ABSTRACT
In prion diseases, abnormal prion protein (PrPSc) is considered as the main component of the infectious agent. Delineation of PrPSc conformation is expected to be a critical factor in understanding properties of prions. However, practical methods to differentiate between conformers of PrPSc are inadequate. Here, we used two PrPSc-specific monoclonal antibodies (mAbs), 3B7 and 3H6, and found that mAb 3H6 detected a limited portion of PrPSc in five mice-adapted prion strains. The quantity of mAb 3H6-precipitated PrPSc was significantly lesser in 22L compared to other strains. This result provides a direct evidence of the conformational heterogeneity of PrPSc within the prion strains. Conformation-specific probes, like these mAbs, have the potential to be powerful tools for investigating conformational variations in PrPSc.
Keywords: abnormal prion protein (PrPSc), conformation, monoclonal antibody
Transmissible spongiform encephalopathy (TSE) is a neurodegenerative disorder in humans and animals, such as scrapie in sheep and goats. An abnormal isoform of prion protein, PrPSc, which is generated by the posttranslational modification of cellular prion protein (PrPC), accumulates in affected animals. PrPSc is believed to be the major, or the only, component of the infectious agent, that is, the prion [18]. Unlike PrPC, PrPSc has many β-sheets [15], and this structure is considered responsible for the aggregation of PrPSc. These characteristics contribute to the relative resistance to proteinase K (PK) digestion [17]. Biochemical detection of the moieties remaining after PK digestion, designated as PrPres, has generally been utilized as a criterion for diagnosing TSE.
Accumulating evidence indicates that PrPSc-specific monoclonal antibodies (mAbs) can be employed to detect conformations of PrPSc [6, 9, 12, 14, 16, 21]. Although their deduced epitopes vary, almost all mAbs seemed to detect PrPSc irrespective of the species-based differences. The mAb 3B7 and 3H6 possessed different reactivities to the PrPSc of several species; this represents their unique characteristics. This differential reactivity enabled us to trace the conformational transition of mouse PrPSc during adaptation in the sheep-to-mice transmission of scrapie [21]. This also indicated the co-existence of heterogeneous PrPSc in early-passaged mice in inter-species transmission of prion. These unique characteristics of mAbs are expected to provide an advantage in the conformational analysis of PrPSc.
TSE can be transmitted across species, and during this transmission, the incidence of different strains exhibiting different disease phenotypes have often been found [7]. The “protein-only” hypothesis suggests that conformational differences in PrPSc determine the strain phenotype [20]. It has been proposed that PrPSc has several conformations and this concept adequately explains the variation in the susceptibility and the emergence of new strains during interspecies transmission [3, 4]. However, direct evidence based on a biochemical approach is limited. The conformational differences in PrPSc have been estimated by the biochemical properties of PrPres during immunoblotting [5, 20]. These analyses are effective in the comparison of PrPSc conformations among strains. However, distinguishing a particular PrPSc from a mixture of PrPSc is difficult, except in case of clearly distinguishing characteristics [1, 2]. Therefore, simple procedures for conformational discrimination of PrPSc in non-denatured condition would be of great value. In this study, we aimed to determine whether heterogeneous PrPSc exists within mouse-adapted prion strains, by using PrPSc-specific mAbs.
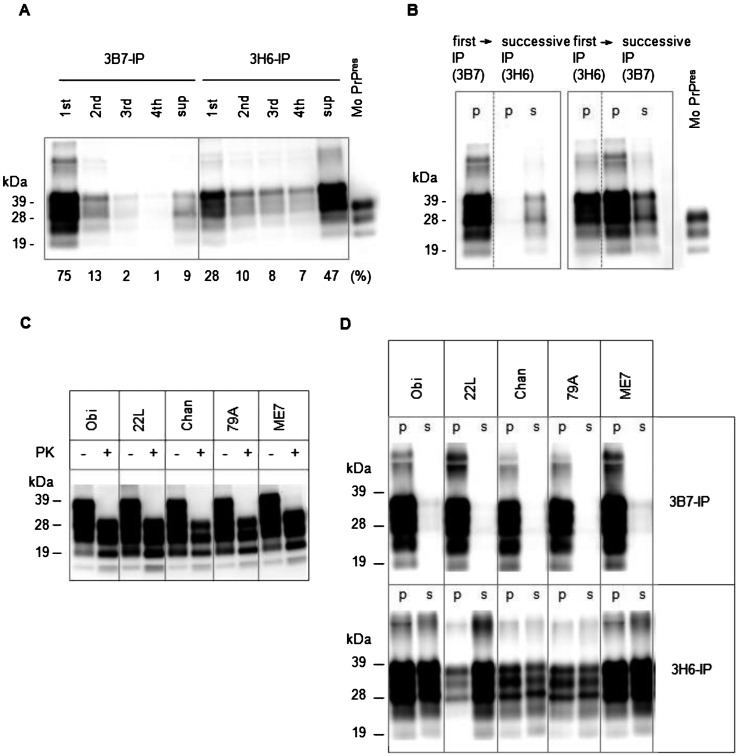
Initially, we examined their reactivity to the scrapie Obihiro [19]-affected mouse brain homogenate by the immunoprecipitation (IP) assay. The mAbs 3B7 and 3H6 were conjugated to Dynabeads M-280 Tosylactivated (Invitrogen, Carlsbad, CA, U.S.A.), in accordance with the manufacturer’s instructions and used in an IP assay as described earlier [21]. In short, 200 µl of 0.025% brain homogenate (equivalent to 50 µg of brain tissue) in 2% Triton X-100 in PBS (Triton/PBS) and 5 µl of mAb-conjugated beads were rotated for 1 hr at room temperature and then washed 4 times with Triton/PBS. The mAb-bound PrPSc was directly eluted into sodium dodecyl sulfate (SDS) sample buffer by heating. The unbound PrP remaining in the supernatant was precipitated using a 2-butanol/methanol solution [11] and resuspended in SDS sample buffer [21]. Both bound and unbound PrPSc were detected by immunoblotting as described previously [21]. Preliminary IP analysis showed that the quantity of PrPSc detected by mAb 3H6 was lower than that detected by mAb 3B7. Additionally, preparatory repetitive IP indicated that most of the mAb 3B7/3H6-precipitated PrPSc was detected by 1st round-IP. However, the supernatant retained 47% of the total PrP after the 4th round of repetitive IP using mAb 3H6 (Fig. 1A). Therefore, we assessed whether the PrPSc remaining in the supernatant after the IP with mAb 3H6 could be detected by mAb 3B7. As shown in Fig. 1B, most of the PrPSc was precipitated with mAb 3B7, and only a small amount remained in the supernatant. In contrast, after IP with mAb 3H6, the supernatant contained a large amount of PrPSc, while most of the remaining PrPSc could be precipitated with mAb 3B7. These results suggested that the mAb 3H6 selectively precipitated a portion of the PrPSc present in the brain homogenate of the Obihiro strain, leaving a substantial amount of PrPSc that could be precipitated by mAb 3B7.
Fig. 1.
Representative immunoblotting results of PrPs after immunoprecipitation (IP) or proteinase K (PK) treatment. PrPs were detected with anti-PrP monoclonal antibody (mAb) T2 [8] in accordance with the standard protocol [21] (n=3 or 4). Intensity of the bands was analyzed with Spot Denso software (AlphaImager, ProteinSimple, Santa Clara, CA, U.S.A.). The signal intensity obtained in independent immunoblots was evaluated by the ratio to the standard material, purified mouse PrPres (Mo PrPres), prepared as described earlier [13]. Mo PrPres was coelectrophoresed with test samples for every PrP immunoblot. The mean intensity of each band in immunoblot of (C, D) is summarized in Table 1. (A) Repetitive IP of PrPSc with mAbs 3B7 and 3H6. The PrPSc that remained in the supernatant (sup) after being precipitated by repeated IP (4 times; 1st to 4th) were detected by immunoblotting. The ratio of each fraction was represented as the percentage of total intensity, which was obtained from 3 independent experiments and is designated underneath immunoblotting panel. (B) Differences in PrPSc of Obihiro precipitated with the mAbs 3B7 and 3H6. The residual PrPSc recovered from the supernatant of the first IP with 1 mAb was detected by successive IP with another mAb. The PrPSc in the precipitate (p) and supernatant (s) of IP were detected. (C) Quantification of PrPres. The mouse brain homogenates were treated (+) or not treated (−) with PK and then detected. PrPres, comprising about 34–52% of total PrP, was present in all examined homogenates. Obi: Obihiro; Chan: Chandler. (D) The result of IP with the mAbs 3B7 and 3H6 of 5 strains. The PrPSc in the precipitate (p) and supernatant (s) of IP were detected.
Next, we determined whether the difference in discrimination of mAb 3H6 was commonly found in other mouse-adapted scrapie strains. The brains of scrapie 22L-, Chandler-, 79A- and ME7-infected mice [10, 22] were examined. We confirmed that all scrapie-affected brains contained approximately similar amounts of PrPres (34–52% of total PrP) by immunoblotting (Fig. 1C and Table 1). Then, IP was performed with both mAbs 3B7 and 3H6, and the ratio of the precipitated PrPSc was calculated as a percentage of total PrP (sum of intensities on the immunoblot of mAb-precipitated PrPSc and that of PrP in the supernatant). We found that mAb 3B7 precipitated approximately 93–96% of the PrPSc from the 22L, Chandler, 79A and ME7 homogenates (Table 1), similar to the results obtained for Obihiro (Fig. 1B), and almost no PrPSc was detected in the remaining supernatant (Fig. 1D). In contrast, the ratio of mAb 3H6-precipitated PrPSc to total PrP was lower: 55% for Chandler, 51% for 79A, 47% for ME7 and 15% for 22L (Table 1). This finding suggested that the partial recognition of PrPSc by mAb 3H6 was not specific to the Obihiro strain, but was also observed in all the prion strains examined. These data suggested that the mAb 3H6 precipitated and unprecipitated PrPSc coexisted in the scrapie-affected mice.
Table 1. Summary of relative intensity of PrPs analyzed by immunoblotting.
| Scrapie strain | WB |
IP |
|
|---|---|---|---|
| PrPres (%)a) | mAb-precipitated PrPSc in brain homogenate (%) |
||
| mAb 3B7b) | mAb 3H6b) | ||
| Obihiro | 51 ± 11 | 96 ± 2 | 43 ± 12 |
| 22L | 52 ± 10 | 95 ± 2 | 15 ± 12* |
| Chandler | 34 ± 8 | 93 ± 3 | 55 ± 3 |
| 79A | 37 ± 11 | 95 ± 3 | 51 ± 5 |
| ME7 | 51 ± 13 | 94 ± 3 | 47 ± 7 |
a) The quantity of PrPres was estimated as the ratio of relative intensity of PrPres (PK+) to that of total PrP (PK–) of immunoblots (n=3). b) The ratio of mAb-precipitated PrPSc to total PrP was calculated according to the intensity obtained by immunoblots (n=3 or 4). The intensity of total PrP was estimated as the sum of the precipitate and the supernatant (p and s in Fig. 1D , respectively). The asterisk indicates significant differences in the mean values analyzed by Student’s t-test between 22L and other 4 strains (P<0.05).
It should be noted that the partial detection of PrPSc by mAb 3H6 was possibly due to its low binding affinity. However, low affinity does not adequately explain the reason for lesser binding to 22L (Fig. 1D and Table 1). Interestingly, these mAbs possessed different species specificity. The mAb 3H6 was considered mouse specific, while mAb 3B7 reacted with the PrPSc of mice, hamsters and deer [21]. This difference in the species specificity of these two mAbs would have an effect on the ratio of mAb-precipitated PrPSc to total PrP. Detailed kinetic analyses of these mAbs need to be carried out in future. Understanding the mechanism underlying the differential reactivity of mAb 3H6 would enable us to clarify the species-specific conformational characteristics of PrPSc.
The findings of this study indicated that the PrPSc of the five strains examined by us consisted of at least two types of PrPSc, viz., the mAb 3H6-precipitated PrPSc and the other PrPSc, even in the strains stabilized by sufficient adaptation. To date, some models of PrPSc conformation at the molecular level have been proposed, which suggest that the PrPSc of an individual strain could be represented as a mixture of several conformations, including intermediate forms [3, 4]. Our data present the direct evidence of heterogeneity of PrPSc in prion-affected mice. It is necessary to compare the conformational characteristics of mAb 3H6-precipitated PrPSc and the other PrPSc in future studies.
In conclusion, our study demonstrated the conformational heterogeneity of PrPSc in mouse-adapted scrapie prions by utilizing the unique specificity of mAb 3H6. A panel of mAbs capable of delineating specific PrPSc conformation could be a powerful tool for further investigation of conformational variation of PrPSc. If other biochemical approaches that focus on the heterogeneity of PrPSc conformation within a strain were to be combined with our mAb-based strategy, it could shed light on the mechanism by which PrPSc conformations may generate various strain phenotypes.
ACKNOWLEDGMENTS
We thank Dr. Y. Kaku, NIID, Japan, for providing comments and suggestions on this article. We also thank the laboratory staff of the NIAH, Japan, for their technical support.
REFERENCES
- 1.Baron T. G., Biacabe A. G.2001. Molecular analysis of the abnormal prion protein during coinfection of mice by bovine spongiform encephalopathy and a scrapie agent. J. Virol. 75: 107–114. doi: 10.1128/JVI.75.1.107-114.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartz J. C., Bessen R. A., Mckenzie D., Marsh R. F., Aiken J. M.2000. Adaptation and selection of prion protein strain conformations following interspecies transmission of transmissible mink encephalopathy. J. Virol. 74: 5542–5547. doi: 10.1128/JVI.74.12.5542-5547.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collinge J.2010. Prion strain mutation and selection. Science 328: 1111–1112. doi: 10.1126/science.1190815 [DOI] [PubMed] [Google Scholar]
- 4.Collinge J., Clarke A. R.2007. A general model of prion strains and their pathogenicity. Science 318: 930–936. doi: 10.1126/science.1138718 [DOI] [PubMed] [Google Scholar]
- 5.Collinge J., Sidle K. C., Meads J., Ironside J., Hill A. F.1996. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature 383: 685–690. doi: 10.1038/383685a0 [DOI] [PubMed] [Google Scholar]
- 6.Curin Serbec V., Bresjanac M., Popovic M., Pretnar Hartman K., Galvani V., Rupreht R., Cernilec M., Vranac T., Hafner I., Jerala R.2004. Monoclonal antibody against a peptide of human prion protein discriminates between Creutzfeldt-Jacob’s disease-affected and normal brain tissue. J. Biol. Chem. 279: 3694–3698. doi: 10.1074/jbc.M310868200 [DOI] [PubMed] [Google Scholar]
- 7.Groschup M., Gretzschel A., Kuczius T.2009. Prion Strains, 1st ed., Walter de Gruyter GmbH & Co., Berlin. [Google Scholar]
- 8.Hayashi H. K., Yokoyama T., Takata M., Iwamaru Y., Imamura M., Ushiki Y. K., Shinagawa M.2005. The N-terminal cleavage site of PrPSc from BSE differs from that of PrPSc from scrapie. Biochem. Biophys. Res. Commun. 328: 1024–1027. doi: 10.1016/j.bbrc.2005.01.065 [DOI] [PubMed] [Google Scholar]
- 9.Horiuchi M., Karino A., Furuoka H., Ishiguro N., Kimura K., Shinagawa M.2009. Generation of monoclonal antibody that distinguishes PrPSc from PrPC and neutralizes prion infectivity. Virology 394: 200–207. doi: 10.1016/j.virol.2009.08.025 [DOI] [PubMed] [Google Scholar]
- 10.Iwamaru Y., Takenouchi T., Ogihara K., Hoshino M., Takata M., Imamura M., Tagawa Y., Hayashi-Kato H., Ushiki-Kaku Y., Shimizu Y., Okada H., Shinagawa M., Kitani H., Yokoyama T.2007. Microglial cell line established from prion protein-overexpressing mice is susceptible to various murine prion strains. J. Virol. 81: 1524–1527. doi: 10.1128/JVI.01379-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwata N., Sato Y., Higuchi Y., Nohtomi K., Nagata N., Hasegawa H., Tobiume M., Nakamura Y., Hagiwara K., Furuoka H., Horiuchi M., Yamakawa Y., Sata T.2006. Distribution of PrPSc in cattle with bovine spongiform encephalopathy slaughtered at abattoirs in Japan. Jpn. J. Infect. Dis. 59: 100–107 [PubMed] [Google Scholar]
- 12.Korth C., Stierli B., Wuthrich K., Oesch B.1997. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature 390: 74–77. doi: 10.1038/36337 [DOI] [PubMed] [Google Scholar]
- 13.Masujin K., Matthews D., Wells G. A., Mohri S., Yokoyama T.2007. Prions in the peripheral nerves of bovine spongiform encephalopathy-affected cattle. J. Gen. Virol. 88: 1850–1858. doi: 10.1099/vir.0.82779-0 [DOI] [PubMed] [Google Scholar]
- 14.Masujin K., Kaku-Ushiki Y., Miwa R., Okada H., Shimizu Y., Kasai K., Matsuura Y., Yokoyama T.2013. The N-terminal sequence of prion protein consists an epitope specific to the abnormal isoform of prion protein (PrPSc). PLoS One 8: e58013. doi: 10.1371/journal.pone.0058013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan K. M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z., Fletterick R. J., Cohen F. E., Prusiner S. B.1993. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U.S.A. 90: 10962–10966. doi: 10.1073/pnas.90.23.10962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paramithiotis E., Pinard M., Lawton T., Laboissiere S., Leathers V. L., Zou W. Q., Estey L. A., Lamontagne J., Lehto M. T., Kondejewski L. H., Francoeur G. P., Papadopoulos M., Haghighat A., Spatz S. J., Head M., Will R., Ironside J., O’rourke K., Tonelli Q., Ledebur H. C., Chakrabartty A., Cashman N. R.2003. A prion protein epitope selective for the pathologically misfolded conformation. Nat. Med. 9: 893–899. doi: 10.1038/nm883 [DOI] [PubMed] [Google Scholar]
- 17.Prusiner S. B.1982. Novel proteinaceous infectious particles cause scrapie. Science 216: 136–144. doi: 10.1126/science.6801762 [DOI] [PubMed] [Google Scholar]
- 18.Prusiner S. B.1998. Prions. Proc. Natl. Acad. Sci. U.S.A. 95: 13363–13383. doi: 10.1073/pnas.95.23.13363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinagawa M., Takahashi K., Sasaki S., Doi S., Goto H., Sato G.1985. Characterization of scrapie agent isolated from sheep in Japan. Microbiol. Immunol. 29: 543–551 [DOI] [PubMed] [Google Scholar]
- 20.Telling G. C., Parchi P., Dearmond S. J., Cortelli P., Montagna P., Gabizon R., Mastrianni J., Lugaresi E., Gambetti P., Prusiner S. B.1996. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science 274: 2079–2082. doi: 10.1126/science.274.5295.2079 [DOI] [PubMed] [Google Scholar]
- 21.Ushiki-Kaku Y., Endo R., Iwamaru Y., Shimizu Y., Imamura M., Masujin K., Yamamoto T., Hattori S., Itohara S., Irie S., Yokoyama T.2010. Tracing conformational transition of abnormal prion proteins during interspecies transmission by using novel antibodies. J. Biol. Chem. 285: 11931–11936. doi: 10.1074/jbc.M109.058859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokoyama T., Kimura K., Tagawa Y., Yuasa N.1995. Preparation and characterization of antibodies against mouse prion protein (PrP) peptides. Clin. Diagn. Lab. Immunol. 2: 172–176 [DOI] [PMC free article] [PubMed] [Google Scholar]