Abstract
TP53 is the most frequently mutated gene in human malignancies; however, de novo somatic mutations in childhood embryonal cancers such as neuroblastoma are rare. We report on the analysis of three independent case–control cohorts comprising 10290 individuals and demonstrate that rs78378222 and rs35850753, rare germline variants in linkage disequilibrium that map to the 3′ untranslated region (UTR) of TP53 and 5′ UTR of the Δ133 isoform of TP53, respectively, are robustly associated with neuroblastoma (rs35850753: odds ratio [OR] = 2.7, 95% confidence interval [CI] = 2.0 to 3.6, P combined = 3.43×10−12; rs78378222: OR = 2.3, 95% CI = 1.8 to 2.9, P combined = 2.03×10−11). All statistical tests were two-sided. These findings add neuroblastoma to the complex repertoire of human cancers influenced by the rs78378222 hypomorphic allele, which impairs proper termination and polyadenylation of TP53 transcripts. Future studies using whole-genome sequencing data are likely to reveal additional rare variants with large effect sizes contributing to neuroblastoma tumorigenesis.
Neuroblastoma is a cancer of the developing sympathetic nervous system that accounts for approximately 10% of all pediatric oncology deaths (1). Familial neuroblastoma is rare (approximately 1%), and most of these cases harbor germline mutations in ALK (2) or PHOX2B (3,4). Through a genome-wide association study of sporadic neuroblastoma, we have reported common single nucleotide polymorphisms (SNPs) within or upstream of CASC15 (5) and FLJ44180 (5), BARD1 (6), LMO1 (7), DUSP12 (8), HSD17B12 (8), DDX4/IL31RA (8), HACE1 (9), and LIN28B (9), along with a common copy number variation within NBPF23 (10), as each being highly associated with neuroblastoma. Collectively, however, these variants still account for only a small portion of the risk for developing neuroblastoma. Recent next-generation sequencing studies of paired tumor-normal genomes in neuroblastoma have revealed a striking paucity of somatic mutations (11–14); we hypothesized that rare variants in the host genome play an important role in tumorigenesis and explain a substantial proportion of the unidentified heritability in this disease.
Disruption or malfunction of the p53 pathway is a near-universal hallmark of cancer (15–17). Germline mutations in TP53 are the cause of Li-Fraumeni syndrome, and individuals who carry these mutations are at increased risk of developing a wide array of cancers at an early age (18). Recently, a rare polymorphism (rs78378222) in the 3΄ UTR of TP53 was found to confer susceptibility to cutaneous basal cell carcinoma, prostate cancer, glioma, and colorectal adenomas, but not melanoma, colon cancer, or breast cancer (19). Somatic mutations of TP53 or other pathway members, such as MDM2, are rare in primary neuroblastomas obtained at diagnosis (14).
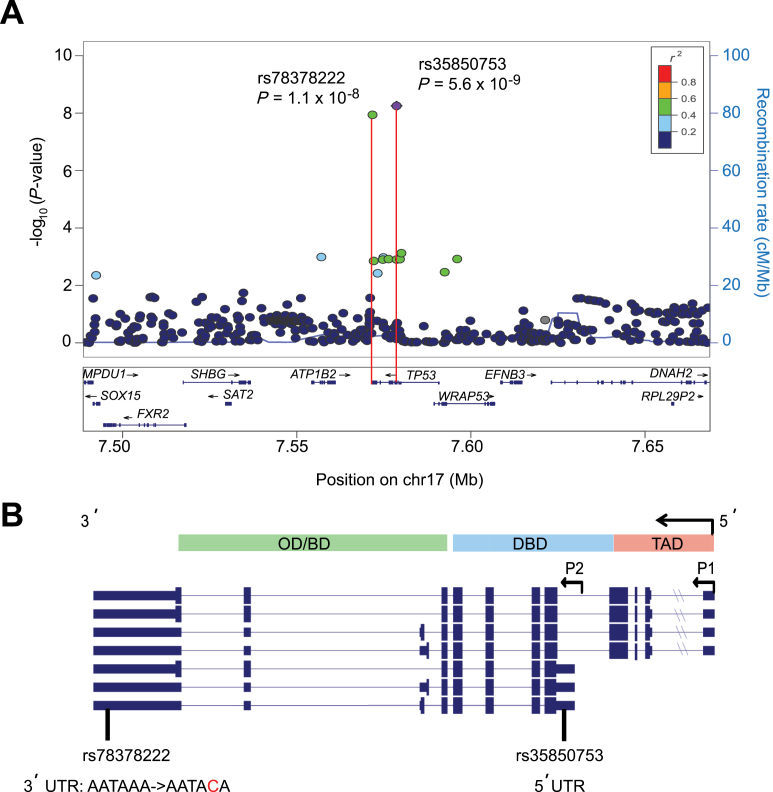
To identify germline variants associated with neuroblastoma at the TP53 locus, we performed genotype imputation on a discovery case series of 2101 neuroblastoma patients accrued and consented through the North American–based Children’s Oncology Group and 4202 control subjects of European ancestry matched genetically and consented through the Children’s Hospital of Philadelphia, as described previously (9). The Ethics Committee at the Children’s Hospital of Philadelphia approved this study. All statistical tests were two-sided, and a P value less than 1.0×10−7 was considered statistically significant for subsequent replication efforts. Imputation was performed using IMPUTE2 (20) with default parameters and Ne equal to 20000, along with a multipopulation reference panel from the worldwide 1000 Genomes Project Phase 1 Interim release. After imputation, SNPs with minor allele frequency (MAF) less than 1% and/or IMPUTE2 info quality score less than 0.8 were removed. The remaining SNPs were tested for association with neuroblastoma using the frequentist association test under the additive model using the “score” method implemented in SNPTEST (21). Although analysis of SNPs directly genotyped on the array revealed only modest evidence for association in the TP53 region (rs8079544: odds ratio [OR] = 1.3; 95% confidence interval [CI] = 1.2 to 1.6; P = 7.2×10−4) (Supplementary Table 1, available online), two imputed SNPs were highly associated with neuroblastoma (rs35850753: OR = 1.9, 95% CI = 1.5 to 2.3, P = 5.6×10−9; rs78378222: OR = 2.0, 95% CI = 1.6 to 2.7, P = 1.1×10−8) (Figure 1A). Imputation probabilities for rs35850753 and rs78378222 were very high (Supplementary Figure 1, available online). To further confirm the accuracy of imputation, we performed polymerase chain reaction (PCR)–based genotyping on 176 case patients and compared these results to the most probable genotype based on imputation. We observed a 96% concordance rate overall (Supplementary Tables 2 and 3, available online).
Figure 1.
Discovery of rare variants within TP53 associated with neuroblastoma. A) Regional association plot of genotyped and imputed single nucleotide polymorphisms (SNPs) at the TP53 locus in a discovery cohort of 2101 case patients and 4202 control subjects of European ancestry. Plot was generated using LocusZoom (25). Y-axes represent the statistical significance of association (-log10 transformed P values) and the recombination rate. SNPs are color-coded based on pair-wise linkage disequilibrium (r 2) with most statistically significant SNP. Most statistically significant SNPs are labeled with P value, and most statistically significant SNP is shown in purple. Allelic P values generated by SNPTEST using score method (two-sided). B) Neuroblastoma-associated SNPs map to the 3΄ untranslated region (UTR) of TP53 and 5΄ UTR of the Δ133 isoform of TP53, respectively. The Δ133 isoform is transcribed by an alternative promoter (P2) and lacks a transactivation domain and part of the DNA binding domain. Shown are known isoforms of TP53 currently reported in the NCBI Reference Sequence database (RefSeq). BD = basic domain; DBD = DNA binding domain; OD = oligomerization domain; TAD = transactivation domain.
The two neuroblastoma-associated SNPs map to UTRs of TP53 and are rare (MAF for rs35850753: 3.6% case patients, 1.9% control subjects; rs78378222: 2.7% case patients, 1.3% control subjects). The most statistically significant SNP, rs35850753, maps to the 5΄ UTR of the Δ133 isoform of TP53, which is transcribed by an alternative promoter, and lacks a trans activation domain (Figure 1B). This isoform has been shown to have a dominant negative effect whereby it inhibits the tumor suppressive functions of full-length TP53 (22). The other SNP, rs78378222, maps to the 3΄ UTR of TP53 and is the same rare variant found by Stacey and colleagues to confer susceptibility to several other cancers (19). It was further demonstrated that this rare allele disrupts the TP53 polyadenylation signal (AATA[A/C]A) resulting in impaired 3′-end processing of TP53 transcripts in blood and adipose tissue (19). We analyzed total RNA from two rs78378222 heterozygous primary neuroblastomas and observed a preference for the wild-type/protective allele in properly terminated and polyadenylated transcripts, whereas improperly terminated “run-on” transcripts were detected almost exclusively from the variant/risk allele (Supplementary Figure 2, available online). Based on data from the 1000 Genomes Phase I European population, rs35850753 and rs78378222 are in linkage disequilibrium (r 2 = 0.52 D΄ = 1). Accordingly, rs35850753 was no longer statistically significant after conditioning on the known functional SNP, rs78378222 (P = 0.07). Neither variant was associated with clinical or biological covariables (Supplementary Table 4, available online); however, given the low MAF of both SNPs, larger sample sizes are required before definitive conclusions can be drawn on subgroup association. Lastly, we tested the TP53 codon 72 Arg/Pro missense variant (rs1042522) for association with overall survival in 1809 neuroblastoma case patients; however, we were unable to replicate the recent report that codon 72 Pro/Pro predicts poor survival in neuroblastoma (23) (Supplementary Figure 3 and Supplementary Table 5, available online).
We next sought to replicate the rs35850753 and rs78378222 associations in an African ancestry cohort of 365 neuroblastoma case patients and 2491 genetically matched control subjects (9). We used IMPUTE2 (20) to infer genotypes in a 6-Kb region around the TP53 locus using data from the 1000 Genomes Project. Similar to the European ancestry samples, imputation probabilities were very high in individuals of African ancestry (Supplementary Figure 4, available online). Using the proportion of African admixture as a covariable to correct for varying degrees of admixture among our samples, we observed highly statistically significant associations at both SNPs (Table 1). Indeed, these were the two most statistically significant SNPs within the 6-Kb region surrounding TP53 (Supplementary Figure 5, available online).
Table 1.
Statistically significantly associated single nucleotide polymorphisms within TP53 at 17p.13*
| SNP | A1/A2 | Cohort† | Frequency A1 case patients | Frequency A1 control subjects | P‡ | OR (95% CI) |
|---|---|---|---|---|---|---|
| rs35850753 | T/C | European ancestry | 0.036 (n = 2101) | 0.019 (n = 4202) | 5.6×10–9 | 1.9 (1.5 to 2.3) |
| African ancestry | 0.012 (n = 365) | 0.004 (n = 2491) | 1.3×10–3 | 3.4 (1.5 to 7.7) | ||
| Italian | 0.025 (n = 338) | 0.005 (n = 781) | 9.0×10–5 | 4.7 (2.0 to 11.1) | ||
| Combined | 3.4×10–12 | 2.7 (2.0 to 3.6) | ||||
| rs78378222 | G/T | European ancestry | 0.027 (n = 2101) | 0.013 (n = 4202) | 1.1×10–8 | 2.0 (1.6 to 2.7) |
| African ancestry | 0.011 (n = 365) | 0.002 (n = 2491) | 4.2×10–5 | 5.1 (2.0 to 12.8) | ||
| Italian | 0.009 (n = 335) | 0.001 (n = 753) | 4.2×10–2 | 4.6 (1.2 to 18.7) | ||
| Combined | 2.0×10–11 | 2.3 (1.8 to 2.9) |
* A1, allele, on forward strand; A2: allele 2, on forward strand; CI, confidence interval; OR, odds ratio with respect to A1; SNP, single nucleotide polymorophism.
† No deviations from Hardy–Weinberg equilibrium were observed (P > .001) in all cohorts.
‡ Allelic P values generated by SNPTEST score method (two-sided); combined P values from METAL using inverse-variance method (two-sided).
Finally, we performed PCR-based genotyping of rs35850753 and rs78378222 in an Italian cohort of 351 neuroblastoma case patients and 780 control subjects as another independent replication effort. Both SNPs showed statistically significant association in the same direction seen in the European and African ancestry samples (Table 1). Meta-analysis of 10290 individuals from the three studies using the inverse-variance method within METAL (24) resulted in highly statistically significant associations with larger effect sizes than seen for common variants identified by genome-wide association studies (rs35850753: OR = 2.7, 95% CI = 2.0 to 3.6, P = 3.43×10−12; rs78378222: OR = 2.3, 95% CI = 1.8 to 2.9, P = 2.03×10−11) (Table 1).
We have used imputation to infer genotypes for variants in the 1000 Genomes Project not directly assayed on the SNP arrays used. It is possible that additional rare variants within or nearby TP53 are associated with neuroblastoma, but were not detected in our study because of limitations in the imputation process.
In conclusion, here we describe the first report of rare germline variation associated with neuroblastoma susceptibility. Our findings add to the complex repertoire of human cancers influenced by the rs78378222 hypomorphic allele, which impairs proper termination and polyadenylation of TP53 transcripts. Future studies using whole-genome sequencing data are likely to reveal additional rare variants contributing to neuroblastoma tumorigenesis.
Funding
This work was supported in part by grants from the National Institutes of Health (R01-CA124709 to JMM; R00-CA151869 to SJD; P30-HD026979 to MD); the Giulio D’Angio Endowed Chair (to JMM); the Alex’s Lemonade Stand Foundation (to JMM); Andrew’s Army Foundation (to JMM); the PressOn Foundation (to JMM); the Abramson Family Cancer Research Institute (to JMM); Fondazione Italiana per la Lotta al Neuroblastoma and Associazione Italiana per la Ricerca sul Cancro (10537 to MC); and the Center for Applied Genomics at the Children’s Hospital of Philadelphia Research Institute (to HH).
S. J. Diskin and J. M. Maris designed the experiment. S. J. Diskin drafted the manuscript, performed SNP association study, and analyzed PCR-based genotyping and sequencing data. M. Devoto and S. J. Diskin replicated SNP associations in the African ancestry cohort. M. Capasso replicated SNP associations in the Italian cohort. M. Diamond performed PCR-based genotype validation in the European and African ancestry cohorts. D. A. Oldridge assisted in evaluation of genotype imputation probabilities. K. R. Bosse assisted with biological interpretation of SNP associations. M. Diamond and M. R. Russell performed 3΄ RACE experiments. K. Conkrite performed TP53 run-on experiments. H. Hakonarson generated and provided all control data for the European and African ancestry cohorts. All authors commented on or contributed to the manuscript.
The study sponsors had no role in the in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
The URLs for data presented herein are as follows: 1000 Genomes Project, http://www.1000genomes.org; LiftOver, http://genome.ucsc.edu/cgi-bin/hgLiftOver; IMPUTE2, http://mathgen.stats.ox.ac.uk/impute/impute_v2; SNPTEST, https://mathgen.stats.ox.ac.uk/genetics_software/snptest; LocusZoom, http://csg.sph.umich.edu/locuszoom; METAL, http://www.sph.umich.edu/csg/abecasis/metal.
References
- 1. Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362(23):2202–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mosse YP, Laudenslager M, Longo L, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455(7215):930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trochet D, Bourdeaut F, Janoueix-Lerosey I, et al. Germline mutations of the paired-like homeobox 2B (PHOX2B) gene in neuroblastoma. Am J Hum Genet. 2004;74(4):761–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mosse YP, Laudenslager M, Khazi D, et al. Germline PHOX2B mutation in hereditary neuroblastoma. Am J Hum Genet. 2004;75(4):727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maris JM, Mosse YP, Bradfield JP, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358(24):2585–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Capasso M, Devoto M, Hou C, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41(6):718–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang K, Diskin SJ, Zhang H, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2010;469(7329):216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nguyen le B, Diskin SJ, Capasso M, et al. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;7(3):e1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diskin SJ, Capasso M, Schnepp RW, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;44(10):1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diskin SJ, Hou C, Glessner JT, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459(7249):987–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheung NK, Zhang J, Lu C, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307(10):1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Molenaar JJ, Koster J, Zwijnenburg DA, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483(7391):589–593 [DOI] [PubMed] [Google Scholar]
- 13. Sausen M, Leary RJ, Jones S, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45(1):12–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pugh TJ, Morozova O, Attiyeh EF, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45(3):279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharpless NE, DePinho RA. p53: good cop/bad cop. Cell. 2002;110(1):9–12 [DOI] [PubMed] [Google Scholar]
- 16. Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310 [DOI] [PubMed] [Google Scholar]
- 17. Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283 [DOI] [PubMed] [Google Scholar]
- 18. Malkin D. p53 and the Li-Fraumeni syndrome. Cancer Genet Cytogenet. 1993;66(2):83–92 [DOI] [PubMed] [Google Scholar]
- 19. Stacey SN, Sulem P, Jonasdottir A, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43(11):1098–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913 [DOI] [PubMed] [Google Scholar]
- 22. Fujita K, Mondal AM, Horikawa I, et al. p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat Cell Biol. 2009;11(9):1135–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cattelani S, Ferrari-Amorotti G, Galavotti S, et al. The p53 codon 72 Pro/Pro genotype identifies poor-prognosis neuroblastoma patients: correlation with reduced apoptosis and enhanced senescence by the p53-72P isoform. Neoplasia. 2012;14(7):634–64322904680 [Google Scholar]
- 24. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]