Abstract
Background
The c-Myc oncoprotein is activated in the majority of colorectal cancers (CRCs), whereas the TGF-β pathway is frequently affected by loss-of-function mutations, for example in SMAD2/3/4 genes. The canonical model places Myc downstream of inhibitory TGF-β signaling. However, we previously demonstrated that Myc also inhibits TGF-β signaling through the miR-17~92 microRNA cluster, raising the question about functional relationships between these two pathways.
Methods
We engineered a series of genetically complex murine and human CRC cell lines in which Myc and TGF-β activities could be manipulated simultaneously. This was achieved through retroviral expression of the Myc–estrogen receptor fusion protein and through Smad4 short hairpin RNA knockdown. Cell lines thus modified were injected subcutaneously in immunocompromised mice, and the resultant tumors (n = 5–10 per treatment group) were analyzed for overall growth and neovascularization. Additionally, the distribution of MYC and TGF-β pathway mutations was analyzed in previously profiled human CRC samples.
Results
In kras-mutated/trp53-deleted murine colonocytes, either Myc activation or TGF-β inactivation increased tumor sizes and microvascular densities approximately 1.5- to 2.5-fold, chiefly through downregulation of thrombospondin-1 and related type I repeat–containing proteins. Combining Myc activation with TGF-β inactivation did not further accelerate tumorigenesis. This redundancy and the negative effect of TGF-β signaling on angiogenesis were also demonstrated using xenografts of human CRC cell lines. Furthermore, the analysis of the Cancer Genome Atlas data revealed that in CRC without microsatellite instability, overexpression of Myc and inactivation of Smads (including acquired mutations in SMAD2) are mutually exclusive, with odds ratio less than 0.1.
Conclusions
In human CRC, gain-of-function alterations in Myc and loss-of-function alterations in TGF-β exhibit a masking epistatic interaction and are functionally redundant.
The c-Myc oncoprotein is central to the pathogenesis of colorectal cancers (CRCs), in which it is frequently overexpressed because of activating mutations in the WNT pathway or, less commonly, through MYC gene amplification (1). To determine how MYC functions in genetically complex CRC, we had previously established a novel syngeneic mouse model based on sequential transformation of p53-null primary colonocytes with mutant Ki-Ras and Myc. In this model, Myc was dispensable for proliferation but conferred upon neoplastic cells a hypervascular phenotype (2). This occurred through downregulation of thrombospondin-1 (Tsp-1) and related antiangiogenic factors of the type I repeat (TSR) superfamily, such as CTGF (3). Of note, their downregulation was not simply promoter based but required upregulation of the miR-17~92 microRNA cluster by Myc (2). Conversely, many antiangiogenic TSR proteins are induced by TGF-β (4–6). This implied that TGF-β signaling could be antiangiogenic in the context of CRC and that Myc needs to counteract these antiangiogenic effects during tumorigenesis.
To test this bipartite hypothesis, we transduced a variety of CRC cell lines with retro- and lentiviruses encoding Myc, its tamoxifen-inducible variant (MycERTAM), and short hairpin RNAs (shRNAs) targeting Smad4, the essential component of TGF-β signaling. We discovered that either Myc activation or TGF-β inactivation, but not both, is needed to promote angiogenesis and tumor growth. This redundancy was further corroborated by Cancer Genome Atlas data showing a surprising mutual exclusivity of gain-of-function mutations in MYC and loss-of-function mutations in various SMAD genes.
Methods
Cell Lines and Treatments In Vitro
RasMycER colonocytes were generated by retroviral infection of p53-null, Ki-Ras–transformed mouse colonocytes using the MigR1-MycER retrovirus and sorted for green fluorescent protein. Propagation and treatment of these and other cell lines are described in the Supplementary Methods (available online).
Virus Infection, shRNA Knockdown, and Rescue
RasMycER cells were infected with the PLKO.1 lentiviruses expressing variously effective shRNAs against mouse Smad4. SH-cont (with a nontargeting hairpin) and SH-Smad4 cell lines were generated. SW837 cells were similarly infected with the PLKO.1 lentiviruses expressing shRNA against human Myc, human Smad4, or firefly luciferase. See the Supplementary Methods (available online) for further details.
Tumor Xenograft Studies
To generate murine carcinomas, 1–2×106 RasMycER colonocytes, HCT116 cells, or SW837 cells resuspended in 100 µL of phosphate-buffered saline were injected subcutaneously into the flanks of NSG (NOD.Cg-Prkdc scid Il2rg tm1Wjl/SzJ) mice aged 8 to 12 weeks. Mice of either sex from the in-house breeding facility were used. When indicated, 1mg of tamoxifen (Sigma, St. Louis, MO) resuspended in corn oil was administered by daily intraperitoneal injection. At least five mice were used per group in all animal studies (see the Supplementary Methods, available online). All animal protocols were approved by the Children’s Hospital of Philadelphia Animal Care and Use Committee (protocol 2012-12-902).
Immunohistochemistry and Vessel Quantitation
Anti-CD34 (Abcam, Cambridge, MA; Ab8158 and Ab81289) staining was performed on paraffin-embedded tumor sections using standard procedures. Microvessel density analysis was performed either using Aperio Positive Pixel Count Algorithm (version 9.1; Leica, Buffalo Grove, IL) or manually by two separate investigators on digitally scanned slide images (see the Supplementary Methods, available online).
Quantitative Real-Time Polymerase Chain Reaction Analysis
RNA extraction, cDNA synthesis, and Sybr green quantitative real-time polymerase chain reaction (qPCR) for CTGF or Taqman qPCR for microRNAs was performed as described in the Supplementary Materials (available online) and previously (5).
Immunoblotting
Cultured cells were lysed in plates, and frozen tumors were crushed to a fine powder then extracted using radioimmunoprecipitation assay (RIPA) buffer. Western blotting was carried out as described in the Supplementary Methods (available online) and previously (2,4).
CRC Patient Studies
Primary data for this analysis have been published previously (1) and are stored in Cancer Genomics Hub (https://cghub.ucsc.edu/, last accessed on 02/26/2014). Mutations, putative copy-number alterations from GISTIC, and/or mRNA expression data (RNASEQ; z score = 2) of complete nonhypermutated patient samples (n = 165) were analyzed using cBioPortal for Cancer Genomics platform (http://www.cbioportal.org/public-portal/, last accessed on 02/26/2014) (7,8). OncoPrints, copy number variation-RNA sequencing (CNV-RNASEQ) correlation plots, and mutual exclusivity odds ratio (ORs) were reported out. Access to Cancer Genome Atlas datasets was approved by the Responsible Data Access Committee of National Center for Biotechnology Information (NCBI) database of Genotypes and Phenotypes (dbGAP) (request 15694–5).
Statistics
Most statistical analyses were performed using one-way analysis of variance (ANOVA) or Tukey’s test. Two-way ANOVA was also performed to determine the interactions between MYC expression and TGF-β signaling. Type III two-way ANOVA was used to analyze experiments with different sample sizes. All statistical tests were two-sided when applicable, and a P value of less than .05 was considered statistically significant.
Results
Myc, Tsp-1, and Angiogenesis in the Mouse Model of CRC
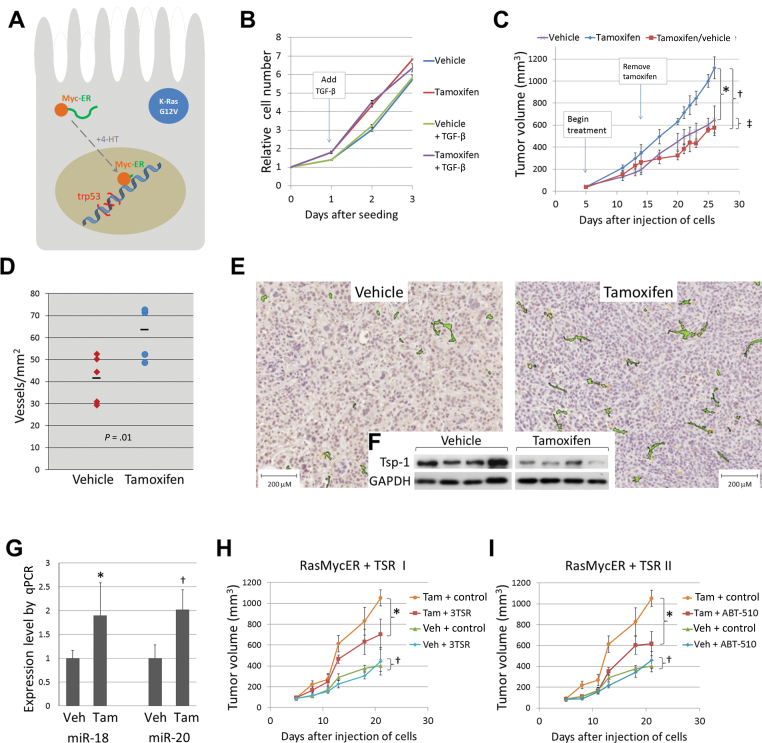
To determine whether transient activation of Myc enhances growth of Ki-Ras–transformed/p53-null mouse colonocytes, we engineered these cells to express the MycERTAM transgene along with green fluorescent protein (Figure 1A). Although activation of Myc caused cells to grow in clumps with less adhesion to the substratum, this difference was suppressed upon treatment with TGF-β (Supplementary Figure 1A, available online). Furthermore, Myc activation did not affect cell accumulation in vitro, either in the presence or in the absence of TGF-β (Figure 1B). RasMycER cells were then injected subcutaneously into the flanks of immunocompromised NSG mice. When tumors became palpable, animals were randomized, with one group receiving vehicle alone, whereas two others received tamoxifen. After 9 days, tamoxifen was discontinued in one of these latter groups. At 3 weeks, all animals were euthanized, and the tumors were weighed and processed for histology. Activation of Myc resulted in a statistically significant enhancement of tumor growth (1.7-fold difference on day 25; P < .001) (Figure 1C, blue vs purple lines). Moreover, Myc hyperactivity was continuously required because tumor growth immediately slowed in mice that stopped receiving tamoxifen (P = .27) (Figure 1C, red vs purple lines; see replicate experiment in Supplementary Figure 1B, available online).
Figure 1.
Effects of transient Myc activation and thrombospondin-1 (Tsp-1) repression on tumor growth. A) Schematic depiction of RasMycER cell harboring activated KRAS (with the G12V mutation) and null for p53. When tamoxifen (4-HT) is added, conformation changes allow the MycER fusion protein to enter the nucleus and regulate transcription. B) Growth of RasMycER cells seeded in 96-well plates in 4-HT or vehicle alone. Where indicated, 0.5ng/mL of TGF-β1 or vehicle was added to media on day 1. Cell accumulation was quantitated using the WST-1 assay and represented by line plots for each condition. The experiment was repeated twice using duplicate wells for each condition. C) Subcutaneous growth of RasMycER tumors in mice treated with vehicle alone, tamoxifen, or tamoxifen then vehicle alone. Statistical significance per one-way analysis of variance: *P < .001; †P < .001; ‡P = .27. Error bars represent standard deviation. D) Microvessel density of RasMycER tumors from mice treated with vehicle alone or tamoxifen in a representative experiment (n = 5 in each treatment group). Dashes represent the average for each group. Statistical significance per one-way analysis of variance: P = 0.01. E) Representative sections of tumors shown in D costained with anti-CD34 antibody and hematoxylin. Positive staining colorized using Aperio ImageScope software. F) Tsp-1 levels in lysates from tumors shown in E. GAPDH serves as loading control for immunoblotting. G) Average relative expression of miR-18 and miR-20 in tumors shown in D as determined by Taqman quantitative real-time polymerase chain reaction (qPCR; n=5, experiment repeated twice). Statistical significance per one-way analysis of variance:*P = .01; †P = .05. Error bars represent standard deviation. H) Subcutaneous growth of RasMycER tumors in mice given daily intraperitoneal injections of vehicle alone (Veh) or tamoxifen (Tam), each combined with either vehicle for TSRs (control) or the Tsp-1 mimetic 3TSR. Statistical significance per one-way analysis of variance:*P = .002; †P = .54. Error bars represent standard deviation. I) Subcutaneous growth of RasMycER tumors in mice given daily intraperitoneal injections of vehicle alone (Veh) or tamoxifen (Tam), each combined with either vehicle for TSRs (control) or the Tsp-1 mimetic ABT-510. Statistical significance per one-way analysis of variance:*P < .001; †P = .22. Error bars represent standard deviation.
To examine whether changes in the tumor microenvironment are responsible for this enhanced growth, we stained sections with an anti-CD34 antibody to identify vascular endothelial cells. Quantitative analysis of vessels within tumors revealed markedly increased microvessel density in tumors with transient Myc activation (1.6-fold difference; P = .01) (Figure 1, D and E). Furthermore, immunoblotting analysis of tumor cell lysates demonstrated consistent downmodulation of Tsp-1 in tumors with Myc activation (Figure 1F). In agreement with our previous findings (2,4), miR-18 and miR-20, representative members of the Tsp-1–targeting miR17~92 microRNA cluster, were markedly elevated in tumors with MycER activation (P = .01 for miR-18; P = .05 for miR-20) (Figure 1G), where they could contribute to the observed reduction in Tsp-1.
To test whether Tsp-1 is causally involved in Myc-dependent tumor growth, we replenished the tumor microenvironment with Tsp-1–based peptides. Mice with palpable subcutaneous RasMycER tumors were treated by daily intraperitoneal injections of the Tsp-1 peptide mimetic 3TSR (9) in the presence and absence of tamoxifen-activated Myc. When animals with MycOFFTSPHIGH neoplasms were treated with 3TSR, there was no effect on tumor volume (P = .54) (Figure 1H, compare blue and green symbols). In contrast, replenishment of Tsp-1 in the microenvironment of MycONTSPLOW tumors had a strong inhibitory effect (compare yellow and red symbols, 1.5-fold difference; P = .002). In fact, it largely cancelled the selective advantage afforded by Myc activation. To corroborate these finding, we used another TSR mimetic, ABT-510 (10). The ABT-510 results obtained with both MycOFFTSPHIGH (P = .22) and MycONTSPLOW (P < .001) tumors were essentially identical to the 3TSR results (1.7-fold difference in Tam-treated mice) (Figure 1I).
Myc and TGF-β Effects in Genetically Complex Murine CRC Cells
Although Tsp-1 was easily detected by immunoblotting in RasMycER tumor lysates, it was not detectable in cultured cells unless they were treated with TGF-β1. However, upon activation of Myc with tamoxifen, TGF-β1 induction of Tsp-1 was almost completely blocked (Figure 2A). We then asked what effect blocking TGF-β signaling would have in the absence or presence of activated Myc.
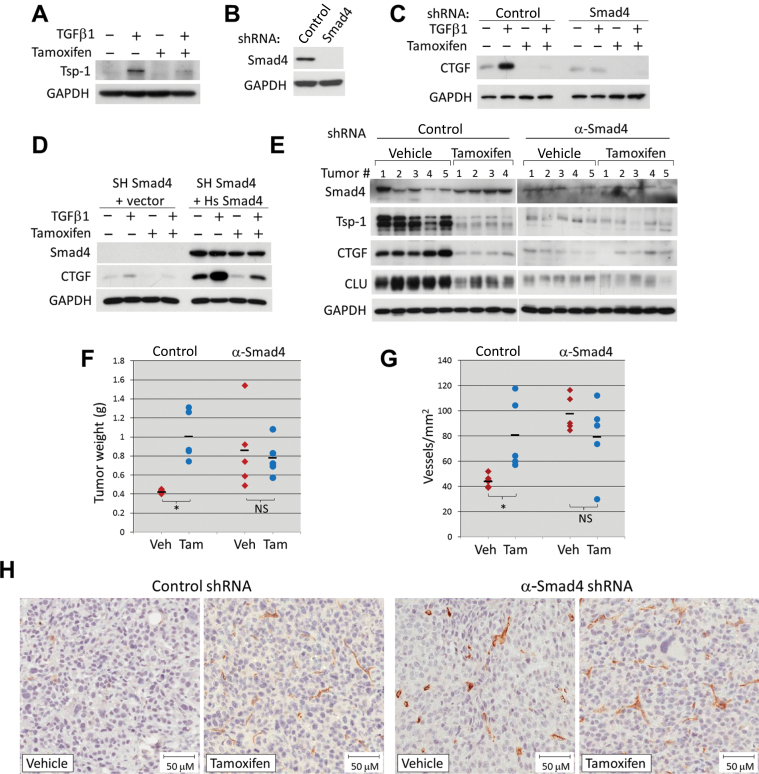
Figure 2.
Effects of Myc and TGF-β on TSR expression in colorectal cancer cells. All proteins were quantified by immunoblotting. Each experiment was repeated at least two times. A) Thrombospondin-1 (Tsp-1) in cultured RasMycER cells pretreated for 24 hours with vehicle (95% ethanol) alone or 250nM 4-hydroxytamoxifen, then with either vehicle or 5ng/mL TGF-β1 for 24 hours. B) Smad4 protein in cultured RasMycER cells stably expressing either a nontargeting short hairpin RNA (shRNA; SH Control) or shRNA against mouse Smad4 (SH Smad4). C) CTGF in cultured RasMycER SH control or SH Smad4 cells each treated as in A. D) Smad4 and CTGF levels in cultured RasMycER SH Smad4 cells infected with empty vector or with human Smad4, each treated as in A. E) TSR proteins in lysates from RasMycER SH control or RasMycER SH Smad4 tumors treated with vehicle alone or tamoxifen in a representative experiment. F) Weights of tumors shown in E. Dashes represent average for each group (n = 5). Pairwise statistical significance per Tukey’s test (in conjunction with two-way analysis of variance):*P = .003; non-statistically significant (NS): P = .65. G) Microvessel density in RasMycER SH control or RasMycER SH Smad4 tumors treated with vehicle (Veh) alone or tamoxifen (Tam). Dashes represent average for each group (n = 5). Statistical significance per Tukey’s test (in conjunction with two-way analysis of variance):*P = .02; non-statistically significant (NS): P = .21; H) Representative sections from tumors shown in G stained with anti-CD34 antibody and hematoxylin.
To this end, we infected RasMycER colonocytes with several lentiviruses expressing shRNAs against mouse Smad4 and obtained stable cell populations through puromycin selection. Among the constructs, one shRNA reduced Smad4 levels very effectively, whereas another, chosen as a negative control, did not (Figure 2B). Treatment of these two cell lines in vitro with TGF-β1 showed a profound dampening of TGF-β1 induction of CTGF by the Smad4-targeting but not the control shRNA. Furthermore, activation of MycER inhibited CTGF induction in shRNA control cells but had little effect in cells with Smad4 shRNA, where induction was already abolished (Figure 2C). To rule out nonspecific effects of the anti-Smad4 hairpin, we rescued Smad4 expression using the human ortholog, which does not contain the short hairpin target sequence. Transduction with this rescue construct increased basal CTGF levels and also restored robust induction of CTGF by TGF-β (Figure 2D). Again, the induction of CTGF was strongly blunted in the presence of Myc (Figure 2D, two rightmost lanes)
Next, RasMycER cells expressing control and α-Smad4 shRNA were used to generate tumors in either tamoxifen- or vehicle-treated mice. Activation of MycER in tumors expressing the control shRNA resulted in markedly reduced levels of all TSR family proteins assayed, including Tsp-1, CTGF, and clusterin, whereas Smad4 levels were maintained (Figure 2E, left panel). In contrast, in Smad4-deficient tumors, the protein levels of Tsp-1, CTGF, and clusterin were greatly reduced even in the absence of tamoxifen because of inactive TGF-β signaling (Figure 2E, right panel). Notably, low TSR protein levels tracked with large tumor sizes (SH-cont vehicle vs tamoxifen: 2.3-fold difference, P = .003; SH-Smad4 vehicle vs tamoxifen: 1.2-fold difference, P = .65) (Figure 2F; see replicate experiment in Supplementary Figure 1C, available online). To determine whether robust vascularity also tracked with low TSR protein levels, we stained tumor sections with the anti-CD34 antibody. Regardless of whether the reduced TSR expression was due to MycER activation or Smad4 knockdown, resultant tumors had markedly higher microvessel densities than tumors with robust TSR expression (SH-cont vehicle vs tamoxifen: 1.8-fold difference, P = .02; SH-Smad4 vehicle vs tamoxifen: 1.2-fold difference, P = .21) (Figure 2, G and H). The concordance between tumor sizes and neovascularization suggested that MYC-driven angiogenesis is limiting to tumor growth.
Activation of MYC and Loss-of-Function Mutations in the TGF-β Pathway in Human CRC Cell Lines
We thus asked whether Myc regulates TGF-β1 induction of Tsp-1 in human CRC cells. Because most commonly used human CRC cell lines are impaired in that pathway because of mutations in either the TGF-β type II receptor and Smad2/3/4 or polymorphisms in type I receptor (11,12), we first tested several human colonic epithelial cell lines that were reported either to have the intact TGF-β signaling pathway (SW837 CRC cells) (13) or to be nontumorigenic (NCM356 and NCM460 cells) (14). Stimulation of each of these three cell lines for 1 hour with 5ng/mL of TGF-β1 resulted in phosphorylation of receptor-regulated Smads (ie, Smad2 and Smad3) (Figure 3A). However, although Smad4 was robustly expressed in SW837 cells, we saw no detectable expression of Smad4 in NCM356 and NCM460 cells. Moreover, 48-hour treatment with TGF-β1 induced expression of both Tsp-1 and CTGF only in SW837 cells (Figure 3A).
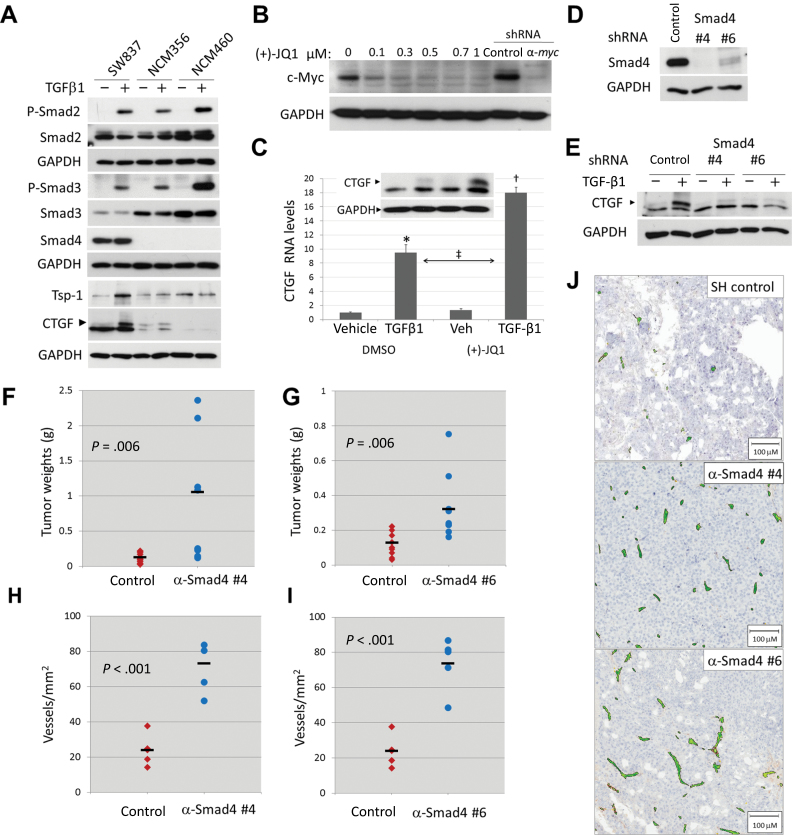
Figure 3.
TGF-β pathway effects on angiogenesis in human colorectal cancer cell lines. All in vitro experiments were repeated at least two times. A) Levels and phosphorylation state of Smad proteins and levels of TSR proteins in cultured SW837, NCM356, and NCM460 cells. All cultures were treated with either vehicle or 5ng/mL TGF-β1 for 1hour (for pSmads Westerns) or 48 hour (for thrombospondin-1 [Tsp-1] and CTGF Westerns). B) Myc expression in cultured SW837 cells treated with dimethyl sulfoxide (DMSO) or increasing concentrations of (+)-JQ1 for 48 hour. For comparison, also shown are Myc levels in SW837 cells stably expressing control and anti-MYC short hairpin RNAs (shRNAs). C) CTGF RNA levels as measured by quantitative real-time polymerase chain reaction in cultured SW837 cells pretreated for 48 hours with DMSO or 0.5 μM (+)-JQ1, then for 8 hours with 0.5ng/mL TGF-β1. Pairwise statistical significance per Tukey’s test in conjunction with two-way analysis of variance: *P < .001; †P < .001; ‡ P < .001. Factor significance per two-way analysis of variance is as follows: MYC inhibition (DMSO vs (+)-JQ1): P < .001; TGF-β activation (vehicle vs TGF-β1): P < .001; interaction between MYC inhibition × TGF-β activation: P < .001. Inset, CTGF protein (arrowhead) under the same conditions except TGF-β1 treatment lasted for 24 hours. D) Smad4 protein in cultured SW837 cells stably expressing either a nontargeting shRNA (SH Control) or two shRNAs against human Smad4 (SH Smad4). E) CTGF protein (arrowhead) in cultured SW837 cell lines shown in D treated for 24 hours either with vehicle or 5ng/mL TGF-β1. F) Weights of subcutaneous tumors derived from SW837 SH control or SH Smad4 #4 cells. Dashes represent the average for each group (n = 10). G) Weights of subcutaneous tumors derived from SW837 SH control or SH Smad4 #6 cells. Dashes represent the average for each group (n = 10). H) Microvessel density in SW837 SH control or SH Smad4 #4 tumors shown in F. Dashes represent average for each group (n = 5). I) Microvessel density in SW837 SH control or SH Smad4 #6 tumors shown in G. Dashes represent average for each group (n = 5). J) Representative sections from tumors shown in H and I stained with anti-CD34 antibody and hematoxylin. Positive staining colorized using Aperio ImageScope software.
We used the SW837 model to determine whether high levels of endogenous Myc protein [presumably due to the APC mutation (15)] were limiting TGF-β1–induced expression of TSR proteins. Using the chemical inhibitor of BET bromodomains JQ1 (16,17), we were able to inhibit c-Myc expression in SW837 cells to the same degree as achieved by infection with a lentivirus expressing an anti-MYC shRNA (Figure 3B). When SW837 cells were treated with a suboptimal (0.5ng/mL) amount of TGF-β1 after pretreatment with JQ1, CTGF was induced more robustly, both at the RNA and the protein levels (P < .001) (Figure 3C). The interaction between TGF-β1 and JQ1 treatments was statistically significant per two-way ANOVA (P < .001). To determine the functional significance of reduced TGF-β signaling for tumor growth, we knocked down Smad4 expression in SW837 cells using two independent hairpins (Figure 3D). Both shRNAs strongly inhibited induction of CTGF by TGF-β (Figure 3E). When implanted in mice, cells with Smad4 knockdown formed larger tumors (shRNA #4 vs. control: 8.2-fold difference, P = .006; shRNA #6 vs. control: 2.5-fold difference, P = .006) (Figure 3, F and G). This was accompanied by a markedly increased vascularization (shRNA #4 vs. control: 3.0-fold difference, P < .001; shRNA #6 vs. control: 3.1-fold difference, P < .001) (Figure 3, H, I, and J).
To directly compare the effects of Myc overexpression and TGF-β inactivation, we chose to use a cell line without the APC mutation and, consequently, with relatively modest basal levels of Myc [despite the activating Ser-45 deletion in CTNNB1, the positive regulator of MYC (15)]. HCT116 cells are known to have a homozygous mutation in the mismatch repair gene hMLH1 on human chromosome 3 and exhibit microsatellite instability leading to inactivation of the TGF-βRII gene (located on the same chromosome) (Figure 4A). However, there is an engineered derivative (HCT116:3–6) in which both hMLH1 and TGF-βRII expression have been restored through the transfer of a single chromosome 3 from normal fibroblasts (Figure 4B) (18,19). We introduced a Myc-expression vector into both HCT116 and HCT116:3–6 cells by means of retroviral transduction (Figure 4C). As anticipated, treatment with TGF-β caused Smad2 and Smad3 phosphorylation in the latter but not the former (Figure 4D). Moreover, Myc overexpression reduced levels of pSmad3 and, to a lesser extent, pSmad2. As we described previously (20), in cultured HCT116 cells, the basal levels of Tsp-1 are very low, making CTGF a more reliable readout. Notably, in HCT116:3–6 cells, Myc strongly blocked induction of CTGF, which is an important functional readout for TGF-β signaling (Figure 4D).
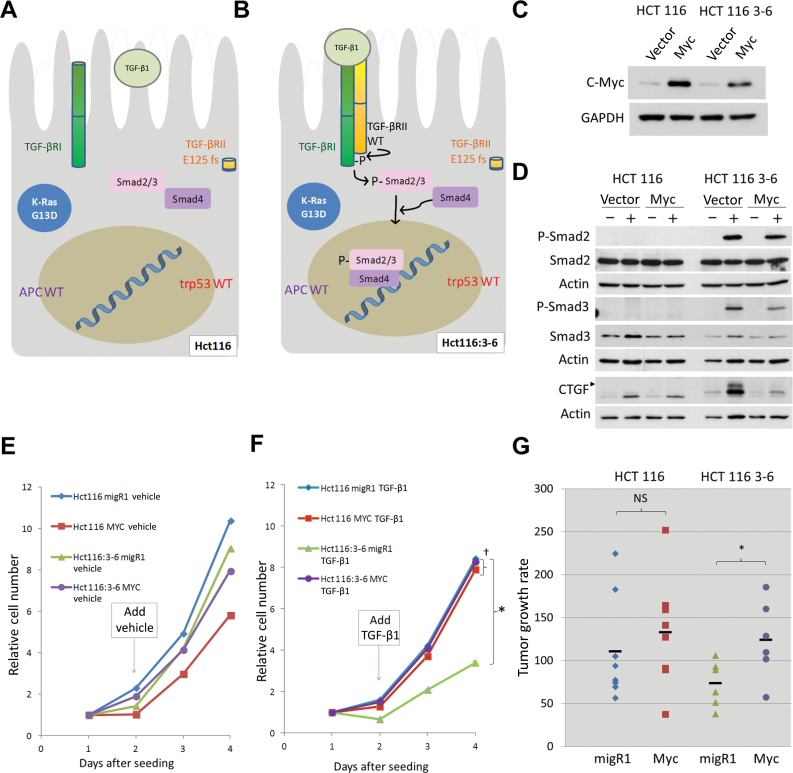
Figure 4.
Interplay between Myc and TGF-β signaling in human colorectal cancer cell lines. All in vitro experiments were repeated at least two times. A) Schematic of HCT116 cell harboring KRAS G13D and TGF-βRII E125fs mutations but possessing WT p53 and APC. B) Schematic of HCT116:3–6 with TGF-βRII restored, resulting in phosphorylation and translocation of Smad proteins into the nucleus. C) Myc protein levels in cultured HCT116 and HCT116:3–6 cells after stable transduction with empty vector (MigR1) or vector encoding human Myc. D) Levels and phosphorylation state of Smad proteins and induction of CTGF in cultured HCT116 cells and HCT116:3–6 cells treated with vehicle or 5ng/mL TGF-β1 for 1 hour (phospho-Smads) or 24 hours (CTGF, arrowhead), with actin serving as loading control. E) HCT116 and HCT116:3–6 with MigR1 or human MYC were seeded in 96-well plates (day = 0). At day 1, vehicle or 5ng/mL TGF-β1 (F) was added to media. Cell accumulation was quantitated every day using the WST-1 assay and represented by line plots for each condition (three technical replicates per sample were used, experiment was repeated twice). Statistical significance per one-way analysis of variance:*P < .001 (HCT116:3-6 MigR1 vs HCT116:3-6 MYC); †P = .63 (HCT116 MigR1 vs HCT116 MYC). Error bars represent standard deviation. G) Subcutaneous tumor growth rates of HCT116 and HCT116:3–6 cells with stably transduced empty vector (MigR1) or with human MYC in mice. Significance per two-sided paired t test: *P = .02; non-statistically significant (NS): P = .23.
Additionally, MYC presence did not affect cell accumulation of either untreated HCT116 or HCT116:3–6 cells in vitro (Figure 4E). However, activating the TGF-β signaling pathway in vector-only HCT116:3–6 reduced cell accumulation, and this reduction was completely rescued by Myc overexpression (P < .001) (Figure 4F, compare green and purple plots). Conversely, omission of chromosome 3 rendered Myc overexpression completely redundant (P = .63) (Figure 4F, compare blue and red plots). To see whether these opposing effects of MYC and TGF-β on cell accumulation would also manifest themselves in vivo, HCT116 and HCT116:3–6 cells, with or without MYC, were used to produce subcutaneous xenografts, which are exposed to TGF-β stimulation (see Figure 2E). Once again, we observed that Myc overexpression enhanced tumor growth only in the presence of functional TGF-β signaling (1.7-fold difference, P = .02) (HCT116:3–6 xenografts in Figure 4G, compare green and purple symbols) and was redundant in the context of TGF-β–deficient HCT116 cells (1.2-fold difference, P = .23) (Figure 4G, compare blue and red symbols; see also Supplementary Figure 2A, available online).
Overexpression of MYC and inactivation of SMADs in primary human CRCs.
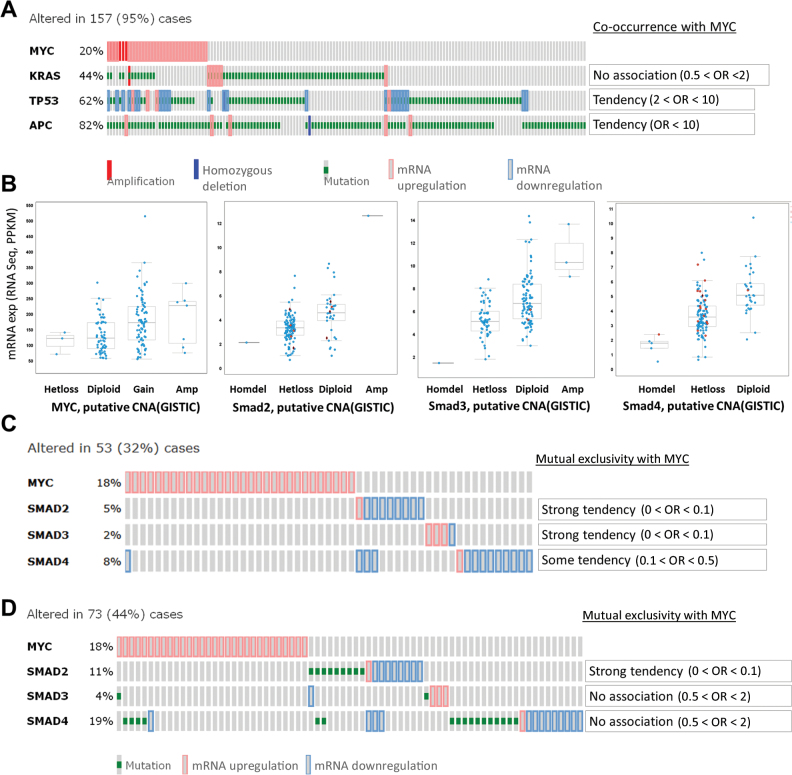
To determine how relevant our syngeneic and xenograft systems were to human cancers, we analyzed the co-occurrence of APC, MYC, RAS, and TP53 mutations in primary human samples profiled in the recent Cancer Genome Atlas study (1). To avoid analyzing samples with hundreds of passenger mutations, we limited our analysis to 165 comprehensively characterized tumors without microsatellite instability. Such tumors do not usually accumulate mutations in TGFBR2 (21) but inactivate one of the Smads, most frequently Smad4 and Smad2 (1). Additionally, to exclude cases with MYC amplification but no overexpression, we used RNASEQ values as a surrogate marker for MYC copy number variations. As predicted (15), there was a strong association (OR > 10) between MYC overexpression and APC mutations. We also observed an association (OR = 2–10) between MYC overexpression and inactivating mutations in TP53 (Figure 5A). Overall, 20% (n = 33 cases) of tumors without microsatellite instability harbor oncogenic mutations in all three CRC genes—APC, KRAS and TP53—further validating our murine RasMycER colonocyte and SW837 models.
Figure 5.
Myc and TGF-β signaling in human colorectal cancer (CRC). A) OncoPrint of MYC, KRAS, TP53, and APC mutations, putative copy-number alterations from GISTIC and mRNA expression data (RNASEQ; z score = 2) of complete nonhypermutated CRC patient samples, which were analyzed using the cBioPortal for Cancer Genomics platform. One hundred fifty-seven (95%) cases had mutations in either or all of the genes analyzed. Odds ratios (ORs), indicating the tendency for MYC amplification/overexpression to co-occur with KRAS, TP53, and APC mutations, putative copy-number alterations, and changes in mRNA expression are shown (right). B) MYC, SMAD2, SMAD3, and SMAD4 mRNA expression data (RNASEQ; z score = 2) of complete nonhypermutated CRC patient samples in A were analyzed against their putative copy-number alterations (CNA, from GISTIC). Missense and nonsense mutations are represented by red circles and black diamonds (red outline), respectively. C) OncoPrint of MYC, SMAD2, SMAD3, and SMAD4 mRNA expression data (RNASEQ; z score = 2) of complete nonhypermutated CRC patient samples in A. Fifty-three (32%) cases had changes in the mRNA expression for the genes analyzed. The odds ratios for MYC overexpression to be mutually exclusive from alterations in SMAD2/3/4 mRNA expression are indicated (right). D) OncoPrint and odds ratio of MYC, SMAD2, SMAD3, and SMAD4 mutations and mRNA expression data (RNASEQ; z score = 2) of complete nonhypermutated CRC patient samples in A. Seventy-three (44%) cases had alterations in the genes of interest. In this analysis, MYC overexpression and Smad2 mutation/mRNA downregulation were mutually exclusive. Statistical significance per Fisher’s exact test: P = .02.
We then asked whether overexpression of MYC and inactivation of SMADs are mutually exclusive, as predicted by our model. Again, to exclude cases with copy number variations but no deregulation at the mRNA levels (Figure 5B), RNASEQ values were used to assess expression levels. For both SMAD2 and SMAD3, we observed strong tendency toward mutual exclusivity with MYC activation (OR < 0.1). A weaker tendency was observed for SMAD4 (OR = 0.1–0.5) (Figure 5C). We then included inactivating point mutations in this analysis. We found that, of the three SMADs, only SMAD2 mutations exhibited strong tendency toward mutual exclusivity (OR < 0.1) (Figure 5D). This was in agreement with the observation that MYC exerts stronger effect on phosphorylation of Smad3 than Smad2 (Figure 4D), perhaps necessitating genetic inactivation in the latter case. Interestingly, when the same analysis was repeated on microsatellite-unstable tumors, we saw no evidence of Myc deregulation, although other CRC cancer genes and components of the TGF-β pathways were mutated as expected (Supplementary Figure 2, B and C, available online).
Discussion
Although CRC patient outcomes have been improving, 5-year survival rates remain at approximately 10% for stage IV CRC with distant metastasis, necessitating major therapeutic advances (22). Much hope is hinging on next-generation sequencing and other systems biology approaches to CRC analysis (23,24). Just 18 months ago, the Cancer Genome Atlas reported genome-scale analysis of 276 samples, including exome sequence, DNA copy number, promoter methylation, and messenger RNA and microRNA expression (1). This in-depth analysis revealed several new therapeutic targets and also underscored the importance of WNT and TGF-β pathways, which ultimately converge on MYC.
Admittedly, TGF-β plays complex and often contradictory roles in disease in general and cancer in particular (25,26). Its tumor suppressive role is most commonly attributed to inhibition of cell proliferation (27). However, in addition to this cell-intrinsic effect, TGF-β clearly plays a role in epithelial–mesenchymal transition, invasion, metastasis, and other cell-extrinsic phenotypes (28–30). This complexity is due, at least in part, to TGF-β–tumor microenvironment interactions, examples of which are beginning to emerge (31). Interestingly, depending on context, TGF-β has been ascribed either pro- or antiangiogenic activities [reviewed in (32)]. The majority of recent articles appear to emphasize proangiogenic effects of TGF-β, to the point of suggesting that ongoing clinical trials of TGF-β inhibitors [eg, fresolimumab (33)] could reveal their antiangiogenic effects (34). Nevertheless, our data argue, at least in the context of CRC, that TGF-β inhibits neovascularization, that antiangiogenesis strongly contributes to the overall tumor-suppressive effects of TGF-β, that MYC must counteract these effects to promote neoplastic growth, and that in the absence of TGF-β signaling the importance of MYC would be greatly diminished.
One limitation of our study is that angiogenesis was tested in subcutaneous xenograft models, in which blood vessels are different in several respects (including poor pericyte coverage) from those found in autochthonous tumors (35). Furthermore, because we wanted to measure tumor growth longitudinally, we implanted CRC cells subcutaneously, not in an orthotopic environment of the cecal wall (2). Finally, although the microvessel density is a commonly used descriptor of tumor vasculature, we have yet to measure the effects of TGF-β on tumor tissue perfusion and hypoxia, which play important roles in tumorigenesis.
Nevertheless, we were able to validate our key finding in spontaneous human tumors by demonstrating that CRCs with very high MYC levels are devoid of common SMAD mutations. This suggested a surprising redundancy between gain-of-function mutations in the MYC pathway and loss-of-function mutations in the TGF-β pathway. The paramount importance of this axis for Myc-driven colon cancer was surprising, given the pleiotropic effects of Myc on neoplastic growth. These findings also have implications for cancer therapeutics. Small molecule inhibitors of bromodomain proteins (eg, I-BET151 and JQ1) have shown efficacy against assorted leukemias and Burkitt’s lymphoma, in large part through profound inhibition of MYC transcription (17,36). These and related compounds are currently being tested in CRC patients (eg, GSK525762 in phase I clinical trial: http://www.cancer.gov/clinicaltrials/search/results?protocolsearchid=10450167, last accessed February 26, 2014). However, it is currently unknown which CRC subtypes will be most susceptible to this therapy.
Of clinical relevance is the fact that most CRCs with microsatellite instability and close to 15% of CRCs with chromosomal instability lack functional TGF-β signaling (1). The evidence for masking epistasis, uncovered in our study, would argue that such patients may not benefit from MYC-targeting drugs as much as patients with TGFBR2/SMAD–sufficient neoplasms and that other therapeutic options should be pursued instead. However, TGF-β deficiency might serve as a biomarker of sensitivity to TSR mimetics (37), which can be construed as an effective TGF-β target replacement therapy.
Funding
This work was supported by grants from the National Institutes of Health (CA122334 to ATT; T32 HL07915 to GST; and CA130895 and NS071197 to JL).
M. Dews and G. S. Tan contributed equally to this work.
The study sponsor had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
We thank the members of our laboratories, especially Drs James Psathas, Elena Sotillo, and Jamie Fox (CHOP) and Mark Duquette (BIDMC) for advice and support. We thank Dr. Andy Minn (University of Pennsylvania) and Yimei Li (CHOP) for their advice regarding statistical analysis of tumor samples and Daniel Martinez at the CHOP Pathology Core for his help with quantitation of tumor vasculatures. Drs Jack Henkin (Northwestern University) and David Boothman (UT Southwestern) are acknowledged for their kind gifts of ABT-510 and HCT116:3–6 cells, respectively, as well as helpful advice regarding the use of these reagents.
References
- 1. Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38(9):1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tucker RP. The thrombospondin type 1 repeat superfamily. Int J Biochem Cell Biol. 2004;36(6):969–974 [DOI] [PubMed] [Google Scholar]
- 4. Dews M, Fox JL, Hultine S, et al. The Myc-mir-17~92 axis blunts TGFb signaling and production of multiple TGFb-dependent antiangiogenic factors. Cancer Res. 2010;70(20):8233–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fox JL, Dews M, Minn AJ, Thomas-Tikhonenko A. Targeting of TGFb signature and its essential component CTGF correlates with improved survival in glioblastoma. RNA. 2013;19(2):177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mestdagh P, Bostrom AK, Impens F, et al. The miR-17–92 microRNA cluster regulates multiple components of the TGF-b pathway in neuroblastoma. Mol Cell. 2010;40(5):762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):l1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miao WM, Seng WL, Duquette M, Lawler P, Laus C, Lawler J. Thrombospondin-1 type 1 repeat recombinant proteins inhibit tumor growth through transforming growth factor-b-dependent and -independent mechanisms. Cancer Res. 2001;61(21):7830–7839 [PubMed] [Google Scholar]
- 10. Haviv F, Bradley MF, Kalvin DM, et al. Thrombospondin-1 mimetic peptide inhibitors of angiogenesis and tumor growth: design, synthesis, and optimization of pharmacokinetics and biological activities. J Med Chem. 2005;48(8):2838–2846 [DOI] [PubMed] [Google Scholar]
- 11. Valle L, Serena-Acedo T, Liyanarachchi S, et al. Germline allele-specific expression of TGFBR1 confers an increased risk of colorectal cancer. Science. 2008;321(5894):1361–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pasche B, Knobloch TJ, Bian Y, et al. Somatic acquisition and signaling of TGFBR1*6A in cancer. JAMA. 2005;294(13):1634–1646 [DOI] [PubMed] [Google Scholar]
- 13. Ijichi H, Ikenoue T, Kato N, et al. Systematic analysis of the TGF-beta-Smad signaling pathway in gastrointestinal cancer cells. Biochem Biophys Res Comm. 2001;289(2):350–357 [DOI] [PubMed] [Google Scholar]
- 14. Stauffer JS, Manzano LA, Balch GC, Merriman RL, Tanzer LR, Moyer MP. Development and characterization of normal colonic epithelial cell lines derived from normal mucosa of patients with colon cancer. Am J Surg. 1995;169(2):190–195 [DOI] [PubMed] [Google Scholar]
- 15. He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281(5382):1509–1512 [DOI] [PubMed] [Google Scholar]
- 16. Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mertz JA, Conery AR, Bryant BM, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koi M, Umar A, Chauhan DP, et al. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N’-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54(16):4308–4312 [PubMed] [Google Scholar]
- 19. Araki S, Eitel JA, Batuello CN, et al. TGF-b1-induced expression of human Mdm2 correlates with late-stage metastatic breast cancer. J Clin Invest. 2010;120(1):290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sundaram P, Hultine S, Smith LM, et al. p53-responsive miR-194 inhibits thrombospondin-1 and promotes angiogenesis in colon cancers. Cancer Res. 2011;71(24):7490–7501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Markowitz S, Wang J, Myeroff L, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268(5215):1336–1338 [DOI] [PubMed] [Google Scholar]
- 22. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2013. 10.1016/S0140-6736(08)61345-8. [DOI] [PubMed] [Google Scholar]
- 23. Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113 [DOI] [PubMed] [Google Scholar]
- 24. Bass AJ, Lawrence MS, Brace LE, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43(10):964–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gordon KJ, Blobe GC. Role of transforming growth factor-beta superfamily signaling pathways in human disease. Biochim Biophys Acta. 2008;1782(4):197–228 [DOI] [PubMed] [Google Scholar]
- 26. Bellam N, Pasche B. Tgf-beta signaling alterations and colon cancer. Cancer Treat Res. 2010;155:85–103 [DOI] [PubMed] [Google Scholar]
- 27. Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFb influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nature Cell Biol. 2001;3(4):400–408 [DOI] [PubMed] [Google Scholar]
- 28. Bierie B, Moses HL. Tumour microenvironment: TGFb: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6(7):506–520 [DOI] [PubMed] [Google Scholar]
- 29. Simms N, Rajput A, Sharratt EA, et al. Transforming growth factor-b suppresses metastasis in a subset of human colon carcinoma cells. BMC Cancer. 2012;12(1):221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu XQ, Rajput A, Geng L, Ongchin M, Chaudhuri A, Wang J. Restoration of transforming growth factor-beta receptor II expression in colon cancer cells with microsatellite instability increases metastatic potential in vivo. J Biol Chem. 2011;286(18):16082–16090 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31. Dong M, How T, Kirkbride KC, et al. The type III TGF-beta receptor suppresses breast cancer progression. J Clin Invest. 2007;117(1):206–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pepper MS. Transforming growth factor-beta: vasculogenesis, angiogenesis, and vessel wall integrity. Cytokine Growth Factor Rev. 1997;8(1):21–43 [DOI] [PubMed] [Google Scholar]
- 33. Lonning S, Mannick J, McPherson JM. Antibody targeting of TGF-beta in cancer patients. Curr Pharm Biotechnol. 2011;12(12):2176–2189 [DOI] [PubMed] [Google Scholar]
- 34. van Meeteren LA, Goumans MJ, Ten DP. TGF-beta receptor signaling pathways in angiogenesis; emerging targets for anti-angiogenesis therapy. Curr Pharm Biotechnol. 2011;12(12):2108–2120 [DOI] [PubMed] [Google Scholar]
- 35. Carmeliet P, Jain RK. Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nat Rev Drug Discov. 2011;10(6):417–27 [DOI] [PubMed] [Google Scholar]
- 36. Dawson MA, Prinjha RK, Dittmann A, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Campbell N, Greenaway J, Henkin J, Petrik J. ABT-898 induces tumor regression and prolongs survival in a mouse model of epithelial ovarian cancer. Mol Cancer Ther. 2011;10(10):1876–1885 [DOI] [PubMed] [Google Scholar]