Abstract
The mammalian target of rapamycin (mTOR) regulates cell growth by integrating nutrient and growth factor signaling and is strongly implicated in cancer. But mTOR is not an oncogene, and which tumors will be resistant or sensitive to new ATP-competitive mTOR inhibitors now in clinical trials remains unknown. We screened a panel of over 600 human cancer cell lines to identify markers of resistance and sensitivity to the mTOR inhibitor PP242. RAS and PIK3CA mutations were the most significant genetic markers for resistance and sensitivity to PP242, respectively; colon origin was the most significant marker for resistance based on tissue type. Among colon cancer cell lines, those with KRAS mutations were most resistant to PP242, while those without KRAS mutations most sensitive. Surprisingly, cell lines with co-mutation of PIK3CA and KRAS had intermediate sensitivity. Immunoblot analysis of the signaling targets downstream of mTOR revealed that the degree of cellular growth inhibition induced by PP242 was correlated with inhibition of phosphorylation of the translational repressor 4E-BP1, but not ribosomal protein S6. In a tumor growth inhibition trial of PP242 in patient-derived colon cancer xenografts, resistance to PP242 induced inhibition of 4E-BP1 phosphorylation and xenograft growth was again observed in KRAS mutant tumors without PIK3CA co-mutation, compared to KRAS WT controls. We show that, in the absence of PIK3CA co-mutation, KRAS mutations are associated with resistance to PP242 and that this is specifically linked to changes in the level of phosphorylation of 4E-BP1.
Keywords: mTOR, colon cancer, PIK3CA, KRAS, xenograft, PP242
Introduction
Clinically approved kinase inhibitors such as imatinib, vemurafenib, and crizotinib show strong anti-tumor responses in patients with mutated forms of their target kinases, BCR-ABL, BRAF V600E, and EML4-ALK, respectively (1–3). The intrinsic sensitivity of cancer cells expressing these mutationally activated kinase alleles provides a template for patient selection and clinical trial design (4). However, many of the protein kinase targets currently being investigated in cancer such as MEK, ERK, AKT, and mTOR are not commonly mutated, but rather lie at critical nodes in conserved cancer signaling pathways. The design of clinical trials for experimental therapeutics that inhibit these targets is challenging as mutations upstream of these nodes may or may not predict sensitivity to inhibition of downstream kinases (5).
Deregulated mammalian target of rapamycin (mTOR) signaling is present in human diseases that alter metabolism, including diabetes and cancer (6). An essential and evolutionarily conserved regulator of cell metabolism, mTOR is the catalytic core of two related heteromeric protein complexes, mTORC1 and mTORC2 (7–9). In cancer, conserved mTOR mutations or gene amplifications have not been identified; instead, mTORC1 is activated by mutations in upstream signaling networks (10,11). The network most implicated in oncogenic mTORC1 signaling is the PI3K/AKT/TSC pathway (12). Enhanced response to mTOR inhibition in patients with rare somatic TSC mutations supports the rationale of targeting this network (13).
Temsirolimus and everolimus, derivatives of the natural product rapamycin, are the only mTOR inhibitors currently approved for the treatment of solid tumors but their activity is limited and mechanism of action debated (14,15). A new and potentially more efficacious class of mTOR inhibitors has been developed specifically to target cancer (16–19). These small molecule drugs competitively target the ATP binding pocket of the mTOR kinase domain and have now entered clinical trials (20). The discovery of these molecules allowed for the division of canonical mTORC1 substrates into classes: those sensitive to inhibition by rapamycin (p70S6 kinase, S6K; and its direct target ribosomal protein S6, rpS6) and those that were relatively insensitive to rapamycin (eIF4E Binding Proteins, 4E-BPs). Inhibition of 4E-BP activity downstream of mTORC1 is responsible for the anti-proliferative effects of PP242 in cell culture models (21). But it remains unclear whether rapamycin sensitive or insensitive targets downstream of mTORC1 are the clinically relevant biomarkers for treatment efficacy.
Colon cancer contains many of the most prevalent aberrations in cancer including KRAS and PIK3CA (phosphatidylinositol 3-kinase catalytic subunit alpha) mutations and loss of PTEN (phosphatase and tensin homolog) expression (22). The frequency of these mutations makes it possible to study how each contributes to resistance and sensitivity to molecularly targeted therapies (23). Here we used a high-throughput cell-screening platform to identify a genetic signature for primary resistance of colon cancer to the ATP-competitive mTOR inhibitor PP242. We validated these observations in cultured cell lines and primary human tumor xenografts and identified a biomarker for PP242 efficacy.
Results
Screening of cancer cell lines
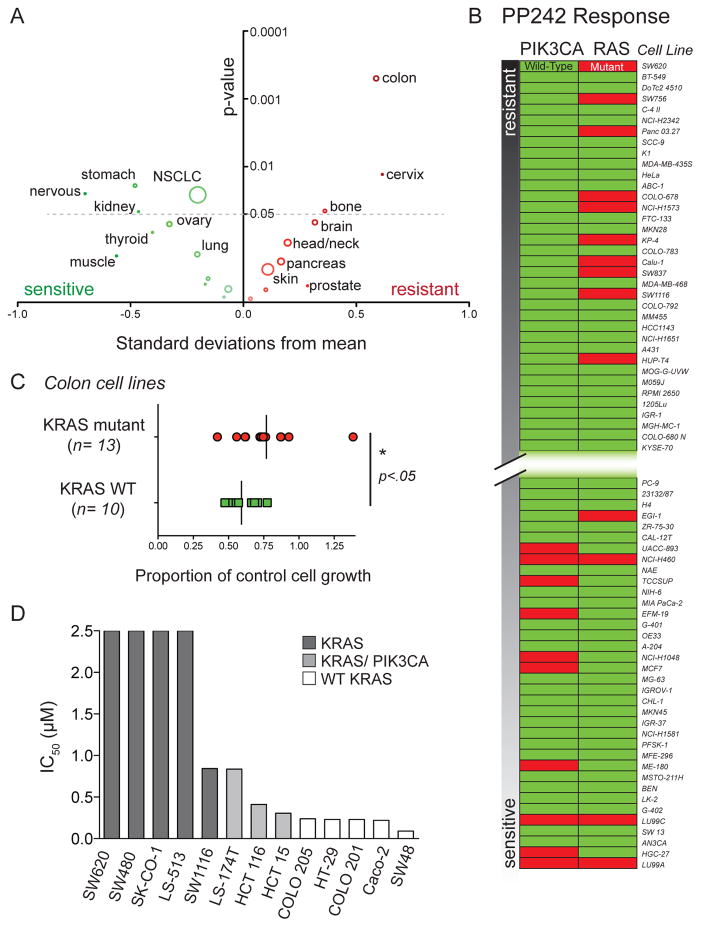
We identified markers of resistance or sensitivity to the ATP-competitive mTOR inhibitor PP242 in solid tumor cell lines. Our approach relied on automated screening of growth of solid tumor cell lines that were annotated for common oncogenic mutations and tissue of origin (Tables S1 and S2) (24). The cell line set (n=666) was treated with 500 nM PP242 and assayed for growth inhibition at 72 hours (The complete data set is available as an online supplement). The PP242 treatment results were normally distributed (Shapiro-Wilk test, p= 0.145) and centered upon 57.9% of untreated control providing maximal sensitivity to detect both resistant and sensitive cell lines. Significant differences were observed in the response of cell lines grouped by tissue of origin (Fig. 1A and Table S1). Several cell types had mean PP242 responses that were greater than 0.5 standard deviations from the complete set. Cell types that were significantly sensitive to PP242 treatment included nervous system, stomach, kidney and non-small cell lung cancer (NSCLC). Only three types were significantly resistant: bone, cervix and colon. Of all significantly resistant and sensitive cell types, colon was distinct for its combination of large magnitude of resistance (0.59 standard deviations higher than the population mean) and the significance of this difference (p=0.005).
Figure 1.
An unbiased cell screen reveals factors leading to resistance and sensitivity to the ATP-competitive mTOR inhibitor PP242 (A) Colon cell origin is a strong predictor for resistance to PP242 treatment. The mean response of each cell type to 500 nM PP242 treatment was plotted as a standard deviation from the population mean. The y-axis indicates the significance of the test-statistic for each independent difference in means (cell type versus population). The size of the circle corresponds to the number of cell lines of each type analyzed. Colon origin (n=39) was the strongest single predictor of resistance or sensitivity to PP242 among all annotated organ types. (B) PIK3CA mutations are prevalent in cell lines sensitive to PP242 while RAS marks cell lines that are resistant to PP242. The set of 357 cell lines with known mutation status for PIK3CA and RAS were assayed for growth inhibition and ranked according to the inhibition results. The 10% most resistant and most sensitive cell lines are shown here in order of increasing response to PP242. (C) KRAS mutant colorectal cells are more resistant to PP242 than cells harboring WT KRAS. Comparison of means was made using Student’s t-test. (D) IC50 values for PP242 in selected colon cancer cell lines. KRAS mutant cell lines with concomitant PIK3CA mutations are more sensitive to PP242 than KRAS mutants alone.
For a subset of cell lines (n=357), we used information from the Sanger COSMIC (http://www.sanger.ac.uk/genetics/CGP/cosmic/) database of cancer cell lines to annotate mutations in 15 of the most common oncogenes (Table S2). We analyzed the role of these mutations in accounting for sensitivity or resistance to PP242 by two independent methods. Multivariate linear regression of all genotypes against PP242 growth response showed that RAS and PIK3CA were significant and independent predictors of resistance and sensitivity, respectively (test statistics from the regression analysis: RAS, p=0.045; PIK3CA, p=0.017). We also tested whether RAS and PIK3CA mutations were significantly enriched in the population of the most sensitive and resistant cell lines (Fig. 1B). PIK3CA mutations were absent in the 10% most PP242 resistant cell lines while enriched in the 10% most sensitive ones (Fisher’s exact test: resistant cell lines, p=0.0048; sensitive cell lines, p=0.013). In the most PP242 sensitive cell lines, the reduced number of RAS mutations was significant (Fisher’s exact test: p=0.030), but the enrichment of mutants in resistant cell lines was not (p=0.35). Compared to prior studies that examined responsiveness to inhibitors targeting a specific genetic lesion (such as lapatinib and the EGFR mutant L858R) (24), the strength of the correlation between either PIK3CA or RAS mutation and resistance or sensitivity to PP242 was modest.
Knowing that RAS was a modest marker of resistance to PP242, we asked if the high frequency of RAS mutations in colon cancer might account for the degree of PP242 resistance observed in this tumor type. KRAS mutant colon cell lines were significantly more resistant than wild-type (WT) colon cancer cell lines (Fig. 1C). Interestingly, the resistance of RAS mutant cell lines to PP242 observed within the screening set appears to be driven by RAS mutant pancreas and colon cancer cell lines; RAS dependent resistance was not observed in NSCLC. To quantify the degree of resistance to PP242 imparted upon colon cancer cell lines by KRAS mutations, we determined the half maximal inhibitory concentration (IC50) for a subset of cell lines (Fig. 1D). The potency (IC50) of PP242 in colon cell lines varied widely from 90 nM to 8 μM (Table 1). KRAS mutant colon cell lines were significantly more resistant to PP242 than WT cell lines (p=0.0036; unpaired t-test). The most PP242 resistant cell line, SW620 (IC50 =8 μM) is mutant for KRAS. Of special note were KRAS mutant cell lines with intermediate sensitivity to PP242. This group of cell lines had both KRAS mutations and PIK3CA mutations and their responsiveness to PP242 was closer to that of sensitive cell lines than the resistant ones. The most sensitive colon cancer cell lines were all WT for KRAS. To identify a mechanism for the varied responsiveness of colon cancer cells to PP242, analysis of the downstream effectors of mTOR was performed.
Table 1.
KRAS and PIK3CA mutations modulate sensitivity to mTOR inhibition
| Cell Line | Mutational Status | PP242 (μM) | ||
|---|---|---|---|---|
| KRAS | PIK3CA | bRAF | ||
| SW620 | G12V | WT | WT | 7.8 |
| SW480 | G12V | WT | WT | 4.6 |
| SK-CO-1 | G12V | WT | WT | 4 |
| LS-513 | G12D | WT | WT | 3.9 |
| SW1116 | G12A | WT | WT | 0.84 |
| LS-174T | G12D | H1047R | WT | 0.84 |
| HCT 116 | G13D | H1047R | WT | 0.41 |
| HCT 15 | G13D | E545K | WT | 0.3 |
| COLO 205 | WT | WT | V600E | 0.24 |
| HT-29 | WT | WT | V600E | 0.23 |
| COLO 201 | WT | WT | V600E | 0.23 |
| Caco-2 | WT | WT | WT | 0.22 |
| SW48 | WT | WT | WT | 0.09 |
mTORC1 substrates 4E-BP1 and rpS6 are differentially inhibited by PP242
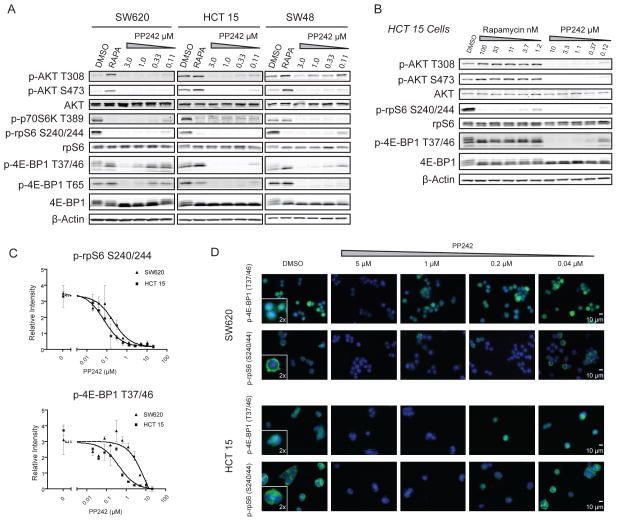
To ascertain the signaling alterations that lead to the spectrum of responses to PP242 in colon cancer cells, we examined whether mTOR substrate phosphorylation was differentially inhibited in resistant versus sensitive cell lines. Three representative cell lines were studied: SW620 (Mut for KRAS), HCT 15 (Mut for both KRAS and PIK3CA) and SW48 (WT for KRAS and PIK3CA) (Fig. 2A). In PP242-sensitive HCT 15 and SW48 cell lines, 1 hour of treatment with PP242 similarly reduced the phosphorylation of mTORC1 substrates S6K and 4E-BP1, as well as mTORC2 substrate AKT S473. We were surprised to observe that in the PP242-resistant cell line SW620, mTORC1 substrates were differentially inhibited by PP242. Phosphorylation of S6K and its effector rpS6 were potently inhibited by PP242, as in the PP242-sensitive cell line HCT 15, but the phosphorylation of 4E-BP1 was poorly inhibited even at high drug concentrations. Expression levels of 4E-BP1 or basal amounts p-4E-BP1 did not correlate with response to PP242 in the cell lines examined (Figs. 2A and S1A). Resistance of 4E-BP1 to dephosphorylation was observed in other KRAS mutant colon cancer cell lines as well (Fig. S1B). We performed the same western blot analysis with 2 additional active site mTOR inhibitors: KU-0063794, which is based on a different chemical scaffold from PP242 (19), and MLN0128 (previously INK128), a clinical derivative of PP242 (25). In both cases, 4E-BP1 phosphorylation was less potently inhibited in SW620 cells compared to HCT 15 cells, as was observed with PP242 (Figs. S1C and S1D).
Figure 2.
mTORC1 substrates are differentially inhibited in PP242 resistant versus sensitive cell lines (A) 4E-BP1 is differentially inhibited in PP242 resistant and sensitive colon cancer cell lines. Representative cell lines SW620, HCT 15 and SW48 were treated with PP242 or rapamycin (20 nM) for 1 hour and analyzed by western blotting. (B) Differential inhibition is reminiscent of incomplete mTORC1 inhibition by rapamycin. In HCT 15 cells, rapamycin only partially inhibits 4E-BP1 phosphorylation after a 1 hour treatment, despite potently blocking rpS6 phosphorylation. (C) Quantification of mTORC1 substrate inhibition shows that inhibition of p-4E-BP1 and not inhibition of p-rpS6 tracks with growth inhibition. Quantification was performed on western blots of lysed cells after treatment for 1 hour with increasing PP242 concentrations in two independent experiments. (D) Immunofluorescence of mTORC1 substrates reveals consistent subcellular localization despite differential inhibition by PP242. SW620 and HCT 15 cells were treated with increasing concentrations of PP242 for 1 hour, formalin fixed and stained for either p-4E-BP1 or p-rpS6 (both green) and counterstained with DAPI (blue). PP242 treatment does not alter the subcellular localization of either phosphorylated substrate.
Differential inhibition of mTOR substrates has previously been observed with rapamycin where inhibition of phosphorylation of mTORC1 substrates bifurcates between sensitive (rpS6) and insensitive (4E-BP1) targets. Rapamycin has no short-term activity against mTORC2 and incompletely inhibits mTORC1; it fails to inhibit 4E-BP1 phosphorylation while paradoxically increasing AKT phosphorylation (26). All of these effects were visible in the KRAS mutant colon cancer cell line HCT 15 (Fig. 2B). Feedback activation of AKT has also been observed in human trials with rapamycin derivatives (27). Unlike rapamycin, PP242 suppressed feedback activation of AKT over 24 hours and fully inhibited cell growth (Figs. S2A and S2B). These findings add to work in colon and other cell types showing that the more complete inhibition of mTOR by PP242 results in greater inhibition of cell growth than that achieved by rapamycin (28–30).
To quantify the sensitivity of 4E-BP1 phosphorylation to inhibition by PP242 and relate it to cell growth, phosphoprotein IC50 curves were constructed (Fig. 2C). The IC50 for inhibition of phosphorylation of rpS6 was similar in HCT 15 and SW620 cell lines (72 and 212 nM respectively, a 3-fold difference). Conversely, the IC50 values for inhibition of 4E-BP1 phosphorylation were 0.43 μM for HCT 15 cells and 10 μM for SW620 cells. The IC50 values for 4E-BP1 phosphorylation and the difference between cell lines (23-fold) correspond closely to the growth IC50 values (0.30 μM and 7.8 μM, a 26-fold difference).
Immunofluorescence imaging of p-4E-BP1 and p-rpS6 in treated SW620 and HCT 15 cells established that rpS6 is excluded from the nucleus in both cell lines and is similarly inhibited (Fig. 2D). p-4E-BP1 partitions between the cytoplasm and nucleus as previously described (31). Differential inhibition of 4E-BP1 phosphorylation by PP242 between cell lines was not correlated with a difference in subcellular localization of 4E-BP1 (Figs. 2D and S3). Single cell analysis showed no PP242 resistant subpopulation within the SW620 cell line that would account for the differences in sensitivity (Fig. S3). The consistent inhibition of S6K target rpS6 in SW620 and HCT 15 cells demonstrates that PP242 enters the cells and binds mTOR with similar efficacies.
MAPK signaling differences do not alter mTORC1 substrate specificity
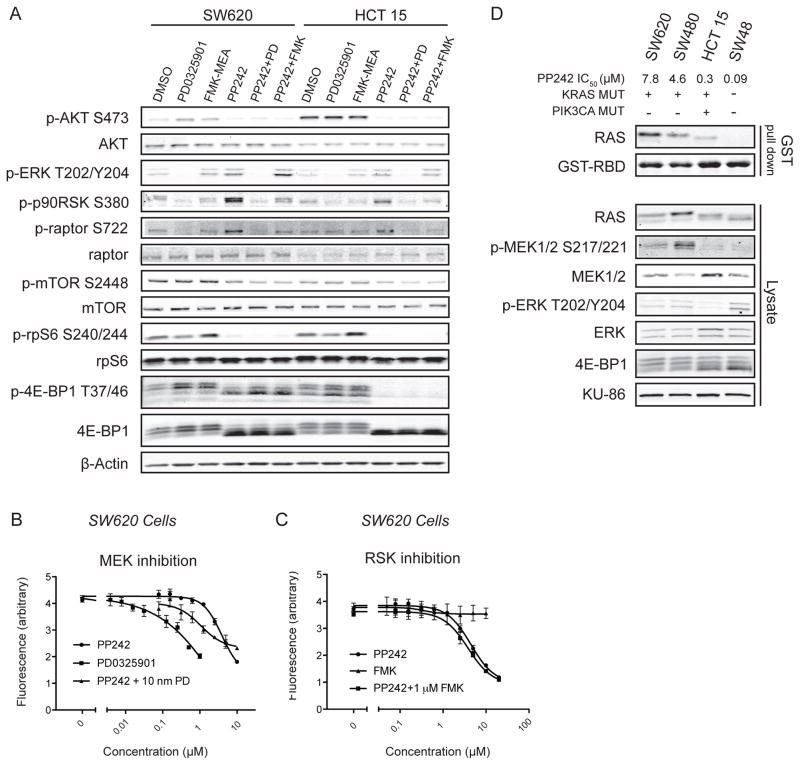
Our finding that KRAS mutant colon cancer cell lines exhibit distinct patterns of substrate inhibition by PP242 led us to question whether outputs of the mitogen activated protein kinase (MAPK) pathway were impacting downstream mTOR substrates. It is known that the mTORC1 component raptor is phosphorylated by the ERK substrate, p90RSK, and we examined whether inhibition of this effect would impact mTOR dependent phosphorylation of 4E-BP1 and rpS6 (32). Immunoblotting revealed that expression of raptor and the phosphorylation of mTOR is greater in the resistant SW620 cell line compared to sensitive HCT 15 cells, however phosphorylation of raptor appeared to be equivalent (Fig. 3A). MAPK pathway activation as assayed by ERK phosphorylation was also comparable between the two KRAS mutant cell lines regardless of PIK3CA mutation status.
Figure 3.
Inhibition of MAPK signaling does not alter mTORC1 substrate phosphorylation (A) Treatment of cell lines with MAPK inhibitors does not sensitize mTOR substrates to PP242 inhibition. Cells were treated with the MEK inhibitor PD0325901 (20 nM), the p90RSK inhibitor FMK-MEA (3 μM), PP242 (1 μM) singly or in combination for 1 hour. Neither FMK-MEA nor PD0325901 sensitize mTOR substrates to PP242 treatment. (B–C) Inhibition of ERK but not p90RSK augments the cell growth arrest induced by PP242. The MEK inhibitor PD0325901 is a potent inhibitor of cell growth, but selective inhibition of ERK substrate p90RSK by FMK-MEA does not recapitulate this phenotype in a three-day cell growth assay measured using a resazurin assay. (D) RAS-GTP loading, but not MAPK pathway activation, correlates with resistance to PP242. Glutathione pull down of RAS-GTP with GST-RBD shows high RAS-GTP levels in unstimulated KRAS mutant PP242 resistant cell lines. Significantly different levels of RAS-GTP activation were observed within KRAS mutant cells.
Pharmacological inhibition of the MAPK pathway by the MEK inhibitor PD0325901 potently inhibited cell growth in KRAS mutant SW620 cells (33), without impacting mTORC1 signaling in either SW620 or HCT15 cells (Fig. 3A and 3B). In contrast, inhibition of phosphorylation of p90RSK by FMK-MEA (34,35), did not affect mTORC1 substrate phosphorylation, (Fig. 3A) or cell growth, even at high concentrations (Fig. 3C). The MAPK pathway does not directly alter the phosphorylation of mTOR substrates in colon cancer.
Combination treatment of PD0325901 and an AKT inhibitor leads to profound growth arrest and synergistic inhibition of 4E-BP1 phosphorylation (36). We asked whether the combination of PP242 and a MAPK inhibitor would similarly lead to enhanced suppression of phosphorylation of 4E-BP1. By itself, acute treatment with PP242 modestly increased phosphorylation of ERK, and significantly increased phosphorylation of p90RSK and consequently raptor, suggesting that MAPK combination therapy could be useful (Fig. 3A). This effect was most prominent in the PP242 resistant SW620 cells. Combination treatment of PP242 and PD0325901 was additive and at least partly mTOR independent (Fig. 3B). However, addition of PD0325901 did not increase the inhibition of phosphorylation of 4E-BP1 beyond what was achieved with PP242 in both resistant and sensitive cell lines (Fig. 3A). There was no additive benefit in treatment combining FMK-MEA with PP242 (Fig. 3C). Thus the enhanced inhibitory effects of combining PP242 with PD0325901 on cell growth are not due to increased inhibition of mTOR substrates but rather ERK substrates that are independent of p90RSK.
MAPK signaling did not explain the differential sensitivity of KRAS mutant colon cancer cell lines to PP242, so we examined RAS directly. Mutations activate RAS by biasing the fraction in the active GTP-bound form. This conformation can be selectively pulled down using a GST tagged RAS binding domain (RBD) taken from c-RAF-1 (37). We observed that the amount of RAS-GTP pulled down differed among KRAS mutant cell lines and was inversely correlated with sensitivity to PP242 (Fig. 3D). KRAS WT and PP242-sensitive cell line SW48 had non-detectable amounts of RAS-GTP in the basal state, yet KRAS mutant cell lines HCT 15, SW480 and SW620 had progressively more RAS-GTP. Our experiment differentiates KRAS mutant cell lines by levels of RAS-GTP and shows a correlation between this activation and resistance to mTOR inhibition by PP242. This was not accompanied by a similar trend in MAPK pathway activation, suggesting that other RAS effectors may be responsible for transmitting the mTOR resistance phenotype observed in high RAS-GTP cells.
Mutant PIK3CA but not PTEN loss leads to mTOR inhibitor sensitization
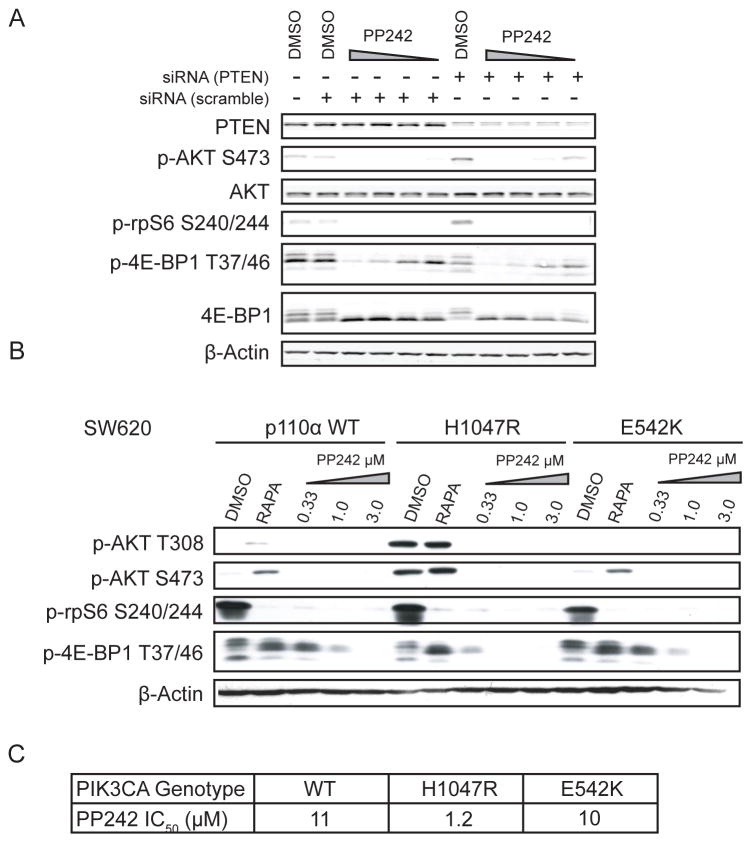
Phosphoinositide signaling in colon cancers is deregulated by both loss of PTEN expression (35%) and activation of PIK3CA (15%) (38,39). Our observation that KRAS mutant cell lines with PIK3CA mutations were sensitive to PP242 led us to investigate how changes in phosphoinositide signaling impact the sensitivity of mTOR to PP242. A previous analysis of PTEN status in breast cancer cell lines observed no correlation between PTEN expression and PP242 sensitivity (40). Our analysis of PTEN in the complete PP242 cell screen similarly showed no relation between PTEN mutations and PP242 sensitivity, but this analysis was confined to the small subset of PTEN mutant tumors whereas loss of expression is more often accomplished via epigenetic mechanisms. All of our cell lines express PTEN, and because loss of PTEN can activate AKT signaling, we wanted to test whether reduction of PTEN would sensitize cells to PP242. Using siRNA directed against PTEN, a 96-hour knockdown was performed and the cells were analyzed for their response to PP242 (Fig. 4A). PTEN knockdown modestly increased p-AKT S473 levels but did not change the dose response on either mTORC1 substrate. Our results are consistent with prior studies showing that PTEN and PIK3CA activate mTOR signaling downstream of AKT in non-redundant ways.
Figure 4.
PIK3CA mutation but not PTEN loss sensitizes KRAS mutant cells to PP242 (A) siRNA against PTEN does not sensitize KRAS mutant CRC cells to PP242. SW620 cells were treated with siRNA against PTEN for 72 hours prior to 1 hour drug treatment with PP242 (3.0, 1.0, 0.33 and 0.1 μM). (B) Addition of mutant PIK3CA to the KRAS mutant cell line SW620 increases sensitivity to PP242. Retroviral insertion of either WT PIK3CA, helical (E542K) or kinase (H1047R) domain mutations only resulted in elevated basal AKT activation in the H1047R case. Cells were treated with rapamycin (20 nM) or PP242 (3.0, 1.0, 0.3 μM) for 1 h before lysis. (C) IC50s for SW620 cell lines engineered to contain additional PIK3CA mutations.
To determine if mutant PIK3CA is sufficient to sensitize SW620 cells to PP242, we transfected SW620 cells with constructs of the most common PIK3CA mutations. Mutations in PIK3CA cluster into two conserved “hotspots”: a kinase domain hotspot, most commonly H1047R, and helical domain mutations, most commonly E542K or E545K (41). As previously shown, the PIK3CA kinase domain mutant was more transforming than the helical domain mutations and conferred a substantial growth advantage to the cells (Fig. S4) (42).
We treated the PIK3CA and KRAS co-mutant isogenic cell lines with PP242 and analyzed phospho-signaling. The basal phosphorylation of both AKT T308 and S473 was significantly higher in the H1047R mutant than either the WT or the helical domain E542K mutant expressing cells (Fig. 4B). The inhibition of phosphorylation of the mTOR substrate 4E-BP1 was only significantly altered in the H1047R mutant expressing cell line. Activated AKT signaling sensitized KRAS mutant cells to mTOR inhibition consistent with the response of mTORC1 substrate 4E-BP1 to acute pharmacological inhibition. To confirm the relationship between 4E-BP1 inhibition and cell growth, SW620 PIK3CA mutant cell growth was assayed and the PP242 IC50 was determined. The values of 10 and 11 μM for the E542K and WT lines were nearly identical to the measured SW620 parental cell line (8 μM) whereas the H1047R cell line was approximately 8-fold more sensitive to PP242 (Fig. 4C).
KRAS mutation status predicts response to PP242 in human primary xenografts
We sought to validate our cell line observations about PP242 sensitivity and KRAS and PIK3CA mutation status in an in vivo model of human colon cancer, patient-derived xenografts. Such xenografts allow patient tumors to be maintained in vivo without undergoing the irreversible changes that occur upon in vitro culture (43). Patient-derived xenografts overcome many of the problems that render standard cell line and cell line derived xenografts models poorly predicative of clinical response (44,45). Their utility in colon cancer was recently demonstrated by the identification of a genetic marker of resistance to the anti-EGFR antibody cetuximab (46).
Xenografts were established from liver metastases of patients with colon cancer resected with curative intent (47) (Table S3). Non-diagnostic portions of removed metastases were implanted, characterized and subsequently passaged in athymic nude mice (Figs. S5A, S5B and S6).
To determine the effects of PP242 in patient-derived xenografts with genetic lesions common in colon cancer, three different patient-derived tumors representing three different combinations of mutant PIK3CA and KRAS were analyzed: WT KRAS and WT PIK3CA (CR 698); Mut KRAS and WT PIK3CA (CR 702); Mut KRAS and Mut PIK3CA (CR 727) (Table S3). Cohorts of single tumor-bearing mice were treated once daily with PP242 or vehicle for 30 days or until (control) tumor burden had reached protocol limits. Treatment was tolerated (Fig. S7).
PP242 slowed tumor growth compared to control (Fig. 5A). In trials with either WT or double mutant tumors (CR 698 and CR 727, respectively), the decrease in tumor growth between treatment and control arms was apparent after seven days. This was in contrast to the more modest effect of PP242 in the KRAS single mutant tumor (CR 702), where the difference in tumor growth was only significant after 28 days. In no trial did PP242 lead to significant tumor regression (>50% in volume) in an individual mouse, but stable disease (final tumor volume of −50% to +20% of starting) was achieved in 26% of mice with CR 698 or CR 727 tumors (and no mice with CR 702 tumors). In PP242 responsive tumors, the growth inhibitory effects were not accompanied by a histological change in tumor characteristics.
Figure 5.
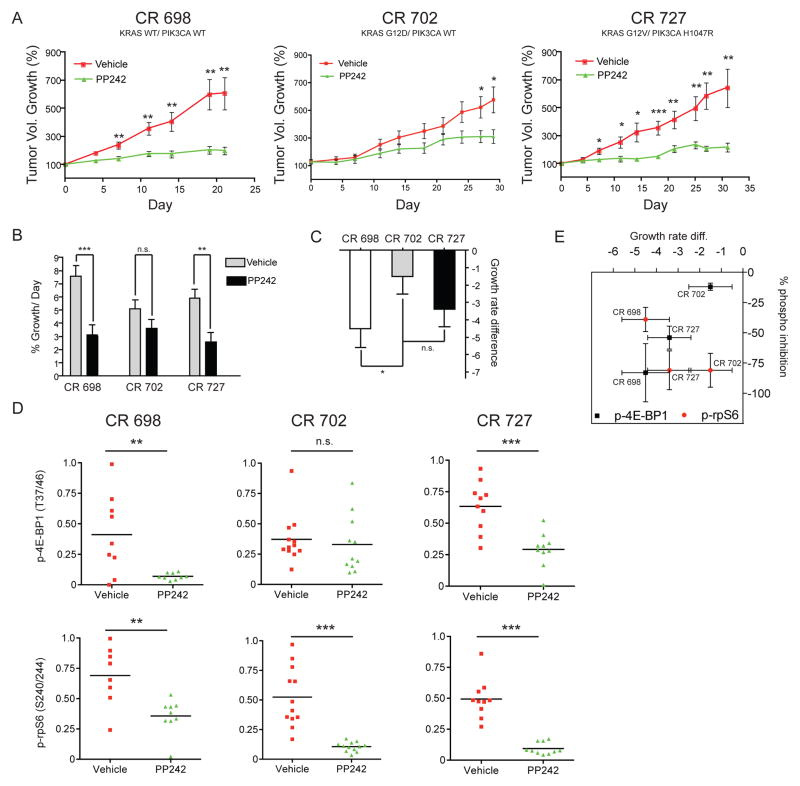
KRAS mutant patient-derived xenografts are resistant to PP242 by incomplete inhibition of 4E-BP1 phosphorylation. (A) Percent growth curves of three xenografts show differences in response to PP242 treatment. KRAS and PIK3CA genotypes are as follows: CR 698 (KRAS WT/PIK3CA WT), CR 702 (KRAS Mut/PIK3CA WT), CR 727 (KRAS Mut/PIK3CA Mut). Mice were given 100 mg/kg PP242 once daily or vehicle for the indicated time. Tumors were normalized to 100 percent at the beginning of dosing and percent growth ±SEM was plotted for each day when tumor volume measurements were taken. Asterisks indicate significant differences in tumor growth at each measurement point as determined by an unpaired t-test (* p< .05, ** p< .01, *** p< .001). (B) Treatment effect is significant in tumors CR 698 and CR 727. Tumor growth rates were calculated using a linear mixed effects model. PP242 led to a significant reduction in growth rate as calculated using a Wald test (asterisks represent the same p values as in A) in the KRAS WT tumor CR 698 and the double mutant tumor CR 727, but not the KRAS single-mutant tumor CR 727. (C) PP242 is most effective at inhibiting growth of the KRAS WT tumor CR 698. Comparison of the growth rate difference calculated from the model shows that PP242 is significantly more effective at inhibiting growth in CR 698 than in CR 702. The growth rate difference is the growth rate of the PP242 treated tumors minus the control growth rate. All other comparisons were not statistically significant. (D) Whole-tumor western blots show that p-4E-BP1 levels were significantly more reduced by PP242 treatment in KRAS WT and KRAS/PIK3CA double mutant tumors but not in KRAS single-mutant tumors. After treatment with either PP242 or vehicle for 30 days, tumors were removed and analyzed by western blot for phosphoprotein analysis. Bands were quantified by fluorescent antibodies and intensities internally normalized to those of β-actin. Intensities are reported as arbitrary normalized fluorescence units. Statistical comparisons were made using two-tailed t-tests as in Figure 3A. (E) Changes in p-4E-BP1 but not p-rpS6 correlate with changes in tumor growth. A plot of the tumor growth rate difference (Figure 5C) versus percent inhibition of p-4E-BP1 and p-rpS6 shows that the efficiency in inhibiting 4E-BP1 phosphorylation correlates linearly with the percent growth defect between treated and untreated tumors. Percent inhibition of p-rpS6 does not vary significantly with genotype or tumor growth defect as calculated from linear mixed effects model.
To directly compare the separate trials and better understand the differences between the resistant KRAS mutant tumor CR 702, and sensitive tumors CR 727 and CR 698, we fit the trial data to a linear mixed effects model. Using the model, we obtained a daily tumor growth rate for each treatment condition and compared the effect of PP242 on tumor growth rate (Fig. 5B). Differences in tumor growth rate between xenografts could not be independently excluded as contributing to the treatment effect, but molecular data strongly suggest that the effects were related to mTOR inhibition. The effect of PP242 treatment on tumor growth was highly significant as determined by a Wald test for both the CR 698 trial (p <0.001) and CR 727 trial (p=0.001) but not for the CR 702 trial (p =0.123). Further comparison of the magnitude of the PP242 treatment effect showed that mTOR inhibition was significantly more effective in the CR 698 trial than the CR 702 trial, (p=0.04) but that the difference in treatment effect was not statistically significant between CR 702 and CR 727 (Fig. 5C).
Inhibition of 4E-BP1 and not rpS6 correlates with anti-tumor effect of PP242
To identify a basis for the differential effect of PP242 on tumor growth, we conducted phospho-signaling analysis by quantitative western blotting (Fig. 5D). Western blots showed an unambiguous inhibition of p-rpS6 in all PP242-treated samples indicating that mTOR was at least partially inhibited in all trials and that PP242 was able to access the tumors equally. The phosphorylation of substrate 4E-BP1 was differentially inhibited among the different trials. p-4E-BP1 was significantly inhibited in the CR 698 and CR 727 trials, but not CR 702. Immunohistochemical staining for p-4E-BP1 was preformed on select tumors, and the results were consistent with the western blotting (Fig. S8). Percent inhibition of p-4E-BP1, not p-rpS6, linearly correlated with tumor growth inhibition (Fig. 5E).
These trials showed a striking primary resistance to PP242 treatment in a KRAS mutant tumor (CR 727) that was not evident in either a tumor with wild type KRAS or a tumor with a PIK3CA mutation in addition to KRAS.
Discussion
The ability to identify determinants of primary resistance to targeted therapies undergoing clinical development has the potential to provide a benefit to patients by guiding patient selection and development of effective biomarkers. We approached this study with the hypothesis that upstream inputs of mTOR that are mutated would affect sensitivity to mTOR inhibitors. Our large cell screen showed that the average sensitivity of different tumors types varied significantly, reflecting the different genetic make-ups of these cancers. Additionally, we were encouraged to find significant positive and negative correlations to PP242 efficacy with mutations in PIK3CA and KRAS, respectively. Newly published data validates our approach showing that PI3KCA H1047R mutations are predictive of response to rapamycin based mTOR inhibitors in patients (48). We chose to examine colon cancer in detail because both mutations implicated in sensitivity and resistance are present and the extreme resistance to mTOR inhibitors in KRAS mutant colon cancer cell lines was unique.
Differences in sensitivity to PP242 among tumors and cell lines were linked to differences in the sensitivity of mTOR substrates to pharmacological inhibition of their phosphorylation. What was unexpected about our findings was the decoupling of inhibition of mTORC1 substrates 4E-BP1 and rpS6 by an ATP-competitive mTOR inhibitor. The phosphorylation status of 4E-BP1 is a better indicator of the functional state of mTOR than S6K and closely correlates with the growth arrest caused by mTOR inhibition. In the single mutant KRAS patient-derived xenograft and cell lines, the inhibition of mTORC1 substrates by PP242 is similar to that of rapamycin, which is now well established to block p-rpS6 but not p-4E-BP1 by a still unknown mechanism. Our observation that in certain cell lines, an ATP-competitive inhibitor could produce these rapamycin like differential inhibitory effects, suggests that a common mechanism may underlie these phenomena.
To test the hypothesis that activated PIK3CA signaling conferred sensitivity to mTOR inhibition, even in a mutant KRAS background, isogenic cell lines were created that differed only by their PIK3CA mutation status. Increased sensitivity to PP242 was only observed in the cell line containing an AKT activating PIK3CA mutation. This result adds to a significant set of data that shows that upstream AKT mutations sensitize tumors to mTOR inhibition. The resistance driven by mutant KRAS is not reversible by short-term MAPK inhibition, leading us to conclude that phosphorylation changes regulating mTORC1 do not affect substrate accessibility. Our observation that resistance is correlated with RAS-GTP loading functionally differentiates cell lines with KRAS mutations, and suggests that the mechanism of resistance is directly KRAS driven but independent of MAPK signaling.
Concurrent to our work on primary resistance to ATP-competitive mTOR inhibitors, other groups have recently reported mechanisms of acquired resistance to these agents. In a study of human mammary epithelial cells treated at sublethal doses of the dual PI3K/mTOR inhibitor BEZ235, Roberts and coworkers identified MYC amplification in one cell line and eIF4E amplification in another cell line (49). Sonenberg and coworkers characterized PP242 resistant E1A/Ras-clones generated by growing the cells for two months in drug (50). The emergent clones exhibited down regulation of 4E-BP1 and 2 or overexpressed eIF4E, highlighting the importance of monitoring the ratio of 4E-BPs and eIF4E in tumors to assess likely responses to ATP-competitive mTOR inhibitors. These two studies on emergent resistance further highlight the importance of translational regulation in response to mTOR inhibitors.
Owing to the lack of direct inhibitors of KRAS, considerable effort has been made to uncover druggable targets (such as kinases TBK1 and STK33) that might act in a synthetic lethal manner with KRAS mutant tumors (51). Our study reveals a different approach to the same goal, that of finding a small molecule that is effective even in a setting of KRAS mutant tumors. We find that the presence of a second oncogene, PI3KCA, provides a signal that sensitizes the KRAS mutant tumors to an ATP-competitive mTOR inhibitor. This result is surprising because the presence of a second lesion typically provides the cancer with a bypass mechanism to avoid kinase inhibitor sensitivity (52). Of note is that 3 of the 4 RAS mutant cell lines that were among the 10% most sensitive to PP242 also harbored a PIK3CA mutation.
In the context of colon cancer, a solid tumor in which small molecule kinase inhibitors have yet to achieve significant clinical utility, our findings suggest subsets of patients who may benefit from targeted anti-mTOR therapy. First, mutations in PIK3CA are likely to be sensitive to mTOR inhibition. The importance of selectively treating mutant PI3KCA colon cancer patients has been highlighted by a retrospective analysis demonstrating that low dose aspirin (by an as yet unclear mechanism) can dramatically prolong survival in those patients (53). Additionally, patients with WT KRAS are likely to be responsive to mTOR inhibitors. Furthermore, we believe that among colon cancer patients with mutant KRAS, those with concomitant hyperphosphorylation of AKT induced by mutant PIK3CA may benefit from ATP-competitive mTOR inhibitors. Finally, although rpS6 remains the default biomarker for mTOR inhibitors, our study shows that the phosphorylation status of 4E-BP1 may be a more relevant biomarker of treatment efficacy for ATP-competitive inhibitors.
Materials and Methods
Inhibitors
The mTOR inhibitors PP242 and MLN0128 were synthesized from commercially available starting materials as previously reported (16,25). Rapamycin and PD0325901 were purchased from EMD-Millipore chemicals (Billerica, MA). FMK-MEA was a gift of Jack Taunton (UC San Francisco, San Francisco, CA).
Cell Screen
The automated cell screen was performed using PP242 (500 nM) as previously described (24). Complete results from the PP242 screen are available as a supplementary file (Data file S1).
Cell culture
Cell lines were purchased from the American Type Tissue Collection (ATCC, Manassas, VA) and cultured according to their recommendations in antibiotic-free media.
Cell proliferation assay
Cells were plated on 96-well plates at densities between 2500 and 5000 cells/well. Cell growth was assayed using resazurin sodium salt (Sigma, Saint Louis, MO) and measured using a Safire bottom-reading fluorescent plate reader with excitation at 530 nm and emission at 590 nm.
Western blot analysis
Cells were grown in 6 or 12-well plates and treated with inhibitor(s) or vehicle (0.1% or 0.2% DMSO for single or combination drug assays, respectively) for 1 h unless otherwise noted. Cells were then lysed in radio-immunoprecipitation assay buffer (RIPA); lysates were normalized for protein content using a Bradford assay (absorbance at 595 nm), resolved by SDS-PAGE, transferred to nitrocellulose and blotted. Phosphorylation-specific antibodies were purchased from Cell Signaling Technology (Danvers, MA), except for phospho-raptor S722 (Millipore) and visualized by HRP or fluorescent secondary antibodies. Quantitative western blotting was accomplished using fluorescent secondary antibodies (800 nm emission) for visualization using an Odyssey IR scanner from LI-COR Biosciences (Lincoln, NE). All reported band intensities were internally normalized to β-Actin and each experiment was done in independent biological duplicates.
Immunofluorescence
Cells were plated on fibronectin treated glass-bottom 12-well plates, drug treated and then fixed and stained following standard protocols (Cell Signaling Technology). An Alexa Fluor 488 conjugated goat anti-rabbit secondary antibody (Life Technologies, Carlsbad, CA) was used to visualize the cells using a Zeiss Axiovert 200M fluorescence microscope. Nuclei were counterstained with Hoechst 33342 dye (Pierce). Image analysis was conducted with the software suite MetaMorph.
GST-RBD Pull Down
Assay was adapted from a protocol published with the Pierce active RAS pull down and detection kit (#16117). Cells were lysed in HEPES lysis buffer (40 mM HEPES pH 7.4, 150 mM NaCl, 0.1% Tx-100). GST Pull down was conducted according to protocol using purified GST-RBD-c-Raf-1 and glutathione beads (GE Healthcare, Pittsburgh, PA). Pan RAS antibody was from Epitomics (Burlingame, CA).
siRNA
Pooled siRNA against PTEN (SMARTpool PTEN) or control scramble siRNA was purchased from Millipore. siRNA was transfected using DharmaFECT 2 reagent according to manufacture’s instructions (Thermo Scientific Dharmacon Products, Lafayette, CO). 96 h after transfection, cells were treated with drugs and western blotted.
Mutant PIK3CA SW620 cell lines
Mutant p110α expressing SW620 lines were created by retroviral infection using a pMIG-p110α plasmid as previously reported (54).
Patient-derived xenografts
The research protocol was approved by the Committee on Human Research of the University of California, San Francisco (UCSF) and patient consent was obtained. All animal studies followed a protocol approved by the UCSF Animal Care and Use Committee.
To establish xenografts, excess tumor removed during hepatic resection was minced under sterile conditions to generate pieces approximately 4–8 mm3. These were dipped into sterile matrigel (BD Biosciences, Sparks, MD) and implanted subcutaneously (SC) onto the flanks of female athymic mice (FOXN1 nude, Harlan, Indianapolis, IN). Non-implanted pieces were flash-frozen in liquid nitrogen and banked. For each tumor, 2–4 pieces were implanted into 2–3 mice to establish the initial xenograft passage. When tumor volume reached ~1,000 mm3, mice were sacrificed; tumors were divided and implanted SC into new animals.
Drug treatment
PP242 was prepared as a 25 mg/mL suspension in 3.1% NMP, 81.6% PVP, 15.3% H2O. 100 μL of PP242 suspension (2.5 mg/dose equal to 100mg/kg) or vehicle alone was given orally once daily. For drug efficacy trials, mice bearing single tumors were treated for 30 days or when tumors reached 3000 mm3 and then sacrificed and tumors were harvested.
Histologic analysis of xenograft tumors
4 μm sections prepared from FFPE tissue were stained with hematoxylin & eosin (H & E) and submitted for review by the pathologist, J.P.S.
Sequencing
DNA extracted from surgical specimens with the Qiagen tissue kit (Qiagen, Valencia, CA) was sequenced using standard Sequenom platform protocols (Sequenom, San Dieg, CA) and colon cancer-specific mutational panel (ColoCarta) (55). iPLEX well 6, containing KRAS-Q61L and HRAS-Q61L primers was omitted.
Statistical analyses
Statistical tests and curve fitting was conducted using the programs Stata and Prism. Univariate and multivariate regressions were conducted using the default analysis models in Stata. Comparison of means was conducted using two-tailed Student’s t tests. Data from drug treatment trials was fit to a linear mixed effects model as previously described (56) using Stata. Wald tests determined whether the slopes differed for different treatments.
Supplementary Material
Acknowledgments
Financial Support: CEA is supported in part by American Cancer Society Postdoctoral Fellowship 11-183-TBG; she also acknowledges support Millennium Pharmaceuticals provided through the Alliance for Clinical Trials in Oncology Foundation. R.S.W is supported in part by a charitable donation from the Littlefield 2000 Trust. K.M.S. is supported by the Howard Hughes Medical Institute and the Waxman Foundation.
We would like to thank Dan Moore for his critical assistance in the statistical analysis, Sandy Devries for FISH analysis, Katherine Pogue-Guile for permission to use the ColoCarta oligonucleotide primer design files and Morris Feldman, Jon Ostrem and Iana Serafimova for reagents. C.E.A. is supported in part by American Cancer Society Postdoctoral Fellowship 11-183-TBG; she also acknowledges support Millennium Pharmaceuticals provided through the Alliance for Clinical Trials in Oncology Foundation. R.S.W is supported in part by a charitable donation from the Littlefield 2000 Trust. K.M.S. is supported by the Howard Hughes Medical Institute and the Waxman Foundation.
Footnotes
Conflicts of Interest: Kevan Shokat is an inventor on patents from UCSF relating to PP242 and MLN0128 licensed to Millennium Pharmaceuticals and is a member of the Millennium Pharmaceuticals Scientific Advisory Board.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Kantarjian H, Sawyers C, Hochhaus A, Guilhot F, Schiffer C, Gambacorti-Passerini C, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002 Feb 28;346(9):645–52. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- 2.Kwak EL, Bang Y-J, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010 Oct 28;363(18):1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011 Jun 30;364(26):2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jänne PA, Gray N, Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat Rev Drug Discov. 2009 Mar 8;8(9):709–23. doi: 10.1038/nrd2871. [DOI] [PubMed] [Google Scholar]
- 5.Joseph EW, Pratilas CA, Poulikakos PI, Tadi M, Wang W, Taylor BS, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci USA. 2010 Aug 17;107(33):14903–8. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laplante M, Sabatini DM. Cell. 2. Vol. 149. Elsevier Inc; 2012. Apr 13, mTOR Signaling in Growth Control and Disease; pp. 274–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarbassov DD, Ali SM, Kim D-H, Guertin DA, Latek RR, Erdjument-Bromage H, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004 Jul 27;14(14):1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 8.Hara KK, Maruki YY, Long XX, Yoshino K-IK, Oshiro NN, Hidayat SS, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002 Jul 26;110(2):177–89. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 9.Loewith RR, Jacinto EE, Wullschleger SS, Lorberg AA, Crespo JLJ, Bonenfant DD, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002 Sep 1;10(3):457–68. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 10.Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2008 Dec 1;27(Suppl 2):S43–51. doi: 10.1038/onc.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T, Nakashima A, Guo L, Coffman K, Tamanoi F. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. 2010 May 6;29(18):2746–52. doi: 10.1038/onc.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006 May 25;441(7092):424–30. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 13.Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome Sequencing Identifies a Basis for Everolimus Sensitivity. Science. 2012 Aug 23; doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008 Aug 9;372(9637):449–56. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 15.Hudes G, Carducci M, Tomczak P, Dutcher J, Figlin R, Kapoor A, et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med. 2007 May 31;356(22):2271–81. doi: 10.1056/NEJMoa066838. [DOI] [PubMed] [Google Scholar]
- 16.Apsel B, Blair JA, Gonzalez B, Nazif TM, Feldman ME, Aizenstein B, et al. Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat Chem Biol. 2008 Oct 12;4(11):691–9. doi: 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, et al. Active-Site Inhibitors of mTOR Target Rapamycin-Resistant Outputs of mTORC1 and mTORC2. Plos Biol. 2009 Feb 1;7(2):e1000038–8. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thoreen C, Kang S, Chang J, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mTOR inhibitor reveals rapamycin-insensitive functions of mTORC1. J Biol Chem. 2009 Jan 15; doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García Martínez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, et al. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009 Jun 12;421(1):29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y-J, Duan Y, Zheng XFS. Targeting the mTOR kinase domain: the second generation of mTOR inhibitors. Drug Discovery Today. 2011 Apr 1;16(7–8):325–31. doi: 10.1016/j.drudis.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dowling RJOR, Topisirovic II, Alain TT, Bidinosti MM, Fonseca BDB, Petroulakis EE, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010 May 28;328(5982):1172–6. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markowitz SD, Bertagnolli MM. Molecular Basis of Colorectal Cancer. N Engl J Med. 2009 Dec 17;361(25):2449–60. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lièvre A, Bachet J-B, Boige V, Cayre A, Le Corre D, Buc E, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008 Jan 20;26(3):374–9. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 24.McDermott U, Sharma SV, Dowell L, Greninger P, Montagut C, Lamb J, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci USA. 2007 Dec 11;104(50):19936–41. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012 Feb 22;:1–10. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Reilly KE, Rojo F, She Q-B, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006 Feb 1;66(3):1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008 Jan 22;5(1):e8. doi: 10.1371/journal.pmed.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blaser B, Waselle L, Dormond-Meuwly A, Dufour M, Roulin D, Demartines N, et al. Antitumor activities of ATP-competitive inhibitors of mTOR in colon cancer cells. BMC Cancer. 2012;12:86. doi: 10.1186/1471-2407-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atreya CE, Ducker GS, Feldman ME, Bergsland EK, Warren RS, Shokat KM. Combination of ATP-competitive mammalian target of rapamycin inhibitors with standard chemotherapy for colorectal cancer. Invest New Drugs. 2012 Jan 24; doi: 10.1007/s10637-012-9793-y. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, et al. Genetic Dissection of the Oncogenic mTOR Pathway Reveals Druggable Addiction to Translational Control via 4EBP-eIF4E. Cancer Cell. 2010 Mar 16;17(3):249–61. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rong L, Livingstone M, Sukarieh R, Petroulakis E, Gingras A-C, Crosby K, et al. Control of eIF4E cellular localization by eIF4E-binding proteins, 4E-BPs. RNA. 2008 May 29;14(7):1318–27. doi: 10.1261/rna.950608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrière A, Cargnello M, Julien L-A, Gao H, Bonneil E, Thibault P, et al. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol. 2008 Sep 9;18(17):1269–77. doi: 10.1016/j.cub.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 33.Barrett SD, Bridges AJ, Dudley DT, Saltiel AR, Fergus JH, Flamme CM, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg Med Chem Lett. 2008 Dec 15;18(24):6501–4. doi: 10.1016/j.bmcl.2008.10.054. [DOI] [PubMed] [Google Scholar]
- 34.Le N-T, Takei Y, Shishido T, Woo C-H, Chang E, Heo K-S, et al. p90RSK Targets the ERK5-CHIP Ubiquitin E3 Ligase Activity in Diabetic Hearts and Promotes Cardiac Apoptosis and Dysfunction. Circ Res. 2012 Feb 17;110(4):536–50. doi: 10.1161/CIRCRESAHA.111.254730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen MS, Zhang C, Shokat KM, Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005 May 27;308(5726):1318–21. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.She Q-B, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010 Jul 13;18(1):39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor SJ, Resnick RJ, Shalloway D. Nonradioactive determination of Ras-GTP levels using activated ras interaction assay. Meth Enzymol. 2001;333:333–42. doi: 10.1016/s0076-6879(01)33067-7. [DOI] [PubMed] [Google Scholar]
- 38.Bohanes P, LaBonte MJ, Winder T, Lenz H-J. Predictive molecular classifiers in colorectal cancer. Semin Oncol. 2011 Aug;38(4):576–87. doi: 10.1053/j.seminoncol.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 39.TCGA Network. Nature. 7407. Vol. 487. Nature Publishing Group; 2012. Jul 10, Comprehensive molecular characterization of human colon and rectal cancer; pp. 330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weigelt B, Warne PH, Downward J. PIK3CA mutation, but not PTEN loss of function, determines the sensitivity of breast cancer cells to mTOR inhibitory drugs. Oncogene. 2011 Jul 21;30(29):3222–33. doi: 10.1038/onc.2011.42. [DOI] [PubMed] [Google Scholar]
- 41.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci USA. 2008 Feb 19;105(7):2652–7. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samuels Y, Diaz LA, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005 Jun 1;7(6):561–73. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009 Apr 15;69(8):3364–73. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talmadge JE, Singh RK, Fidler IJ, Raz A. Murine models to evaluate novel and conventional therapeutic strategies for cancer. Am J Pathol. 2007 Mar 1;170(3):793–804. doi: 10.2353/ajpath.2007.060929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res. 2003 Sep 15;9(11):4227–39. [PubMed] [Google Scholar]
- 46.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, et al. A Molecularly Annotated Platform of Patient-Derived Xenografts (“Xenopatients”) Identifies HER2 as an Effective Therapeutic Target in Cetuximab-Resistant Colorectal Cancer. Cancer Discovery. 2011 Nov 1;1(6):508–23. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 47.House MG, Ito H, Gönen M, Fong Y, Allen PJ, DeMatteo RP, et al. Survival after hepatic resection for metastatic colorectal cancer: trends in outcomes for 1,600 patients during two decades at a single institution. J Am Coll Surg. 2010 May;210(5):744–52. 752–5. doi: 10.1016/j.jamcollsurg.2009.12.040. [DOI] [PubMed] [Google Scholar]
- 48.Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek V, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early phase clinical trials. Cancer Res. 2012 Oct 12; doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci USA. 2011 Aug 29; doi: 10.1073/pnas.1108237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, et al. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res. 2012 Oct 24; doi: 10.1158/0008-5472.CAN-12-2395. [DOI] [PubMed] [Google Scholar]
- 51.Brough R, Frankum JR, Costa-Cabral S, Lord CJ, Ashworth A. Searching for synthetic lethality in cancer. Curr Opin Genet Dev. 2011 Feb 1;21(1):34–41. doi: 10.1016/j.gde.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 52.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010 Feb 1;10(2):130–7. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, et al. Aspirin Use, Tumor PIK3CAMutation, and Colorectal-Cancer Survival. N Engl J Med. 2012 Oct 25;367(17):1596–606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zunder ER, Knight ZA, Houseman BT, Apsel B, Shokat KM. Discovery of drug-resistant and drug-sensitizing mutations in the oncogenic PI3K isoform p110 alpha. Cancer Cell. 2008 Aug 12;14(2):180–92. doi: 10.1016/j.ccr.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fumagalli D, Gavin PG, Taniyama Y, Kim S-I, Choi H-J, Paik S, et al. A rapid, sensitive, reproducible and cost-effective method for mutation profiling of colon cancer and metastatic lymph nodes. BMC Cancer. 2010 Mar 16;10(1):101–1. doi: 10.1186/1471-2407-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinheiro JC, Bates DM. Mixed Effects Models in S and S-Plus. Springer; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.