Abstract
Combined treatment with interferon alpha (IFN-α) and ribavirin (RBV) can effectively cure HCV infection in a significant proportion of patients, but effects of this regimen on cellular reservoirs for human immunodeficiency virus type 1 (HIV-1) are unknown. Here, we show that treatment with IFN-α/RBV led to a moderate but significant and sustained decline of HIV-1 DNA in CD4 T cells from HIV-1/hepatitis C virus–coinfected patients receiving highly active antiretroviral therapy (n = 12). However, in vitro experiments failed to demonstrate an effect of pharmacological doses of IFN-α on HIV-1 reactivation. Together, these data suggest that treatment with IFN-α/RBV can moderately reduce the reservoir of HIV-1–infected CD4 T cells that persists despite suppressive antiretroviral therapy.
Keywords: HIV-1 reservoir, interferon-α, ribavirin, HIV-1/HCV coinfection
Interferon alpha (IFN-α) is a type I interferon with potent inhibitory activities against a range of viral pathogens. When used in combination with ribavirin (RBV), pharmacological dosages of IFN-α can cure hepatitis C virus (HCV) infection in a significant proportion of patients, although treatment is in many cases complicated by toxicities [1]. IFN-α also exerts active antiviral activities against human immunodeficiency virus type 1 (HIV-1) in vitro [2], and pharmacological administration of IFN-α can reduce HIV-1 RNA loads in otherwise untreated patients by approximately 5- to 10-fold [3]. Reduction of HIV-1 replication is also observed after administration of IFN-α receptor agonists in animal models of simian immunodeficiency virus infection [4]. Such antiviral activities against actively replicating HIV-1 are likely related to the IFN-mediated upregulation of a group of host genes that can restrict individual viral replication steps in viral target cells [5].
Effects of IFN-α against HIV-1–infected cells in patients receiving suppressive antiretroviral combination therapy remain less well defined. In such patients, the majority of HIV-1–positive cells are latently infected, and do not actively transcribe HIV-1 genes, although they can harbor replication-competent virus in a transcriptionally silent form that contributes to viral rebound after treatment discontinuation [6]. Treatment modalities that can specifically target such latently infected CD4 T cells are of critical interest for inducing a drug-free remission of HIV-1 infection in a larger number of HIV-1 patients. In a recent clinical trial, a reduction of integrated HIV-1 DNA in CD4 T cells was noted in a group of patients after receiving treatment with IFN-α [7], raising the possibility that IFN-α may, at least in specific patients, reduce the quantity of latently infected CD4 T cells. Here, we analyzed changes of the HIV-1 CD4 T-cell reservoir in a cohort of HIV/HCV-coinfected patients receiving highly active antiretroviral therapy (HAART) and undergoing HCV treatment with IFN-α and RBV.
METHODS
Patients
Peripheral blood mononuclear cell (PBMC) samples were obtained from HIV/HCV-coinfected patients who received suppressive HAART and HCV treatment with IFN-α (180 μg/week of pegylated IFN-α 2a [Roche]) and RBV (600 mg twice daily for ≥75 kg body weight, 500 mg twice daily for ≥75 kg body weight) at the Virgen del Rocio Hospital, Sevilla, Spain. All patients had undetectable HIV-1 RNA in commercial polymerase chain reaction (PCR) assays for the entire time during which samples were collected for this analysis. Clinical and demographical characteristics of the HIV/HCV-coinfected study patients are summarized in Table 1. For viral reactivation assays, PBMCs were collected from HAART-treated HIV-1–infected persons at Massachusetts General Hospital; these individuals had suppressed viremia (<50 copies/mL) for a minimum of 1 year. All patients gave written consent to donate samples for research purposes. The study was approved by the local institutional review boards in Sevilla and Boston, and conducted in agreement with the Declaration of Helsinki.
Table 1.
Clinical and Demographic Characteristics of HIV/Hepatitis C Virus–Coinfected Study Patients
| Patient | Age, y | Sex | Ethnicity | HCV Treatment Duration, wk | HCV Treatment Response | HCV Load |
IL-28B | HCV Genotype | CD4 Count, per µL |
HAART Regimen | Interval Duration, wk |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Onset of IFN-α/RBV | End of IFN-α /RBV | Onset of IFN-α /RBV | End of IFN- α/RBV | Before HCV Treatment | After HCV Treatment | |||||||||
| 1 | 48 | Female | White | 52 | NRa | 1 573 549 | 515 941 | CT | 1A | 466 | 189 | FTC, TDF, NVP | 52 | 21 |
| 2 | 41 | Female | White | 62 | SVRb | 6 082 084 | NDc | CT | 1B | 336 | 327 | DRV/r | 76 | 32 |
| 3 | 54 | Male | White | 67 | NR | 12 549 350 | 280 | CC | 1B | 982 | 673 | 3TC, ABC, NVP | 62 | 35 |
| 4 | 44 | Male | White | 48 | SVR | 5 166 727 | ND | CT | 1A | 456 | 404 | ETV, FTC, TDF | 45 | 46 |
| 5 | 67 | Male | White | 9 | NR | 23 500 000 | 9000 | CT | 1B | 656 | 174 | LPV/r, NVP | 34 | 41 |
| 6 | 38 | Male | White | 25 | NR | 4 910 000 | 3000 | CT | 1B | 653 | 295 | 3TC, d4 T, EFV | 29 | 36 |
| 7 | 50 | Male | White | 25 | SVR | 181 886 | ND | CC | 3A | 519 | 477 | 3TC, ABC, NVP | 66 | 24 |
| 8 | 42 | Male | White | 13 | NR | 4 988 745 | 600 102 | CT | 1A | 447 | 422 | FTC, TDF, LPV/r | 71 | 84 |
| 9 | 41 | Male | White | 16 | NR | 24 612 | 29 570 | CT | 4 | 334 | 255 | DRV/r, MRV, RAL | 65 | 28 |
| 10 | 46 | Male | White | 82 | SVR | 19 770 008 | ND | CC | 1A | 481 | 228 | 3TC, ABC, RAL | 26 | 67 |
| 11 | 43 | Female | White | 24 | NR | 885 381 | NDc | CC | 3A | 659 | 433 | ABC, D4T, NVP | 28 | 44 |
| 12 | 47 | Male | White | 49 | SVR | 96 000 | ND | CC | 1A | 506 | 507 | D4T, EFV, NVP | 51 | 27 |
Abbreviations: 3TC, lamivudine; ABC, abacavir; d4T, stavudine; DRV/r, darunavir/ritonavir; EFV, efavirenz; ETV, etravirine; FTC, emtricitabine; HAART, highly active antiretroviral therapy; HCV, hepatitis C virus; IFN, interferon; IL-28B, interleukin 28B; LPV/r, lopinavir/ritonavir; MRV, maraviroc; NVP, nevirapine; RAL, raltegravir; RBV, ribavirin; TDF, tenofovir.
a NR: no-response, detectable HCV load 24 weeks after the end of HCV therapy.
b SVR: sustained virological response, undetectable HCV load 24 weeks after the end of HCV therapy.
c ND: Not detected using commercial polymerase chain reaction assays.
Assessment of the Interleukin 28B Polymorphism
The interleukin 28B (IL-28B) polymorphism (rs12979860) was analyzed according to standard protocols in commercial laboratories.
Isolation of CD4 T Cells
CD4+ T cells were isolated by immunomagnetic enrichment from 10 million PBMCs using an autoMACS Pro Separator (Miltenyi) according to the manufacturer's instructions. The purity of the CD4+ T cells was >95% as assessed by flow cytometry (data not shown).
Analysis of HIV-1 DNA
Isolated CD4 T cells were digested to extract cell lysates. We amplified total HIV-1 DNA with primers and probes previously described [8]. Integrated HIV-1 DNA was detected using nested PCR with Alu-1/Alu-2 primers and HIV-1 LTR primer L-M667 for the first-round PCR, and LTR primer AA55M, Lambda T primers, and MH603 probe for the second-round quantitative PCR, as described previously [9]. Serial dilutions of HIV-1 DNA from cell lysates of the HIV-1–infected cell line 293 T (provided by F. Bushman, University of Pennsylvania, Philadelphia) were used for reference purposes.
HIV-1 Reactivation Assays
PBMCs from HIV-1 patients on suppressive antiretroviral therapy were seeded at a density of 1 × 106 cells/mL and incubated with IFN-α (1000 units/mL), RBV (1 μg/mL), or panobinostat (0.1 μM). Raltegravir (0.5 μM) and zidovudine (1 μM) were added to the media to prevent further rounds of infection. RNA from isolated CD4 T cells was harvested after 48 hours and analyzed for induction of cell-associated HIV-1 RNA. For this purpose, RNA was extracted (mirVana RNA isolation kit, Life Technologies) and reverse transcribed according to standard protocols. HIV-1 complementary DNA was amplified using TaqMan-based quantitative PCR as previously described [10]; transcripts encoding β-actin were used as reference genes, and the final data were plotted as fold change compared to unstimulated controls. Simultaneously, transcripts encoding for IRF-7 were amplified using commercial gene expression assays (TaqMan, Life Technologies). To analyze cell-associated HIV-1 DNA, total cellular DNA was harvested after 5 days of culture, and analyzed for total and integrated HIV-1 DNA as described above.
Statistical Analysis
Differences were tested for statistical significance with paired Wilcoxin signed rank test. Correlations were analyzed using Spearman correlation coefficient.
RESULTS
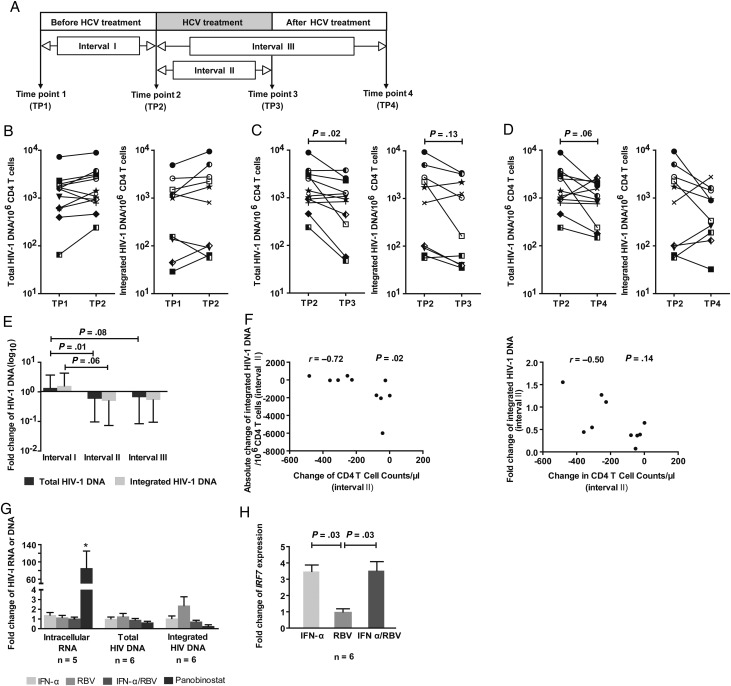
To investigate effects of IFN-α/RBV treatment on the HIV-1 reservoir in CD4 T cells, we retrospectively analyzed total and integrated cell-associated HIV-1 DNA in 12 HIV/HCV-coinfected patients receiving therapy for HCV infection while being treated with suppressive antiretroviral combination therapy. Patients' clinical and demographic characteristics are summarized in Table 1. Changes in HIV-1 DNA in isolated CD4 T cells were analyzed in a time interval immediately preceding IFN-α/RBV treatment (median of 51 weeks [range, 26–71 weeks]), during the time of IFN-α/RBV exposure (median duration of 36.5 weeks [range, 9–82 weeks]), and during a time period after completion of HCV treatment (median duration of 35.5 weeks [range, 21–84 weeks]) (Figure 1A). Whereas HIV-1 DNA levels remained essentially stable during the time preceding IFN-α/RBV therapy (Figure 1B), there was a statistically significant, approximately 2-fold decrease of cell-associated total and integrated HIV-1 DNA in CD4 T cells during IFN-α/RBV therapy (Figure 1C and 1E). This decrease in the viral CD4 T-cell reservoir persisted after discontinuation of IFN-α/RBV therapy, but no further reduction of viral CD4 T-cell–associated DNA was noticed when IFN-α/RBV treatment was stopped (Figure 1D and 1E). The ratio between total and integrated HIV-1 DNA, possibly indicative of ongoing HIV-1 replication [11], remained unaffected by HCV treatment with IFN-α/RBV (data not shown). Decreases of HIV-1 DNA during treatment with IFN-α/RBV were highly variable among the analyzed study subjects, but the degree of viral DNA reduction during IFN-α/RBV treatment was not correlated to the duration of HCV treatment, to the duration of prior therapy with antiretroviral agents, to HCV treatment responses, or to IL-28B polymorphisms. We observed a trend for an inverse relation between the reduction of HIV-1 DNA in CD4 T cells and corresponding declines of absolute CD4 T cells during treatment with IFN-α/RBV, indicating that more pronounced decreases of HIV-1 DNA preferentially occurred in patients who did not develop substantial treatment-associated lymphopenia (Figure 1F); however, our retrospective cohort study was too small to more definitively determine factors associated with or predictive of the effects of IFN-α/RBV on the HIV-1 reservoir in HAART-treated patients. Together, these findings suggest that pharmacological administration of IFN-α/RBV for HCV therapy can cause a moderate but significant decrease of HIV-1 DNA in CD4 T cells from HIV/HCV-coinfected patients.
Figure 1.
Reduction of CD4 T-cell–associated human immunodeficiency virus type 1 (HIV-1) DNA during interferon alpha (IFN-α)/ribavirin (RBV) treatment in HIV-1/hepatitis C virus (HCV)–coinfected patients. A, Time intervals during which changes in HIV-1 DNA were evaluated in HIV-1/HCV-coinfected patients. B–D, Changes in total (left panels) and integrated (right panels) HIV-1 DNA in CD4 T cells from HIV-1/HCV-coinfected study patients during time intervals I (B), II (C), and III (D). Amplification of integrated HIV-1 DNA was unsuccessful in 2 study subjects. Patient 1:  ; Patient 2:
; Patient 2:  ; Patient 3:
; Patient 3:  ; Patient 4:
; Patient 4:  ; Patient 5:
; Patient 5:  ; Patient 6:
; Patient 6:  ; Patient 7:
; Patient 7:  ; Patient 8:
; Patient 8:  ; Patient 9:
; Patient 9:  ; Patient 10:
; Patient 10:  ; Patient 11:
; Patient 11:  ; Patient 12:
; Patient 12:  . E, Fold change of HIV-1 DNA in indicated time intervals. Data reflect median and range. F, Inverse association between maximum CD4 T-cell changes during HCV therapy and corresponding absolute and relative changes in CD4 T-cell–associated integrated HIV-1 DNA. Spearman correlation coefficients are shown. G, Analysis of HIV-1 RNA and total or integrated HIV-1 DNA in primary CD4 T cells from highly active antiretroviral therapy–treated patients after in vitro exposure to IFN-α, IFN-α + RBV, or panobinostat. *P = .06 in comparison to untreated control. H, IRF7 mRNA expression in CD4 T cells after in vitro exposure to IFN-α, RBV or IFN-α + RBV.
. E, Fold change of HIV-1 DNA in indicated time intervals. Data reflect median and range. F, Inverse association between maximum CD4 T-cell changes during HCV therapy and corresponding absolute and relative changes in CD4 T-cell–associated integrated HIV-1 DNA. Spearman correlation coefficients are shown. G, Analysis of HIV-1 RNA and total or integrated HIV-1 DNA in primary CD4 T cells from highly active antiretroviral therapy–treated patients after in vitro exposure to IFN-α, IFN-α + RBV, or panobinostat. *P = .06 in comparison to untreated control. H, IRF7 mRNA expression in CD4 T cells after in vitro exposure to IFN-α, RBV or IFN-α + RBV.
To explore mechanisms underlying the reduction of HIV-1 DNA during HCV treatment, we used in vitro experiments to test whether pharmacological doses of IFN-α can effectively reactivate HIV-1 replication in CD4 T cells. For this purpose, we analyzed HIV-1 RNA transcription in CD4 T cells from HAART-treated patients after ex vivo exposure to IFN-α, IFN-α/RBV, or the potent histone deacetylase inhibitor panobinostat. We observed that exogenous IFN-α upregulated the interferon-stimulated gene IRF7, which is indicative of cellular responsiveness to IFN-α, but failed to reactivate HIV-1 gene expression under conditions in which the histone deacetylase inhibitor panobinostat was effective in increasing HIV-1 gene expression (Figure 1G and 1H). We also failed to notice an effect of IFN-α or IFN-α/RBV on HIV-1 DNA levels during in vitro culture of CD4 T cells from HAART-treated patients (Figure 1G). Together, these findings suggest that the reduction of HIV-1 DNA during pharmacological treatment with IFN-α/RBV is not due to direct effects of IFN-α or IFN-α/RBV on HIV-1 gene transcription or viral reactivation from latency.
DISCUSSION
To our knowledge, this work represents the first analysis of changes in CD4 T-cell–associated HIV-1 DNA in HAART-treated patients undergoing therapy with IFN-α and RBV. Our data indicate that during combined treatment with IFN-α and RBV, CD4 T-cell–associated total and integrated HIV-1 DNA in HAART-treated patients can decline by approximately 2-fold. Notably, these changes occurred selectively during HCV treatment, and not during time periods immediately before or after treatment with IFN-α and RBV, indicating that the observed effects are associated with HCV treatment and do not reflect a general decline of HIV-1 DNA that typically occurs during early stages of suppressive antiretroviral therapy. Moreover, the modest decline of HIV-1 DNA during IFN-α/RBV treatment in our relatively small study cohort is unlikely to contribute to HIV-1 eradication in a clinically significant way. In fact, recent estimates suggest that reduction of CD4 T-cell–associated HIV-1 DNA by 100- to 1000-fold will likely be necessary to approach viral eradication and induce an increased ability to maintain a drug-free remission of HIV-1 infection after discontinuation of HAART [12]. Due to limited sample availabilities, we were unable to evaluate the frequency of cells harboring replication-competent virus before and after HCV therapy; however, total and integrated HIV-1 DNA in CD4 T cells are acceptable biomarkers for the HIV-1 reservoir size in clinical studies [13].
The mechanism of IFN-α–induced reduction of HIV-1 DNA in CD4 T cells from HAART-treated patients remains uncertain. Notably, it is well recognized that IFN-α treatment is associated with decreasing CD4 T-cell counts, raising the possibility that reductions of HIV-1 DNA during IFN-α/RBV therapy may simply result from unspecific lymphocellular toxicity of this treatment regimen. However, our findings seem to imply that during IFN-α/RBV treatment, HIV-1–positive CD4 T cells decline more substantially than HIV-1–negative CD4 T cells, consistent with some degree of selective pharmacological activity against HIV-1–infected cells. In addition, reductions of HIV-1 DNA in CD4 T cells were most pronounced in patients with limited treatment-associated lymphopenia, suggesting that effective reduction of HIV-1–infected cells during IFN-α/RBV therapy may depend on preserved immune function of antiviral effector cells. An IFN-α–dependent reactivation of HIV-1 gene expression that would subsequently translate into selective death of cells in which reactivation occurred is a less likely scenario, given that we failed to observe any effect of IFN-α or IFN-α/RBV on HIV-1 RNA reactivation in in vitro assays. However, it is possible that IFN-α–mediated changes in CD4 T-cell immune activation or gene expression may have contributed to the observed findings. For instance, IFN-α is known to upregulate intracellular sensors of microbial RNA such as RIG-I, which can recognize HIV-1 RNA [14], and induce apoptosis in cells upon binding to target viral RNA [15]. Therefore, treatment with IFN-α may increase death rates in cells in which HIV-1 reactivation occurs naturally. Further investigation of the mechanisms underlying the reduction in HIV-1 DNA during IFN-α/RBV therapy may be helpful for designing improved clinical strategies to target HIV-1 reservoirs that persist despite HAART.
Notes
Financial support. This work was supported by Redes Telemáticas de Investigación Cooperativa en Salud (grant RETICS; 2012, Red de SIDA RD12/0017/0029 and RD12/0017/0037 to M. Le.) and by Fondo de Investigación Sanitaria (contracts P08/00172 to E. R.-M. and CD10/00382 to S. F.-M.). M. B. is supported by a fellowship award from the European Molecular Biology Organization, and by the Tosteson fellowship award from Massachusetts General Hospital. M. Li. is supported by the US National Institutes of Health (grant numbers AI093203 and AI098487), by the Clinical Scientist Development Award from the Doris Duke Charitable Foundation (grant number 2009034), and by the American Foundation for AIDS Research (grant number 108302-51-RGRL). X. G. Y. is supported by the US National Institutes of Health (grant number AI089339).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Hoofnagle JH, Seeff LB. Peginterferon and ribavirin for chronic hepatitis C. N Engl J Med. 2006;355:2444–51. doi: 10.1056/NEJMct061675. [DOI] [PubMed] [Google Scholar]
- 2.Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe. 2007;2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tavel JA, Huang CY, Shen J, et al. Interferon-alpha produces significant decreases in HIV load. J Interferon Cytokine Res. 2010;30:461–4. doi: 10.1089/jir.2009.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanderford TH, Slichter C, Rogers KA, et al. Treatment of SIV-infected sooty mangabeys with a type-I IFN agonist results in decreased virus replication without inducing hyperimmune activation. Blood. 2012;119:5750–7. doi: 10.1182/blood-2012-02-411496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillai SK, Abdel-Mohsen M, Guatelli J, et al. Role of retroviral restriction factors in the interferon-alpha-mediated suppression of HIV-1 in vivo. Proc Natl Acad Sci U S A. 2012;109:3035–40. doi: 10.1073/pnas.1111573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coiras M, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol. 2009;7:798–812. doi: 10.1038/nrmicro2223. [DOI] [PubMed] [Google Scholar]
- 7.Azzoni L, Foulkes AS, Papasavvas E, et al. Pegylated interferon alfa-2a monotherapy results in suppression of HIV type 1 replication and decreased cell-associated HIV DNA integration. J Infect Dis. 2013;207:213–22. doi: 10.1093/infdis/jis663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liszewski MK, Yu JJ, O'Doherty U. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods. 2009;47:254–60. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brussel A, Delelis O, Sonigo P. Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. Methods Mol Biol. 2005;304:139–54. doi: 10.1385/1-59259-907-9:139. [DOI] [PubMed] [Google Scholar]
- 10.Palmer S, Wiegand AP, Maldarelli F, et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2003;41:4531–6. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mexas AM, Graf EH, Pace MJ, et al. Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. AIDS. 2012;26:2295–306. doi: 10.1097/QAD.0b013e32835a5c2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill A, Rosenbloom D, Nowak MA, Siliciano R. Predicting outcomes of treatments to eradicate the HIV latent reservoir. Program and Abstracts of the 20th Conference on Retroviruses and Opportunistic Infections,; Atlanta, GA. 2013. [Google Scholar]
- 13.Eriksson S, Graf EH, Dahl V, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaitin DA, Schreiber G. Upregulation of a small subset of genes drives type I interferon-induced antiviral memory. J Interferon Cytokine Res. 2007;27:653–64. doi: 10.1089/jir.2006.0162. [DOI] [PubMed] [Google Scholar]
- 15.Besch R, Poeck H, Hohenauer T, et al. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J Clin Invest. 2009;119:2399–411. doi: 10.1172/JCI37155. [DOI] [PMC free article] [PubMed] [Google Scholar]