [PubMed] [Google Scholar]
- Flutre T, Duprat E, Feuillet C, Quesneville H. (2011) Considering transposable element diversification in de novo annotation approaches. PLoS ONE 6: e16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. (2009) Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60: 433–453 [DOI] [PubMed] [Google Scholar]
- Gao F, Gao Q, Duan X, Yue G, Yang A, Zhang J. (2006) Cloning of an H+-PPase gene from Thellungiella halophila and its heterologous expression to improve tobacco salt tolerance. J Exp Bot 57: 3259–3270 [DOI] [PubMed] [Google Scholar]
- Gebert M, Meschenmoser K, Svidová S, Weghuber J, Schweyen R, Eifler K, Lenz H, Weyand K, Knoop V. (2009) A root-expressed magnesium transporter of the MRS2/MGT gene family in Arabidopsis thaliana allows for growth in low-Mg2+ environments. Plant Cell 21: 4018–4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisel N. (2011) Constitutive versus responsive gene expression strategies for growth in changing environments. PLoS ONE 6: e27033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Li P, Ma S, Indu Rupassara S, Bohnert HJ. (2005) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J 44: 826–839 [DOI] [PubMed] [Google Scholar]
- Grant GR, Farkas MH, Pizarro AD, Lahens NF, Schug J, Brunk BP, Stoeckert CJ, Hogenesch JB, Pierce EA. (2011) Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM). Bioinformatics 27: 2518–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH. (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Krämer U. (2008) Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453: 391–395 [DOI] [PubMed] [Google Scholar]
- Haraksingh RR, Snyder MP. (2013) Impacts of variation in the human genome on gene regulation. J Mol Biol 425: 3970–3977 [DOI] [PubMed] [Google Scholar]
- Harewood L, Chaignat E, Reymond A. (2012) Structural variation and its effect on expression. Methods Mol Biol 838: 173–186 [DOI] [PubMed] [Google Scholar]
- Harewood L, Schütz F, Boyle S, Perry P, Delorenzi M, Bickmore WA, Reymond A. (2010) The effect of translocation-induced nuclear reorganization on gene expression. Genome Res 20: 554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, Forczek E, Joly-Lopez Z, Steffen JG, Hazzouri KM, et al. (2013) An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet 45: 891–898 [DOI] [PubMed] [Google Scholar]
- Helvaci C, Mordogan H, Colak M, Gundogan I, Çolak M. (2004) Presence and distribution of lithium in borate deposits and some recent lake waters of west-central Turkey. Int Geol Rev 46: 177–190 [Google Scholar]
- Ingle RA, Mugford ST, Rees JD, Campbell MM, Smith JAC. (2005) Constitutively high expression of the histidine biosynthetic pathway contributes to nickel tolerance in hyperaccumulator plants. Plant Cell 17: 2089–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108 [DOI] [PubMed] [Google Scholar]
- Jiang WK, Liu YL, Xia EH, Gao LZ. (2013) Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants. Plant Physiol 161: 1844–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger TE, Sen S, Bray E, Stahl E, Wayne T, McKay J, Richards JH. (2010) Exploring genetic and expression differences between physiologically extreme ecotypes: comparative genomic hybridization and gene expression studies of Kas-1 and Tsu-1 accessions of Arabidopsis thaliana. Plant Cell Environ 33: 1268–1284 [DOI] [PubMed] [Google Scholar]
- Kato Y, Miwa K, Takano J, Wada M, Fujiwara T. (2009) Highly boron deficiency-tolerant plants generated by enhanced expression of NIP5;1, a boric acid channel. Plant Cell Physiol 50: 58–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling CI, Weisshaar S, Lin RPC, Bohlmann J. (2008) Functional plasticity of paralogous diterpene synthases involved in conifer defense. Proc Natl Acad Sci USA 105: 1085–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. (2011) Adaptive seeds tame genomic sequence comparison. Genome Res 21: 487–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Osbourn A. (2012) Making new molecules: evolution of pathways for novel metabolites in plants. Curr Opin Plant Biol 15: 415–423 [DOI] [PubMed] [Google Scholar]
- Kondrashov FA. (2012) Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci 279: 5048–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronzucker HJ, Britto DT. (2011) Sodium transport in plants: a critical review. New Phytol 189: 54–81 [DOI] [PubMed] [Google Scholar]
- Lamdan NL, Attia Z, Moran N, Moshelion M. (2012) The Arabidopsis-related halophyte Thellungiella halophila: boron tolerance via boron complexation with metabolites? Plant Cell Environ 35: 735–746 [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xiao J, Wu J, Duan J, Liu Y, Ye X, Zhang X, Guo X, Gu Y, Zhang L, et al (2012) A tandem segmental duplication (TSD) in green revolution gene Rht-D1b region underlies plant height variation. New Phytol 196: 282–291 [DOI] [PubMed] [Google Scholar]
- Loqué D, Ludewig U, Yuan L, von Wirén N. (2005) Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol 137: 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJM. (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12: 250–258 [DOI] [PubMed] [Google Scholar]
- Maathuis FJM, Filatov V, Herzyk P, Krijger GC, Axelsen KB, Chen S, Green BJ, Li Y, Madagan KL, Sánchez-Fernández R, et al. (2003) Transcriptome analysis of root transporters reveals participation of multiple gene families in the response to cation stress. Plant J 35: 675–692 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA. (2008) Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae). Plant Cell 20: 2559–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron LG, Guimarães CT, Kirst M, Albert PS, Birchler JA, Bradbury PJ, Buckler ES, Coluccio AE, Danilova TV, Kudrna D, et al. (2013) Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc Natl Acad Sci USA 110: 5241–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massouras A, Waszak SM, Albarca-Aguilera M, Hens K, Holcombe W, Ayroles JF, Dermitzakis ET, Stone EA, Jensen JD, Mackay TF, et al (2012) Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS Genet 8: e1003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa K, Fujiwara T. (2010) Boron transport in plants: co-ordinated regulation of transporters. Ann Bot (Lond) 105: 1103–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Tester M. (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Muñoz-Amatriaín M, Eichten SR, Wicker T, Richmond TA, Mascher M, Steuernagel B, Scholz U, Ariyadasa R, Spannagl M, Nussbaumer T, et al. (2013) Distribution, functional impact, and origin mechanisms of copy number variation in the barley genome. Genome Biol 14: R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilhan TG, Emre YA, Osman K. (2008) Soil determinants for distribution of Halocnemum strobilaceum Bieb. (Chenopodiaceae) around Lake Tuz, Turkey. Pak J Biol Sci 11: 565–570 [DOI] [PubMed] [Google Scholar]
- Nitcher R, Distelfeld A, Tan C, Yan L, Dubcovsky J. (2013) Increased copy number at the HvFT1 locus is associated with accelerated flowering time in barley. Mol Genet Genomics 288: 261–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Dassanayake M, Bohnert HJ, Cheeseman JM. (2012) Life at the extreme: lessons from the genome. Genome Biol 13: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Dassanayake M, Haas JS, Kropornika A, Wright C, d’Urzo MP, Hong H, Ali S, Hernandez A, Lambert GM, et al. (2010) Genome structures and halophyte-specific gene expression of the extremophile Thellungiella parvula in comparison with Thellungiella salsuginea (Thellungiella halophila) and Arabidopsis. Plant Physiol 154: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Leidi E, Zhang Q, Hwang SM, Li Y, Quintero FJ, Jiang X, D’Urzo MP, Lee SY, Zhao Y, et al. (2009) Loss of halophytism by interference with SOS1 expression. Plant Physiol 151: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini F, D’Urzo MP, Inan G, Serra S, Oh DH, Mickelbart MV, Consiglio F, Li X, Jeong JC, Yun DJ, et al. (2010) A comparative study of salt tolerance parameters in 11 wild relatives of Arabidopsis thaliana. J Exp Bot 61: 3787–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Li J, Pittman JK, Berkowitz GA, Yang H, Undurraga S, Morris J, Hirschi KD, Gaxiola RA. (2005) Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc Natl Acad Sci USA 102: 18830–18835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz JA, Gould TD, Manji HK. (2004) Molecular effects of lithium. Mol Interv 4: 259–272 [DOI] [PubMed] [Google Scholar]
- Schlattl A, Anders S, Waszak SMS, Huber W, Korbel JO. (2011) Relating CNVs to transcriptome data at fine resolution: assessment of the effect of variant size, type, and overlap with functional regions. Genome Res 21: 2004–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu KK, Purugganan MD. (2005) Evolutionary and ecological genomics of Arabidopsis. Plant Physiol 138: 578–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. (2007) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Miwa K, Fujiwara T. (2008) Boron transport mechanisms: collaboration of channels and transporters. Trends Plant Sci 13: 451–457 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Wallace IS, Takano J, Roberts DM, Fujiwara T. (2008) NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 20: 2860–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hoopen F, Cuin TA, Pedas P, Hegelund JN, Shabala S, Schjoerring JK, Jahn TP. (2010) Competition between uptake of ammonium and potassium in barley and Arabidopsis roots: molecular mechanisms and physiological consequences. J Exp Bot 61: 2303–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77 [DOI] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. (2010) Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat Genet 42: 260–263 [DOI] [PubMed] [Google Scholar]
- van de Mortel JE, Almar Villanueva L, Schat H, Kwekkeboom J, Coughlan S, Moerland PD, Ver Loren van Themaat E, Koornneef M, Aarts MGM. (2006) Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol 142: 1127–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MY, Siddiqi MY, Glass DM. (1996) Interactions between K+ and NH4+: effects on ion uptake by rice roots. Plant Cell Environ 19: 1037–1046 [Google Scholar]
- Wang Y, Wu WH. (2013) Potassium transport and signaling in higher plants. Annu Rev Plant Biol 64: 451–476 [DOI] [PubMed] [Google Scholar]
- Wu HJ, Zhang Z, Wang JY, Oh DH, Dassanayake M, Liu B, Huang Q, Sun HX, Xia R, Wu Y, et al. (2012) Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc Natl Acad Sci USA 109: 12219–12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360 [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Du Z, Su Z. (2013) PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res 41: W98–W103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Loqué D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wirén N. (2007) The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19: 2636–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.