Bacteria that cause late-onset neonatal sepsis often reside in the infant gut before they invade the bloodstream. They are rarely found in infants without sepsis. Screening, augmented hygiene, and specific pathogen decolonization might prevent bloodstream infections in premature infants.
Keywords: Escherichia coli, group B streptococci, Serratia marcescens, septicemia, whole-genome sequencing
Abstract
Background. Late-onset sepsis is a major problem in neonatology, but the habitat of the pathogens before bloodstream invasion occurs is not well established.
Methods. We examined prospectively collected stools from premature infants with sepsis to find pathogens that subsequently invaded their bloodstreams, and sought the same organisms in stools of infants without sepsis. Culture-based techniques were used to isolate stool bacteria that provisionally matched the bloodstream organisms, which were then genome sequenced to confirm or refute commonality.
Results. Of 11 children with late-onset neonatal bloodstream infections, 7 produced at least 1 stool that contained group B Streptococcus (GBS), Serratia marcescens, or Escherichia coli before their sepsis episode with provisionally matching organisms. Of 96 overlap comparison subjects without sepsis temporally associated with these cases, 4 were colonized with provisionally matching GBS or S. marcescens. Of 175 comparisons of stools from randomly selected infants without sepsis, 1 contained a GBS (this infant had also served as an overlap comparison subject and both specimens contained provisionally matching GBS). Genome sequencing confirmed common origin of provisionally matching fecal and blood isolates. The invasive E. coli were present in all presepticemic stools since birth, but gut colonization with GBS and S. marcescens occurred closer to time of bloodstream infection.
Conclusions. The neonatal gut harbors sepsis-causing pathogens, but such organisms are not inevitable members of the normal microbiota. Surveillance microbiology, decolonization, and augmented hygiene might prevent dissemination of invasive bacteria between and within premature infants.
Late-onset sepsis, defined as bloodstream infections occurring after 72 hours of age, remains a major threat to premature infants [1, 2]. Whereas early-onset sepsis is caused by pathogens acquired from the amnion or birth canal [3], the origin of bloodstream infections in premature infants that occur between 72 hours of age and discharge from neonatal intensive care units (NICUs) is less well established. Staphylococcal bloodstream infections probably originate from the skin [4], but the assumption that the gut harbors pathogens that cause episodes of late-onset sepsis that are unrelated to line infections has not been proven using whole-genome sequencing.
Here, we combined culture-based microbiology and whole-genome sequencing to determine how often stools collected before episodes of sepsis contain the organisms that later invade the bloodstream. We also sought to determine if such organisms are appreciably prevalent among premature infants who do not have sepsis.
METHODS
Study Design
This prospective observational case-cohort study was approved by the Washington University Human Research Protection Office. Subjects, who were enrolled after parents provided informed consent, were participants in a study of risk factors for necrotizing enterocolitis. The present study had 3 prespecified analyses: First, we sought to test the hypothesis that infants with late-onset sepsis harbored organisms in their stool that provisionally matched their bloodstream isolate. A pair of isolates was considered to be provisionally matching if the members of the pair share traits appropriate to their species, namely, serotypes (group B Streptococcus [GBS]), sequence types (STs) (GBS and Escherichia coli), and random amplified polymorphic DNA (RAPD) patterns (E. coli). Second, we genome sequenced all blood and a subset of provisionally matching stool isolates from cases (see definition below), and measured pairwise degrees of strain identity by calculating operational variation indices (OVIs) (Supplementary Appendix). The OVI accounts for intrinsic (false) assembly variation, and identifies operationally isogenic bacteria. Third, we determined the frequency with which infants without sepsis in this study cohort harbored in their guts bacteria that provisionally matched those that caused bloodstream infections in subjects with sepsis.
Inclusion criteria were (1) birth during a 632-day interval starting in 2009, (2) birth weight ≤1500 g, and (3) admission to the St Louis Children's Hospital NICU. Infants not expected to survive their first week of life were excluded. All infant stools were collected, stored briefly (4°C), and frozen (−80°C) until analyzed. Stool samples from subjects' mothers were also collected, if available, while the mothers were still in hospital. Cases were defined as infants whose first episode of late-onset sepsis was diagnosed by blood culture drawn after 72 hours of life, if that culture yielded bacteria generally considered to be pathogenic [5]. Subjects were excluded from analysis if their bloodstream isolates were of uncertain virulence (eg, coagulase-negative staphylococci were excluded) or posed logistic difficulties in differentiating them from endogenous fecal strains (eg, enterococci), or if the subjects did not produce sufficient stools before the day of sepsis (Figure 1). The day of sepsis was defined as the collection date of the first blood culture that yielded the qualifying pathogen. All stools that had been collected from all cases before such episodes of qualifying sepsis were subjected to species-appropriate techniques to isolate the organism that matched the bloodstream pathogen (Supplementary Table 1). Cases who produced stools before the onset of sepsis that contained an organism that provisionally matched the bloodstream isolate were termed “precolonized.”
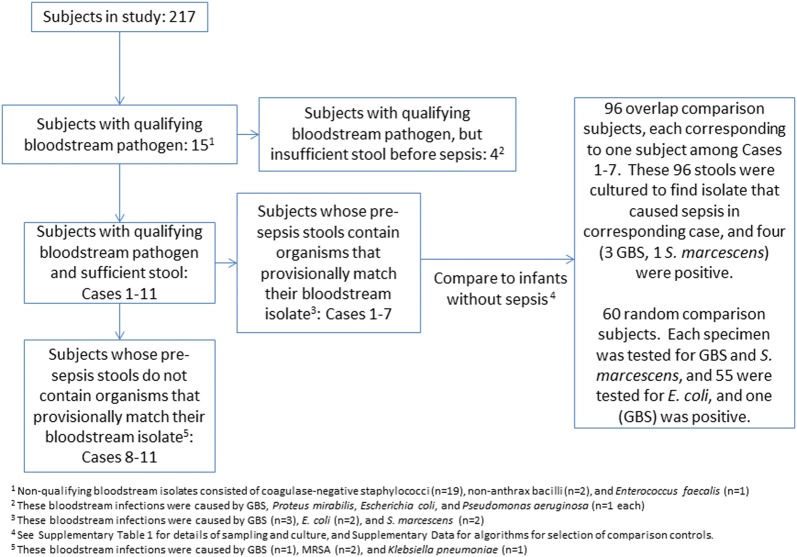
Figure 1.
Study flow chart. Abbreviations: GBS, group B Streptococcus; MRSA, methicillin-resistant Staphylococcus aureus.
Next, we assembled 2 comparison groups of infants without sepsis (Supplementary Data). The first, an overlap comparison group, consisted of study subjects who resided in the NICU at the same time that an episode of sepsis occurred in a precolonized case. This overlap group enabled us to determine the frequency of asymptomatic colonization with a sepsis-causing strain among infants in proximity to such cases. The second, a random comparison group, consisted of a sample of all study subjects throughout the entire NICU without sepsis, to determine the overall rate of asymptomatic colonization with the pathogens of interest. For cases and comparison groups, the days of events of interest were the day of sepsis or the day the analyzed stool was produced, respectively.
Laboratory workers who sought GBS or E. coli in stools were blinded to the septicemic or nonsepticemic status of the subjects from whom specimens originated. Genome Institute staff members were blinded to the subject status of all stool isolates.
Statistical Methods
We used the Fisher exact or Wilcoxon tests for proportions and medians, respectively. Two-tailed P values <.05 were considered significant. Data were collected and managed using REDCap tools at Washington University [6].
RESULTS
Characteristics of Cases Whose Stools Were Colonized Before Sepsis With Bloodstream Isolate
Of the 217 subjects studied, 15 met the case definition of sepsis with a qualifying pathogen, and 11 had sufficient presepsis stools for study (Figure 1). Blood cultures were drawn because of apnea, respiratory failure requiring initiation of, or increased, ventilator assistance, bradycardia, fever, tachypnea, leukocytosis, leukopenia, and concerns about necrotizing enterocolitis. Cases 1–7 produced at least 1 stool before their episode of sepsis that contained an organism (GBS, S. marcescens, or E. coli) that provisionally matched their bloodstream isolate, whereas the presepsis stools of cases 8–11 did not contain organisms that provisionally matched those recovered from their blood (discussed below) (Tables 1 and 2). Bacteria provisionally matching the corresponding bloodstream isolate were recovered from 24 of the 89 (27%) stools produced by cases 1–7 before sepsis occurred, and from 20 of the 34 (54%) stools collected from these 7 infants during the 10 days immediately before the episode of sepsis (Tables 1 and 2, Figure 2).
Table 1.
Characteristics of Cases 1–7 and 2 Comparison Groups
| Characteristics | Precolonized Cases | Overlap Comparison | Random Comparison |
|---|---|---|---|
| Subjects, No. | 7 | 96 | 60 |
| Subjects with positive stools, No. | 7 | 4 | 1 |
| Stools with pathogen/stools tested, No. | 24/89a,20/37b | 4/96 | 1/175 (P < .0001)c |
| Birth weight, g | 800 (735–8159) | 995 (829–1120) (P = .03) | 960 (805–1137) (P = .04) |
| Gestational age at birth, wk | 26.0 (24.1–26.4) | 27.4 (25.2–29) (P = .04) | 27.2 (25.3–28.7) |
| Female sex, No. | 4 | 44 | 33 |
| Race/ethnicity, No. | |||
| African American | 6 | 56 | 33 |
| White | 1 | 38 | 25 |
| Asian | 0 | 2 | 2 |
| Hispanic | 0 | 2 | 1 |
| Singleton births, No. | 6 | 84 | 51 |
| Cesarean delivery, No. | 3 | 64 | 44 |
| Apgar score, 1 min/5 min | 5 (1–6.5)/4 (2–5.5) | 5 (2–6)/7 (5–8) (P = .03)d | 4 (1.5–6)/7 (6–8) (P = .03)d |
| Day of life specimen of interest obtainede | 34 (18–52) | 35 (21–49) | 33 (22–45) |
| Percentage of days of life antibiotics administeredf | 53 (41–86) | 31 (16–48) (P = .02) | 25 (13–41) (P = .003) |
| Most recent antibiotic administered, dg | 14 (6–23) | 18 (5–32) | 15 (6–25) |
| First antibiotic-free day of life | 11 (9–23) | 5 (4–9) (P = .03) | 5 (4–9) |
| Intubated in first 2 days of life, No. | 7 | 91 | 55 |
| No. of days intubatedh | 21 (8–25) | 1 (1–8) | 1 (1–11) |
| Developed NEC, No. | 2 | 7 | 3 |
| Discharged alive, No. | 6 | 93 | 60 |
| Day of life discharged homei | 123 (105–150) | 83 (66–104) (P = .006)i | 85 (65–114) (P = .02)i |
For all categorical variables, values represent number of subjects for whom the variable applies, with the denominator being the number of subjects in the respective columns. All other values are median (interquartile range). P values relate comparison group subjects to cases 1–7, and are provided only if < 0.05.
Abbreviation: NEC, necrotizing enterocolitis.
a All stools produced by cases 1–7 prior to sepsis, including on day of sepsis if stool was produced before blood culture was drawn.
b All stools produced by cases 1–7 in 10 days prior to sepsis, including on day of sepsis if stool was produced before blood culture was drawn.
c The positive specimen rate for the 175 tests performed on the 60 random comparison specimens for GBS, Serratia marcescens, and Escherichia coli were compared with rates of positivity for these pathogens for all presepsis stools, and those collected in the 10 days before sepsis, among the cases. All comparisons were highly significant (P < .001). The overlap comparison group was not subjected to such analysis, because the groups were not microbiologically independent, and the overlap comparison group was used only to identify the extent of asymptomatic colonization in close proximity to cases.
d Statistically significant values were obtained only for the 5-minute Apgar scores.
e Specimen of interest was blood culture that yielded a qualifying pathogen (cases 1–7), or stool (comparison groups).
f Percentage of all days of life prior to collection of specimen of interest on which antibiotics were administered.
g No. of consecutive antibiotic-free days immediately preceding collection of specimen of interest.
h No. of days intubated prior to collection of specimen of interest.
i Among the subjects discharged home alive.
Table 2.
Summary of Cases and of Positive Specimens
| Subjecta | Isolateb | Stools Testedc |
|---|---|---|
| Case 1, DOL 61 (98) | GBS V ST1 | 15, 17, 19, 20, 21, 24, 26, 27, 28, 31, 33, 34, 39, 44, 47, 50, 53, 54, 55, 58, 60 |
| Case 2, DOL 14 (353) | Serratia marcescens | 7, 8, 10, 13 |
| Case 3, DOL 21 (414) | Escherichia coli, ST69 | 8, 11, 12, 13, 21 |
| Case 4, DOL 42 (449) | GBS 1A ST23 | 19, 20, 23, 29, 33, 35, 36, 37, 39, 40 |
| Case 5, DOL 34 (491) | E. coli ST70 | 19, 20, 21, 23, 24, 25, 26, 27, 28, 29, 30, 32, 33 |
| Case 6, DOL 82 (583) | GBS II ST22 | 10, 19, 23, 25, 28, 29, 30, 32, 33, 34, 36, 38, 39, 40, 42, 44, 45, 47, 50, 51, 54, 57, 59, 60, 62, 63, 68, 71, 74, 75, 77, 79, 80 |
| Case 7, DOL 15 (609) | S. marcescens | 7, 11, 14 |
| Case 8, DOL 22 (207) | Klebsiella pneumoniae | 12, 13, 14 |
| Case 9, DOL 27 (217) | MRSA | 17, 18, 19, 20, 21, 23, 25, 26, 27 |
| Case 10, DOL 28 (225) | MRSA | 10, 22, 27 |
| Case 11, DOL 51 (245) | GBS III ST19 | 5, 9, 10, 14, 17, 19, 23, 26, 31d, 32d, 37, 38d, 40d, 42, 44, 46d, 47d, 48, 50d, 51, 53d, 68, 72d |
| Subjects without sepsis (overlap and random comparison) whose stools contained pathogens that provisionally matched bloodstream isolates from cases | ||
| Overlap 1, DOL 63 (98) | GBS V ST1 | 63 |
| Overlap 2, DOL 48 (583) | GBS II ST22 | 48 |
| Overlap 3e, DOL 13 (583) | GBS II ST22 | 13 |
| Random 1e, DOL 22 (592) | GBS II ST22 | 22 |
| Overlap 4, DOL 33 (606) | S. marcescens | 33 |
| Mother of case 3f | E. coli, ST69 | 35 |
Cases 1–7 produced 1 or more stools before sepsis that contained an organism that provisionally matched the subsequent bloodstream isolate. The presepsis stools of cases 8–11 did not contain pathogens that provisionally matched the bloodstream isolates.
Abbreviations: DOL, day of life that specimen was obtained; GBS, group B Streptococcus; MRSA, methicillin-resistant Staphylococcus aureus; ST, sequence type.
a Subjects are categorized as cases or as overlap or random comparison, as defined in text. The specimen of interest was the blood culture that yielded a qualifying pathogen (cases 1–11), or stool that produced a corresponding pathogen (comparison subjects). Number in parentheses denotes day of study specimen was obtained.
b Organism from blood (cases) or stool (comparison subjects), with serotypes (Roman numerals) and/or ST (GBS and E. coli). Underlined organisms were genome sequenced.
c DOL stools were produced that were tested for bloodstream pathogen (all cases), or were tested and positive (comparison subjects). Bolded numbers indicate that stool contained an organism that provisionally matched the corresponding bloodstream isolate. Underlined numbers indicate that organism underwent whole-genome sequencing. Specimens obtained on same DOL as sepsis are listed if stool was produced before blood culture was obtained.
d These stools contained GBS II ST88.
e Overlap comparison subject 3 and random comparison subject 1 are the same individual, sampled 9 days apart.
f Stool from mother of case 3, obtained 2 days after delivery (corresponds to DOL 2 in infant, and day of study 395), contained an E. coli that provisionally matched infant's bloodstream isolate.
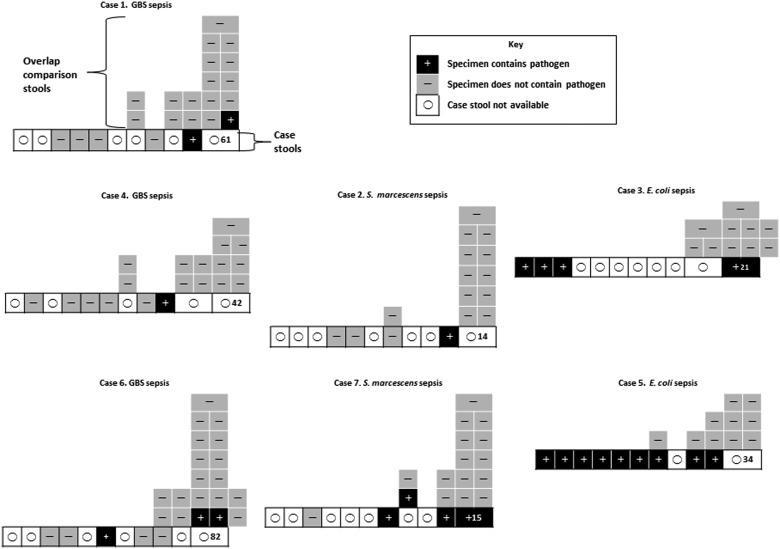
Figure 2.
Precolonized case and overlap comparison group stool culture results. Case 1 annotations apply to all cases. Boxes in bottom rows represent 10 days before sepsis (including day of sepsis if stool was produced before blood culture was obtained). Box with number is day of sepsis; number denotes day of life the episode of sepsis occurred. Boxes above bottom row represent all overlap comparison group specimens relative to day of sepsis in corresponding case, including those collected on the day after sepsis (cases 3 and 6). Inset describes key to positive, negative, and nontested specimens. Abbreviation: GBS, group B Streptococcus.
Escherichia coli with RAPD profiles indistinguishable from the corresponding bloodstream E. coli from cases 3 and 5 were present in all stools produced by these 2 cases before sepsis (Table 2). We interpret these data to mean that the bloodstream pathogens occupied the guts of these infants at the time of their first bowel movement, well in advance of sepsis. In contrast, gut colonization occurred closer to the days on which sepsis occurred in the 5 other cases whose bloodstream infections were caused by GBS and S. marcescens.
Characteristics of Nonsepticemic Infants
Members of both comparison groups weighed more at birth, had greater 5-minute Apgar scores, received antibiotics on fewer days before the specimen of interest was obtained, and were discharged home at younger ages than the precolonized cases. The overlap comparison group also had a greater gestational age at birth and achieved the first antibiotic-free day of life at an earlier age than the precolonized cases.
Of the 96 overlap comparison group subjects, 3 (3%) and 1 (1%) produced stools that contained GBS and S. marcescens, respectively, that provisionally matched bloodstream isolates from their corresponding cases (Tables 1 and 2, Figures 1 and 2). Moreover, each of these 4 subjects resided in bed spaces in close proximity to their corresponding cases in the same multipatient rooms. Only 1 random comparison group stool contained a pathogen of interest. This infant 9 days earlier had served as an overlap comparison subject for case 6, and both of the tested stools contained GBS that provisionally matched the case 6 bloodstream isolate.
Maternal Stools
Maternal stools were available only from the mothers of precolonized cases 3, 6, and 11 for testing in our laboratory, and the only such specimen to yield an organism that provisionally matched a bloodstream isolate was the stool of the mother of case 3. The mothers of cases 1 and 4 were tested for peripartum GBS colonization per their obstetric records, and were negative.
Characteristics of Infants With Bloodstream Infections Whose Stools Did Not Yield the Bloodstream Isolate
Cases 8–11 had bloodstream infections caused by methicillin-resistant Staphylococcus aureus (MRSA) (n = 2), Klebsiella pneumoniae (n = 1), or serotype III ST19 GBS (GBS III ST19) (n = 1), but their presepsis stools did not contain these organisms. However, multiple stools from case 11 contained GBS II ST88 before and after the day of sepsis caused by GBS III ST19, and 1 stool obtained 17 days after the day of sepsis contained GBS III ST19 (Table 2).
Whole-Genome Sequencing
Genome sequencing details are provided in Supplementary Table 2. The following pairings of sequenced isolates demonstrated operational isogenicity: within-host bloodstream and provisionally matching fecal GBS, E. coli, and S. marcescens from cases 1–7 (median OVI, 0.26; interquartile range [IQR], 0.18–0.34); bloodstream isolates from cases 1, 3, 6, and 7 and provisionally matching fecal isolates from their 4 overlap comparison subjects, the random comparison subject who also was an overlap comparison subject for case 6, and the mother of case 3 (median OVI, 0.63; IQR, 0.24–1.2; P = .42 vs median intrahost OVIs). In contrast, individual bloodstream GBS and E. coli were quite distinct from each other, consistent with their different serotypes and STs (median OVI, 589 for all pairwise comparisons; IQR, 512–935; P < .0001 vs within-host pairings for these particular species). However, case 2 and 7 bloodstream S. marcescens, recovered 256 days apart, were operationally isogenic (OVI, 0.24).
The bloodstream GBS III ST19 from case 11 was much less similar to the fecal GBS II ST88 strains from that case (median OVI, 413; IQR, 411–414) than to the fecal GBS III ST19 isolated from the stool on the 17th day after sepsis (OVI, 0.48).
DISCUSSION
In this prospective cohort study, we documented enteric colonization with pathogens that subsequently invaded the bloodstream of premature infants. Our application of whole-genome sequencing to within-host tracking of pathogens that are largely susceptible to antibiotics and that caused infections in a nonoutbreak setting extends this powerful technology beyond antibiotic-resistant hospital infections [7–10]. The high degree of operational isogenicity between corresponding isolates provides an unprecedented level of verification of identity between enteric and bloodstream bacteria, as has been suggested by less precise typing methods, usually antimicrobial resistance [11–15]. Furthermore, our demonstration of intestinal colonization with operationally isogenic pathogens in infants in temporal and spatial proximity to precolonized cases of late-onset sepsis strongly suggests interpatient transmission within NICUs. Our data probably underestimate the number and magnitude of such time-space colonization microclusters and interinfant flow of such pathogens. Specifically, our cohort was limited to infants with birth weights ≤1500 g, so stools from additional children at risk for colonization were not analyzed. Härtel et al recently described microclusters of late-onset neonatal sepsis in nurseries [16]. We expand this concept to include microclusters of asymptomatic colonization associated with single cases of bloodstream infections.
Current efforts to prevent late-onset sepsis focus on catheter care and hand hygiene, but our data draw attention to pathogens harbored in the gut. Microbiologic surveillance of stool might detect infants whose stools contain organisms with potential to invade the bloodstream, which then might prompt augmented hygienic measures to limit the size of colonization microclusters. Sepsis does not generally obligate contact precautions, but it is concerning that case 11 shed potentially invasive GBS of 2 serotypes for at least 21 days after parenteral antibiotics were started. Also, microbiologic assessment of stools could be used to promote more rational use of antibiotics in premature infants [17]. For example, knowledge of the identity and antibiotic susceptibilities of bacteria that colonize the gut has been used to inform the choice of empiric antibiotics in adult intensive care units [15].
By identifying enteric colonization with high-risk organisms, strategies might be developed to prevent bloodstream infections. One tactic worth considering would be specific pathogen decolonization guided by microbiologic diagnosis to reduce the risk of sepsis among the colonized. At the very least, identification of a colonized infant could heighten vigilance for the earliest signs of sepsis in that child. Indeed, prepartum screening [18] and intrapartum antibiotics [19–21] have dramatically reduced the incidence of early-onset GBS disease, but the incidence of late-onset GBS sepsis, which, like early-onset GBS disease has major sequelae [5, 22], remains constant [19, 21, 23]. We recognize that identifying the subsets of fecal E. coli that are potentially invasive presents challenges because of the high rate of colonization with this diverse species (most of the members of which are commensals) and the multiple different pathotypes of E. coli that cause invasive neonatal disease [24]. However, the presence of extended-spectrum β-lactamase–producing gram-negative bacilli in stools of hospitalized adults is strongly associated with subsequent sepsis [13, 14], and E. coli K1 has enhanced ability to cause neonatal sepsis [25]. Hence, it is conceivable that E. coli with traits associated with invasion might be detectable in stool before dissemination ensues. In any event, surveillance strategies for asymptomatically colonized infants, and responses to positive cultures, will need careful development and likely be organism-specific. For example, our data raise the possibility that the presence in feces of GBS or S. marcescens might by itself be sufficient to predict a high likelihood of subsequent bloodstream infection.
Our findings expand our understanding of the biology of late-onset sepsis. The presence of GBS in stools in advance of sepsis in infants who were not in respiratory distress suggests that in late-onset infections, GBS invades hosts from the gut. The E. coli that caused late-onset sepsis in cases 3 and 5 were probably acquired at or soon after birth, because these infants shed this pathogen in each of their stools. The colonization of the mother of case 3 suggests vertical transmission, as this infant was delivered vaginally. In contrast, gut colonization with GBS and S. marcescens seemed in this small cohort to be acquired later in life, from unknown sources.
Case 11, whose bloodstream GBS III ST19 was found in stool only after sepsis, whereas multiple presepsis stools contained GBS of a different serotype and ST, warrants comment. We might have overlooked scarce GBS III ST19 among abundant GBS II ST88 in the presepsis stools; this sepsis episode could have been caused by GBS belonging to 2 different lineages with similar colony morphology and we stored only 1; or GBS III ST19 might have seeded the stool during the episode of sepsis. In any event, this case demonstrates that premature infants can harbor multiple GBS strains for prolonged periods, and also suggests that parenteral antibiotics fail to rapidly clear GBS from the gastrointestinal tact.
Modern sequencing technology, although increasingly economical, yields multiple apparent genomic variants in interstrain comparisons, probably related to data alignment issues. Because the effort required to validate or refute the presence of these variants would be considerable, we developed the easily calculable OVI, which addresses the intrinsic false variant rate by taking into account the number of intraisolate variants generated when a sequence is aligned to itself. The validity of this metric is suggested in case 11: The OVI for the serotype III ST19 GBS from blood and postsepsis stool was nearly 1000-fold less than the OVIs for the genomic alignments of the isolate and the serotype II ST88 GBS fecal strains. The extent to which isogenicity can be inferred from genome data depends on context, including the genomic plasticity of the particular organisms being compared, and the sequencing and alignment tools used. However, a low OVI provides, at least for the purpose of a study such as ours, a high degree of certainty that provisionally matching isolates are operationally isogenic.
This study has several limitations. First, bacteria might lose viability during freezing and thawing of stools prior to culture, leading to falsely negative specimens, and we do not know the precise sensitivity of detection of the target organisms using our culture protocol. However, any lack of sensitivity should apply to cases with sepsis and to subjects without sepsis, so this should not bias comparisons between these groups. We are planning sequence-based methods to measure pathogen density in our samples, which might also shed light on the intestinal microbiome as a risk factor for sepsis [26, 27]. Second, pathogens such as S. aureus and K. pneumoniae might have extraintestinal venues before they invade the bloodstream that were not cultured in this study. Third, there are challenges inherent to the selection of appropriate comparison specimens and subject groups, which should be borne in mind in future efforts to calculate the relative risk of bloodstream infection among colonized neonates. Specifically, groups of compared infants will need to be of sufficient sizes to address the many different host factors that might predispose to sepsis, and the possibility that colonization and invasion risk might vary between species. Also, for organisms that colonize the gut only briefly before they invade the bloodstream, the most appropriate comparison might be between specimens obtained immediately before sepsis and those obtained earlier in life, ie, an acute infection model. Nonetheless, the rarity of sepsis-causing bacteria in the comparison specimens suggests that the risk of dissemination after the gut is colonized is substantial. Fourth, we did not seek coagulase-negative staphylococci in the stool. In premature infants, coagulase-negative staphylococci can be bona fide bloodstream pathogens, but their presence in blood cultures could also reflect skin contamination of the media [28]. Future research to address colonization with actionable coagulase-negative staphylococci would be worthwhile [29], but the study design will need to take into account the very high rate of cutaneous colonization with this species, which could complicate analysis. A final limitation of our study is that we had few maternal stools with which to determine the extent of possible vertical transmission.
In summary, late-onset sepsis is preceded by adventitious colonization of the gastrointestinal tracts of premature infants by specific organisms. These infections are quite consequential: Late-onset sepsis caused by the types of bacteria we recovered from cases 1–7 has mortality rates in excess of 20% [5, 30], and the incidence of late-onset sepsis might be rising [31]. Hence, there is a compelling need for new strategies to prevent the dissemination of bloodstream pathogens between and within premature infants, and our findings suggest that efforts should focus on gut microbes. Expansion of our study to larger cohorts will determine the generalizability of our findings and facilitate construction of predictive models. Novel strategies to consider to control late-onset sepsis include surveillance and reactive (to positive specimens) and selective (pathogen-specific) decontamination. However, the evaluation of such interventions will require careful planning, validation, and implementation.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://cid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Patricia Sellenriek and Pallavi Singh, PhD, for technical advice and assistance; Chad Tomlinson, Wes Warren, and Michael Kiwala for sequence data management and submission; Julie Hoffmann and Laura Linneman for enrollment and specimen collection; Drs Steve Moseley and Robert Rothbaum for helpful editorial comments; and Ms Ariana Jasarevic for administrative support and assistance in manuscript preparation.
Financial support. This work was supported by the National Institutes of Health (grant numbers UH3AI083265, U54 HG004968, AFRI-AI10903, P30DK052574 [Biobank Core]); the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (grant 1 I01 CX000192 01); the Infectious Diseases Society of America Medical Scholars Program (M. A. C.); the Global Alliance to Prevent Prematurity and Stillbirth, an initiative of Seattle Children's Hospital and the Bill & Melinda Gates Foundation (S. D. M.); and the Melvin E. Carnahan Professorship (P. I. T.).
Potential conflicts of interest. P. I. T. has received an honorarium for a lecture at the headquarters of Cepheid, Inc. C.-A. D. B. has received research funding from bioMérieux and Cepheid. G. M. W. is on the scientific advisory boards of RTG and OpGen. J. R. J. has received research grants or contracts from Merck and Syntiron, and has patent applications for tests to detect specific strains of E. coli. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Polin RA, Denson S, Brady MT; Committee on Fetus and Newborn, Committee on Infectious Diseases. Strategies for prevention of health care-associated infections in the NICU. Pediatrics. 2012;129:e1085–93. doi: 10.1542/peds.2012-0145. [DOI] [PubMed] [Google Scholar]
- 2.Boghossian NS, Page GP, Bell EF, et al. Late-onset sepsis in very low birth weight infants from singleton and multiple gestation births. J Pediatr. 2013;162:1120–24.e1. doi: 10.1016/j.jpeds.2012.11.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polin RA; Committee on Fetus and Newborn. Management of neonates with suspected or proven early-onset bacterial sepsis. Pediatrics. 2012;129:1006–15. doi: 10.1542/peds.2012-0541. [DOI] [PubMed] [Google Scholar]
- 4.Darmstadt GL, Saha SK, Choi Y, et al. Population-based incidence and etiology of community-acquired neonatal bacteremia in Mirzapur, Bangladesh: an observational study. J Infect Dis. 2009;200:906–15. doi: 10.1086/605473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoll BJ, Hansen N, Fanaroff AA, et al. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics. 2002;110:285–91. doi: 10.1542/peds.110.2.285. [DOI] [PubMed] [Google Scholar]
- 6.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–81. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koser CU, Holden MT, Ellington MJ, et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med. 2012;366:2267–75. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snitkin ES, Zelazny AM, Thomas PJ, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3004129. 148ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris SR, Cartwright EJ, Torok ME, et al. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis. 2013;13:130–6. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherry NL, Porter JL, Seemann T, Watkins A, Stinear TP, Howden BP. Outbreak investigation using high-throughput genome sequencing within a diagnostic microbiology laboratory. J Clin Microbiol. 2013;51:1396–401. doi: 10.1128/JCM.03332-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith A, Saiman L, Zhou J, Della-Latta P, Jia H, Graham PL., 3rd Concordance of gastrointestinal tract colonization and subsequent bloodstream infections with gram-negative bacilli in very low birth weight infants in the neonatal intensive care unit. Pediatr Infect Dis J. 2010;29:831–5. doi: 10.1097/INF.0b013e3181e7884f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham PL, 3rd, Della-Latta P, Wu F, Zhou J, Saiman L. The gastrointestinal tract serves as the reservoir for gram-negative pathogens in very low birth weight infants. Pediatr Infect Dis J. 2007;26:1153–6. doi: 10.1097/INF.0b013e31814619d4. [DOI] [PubMed] [Google Scholar]
- 13.Harris AD, McGregor JC, Johnson JA, et al. Risk factors for colonization with extended-spectrum beta-lactamase-producing bacteria and intensive care unit admission. Emerg Infect Dis. 2007;13:1144–9. doi: 10.3201/eid1308.070071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy P, Malczynski M, Obias A, et al. Screening for extended-spectrum beta-lactamase-producing Enterobacteriaceae among high-risk patients and rates of subsequent bacteremia. Clin Infect Dis. 2007;45:846–52. doi: 10.1086/521260. [DOI] [PubMed] [Google Scholar]
- 15.Blot S, Depuydt P, Vogelaers D, et al. Colonization status and appropriate antibiotic therapy for nosocomial bacteremia caused by antibiotic-resistant gram-negative bacteria in an intensive care unit. Infect Control Hosp Epidemiol. 2005;26:575–9. doi: 10.1086/502575. [DOI] [PubMed] [Google Scholar]
- 16.Härtel C, Faust K, Avenarius S, et al. Epidemic microclusters of blood-culture proven sepsis in very-low-birth weight infants: experience of the German Neonatal Network. PLoS One. 2012;7:e38304. doi: 10.1371/journal.pone.0038304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandora TJ, Goldmann DA. Preventing lethal hospital outbreaks of antibiotic-resistant bacteria. N Engl J Med. 2012;367:2168–70. doi: 10.1056/NEJMp1212370. [DOI] [PubMed] [Google Scholar]
- 18.Verani JR, McGee L, Schrag SJ; Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention. Prevention of perinatal group B streptococcal disease—revised guidelines from CDC, 2010. MMWR Recomm Rep. 2010;59:1–36. [PubMed] [Google Scholar]
- 19.Phares CR, Lynfield R, Farley MM, et al. Epidemiology of invasive group B streptococcal disease in the United States, 1999–2005. JAMA. 2008;299:2056–65. doi: 10.1001/jama.299.17.2056. [DOI] [PubMed] [Google Scholar]
- 20.Schrag SJ, Zywicki S, Farley MM, et al. Group B streptococcal disease in the era of intrapartum antibiotic prophylaxis. N Engl J Med. 2000;342:15–20. doi: 10.1056/NEJM200001063420103. [DOI] [PubMed] [Google Scholar]
- 21.Bauserman MS, Laughon MM, Hornik CP, et al. Group B Streptococcus and Escherichia coli infections in the intensive care nursery in the era of intrapartum antibiotic prophylaxis. Pediatr Infect Dis J. 2013;32:208–12. doi: 10.1097/INF.0b013e318275058a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Libster R, Edwards KM, Levent F, et al. Long-term outcomes of group B streptococcal meningitis. Pediatrics. 2012;130:e8–15. doi: 10.1542/peds.2011-3453. [DOI] [PubMed] [Google Scholar]
- 23.Jordan HT, Farley MM, Craig A, et al. Revisiting the need for vaccine prevention of late-onset neonatal group B streptococcal disease: a multistate, population-based analysis. Pediatr Infect Dis J. 2008;27:1057–64. doi: 10.1097/INF.0b013e318180b3b9. [DOI] [PubMed] [Google Scholar]
- 24.Mahjoub-Messai F, Bidet P, Caro V, et al. Escherichia coli isolates causing bacteremia via gut translocation and urinary tract infection in young infants exhibit different virulence genotypes. J Infect Dis. 2011;203:1844–9. doi: 10.1093/infdis/jir189. [DOI] [PubMed] [Google Scholar]
- 25.Robbins JB, McCracken GH, Jr, Gotschlich EC, Orskov F, Orskov I, Hanson LA. Escherichia coli K1 capsular polysaccharide associated with neonatal meningitis. N Engl J Med. 1974;290:1216–20. doi: 10.1056/NEJM197405302902202. [DOI] [PubMed] [Google Scholar]
- 26.Madan JC, Salari RC, Saxena D, et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonatal Ed. 2012;97:F456–62. doi: 10.1136/fetalneonatal-2011-301373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mai V, Torrazza RM, Ukhanova M, et al. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS One. 2013;8:e52876. doi: 10.1371/journal.pone.0052876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Healy CM, Palazzi DL, Edwards MS, Campbell JR, Baker CJ. Features of invasive staphylococcal disease in neonates. Pediatrics. 2004;114:953–61. doi: 10.1542/peds.2004-0043. [DOI] [PubMed] [Google Scholar]
- 29.Soeorg H, Huik K, Parm U, et al. Genetic relatedness of coagulase-negative Staphylococci from gastrointestinal tract and blood of preterm neonates with late-onset sepsis. Pediatr Infect Dis J. 2013;32:389–93. doi: 10.1097/INF.0b013e3182791abd. [DOI] [PubMed] [Google Scholar]
- 30.Didier C, Streicher MP, Chognot D, et al. Late-onset neonatal infections: incidences and pathogens in the era of antenatal antibiotics. Eur J Pediatr. 2012;171:681–7. doi: 10.1007/s00431-011-1639-7. [DOI] [PubMed] [Google Scholar]
- 31.Bizarro MJ, Dembry L-M, Baltimore RS, Gallagher PG. Changing patterns in neonatal Escherichia coli sepsis and ampicillin resistance in the era of intrapartum antibiotic prophylaxis. Pediatrics. 2008;121:689–96. doi: 10.1542/peds.2007-2171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.