Abstract
Inefficient and inaccurate repair of DNA damage is the principal cause of DNA mutations, chromosomal aberrations, and carcinogenesis. Numerous multiple-step DNA repair pathways exist whose deployment depends on the nature of the DNA lesion. Common to all eukaryotic DNA repair pathways is the need to unravel the compacted chromatin structure to facilitate access of the repair machinery to the DNA and restoration of the original chromatin state afterward. Accordingly, our cells utilize a plethora of coordinated mechanisms to locally open up the chromatin structure to reveal the underlying DNA sequence and to orchestrate the efficient and accurate repair of DNA lesions. Here we review changes to the chromatin structure that are intrinsic to the DNA damage response and the available mechanistic insight into how these chromatin changes facilitate distinct stages of the DNA damage repair pathways to maintain genomic stability.
Introduction
Over the last 20 years, a clear picture has emerged of how the cell repairs lesions on naked DNA (Friedberg et al. 2006). The current challenge is to understand how DNA repair occurs in its physiological context within the tightly packaged nucleoprotein structure known as chromatin. Within the confines of chromatin, our DNA is rendered highly inaccessible to the central machineries that mediate genomic processes, implying that there are additional layers of complexity for the repair of DNA damage within our cells.
The basic repeating unit of chromatin, the nucleosome, consists of approximately 146 bp of DNA wound around an octamer of histone proteins, comprised of two molecules each of histones H3, H4, H2A, and H2B (Luger et al. 1997). Nucleosomes coat our entire genome, separated only by short internucleosomal linker DNA regions to form arrays of nucleosomes, referred to as the 10-nm fiber or beads on a string form of chromatin. The binding of linker histones and additional non-histone chromatin proteins promotes the compaction of the chromatin into the 30-nm fiber and higher-order chromatin forms in which the DNA is deeply buried. Accordingly, our cells have developed specialized mechanisms to alter the chromatin structure to facilitate and regulate genomic processes.
Physiological chromatin alterations can be divided into five main classes: (1) Enzymes can add and remove post-translational modifications (PTMs) of the histone proteins, including acetylation, methylation, and phosphorylation on specific lysines. Histone PTMs usually function to recruit specific proteins to the chromatin, the identity of which is determined by the type of PTM and the specific histone residue that is modified (Suganuma and Workman 2011). (2) Histone–DNA interactions within the nucleosomes can be transiently broken by ATP-dependent nucleosome remodelers (Hargreaves and Crabtree 2011). (3) The canonical histones can be exchanged for histone variants that have altered properties (Yuan and Zhu 2011). (4) The DNA can be fully exposed by total removal of histones from the DNA, and this is mediated by histone chaperones (Elsässer and D’Arcy 2012). (5) Additional non-histone architectural proteins, such as linker histones, HMG proteins, or hetero-chromatin protein 1 (HP1) can be added to the chromatin to achieve higher levels of compaction (Li and Reinberg 2011), or they can be removed to promote lower-order forms of chromatin such as the 10-nm fiber. These five general mechanisms are often used in a coordinated fashion by the cell to alter the chromatin structure and collectively impart regulation and accuracy on genomic processes.
Although there are many types of DNA lesions and numerous repair mechanisms for them, the role of chromatin structure during DNA repair has been most highly studied in response to UV irradiation, ionizing radiation (IR), or endonuclease-induced double-strand breaks. UV irradiation occurs naturally in response to sunlight exposure, and the resulting DNA intrastrand crosslinks must be rapidly removed by the process of nucleotide excision repair (NER) to prevent inaccurate DNA replication. DNA double-strand breaks (DSBs) arise naturally as a result of genotoxic stress or replication failure, during programmed events such as meiosis and V(D)J recombination, as well as in response to IR and ectopically introduced endonucleases (Hiom 2010). DSBs are repaired by non-homologous end joining (NHEJ) or by homologous recombination (HR).
The physical repair of the DNA lesion per se in the cell is intertwined with a complex network of signal transduction pathways known as the DNA damage response (DDR). The DDR is mainly mediated by the phosphatidylinositol 3-kinase-like protein kinase family members ataxia telangiectasia mutated (ATM), ataxia telangiectasia and Rad3-related (ATR), and the DNA-dependent protein kinase (DNA-PK) (Smith et al. 2010). After recognition of the DNA breaks by damage sensors, ATM and ATR phosphorylate mediator proteins, which act to recruit additional ATM/ATR substrates to sites of DNA damage within discrete nuclear foci, in turn amplifying the DDR. The exact order and timing of DDR factor recruitment to the repair foci is highly dynamic and tightly controlled, and this provides the cell with opportunities to choose between various repair pathways (Bekker-Jensen et al. 2006). Dynamic chromatin modifications such as histone acetylation, phosphorylation, ADP-ribosylation, methylation, and ubiquitylation all play a critical role in DDR factor recruitment, as discussed below in detail. In addition to repair of the DNA lesion itself, the sensor kinases also phosphorylate many effector proteins, either directly or indirectly, via phosphorylating checkpoint 1 (CHK1) and checkpoint 2 (CHK2) kinases to mobilize the effector proteins and bring about the cellular responses to DNA damage including transcriptional upregulation, cell-cycle arrest, senescence, or apoptosis (Lazzaro et al. 2009).
Here, we will review the highly orchestrated array of chromatin changes that are fundamental to DNA repair and the DDR. Distinct chromatin changes occur directly at the site of the DNA lesion, in chromatin domains adjacent to the DNA lesion, as well as global changes that propagate across the entire genome. Our purpose is not to exhaustively cover the details of all DNA repair pathways and the DNA damage response, as this has been done recently elsewhere (Ciccia and Elledge 2010; Polo and Jackson 2011). Instead, we will give only enough information on the DDR and DNA repair to put the chromatin changes in context. Specifically, we will review the current knowledge of the chromatin changes that occur during the processes of NER and DSB repair. The chromatin changes will be described in the temporal order in which they occur in the DNA repair pathway rather than on a modification-by-modification basis (which has been done elsewhere recently in an excellent review (Luijsterburg and van Attikum 2011)). Emphasis will be placed here on relating each specific chromatin change to its apparent or predicted molecular function in the DDR or repair pathway. Other histone PTMs that occur during DNA repair and/or are important for DNA repair but have no currently known molecular function are listed in Table 1 and will not be further discussed. An emerging theme from recent studies is that specific histone modifications occur in a tightly controlled temporal manner to coordinate and amplify the DNA damage response.
Table 1.
Summary of histone modifications that function in the DDR
| Histone | Modification | Enzyme | Role in DDR |
|---|---|---|---|
| H2A | K119ub/K119ub2 | RNF8 | Helps accumulation of BRCA1 and 53BP1, for DSB repair |
| K119poly-ub | RNF168 | ||
| H2AX | K5ac | TIP60 | Facilitates subsequent K119ub of H2AX, as well as H2AX removal from chromatin |
| K36ac | CBP/p300 | Recruits Ku during NHEJ | |
| K119ub | RNF2 | Help accumulate DDR proteins; similar to H2A K119ub (see above) | |
| K119ub2 | RNF8 | ||
| K119poly-ub | TIP6—UBC13 | ||
| S139p | ATM, ATR, DNA-PKcs | Recruits and accumulates DDR proteins, amplifies DDR, mediates IRIF formation through MDC1 recruitment, and triggers dynamic histone acetylation and chromatin remodeling at DSB site (H2A S129p in yeast) | |
| T142p | WSTF | Recruits activated ATM and MDC1 and promotes repair vs apoptosis (through dephosphorylation by EYA1/3) | |
| H2B | K120ub | RNF20-RNF40 | Promotes NHEJ through XRCC4 and Ku recruitment and promotes HR through BRCA1 and RAD51 recruitment |
| S14p | Unknown, this modification is also seen in apoptosis | ||
| H3 | K4me3 | SET1 | Stimulates V(D)J recombination via the RAG complex |
| K9ac | GCN5 | Recruits SWI/SNF complex, promotes spreading of γH2AX domain, and facilitates NER through XPC recruitment | |
| K9me3 | Suv3-9H1/Suv3-9H2 | In undamaged chromatin, retains HP1β; damage-induced phosphorylation of HP1β leads to its dissociation and activation of TIP60 | |
| K14ac | GCN5 | Recruits SWI/SNF complex, promotes spreading of γH2AX domain, and regulates ATM activity through HMGN1-dependent mechanism | |
| K18ac | CBP/p300; GCN5 | Recruits SWI/SNF complex and Ku, promotes NHEJ, and promotes spreading of γH2AX domain | |
| K23ac | GCN5 | Recruits SWI/SNF complex; spreading of γH2AX domain | |
| K36me2 | Metnase/SETMAR | Accumulates NBS1 and Ku to stimulate NHEJ | |
| T11p | CHK1 | Constitutive PTM lost following UV damage, which represses gene expression | |
| K56ac | CBP/p300; RTT109 (yeast) (on free histones) | Decreases after IR to promote NHEJ; localized, transient enrichment of K56ac after HR and UV repair due to chromatin assembly. May function in checkpoint recovery/adaptation | |
| K79me3 | DOT1 | Recruits RAD9 in budding yeast | |
| H4 | K5ac | TIP60-TRRAP; CBP/p300 | Recruits DDR and repair proteins and recruits SWI/SNF nucleosome remodeling complex |
| K8ac | TIP60-TRRAP; CBP/p300 | Recruits DDR and repair proteins and recruits SWI/SNF nucleosome remodeling complex | |
| K12ac | TIP60-TRRAP; CBP/p300 | Recruits DDR and repair proteins and recruits SWI/SNF nucleosome remodeling complex | |
| K16ac | TIP60-TRRAP; CBP/p300; hMOF-1 | Recruits DDR and repair proteins, recruits SWI/SNF nucleosome remodeling complex, accumulates IRIF proteins, and inhibits formation of 30-nm chromatin fibers | |
| K20me2 | Suv4-20H1/Suv4-20H2; MMSET | Recruits 53BP1 to promote NHEJ and inhibits HR via recruitment of 53BP1 | |
| K91ub | BBAP | Constitutive PTM that induces H4K20me and 53BP1 recruitment | |
| S1p | CK2 | Promotes NHEJ |
Chromatin changes that promote DSB repair
Brief overview of DSB repair
The severe dangers posed to a cell harboring a DSB has resulted in highly effective and conserved repair mechanisms that are broadly separated into two categories: (1) HR that employs the undamaged sister chromatid to accurately restore the missing genetic information and (2) NHEJ, which re-ligates the broken ends of the chromosome. Accordingly, mammalian cells have evolved mechanisms to channel the repair of DSBs into the NHEJ pathway in G1 phase before the sister chromatid has been generated. Meanwhile, the cell channels the repair of DSBs into the HR pathway during S and G2 phase (Warmerdam and Kanaar 2010). Briefly, during NHEJ in mammals, the broken DNA ends are bound by the KU70/80 heterodimer, which recruits DNA-PK and keeps the ends in close proximity to each other. If necessary, DNA ends are processed to identify regions of microhomology, which are then ligated together by the XRCC4-DNA Ligase IV (Lieber et al. 2010). During HR in mammals, the Mre11–Rad50–Nbs1 (MRN) complex binds to DNA ends and, CtIP initiates the process of DNA resection to form single-strand overhangs (You and Bailis 2010). Next, the ssDNA is coated with RPA, and RAD51 replaces RPA to form nucleoprotein filaments for strand invasion into an undamaged homologous DNA region, creating a Holliday junction. DNA is resynthesized by polymerases to fill the gaps and the Holliday junction is resolved (Holthausen et al. 2010). Regardless of the DSB repair pathway employed, all DNA repair necessitates rapid recognition of the lesion and coordinated mechanisms for recruitment of the repair machinery in the context of the chromatin template.
Overview of chromatin’s role in DSB repair
It has been known for over 20 years that DNA damage induces chromatin structure alteration. For example, damaged DNA renders chromatin more sensitive to digestion by micrococcal nuclease (Telford and Stewart 1989). We now know that DSB generation causes global and local decon-densation of the chromatin (Dellaire et al. 2009; Kruhlak et al. 2006), which is essential for efficient DNA repair and activation of the DDR.
The growing body of data indicates that DSBs induce myriad changes in the chromatin structure. Nucleosomes around the damage sites are modified by ATP-dependent chromatin remodeling. Meanwhile, histone PTMs are dynamically generated and removed to transiently mark DSBs for recruitment of DDR and repair proteins. Histones are removed from the DNA concomitant with DNA processing, while other histone PTMs serve to trigger the subsequent displacement of repair proteins from the vicinity of the DSB. Furthermore, restoration of the chromatin structure after repair facilitates inactivation of the DNA damage checkpoint and cell cycle re-entry. Strikingly, histone modifications also help determine the location of DSBs made during programmed events such as V(D)J recombination and meiosis. Below we discuss each specific chromatin change in the context of the stage of DSB repair that is most directly promoted by the chromatin change. Needless to say, all upstream chromatin alterations indirectly promote all subsequent events in the DNA repair process.
Chromatin modifications that stimulate DSB formation
The genomic location of DSBs generated during developmentally programmed events is directly stimulated by histone PTMs. The clearest example of this is the formation of DSBs by the RAG1–RAG2 endonucleases during V(D)J recombination in lymphocytes. RAG1–RAG2 are targeted to the V(D)J segments via a direct interaction between the PhD finger of RAG2 and trimethylated histone H3 on lysine 4 (H3 K4me3) (Matthews et al. 2007). Furthermore, the enzymatic activity of the RAG complex itself is stimulated by binding to H3 K4me3 (Grundy et al. 2010; Shimazaki et al. 2009). The physiological relevance of H3 K4me3 in V (D)J recombination is underscored by defective V(D)J recombination that results from reduced H3 K4me3 levels (Matthews et al. 2007).
Meiotic crossover formation occurs in response to programmed DSBs generated at meiotic hotspots by a meiosis-specific endonuclease. Recent studies investigating the rationale for the selection of sites of meiotic DSBs have uncovered a unique role for chromatin in this process. Specifically, H3 K4me3 marks the majority of the meiotic hotspots in yeast and mice (Borde et al. 2009; Buard et al. 2009). In worms, acetylated H2A K5 (H2A K5Ac) modulates the sites of meiotic crossovers (Wagner et al. 2010). Exactly how histone PTMs dictate sites of meiotic crossover is currently unknown, but further studies will undoubtedly reveal the basis for their fundamental role in driving genetic diversity.
The damage sensor PARP1 promotes chromatin alteration during alternative NHEJ and SSBR
Poly(ADP-ribose) polymerase (PARP) family proteins PARP1 and PARP2 have the ability to detect single- and double-strand DNA breaks (Schreiber et al. 2006). In mammalian cells, PARP1 and PARP2 appear to function at a very early stage of single-strand break repair and an alternative NHEJ pathway (Huber et al. 2004). This is because PARP1 and PARP2 are catalytically activated by the DNA breaks, resulting in the local addition of poly(ADP-ribose) chains onto various proteins, including itself, at the site of the DNA break. These poly(ADP-ribosylated) proteins recruit other proteins including the ATP-dependent chromatin remodeling complex known as amplified in liver cancer 1 and CHD1L, which presumably stimulates nucleosome sliding at the site of DNA damage (Ahel et al. 2009). Among PARP1’s many substrates are the histone tails (Messner et al. 2010), but whether this plays a role in the DDR is unknown. Poly(ADP ribose) is also responsible for recruiting repressive polycomb group proteins and the ATP-dependent nucleosome remodeler NuRD (which contains CHD4) to sites of DNA damage (Chou et al. 2010). In this study, they also found that PARP contributes to the removal of nascent RNA and elongating RNA polymerase II from the sites of DNA damage, suggesting that PARP may promote a transcriptionally repressive local environment to facilitate DNA repair (Chou et al. 2010). Clearly, much more research on the role of PARP1/2 and poly(ADP ribose) in the DDR is warranted, especially in light of the fact that PARP1 inhibitors are promising treatments for cancer patients with defective HR pathways, for example, due to BRCA1 or BRCA2 mutations (Sandhu et al. 2011).
Chromatin changes that facilitate the initial activation of the DDR kinase ATM
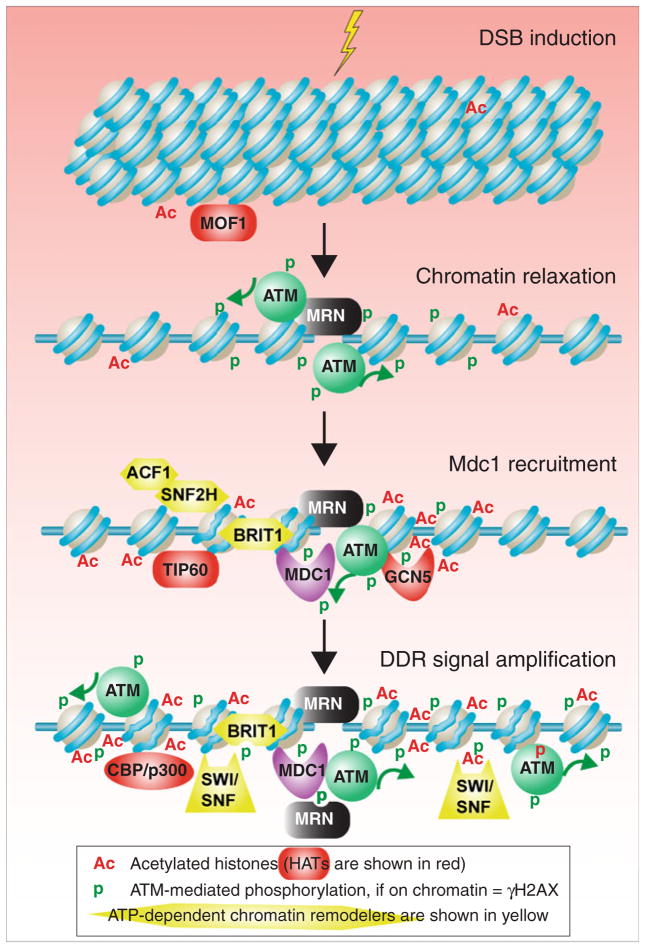
Activated ATM kinase is one of the first proteins to be recruited to a DSB (Fig. 1), where activation of ATM is a consequence of ATM-mediated autophosphorylation (Bakkenist and Kastan 2003). Once recruited to the DNA break, ATM phosphorylates many proteins including a histone H2A variant called H2AX on serine 139 (S139) to form γH2AX. As such, appearance of γH2AX is one of the very early post-DSB events in the mammalian cell, appearing within 1 min of DSB induction (Bewersdorf et al. 2006; Celeste et al. 2002; Rogakou et al. 1998, 1999). Phosphorylation of H2AX is mediated not only by activated ATM kinase but also by the related kinases ATR (which is recruited to single-strand DNA after DNA processing) and DNA-PK (which functions downstream of ATM only in NHEJ) (Pinto and Flaus 2010). Proteins involved in DNA repair and the DDR localize together with DSBs into repair foci that are apparent by immunoflourescence analysis. As such, the appearance of γH2AX foci is a widely used and convenient marker of both ATM activation and DSB breaks.
Fig. 1.
Role of chromatin in amplification of the DDR at a DSB site. As discussed in the text, in response to a DSB, the chromatin structure locally relaxes, facilitating the activation and recruitment of the ATM kinase. Phosphorylation of H2AX by ATM enables recruitment of many proteins including MDC1. ATM-mediated phosphorylation of MDC1 enables further amplification of the DNA damage response. DNA is shown in blue and histones are beige, and the wiggly DNA in nucleosomes around the DSB depict those that have been acted upon by ATP-dependent nucleosome remodelers, to facilitate access to the DNA
Mutations or drugs that alter the global chromatin state in the cell have been shown to facilitate or block activation of ATM, depending on whether they open up or condense the chromatin structure, respectively. H2AX is phosphorylated along megabase-long tracts of chromatin surrounding the DSB following the pattern of ATM spreading in humans (Rogakou et al. 1999). It has also been reported that λ H2AX forms preferentially in euchromatin (open chromatin) (Cowell et al. 2007; Kim et al. 2007b), suggesting that a closed chromatin structure hinders ATM activation. Furthermore, the absence of the histone acetyl transferase (HAT) hMOF-1 delays formation of γH2AX foci (Sharma et al. 2010). hMOF-1 mediates acetylation of histone H4 lysine 16 (H4 K16Ac), which is a unique histone modification in that it abrogates the formation of the compact 30-nm fibers of chromatin (Shogren-Knaak et al. 2006). As such, cells lacking hMOF-1 have globally a more compact chromatin structure that hinders ATM activation. Conversely, global activation of ATM occurs in the absence of DNA damage following treatment with class I and class II histone deacetylase (HDAC) inhibitors, which opens up the chromatin structure by increasing histone acetylation (Bakkenist and Kastan 2003). HMGN1 is a structural component of chromatin that directly or indirectly stimulates H3 K14Ac but is not itself recruited to sites of DNA damage, yet cells lacking HMGN1 show defective ATM induction (Kim et al. 2009). Meanwhile, ATM activation is restored in cells lacking HMGN1 upon chromatin relaxation using histone deacetylase inhibitors. Tellingly, these studies indicate that the preexisting chromatin state in the cell influences, directly or indirectly, activation of ATM and presumably all subsequent events in the repair pathway and DDR.
Although the studies discussed above used experimental manipulations to open up the global chromatin structure to facilitate ATM activation, it appears that our cells naturally open up the chromatin structure locally to bring about the initial round of ATM activation in response to a DSB. Specifically, electron microscopy has revealed rapid local chromatin decondensation around the DSB prior to ATM activation (Kruhlak et al. 2006). This is supportive of the idea that ATM is activated in response to architectural changes in the chromatin structure itself at the DSB site. Such an architectural change could be the DNA break induced relaxation of the torsional stress that is held within a chromatin domain (Fig. 1). Consistent with this idea, the length of the stretch of λ H2AX induced in response to a single DSB corresponds to the length of a topologically constrained chromatin loop domain (Rogakou et al. 1999).
Chromatin modifications that amplify the DDR and facilitate formation of DNA repair foci
γH2AX facilitates recruitment and retention of DDR proteins to repair foci and in so doing amplifies the DDR (Yuan et al. 2010) (Fig. 1). One protein that is directly recruited by γH2AX to repair foci is mediator of DNA damage checkpoint protein 1 (MDC1). MDC1 binds to γH2AX via its BRCT domain and accumulation of MDC1 on the chromatin is essential for formation of ionizing radiation-induced foci (IRIF) (Jungmichel and Stucki 2010; Stucki et al. 2005) (Fig. 1). Formation of γH2AX foci is a self-perpetuating and dynamic process. γH2AX-recruited MDC1 recruits more activated ATM, thus establishing the positive feedback loop of γH2AX expansion along the DNA (Lou et al. 2006; Stucki et al. 2005). MDC1 in turn is phosphorylated by both ATM and casein kinase 2, the latter of which phos-phorylates a series of repeated motifs required for the Mdc1/ Nbs1 interaction that retains MRN in IRIFs along the chromatin (Chapman and Jackson 2008; Melander et al. 2008; Spycher et al. 2008; Wu et al. 2008). MRN and end processing of the DNA then recruits more ATM (Lee and Paull 2007). As such, there are multiple positive feedback loops mediated by histone PTMs and nucleosome remodeling that promote amplification of the DDR and further activation of ATM (Fig. 1).
The binding of MDC1 to γH2AX is modulated by other histone modifications, in addition to its direct interaction with phosphorylated H2AX. MDC1 recruitment to γH2AX additionally requires, directly or indirectly, MOF1-dependent H4 K16 acetylation (Li et al. 2010). MDC1 recruitment is also regulated by phosphorylation of Y142 within the same C-terminal tail of H2AX. MDC1 only binds to unphosphorylated γH2AX Y142, whose dephosphorylation is mediated by EYA1/3 (Cook et al. 2009; Xiao et al. 2009). Interestingly, the phosphorylation status of Y142 of γH2AX appears to determine the choice between activation of the DDR or cell death because apoptotic factors bind to phosphorylated Y142 within γH2AX (Cook et al. 2009).
In addition to recruiting MDC1, γH2AX triggers dynamic acetylation and chromatin remodeling at the site of the DSBs. DNA damage is known to induce H3 acetylation on the same nucleosomes that contain γH2AX flanking the DSB. This is presumably a consequence of the HAT GCN5 binding to γH2AX nucleosomes (Botuyan et al. 2006), leading to its nearby H3 acetylation. The acetylated H3 is bound directly by the bromodomain of the BRG1 subunit of the SWI/SNF ATP-dependent nucleosome remodeler specifically in nucleosomes containing γH2AX (Lee et al. 2010a) and presumably functions to increase the accessibility of the DNA. The recruitment of SWI/SNF in turn stimulates further ATM-mediated H2AX phosphorylation (Botuyan et al. 2006; Park et al. 2006) (Fig. 1).
Independent of H2AX phosphorylation (H2A S129p in yeast), the yeast ATP-dependent nucleosome remodeler RSC is recruited to sites of DSB as a very early event. This is required for subsequent activation of the yeast counterparts of ATM and ATR, which are Tel1 and Mec1, respectively (Chai et al. 2005; Liang et al. 2007; Shim et al. 2005). In mammalian cells, additional ATP-dependent remodelers, SNF2L, SNF2H (Erdel et al. 2010), and ACF1, are recruited to sites of DSBs as very early events (Lan et al. 2010). The recruitment of SNF2H to sites of DNA damage appears to depend on ACF1. Knockdown of components of these complexes leads to defects in both HR and NHEJ as a consequence of defective recruitment of RPA, BRCA1, RAD51, and KU (Lan et al. 2010; Nakamura et al. 2011). More recently, it has been shown that cells lacking ACF1 have a clear reduction in the amount of γH2AX signal at early times following IR exposure (Sánchez-Molina et al. 2011). However, whether ACF1’s recruitment and role at a DSB is dependent on ATM/ATR has not yet been tested, so it is not yet known if ACF1 is required for amplification of the DDR signal or for initial activation of the DDR.
Akin to the situation with GCN5, the H3 and H4 HATs p300 and CBP are also recruited to sites of DSBs. Once recruited, p300/CBP-mediated acetylation works in parallel with the BRM and BRG1 subunits of the SWI/SNF ATP-dependent nucleosome remodeler to promote KU recruitment and NHEJ (Ogiwara et al. 2011). In addition to the interaction between the BRM and BRG1 subunits of the SWI/SNF ATP-dependent nucleosome remodeler with acetylated histones, another ATP-dependent nucleosome remodeler BRIT1 also helps with the accumulation of SWI/SNF at IRIF (Peng and Lin 2009; Rai et al. 2006). The interaction between BRIT1 and SWI/SNF is stimulated by ATM-dependent phosphorylation of the SWI/SNF subunit BAF170 (Peng and Lin 2009). BRIT1 (also known as microcephalin 1) is recruited to IRIF through interactions between its C-terminal BRCT domains and γH2AX (Wood et al. 2007), where it plays a critical role in the DDR. Specifically, BRIT1 depletion impairs accumulation of many downstream DDR proteins including MDC1, KU70, RPA, RAD51, NBS1, and BRCA1 (Lin et al. 2005; Peng and Lin 2009; Rai et al. 2006), suggesting that it might be functioning at quite an early step in the DDR.
Yet another function of γH2AX is to facilitate histone acetylation of H4 and H2B by binding to the Arp4 subunit of the NuA4 HAT complex in yeast (Bird et al. 2002; Downs et al. 2004). Its counterpart is TIP60 in mammals, which also mediates H4 acetylation in response to DSBs that can extend several kilobases on each side of a DSB (Murr et al. 2006). TIP60 recruitment to DSBs promotes the efficient subsequent recruitment of DDR proteins including MDC1, 53BP1, BRCA1, and RAD51 and promotes efficient HR (Murr et al. 2006). The contribution of TIP60 to HR can be partially overcome by chemical relaxation of the chromatin, suggesting that the function of TIP60-mediated histone acetylation is to promote access of DDR proteins to the site of the DSB (Murr et al. 2006). Similarly, elimination of NuA4 HAT activity results in a DSB repair defect, but this defect is abrogated in cells in which chromatin was relaxed chemically (Murr et al. 2006). These results may be related to the fact that relaxation of chromatin by the NuA4/TIP60 complex is required to allow histone ubiquitination that is in turn necessary for recruitment of BRCA1 and 53BP1 (Murr et al. 2006; Xu et al. 2010). In summary, γH2AX plays a critical role in recruiting not only MDC1 to the DSB but also in recruiting multiple different HATs and ATP-dependent nucleosome remodelers to the site of the DSB, which aid in amplification of the DDR (Fig. 1).
The role of histone modifications in recruitment of DNA damage response mediator 53BP1
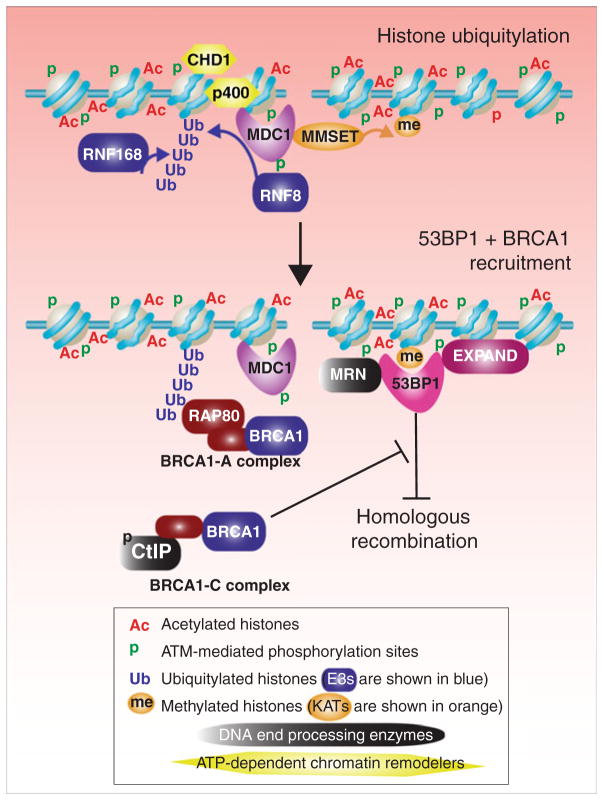
After histone acetylation in the vicinity of the DSB (Fig. 1), histone ubiquitylation occurs (Fig. 2). The ubiquitin ligase ring finger 8 (RNF8) is recruited to the site of the lesion by its direct interaction with ATM-phosphorylated MDC1 (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007). RNF8 adds K63-linked ubiquitin to H2A and H2AX, which creates a docking site for a second E3 ligase RNF168 which makes a polyubiquitin chain on H2A (Doil et al. 2009; Stewart et al. 2009; Zhao et al. 2007). Another ubiquitin ligase, RNF2, also contributes to DNA damage inducible γH2AX ubiquitylation (Chou et al. 2010; Ginjala et al. 2011; Ismail et al. 2010). RNF2 is a component of the polycomb repressive complex 1 (PRC1) that is recruited to sites of damage by binding to the BRCT domain of NBS1 to perform monoubiquitylation of γH2AX (Ismail et al. 2010). There is even evidence that di-ubiquitylation of γH2AX by RNF8 depends on earlier monoubiquitylation by PRC1 (Ginjala et al. 2011), followed by polyubiquitylation by RNF168. Regardless of the mechanism of histone ubiquitylation, 53BP1 recruitment is dependent on this H2A/H2AX polyubiquitylation (Bekker-Jensen et al. 2005; Mailand et al. 2007). However, 53BP1 does not bind directly to ubiquitin chains; rather, it binds via tandem TUDOR domains to dimethylated lysine K20 on histone H4 (Botuyan et al. 2006; Huyen et al. 2004). How histone H2A ubiquitylation facilitates the 53BP1/dimethylated histone interaction is not precisely defined because downregulation of RNF8 does not affect H4 K20 methylation (Pei et al. 2011). Clearly the situation is quite complicated because down-regulation of monoubiquitylation of H4 K91 also reduces H4 K20 methylation and 53BP1 recruitment to damage sites (Yan et al. 2009).
Fig. 2.
Role of chromatin in recruitment of mediators of the DDR to DSB sites. The steps shown in this figure follow on from those in Fig. 1. MDC1 promotes the dimethylation of H4 K20 via recruiting MMSET which in turn is recognized by the 53BP1 checkpoint mediator protein. MDC1 also promotes ubiquitinylation of H2A by recruiting the p400 ATP-dependent nucleosome remodeler and the RNF8 E3 ligase. RNF168 acts after RNF8 to generate a polyubiquitin tail on H2A, which in turn serves to recruit the BRCA1-A checkpoint mediator complex to the DSB site. 53BP1 favors NHEJ over homologous recombination, while the BRCA1-C complex containing the phosphorylated CtIP endonuclease overcomes the repressive effect of 53BP1 on homologous recombination
Like the step-wise action of E3 enzymes to ubiquitylate H2A, the step-wise action of histone methyltransferases is used to generate H4 K20me2 for recruitment of 53BP1 for repair. First, monomethylated H4 K20, generated by PR-SET7, must preexist on the chromatin near the DSB (Botuyan et al. 2006; Oda et al. 2009). This is a prerequisite for MMSET-mediated generation of H4 K20me2. H4 K20me2 and MMSET are locally enriched at the sites of damage due to MMSET recruitment by MDC1 (Pei et al. 2011). MMSET phosphorylation is induced after DNA damage, and this promotes its recruitment to DSB sites via a specific interaction between phosphorylated MMSET and the BRCT domain of MDC1 that is already in the vicinity of the DSB (Pei et al. 2011). While the stable accumulation of 53BP1 into repair foci depends on MDC1 and the RNF8–RNF168 pathways, 53BP1 is still transiently recruited in H2AX deficient cells (Celeste et al. 2003). Indeed, the role of 53BP1 in NHEJ is H2AX-independent (Xie et al. 2007), and this may be related to the fact that the interaction between 53BP1 and monomethylated H4 K20 (remember the dimethylation of H4 K20 requires γH2AX) promotes NHEJ while suppressing HR (Sanders et al. 2004; Xie et al. 2007). While the fission yeast counterpart of 53BP1, Crb2, is also recruited to damage sites via binding to H4 K20me2 (Sanders et al. 2004), budding yeast lacks this histone modification. Instead, the budding yeast counterpart of 53BP1, Rad9, is recruited via interaction with H3 K79me3 (Giannattasio et al. 2005; Wysocki et al. 2005).
As a mediator protein of the DDR, the functional role of 53BP1 is to accumulate in chromatin surrounding the DSB and facilitate checkpoint signaling. Though indirect, 53BP1 ultimately impairs CHK2 phosphorylation (Fernandez-Capetillo et al. 2002; Wang 2002; Ward et al. 2003). In vitro, Lee et al. (2010b) showed that the effect of 53BP1 on CHK2 phosphorylation results from 53BP1/MRN complex interactions that stimulate ATM activity. Investigations into the effects of 53BP1 depletion on the DDR have yielded somewhat inconsistent results ranging from a lack of G2/M arrest altogether to an apparently intact DDR response (FitzGerald et al. 2009). This could be explained, at least in part, by redundant activities of mediator proteins including BRCA1 and MDC1 that may take on somewhat varied roles in different circumstances (Mochan et al. 2003; Peng and Chen 2003; Wilson and Stern 2008). Another interesting possibility is that the observed inconsistent effects of 53BP1 on the DDR are related to its unique role in a special kind of DSB repair: NHEJ of distant ends demonstrated in deprotected telomeres (Dimitrova et al. 2008), Igh class-switch recombination (Manis et al. 2004; Ward et al. 2004), and long-range V(D)J recombination (Difilippantonio et al. 2008). In these scenarios, 53BP1 increases chromatin mobility, allowing for intra- and inter-molecular interactions that may otherwise not occur. Thus, it could be reasoned that the varied impact of 53BP1 on the DDR in earlier studies was affected by the nature of the lesion and its chromatin context. A recent search for 53BP1-interacting proteins identified the previously uncharacterized EXPAND1/MUM protein that mediates DNA damage-induced chromatin decon-densation (Sy and Huen 2010). This chromatin relaxation function of EXPAND is presumably mediated through its interaction with nucleosomes via its PWWP domain, a domain commonly found in chromatin-binding proteins (Huen et al. 2010). Future studies no doubt will reveal how EXPAND relaxes the chromatin structure to promote DNA repair.
The role of histone ubiquitylation in recruitment of BRCA1
The ubiquitylated histones play a critical role in recruitment of BRCA1 to the DSB (Fig. 2). The polyubiquitin chains on histone H2A (discussed above) directly recruit the RAP80 subunit of the BRCA1-containing complex BRCA1-A (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007; Yan et al. 2007). The monoubiquitylation of H2A by RNF8 itself is enhanced by the chromatin remodeling activities of the ATP-dependent nucleosome remodelers p400 and CHD4 (Larsen et al. 2010; Smeenk et al. 2010), presumably by generating a chromatin state that is more open and more accessible for ubiquitylation as they are dispensable for RNF8 recruitment (Xu et al. 2010). Like RNF8 recruitment, p400 recruitment to the DSB is dependent on MDC1 (Xu et al. 2010). Once RNF168 has generated the polyubiquitin chains on H2A, the tandem ubiquitin interacting motifs of the RAP80 subunit of the BRCA1-A complex bind to the polyubiquitin chains on H2A (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007). As such, the ubiquitin chains on H2A are responsible for recruitment of BRCA1-A to sites of DNA repair.
BRCA1 is a component of multiple repair-related protein complexes. These complexes include BRCA1-A, BRCA1-B, and BRCA1-C, named for the ATM- or ATR-phosphorylated proteins Abraxas, BACH1, and CtIP, respectively, with which the C-terminal BRCT repeats of BRCA1 bind (Manke et al. 2003; Wang et al. 2007; Yu and Chen 2004; Yu et al. 2003). How the BRCA1-B and C complexes are recruited to sites of DSBs is not so clear, nor is their function. BRCA1 itself is an E3 ligase, although its substrates are unknown, but BRCA1 promotes recruitment of BRCA2, which appears in turn to promote recruitment of RAD51 (Qing et al. 2011) for homologous recombination.
Chromatin also plays a role in the choice between using the NHEJ and HR pathways for DSB repair. A key protein in this choice appears to be BRCA1 and the associated CtIP endonuclease, presumably within the context of the BRCA1-C complex. Phosphorylation of CtIP occurs as cells enter S phase, enabling it to function in DNA end processing, thus promoting HR during S and G2 phase (Yun and Hiom 2009). Meanwhile in G1 phase, CtIP is not phosphorylated, and its ability to perform DNA end processing is inhibited by 53BP1 bound at the site of the DSB. In fact, CtIP-mediated (Sae2 in yeast) DNA end processing is required for HR (You and Bailis 2010), but only in the presence of 53BP1, as concurrent loss of 53BP1 in BRCA1-deficient cells restores efficient HR (Bouwman et al. 2010; Bunting et al. 2010). Thus, it appears that end processing by phosphorylated CtIP in the BRCA1-C complex effectively removes methylated and ubiquitylated his-tones at the DSB that accumulate an otherwise potent inhibitor of HR, 53BP1 (Lowndes 2010) (Fig. 2).
In addition to H2A and H2AX ubiquitylation, histone H2B ubiquitylation (on K120) also promotes DSB repair (Moyal et al. 2011; Nakamura et al. 2011). H2B ubiquitylation is performed by a complex of RNF20/RNF40 (Pavri et al. 2006). RNF20/RNF40 is recruited to DSBs, although the mechanism for its recruitment is unknown (Moyal et al. 2011). Noteworthy, the ability of RNF20/RNF40 to ubiquitylate H2B requires its ATM-mediated phosphorylation (Moyal et al. 2011). The exact step during DNA repair at which ubiquitylated H2B functions is not known, but its absence results in reduced recruitment of XRCC4 and KU (during NHEJ) and reduced recruitment of BRCA1/2 and RAD51 during HR (Moyal et al. 2011; Nakamura et al. 2011). In yeast, H2B ubiquitylation (on K123) by Rad6/ Bre1 has been tied directly to the DDR, as deletion of either RAD6 or BRE1 activity resulted in defective activation of Rad53 kinase after DNA damage, especially when cells were damaged in G1 (Giannattasio et al. 2005).
Repair of DSBs within heterochromatin requires unique chromatin dynamics
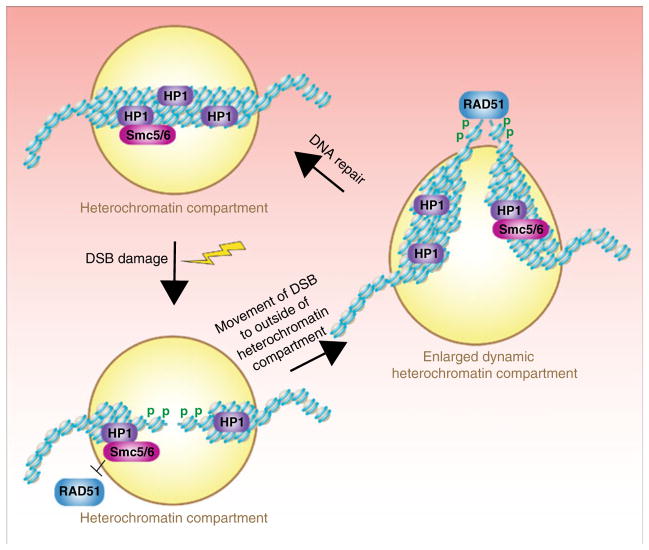
Not all DSBs are created equal, as the chromatin context surrounding the DSB influences the cell’s response to the lesion. Heterochromatin is a highly compacted, transcriptionally inactive chromatin structure that is characterized by the enrichment of specific histone modifications (for example, H3 K9me2 and H3 K9me3) and the proteins that specifically bind to these histone PTMs, such as HP1. It has been suggested that DSBs within heterochromatin are detected at lower frequency and are resolved with slower kinetics (Goodarzi et al. 2008; Ayoub et al. 2008). Furthermore, it has been proposed that delocalization of HP1beta and its binding partner KAP1 from sites of DSBs facilitates repair (Ayoub et al. 2008; Goodarzi et al. 2008). However, a recent careful time-lapse study in Drosophila shows that DSBs within heterochromatin are recognized and processed as efficiently as those in euchromatin (Chiolo et al. 2011) (Fig. 3). Strikingly, this study showed that the recruitment of proteins involved in early repair events makes the heterochromatin rapidly expand and become dynamic, causing the repositioning of the DSB outside of the heterochromatin compartment in order to complete repair (Chiolo et al. 2011). Such a scenario could also explain the difficulty to detect repair foci within heterochromatin and the apparent displacement of HP1-beta from sites of DSBs even in the absence of changes in H3 K9me3 levels (Goodarzi et al. 2008; Ayoub et al. 2008). Like the situation in Drosophila, DNA breaks within heterochromatin in mammalian and human cells also elicit rapid recruitment of repair proteins, and the DSBs are relocalized to outside the heterochromatin compartment presumably for the completion of DNA repair (Falk et al. 2007; Jakob et al. 2011). Despite the differences in chromatin structure and genomic organization between flies, murine, and human cells, DSB movement to the periphery of heterochromatic compartments for the rapid repair of heterochromatic DNA lesions appears to be a general phenomenon.
Fig. 3.
Chromatin dynamics during repair of heterochromatinized DSBs. DSBs within heterochromatin are efficiently and rapidly recognized by the DNA damage checkpoint machinery leading to phosphorylation of H2AX (shown by the green p). The chromatin within the heterochromatin compartment next becomes highly mobile and dynamic, resulting in the movement of the DSB to the periphery of the heterochromatin compartment. Once at the periphery, homologous recombination can occur due to the recruitment of proteins such as Rad51, which are blocked from association with the DSB within the heterochromatin compartment by the Smc5/6 proteins
Functionally, the purpose of the repositioning of DSBs made within heterochromatin to regions away from heterochromatin compartment is to prevent inaccurate homologous recombinational repair with the many identical repeated sequences within heterochromatin (Chiolo et al. 2011). This repositioning of DSBs within heterochromatin to outside of the heterochromatin compartment occurs after DNA processing but before Rad51 recruitment (Chiolo et al. 2011). This DSB movement also required HP1alpha dependent binding of the Smc5/6 complex to the heterochromatin, which served to block Rad51 association with the DSB while it was within the heterochromatin compartment. It will be interesting to learn how Smc5/6 suppresses recruitment of Rad51 and whether HP1alpha and the interacting Smc5/6 proteins are removed from the vicinity of the DSB when it relocates outside the heterochromatin compartment. Also, we know virtually nothing about the mechanism whereby the heterochromatin expands and becomes dynamic in response to a DSB, but it is promoted by the DDR, particularly ATR (Chiolo et al. 2011). Phosphorylation of the heterochromatin protein KAP1 by ATM leads to global chromatin relaxation following DSB damage (Goodarzi et al. 2008; Ziv et al. 2006), but whether this is related to the heterochromatin relaxation that occurs upon damage is not clear. Also, how KAP1 phosphorylation relaxes chromatin is unknown but loss of the ATP-dependent nucleosome remodeler CHD3 from DSBs is implicated in the mechanism (Goodarzi et al. 2011).
Contrary to the reported loss of HP1 beta and KAP1 from DSBs in heterochromatin (Ayoub et al. 2008; Goodarzi et al. 2008), all three isoforms of HP1 (alpha, beta, and gamma) have been shown to be recruited to DSBs and UV lesions (Ayoub et al. 2009; Luijsterburg et al. 2009; Zarebski et al. 2009). In the case of HP1alpha, its recruitment to DSBs occurs independent of H3 K9me3, but it does require its physical interaction with the large subunit of the histone chaperone CAF-1, which also localizes to laser-induced DNA lesions (Baldeyron et al. 2011). Indeed, CAF-1 recruits both HP1alpha and KAP1 to DSBs in mammalian cells (Baldeyron et al. 2011). Functionally, the HP1alpha appears to be required for promoting HR at the end processing stage and for the recruitment of 53BP1, BRCA1, and RAD51 (Baldeyron et al. 2011). The recruitment of KAP1 and HP1alpha to DSBs was short-lived as they rapidly disappeared, while CAF-1 remains at damage sites longer (Baldeyron et al. 2011). These studies have been important in demonstrating that heterochromatin and heterochromatin proteins are not an obstacle for DNA repair, but rather they promote the efficiency and accuracy of DNA repair.
Chromatin changes that facilitate DNA end processing
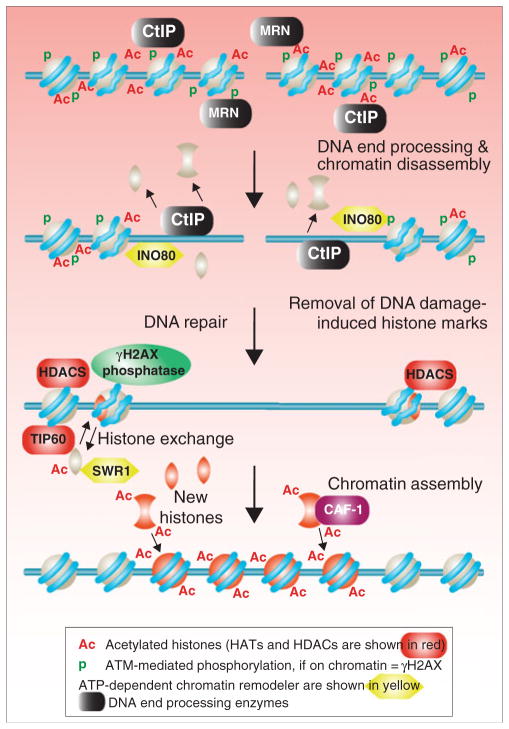
It makes structural sense for the histone proteins to be at least transiently removed from the vicinity of a DSB (Fig. 4). This would allow the DNA repair machinery to gain the intimate contacts that are required for DNA end processing and subsequent events in the DNA repair processes. Indeed, the processes of histone removal and end processing are tightly temporally linked. For example, the MRN end processing complex is required for nucleosome eviction at a DSB site in mammalian cells (Berkovich et al. 2007). Meanwhile, the yeast ATP-dependent nucleosome remodeler INO80 promotes histone removal from the site of a DSB, and Ino80 is required for efficient end processing (Chen et al. 2008; Morrison et al. 2004; van Attikum et al. 2004). Yeast Ino80 is recruited to sites of DSB via interaction between γH2AX and its Arp4 subunit, the same subunit found in the NuA4 HAT that mediated its recruitment for histone acetylation (Cairns 2004; Downs et al. 2004; van Attikum et al. 2004). Indeed, upon formation of DSBs, nucleosome loss is observed up to a few kilobases flanking the damaged sites (Chen et al. 2008; Tsukuda et al. 2005). None of the yeast histone chaperones that have been tested to date are required for removal of histones from the site of a DSB (Chen et al. 2008).
Fig. 4.
Reestablishment of chromatin after DSB repair. DNA resection in yeast is accompanied by loss of the histones from the DNA being processed. Phosphorylated H2AX either gets dephosphorylated or those histones get removed from the chromatin and replaced with unphosphorylated histones (termed histone exchange). Following DNA repair, histone chaperones such as CAF-1 bring in new histone carrying the H3 K56Ac mark onto the repaired DNA, serving as a signal to the DNA damage checkpoint that repair is now complete
While the yeast INO80 ATP-dependent nucleosome remodeler contributes to nucleosome eviction around a DSB, that is not the case with another ATP-dependent nucleosome remodeler SWR1 (Morrison and Shen 2009; van Attikum et al. 2007). Like INO80, SWR1 is recruited to DSBs in a manner dependent on phosphorylated H2A but, unlike INO80, SWR1 does not function to evict nucleosomes (van Attikum et al. 2007). Rather, SWR1 functions to remove canonical H2A and replace it with the histone variant H2AZ. Accordingly, H2AZ is transiently enriched at DSB sites due to the action of SWR1. The absence of H2AZ results in a defect in Rad51 recruitment, although the molecular mechanism as to how H2AZ helps promote Rad51 recruitment is not clear (Kalocsay et al. 2009).
Another histone PTM that may promote DNA end processing is H3 K36me2. This modification is mediated by the Metnase/SETMAR methyltransferase. The function of H3 K36me2 during NHEJ is to recruit NBS1 and the KU complex, although whether this function is direct or indirect is unknown (Fnu et al. 2011). However, given its requirement for NBS1 recruitment, H3 K36me2 is likely to stimulate end processing.
Histone PTMs that prevent promiscuous DNA processing
Binding of the budding yeast Rad9 to H3 K79me2/3 appears to slow down DNA processing. The evidence for this includes the rapid accumulation of ssDNA at DSBs in yeast deleted for RAD9 or the H3 K79 methyltransferase DOT1 whose modification helps recruit Rad9 to breaks (Lazzaro et al. 2008). The human counterpart of Rad9, 53BP1, similarly inhibits promiscuous end resection and plays a critical role in genome stability (Bothmer et al. 2011; Bothmer et al. 2010; Bunting et al. 2010). γH2AX also plays a role in preventing promiscuous end processing during VDJ recombination. DSB ends generated during VDJ recombination are recognized and cleaved by Artemis. As discussed above, γH2AX recruits MDC1 flanking a DSBs, which in turn inhibits CtIP-mediated processing to facilitate only Artemis-mediated processing (Helmink et al. 2011). This recently uncovered function of γH2AX in limiting end processing likely explains the earlier observed requirement for ATM in maintaining DNA ends during lymphocyte antigen receptor gene assembly (Bredemeyer et al. 2006). This function of ATM is likely to provide the molecular explanation for the increase in lymphoid tumors with translocations involving antigen receptor loci associated with ataxia telangiectasia (Bredemeyer et al. 2006).
Chromatin changes that promote recruitment of the late acting DNA repair machinery
In addition to the histone acetylations that have already been discussed above, many other dynamic histone acetylation and deacetylation events occur during the DDR. During the process of homologous recombination in yeast, the levels of many H3 and H4 histone N-terminal tail acetylations increase rapidly on the chromatin in the vicinity of a DSB and are then deacetylated concomitant with DNA repair (Tamburini and Tyler 2005). This is presumably mediated by the dynamic recruitment of the responsible HAT and HDAC enzymes to the local vicinity of the DSB (Tamburini and Tyler 2005). Yet other acetylation marks, for example H3 K56Ac and H4 K16Ac, decrease initially after DNA damage and increase at later times after DNA repair (Das et al. 2009; Miller et al. 2010; Tjeertes et al. 2009; Vempati et al. 2010). This initial decrease in H3 K56Ac and H4 K16Ac is mediated by CHD4-dependent recruitment of HDAC1 and HDAC2 and is required for efficient disassembly of the NHEJ repair factors Artemis and KU (Miller et al. 2010; Polo et al. 2010). At least in the case of KU, its removal from sites of DNA repair has been proposed to facilitate post-repair processes (Postow et al. 2008).
Chromatin modifications also promote the stabilization of some of the very last DNA repair proteins at the DSB. For example, the retention of the XRCC4–ligase IV complex at NHEJ sites is enhanced by the histone chaperone APLF (Rulten et al. 2011). The functional consequence of this is apparent because the absence of APLF renders cells sensitive to DSBs (Mehrotra et al. 2011). Although the functional relevance is not clear, the mammalian INO80 ATP-dependent nucleosome remodeling complex binds to four-way junctions that mimics Holliday junctions in vitro (Wu et al. 2007). It is tempting to speculate that this may represent a mechanism to recruit machinery to help disassemble chromatin ahead of the DNA polymerase during branch migration.
Chromatin changes that promote turning off the DDR after DNA repair
Chromatin structure has to be altered in two general ways after DNA repair. Histones have to be reassembled to repackage the naked newly repaired DNA into chromatin (Fig. 4). In addition, the histone modifications that served to recruit DDR proteins, including γH2AX, have to be lost from the regions flanking the site of repair in order to allow inactivation of the DDR and cell cycle re-entry.
Histone reassembly onto the DNA following DSB repair is tightly coupled to DNA synthesis. In yeast, the assembly of histones H3 and H4 at the site of DSB repair occurs in a manner that requires the histone chaperone CAF-1 (J.K.T and Cc.C., unpublished observation) and another histone chaperone Asf1 (Chen et al. 2008) (Fig. 4). The role of Asf1 in chromatin assembly during DSB repair is indirect, in that Asf1 helps the HAT Rtt109 to acetylate free histones on H3 lysine 56, which then drives their subsequent assembly onto the DNA (Chen et al. 2008). This is presumably a consequence of the increased affinity of histones carrying the H3 K56Ac mark for CAF-1, which itself is physically tethered to sites of DNA synthesis via its interaction with PCNA (Li et al. 2008). In agreement with the situation in yeast, in human cells, CAF-1 is required for the assembly of newly synthesized canonical histone H3.1 at sites of laser irradiation (Polo et al. 2006). Additionally, in humans, the H3 K56Ac mark is enriched at sites of DSB repair after repair is complete, providing a transient memory of the assembly of newly synthesized histones over the repair site (Das et al. 2009; Vempati et al. 2010). Perhaps most interestingly, mutant yeast strains that fail to reassemble chromatin even though they do complete DSB repair are unable to turn off their DNA damage checkpoint (Chen et al. 2008). This result indicates that repair of the DNA break itself is not sufficient information to end the DDR. Instead, assembly of the chromatin and/or the local enrichment of H3 K56Ac over the DNA repair site is somehow involved in signaling to inactivate the DDR machinery (Fig. 4). Exactly how this happens clearly requires more research and should help us better understand the rather vague process of DNA damage checkpoint inactivation or recovery.
The disassembly of histones from the immediate vicinity of the DSB during DNA resection would effectively remove many of the histone PTMs from the DNA. Noteworthy, DNA resection either does not occur or is very limited during NHEJ. As such, removal of histones PTMs from sites of NHEJ repair would either require histone exchange or the action of enzymes that reverse the histone modification (such as histone deacetylases, histone demethylases, deubiquitylases (DUB)). There are many examples of HDACs being recruited to sites of DNA repair (as discussed above), and the BRCA-1A complex actually contains a DUB, Brcc36, that can deubiquitinate histones at least in vitro (Sobhian et al. 2007).
Specialized machinery appears to be involved in removal of γH2AX from the site of the DSB. Indeed, in yeast, chromatin immunoprecipitation (ChIP) analyses have shown that γH2AX is removed from the vicinity of DNA damage even before DNA repair has occurred (Chen et al. 2008; Kim et al. 2007b). Whether this is the case over the whole domain is not clear. The removal of γH2AX from the DNA is potentiated by additional PTMs that are stimulated by its phosphorylation, which promote histone exchange (Heo et al. 2008; Ikura et al. 2007). Whether γH2AX is replaced with another molecule of unphosphorylated H2AX or a different histone monomer depends on the ATP-dependent nucleosome remodeling enzyme involved in exchange and may play a critical role in checkpoint signaling and/or adaptation (Kusch et al. 2004; Papamichos-Chronakis et al. 2006). Specifically, in Drosophila cells, the TIP60 acetyltransferase, a component of NuA4 complex, acetylates γH2AX and promotes SWR1-mediated exchange with unmodified H2A near the DSB sites (Kusch et al. 2004). Once γH2AX has been removed from the DNA in yeast, it is dephosphorylated by the yeast Pph3 phosphatase (Keogh et al. 2006). Furthermore, physical removal of γH2AX from the chromatin in yeast is not sufficient to lead to inactivation of the DDR because dephosphorylation of free γH2AX by Php3 is required for checkpoint inactivation. It is hard to envision how soluble γH2AX maintains an active DDR, given that the function of γH2AX in amplifying the DDR is to recruit and concentrate proteins to the location of the DSB. In contrast to yeast, dephosphorylation of human γH2AX by the human counterpart phosphatase PPC4 occurs on the chromatin (Chowdhury et al. 2008). Clearly the regulation of γH2AX removal from the DNA and/or its dephosphorylation must be very tightly regulated to enable checkpoint inactivation to occur.
Removal of unphosphorylated H2AX from the DNA requires histone acetylation. H2AX is acetylated at K5 by the HAT TIP60 in response to IR, which in turn leads to ubiquitylation of H2AX on K119 (Ikura et al. 2007). This acetylation and ubiquitylation of H2AX together trigger its removal from nucleosomes around the DSB in a manner independent of S139 phosphorylation of H2AX (Ikura et al. 2007). Why and how unphosphorylated H2AX is removed from the DNA and why this requires its acetylation and ubiquitylation are not clear. It is possible that this may just be the mechanism to remove H2AX from the DSB site to enable repair. Alternatively, it may be important in adaptation to an unrepairable DNA break because the removal of H2AX from the chromatin would prevent further phosphorylation of H2AX.
In addition to recruitment of HDACs to the site of DSB repair, there are active mechanisms to prevent histone acetylation after repair. For example, H4 S1p levels increase at a late stage of DSB repair in yeast (Cheung et al. 2005). This modification prevents NuA4 from acetylating the adjacent lysines (Utley et al. 2005), maintaining the chromatin over the repaired DNA in an underacetylated state to favor a more compact and less accessible conformation.
Chromatin changes that promote NER
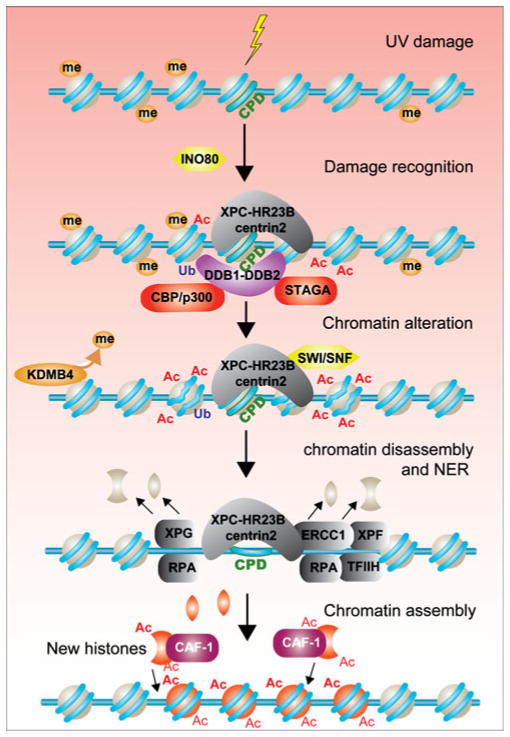
NER is the major mechanism used in cells to remove DNA damage induced by UV irradiation, such as cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6–4) pyrimidone photoproducts (6–4PPs). The importance of this repair mechanism is evidenced by the fact that several severe human diseases are genetically associated with mutations of NER proteins, such as Xeroderma pigmentosum and Cockayne’s syndrome. The detailed molecular mechanism of the NER pathway has been established from in vitro studies (Cleaver et al. 2009; Costa et al. 2003). DNA adducts formed by UV irradiation are initially recognized by the XPC-HR23B-centrin-2 complex and/or the DDB1–DDB2 heterodimer. Next, the DNA duplex is unwound by TFIIH, and this is facilitated by the helicase activity and ATPase activity of the XPD and XPB subunits of TFIIH, respectively. The single-strand DNA is then coated by RPA, as in DSB repair. XPG and ERCC1-XPF cleave the 3′ and 5′ ends of the damaged DNA strand, and the gap is filled by DNA synthesis. The mechanisms responsible for activation of the DDR by UV lesions have been described elsewhere (Giannattasio et al. 2010; Sertic et al. 2011), and we will only focus on the intersection of chromatin and NER here.
Both CPDs and 6–4PPs induce a bend or kink in the DNA, and this is what is recognized by the NER machinery (Kim et al. 1995). While 6–4PPs are located primarily within the linker DNA region, CPDs can also occur within the DNA wrapped around the histone octamer (Gale et al. 1987; Suquet and Smerdon 1993). CPDs within a nucleosome change the dynamic equilibrium of DNA unwrapping and rewrapping around the nucleosome by favoring spontaneous unwrapping of DNA (Duan and Smerdon 2010), thus increasing the accessibility of lesions. Besides the DNA distortion, other factors also actively contribute to reveal and mark the lesion sites for recruitment of the repair machinery. Two E3 ligase complexes, DDB1–DDB2–CUL4A and DDB1–DDB2–CUL4B, can recognize and bind to UV-damaged DNA respectively (Guerrero-Santoro et al. 2008) and monoubiquitylate H2A at these sites (Fig. 5). The ubiquitin ligase Ring2 has also been suggested to mediate UV-induced monoubiquitylation of H2A (Bergink et al. 2006). Furthermore, these DDB1–DDB2 containing E3 complexes have also been shown to weaken DNA–histone interaction by ubiquitylating H3 and H4, facilitating recruitment of XPC to the damage sites (Wang et al. 2006). The function of this histone ubiquitylation is to promote dissociation of DDB2 from the DNA damage site, enabling the recruitment of subsequent NER factors to the lesion (Takedachi et al. 2010).
Fig. 5.
Role of chromatin in NER. DNA distortion that is caused by UV-induced DNA damage (a CPD is shown for example) is recognized by the XPC–HR23B–centrin2 and DDB1–DDB2 complexes. DDB1–DDB2 recruits histone acetyl transferases (shown in red) to the site of the DNA lesion, inducing histone acetylation to further loosen the chromatin structure. For lesions within heterochromatin, H3 K9 is demethylated by KDMB4 to facilitate NER. Histone loss presumably accompanies the excision and removal of the damaged DNA strand, and this is followed by CAF-1 mediated chromatin assembly over the repaired DNA region
Like DSB repair, ATP-dependent nucleosome remodelers play a key role in NER. In yeast, UV irradiation enhances the interaction between Rad4–Rad23 (XPC-HR23B in humans) and the SWI/SNF ATP-dependent nucleosome remodeling complex (Gong et al. 2006), where the SWI/ SNF complex stimulates removal of CPDs (Hara and Sancar 2003; Zhao et al. 2009). Human SWI/SNF is recruited to sites of UV damage via its interaction with XPC and DDB2. This is consistent with the observation that inactivation of SWI/SNF has no effect on recruitment of DDB2 and XPC to CPDs, whereas the recruitment of downstream NER factors such as XPG and PCNA is deficient (Zhao et al., 2009). As such, it would appear that the role of SWI/SNF is to relax the nucleosome structure after damage recognition to facilitate access by the later acting NER factors.
INO80, another ATP-dependent chromatin remodeling complex, also regulates efficient repair of UV-induced photodamage in mammalian cells (Jiang et al. 2010). Like SWI/ SNF, INO80 functions to facilitate the access of NER factors to the sites of UV-induced damage. INO80 is recruited to DNA lesions independently of XPC and XPA, suggesting that it may have an early role in DNA repair. Furthermore, INO80 interacts with DDB1 and may play a role in its recruitment to the sites of UV-induced damage. A more recent study suggests that the role of yeast INO80 is to restore the chromatin structure after UV repair (Sarkar et al. 2010).
UV irradiation induces histone PTMs and chromatin relaxation. Hyperacetylated nucleosomes have been shown over 20 years ago to enhance NER efficiency (Ramanathan and Smerdon 1989). UV irradiation induces hyperacetylation of H3 K9 and K14, and H4 K16, and inactivation of the histone acetyltransferase GCN5 impairs the repair of UV-induced damage (Guo et al. 2011; Yu et al. 2005). In yeast, constitutive hyperacetylation of histone H3 can bypass the requirement for some of the NER machinery, suggesting that their normal function is to promote opening up on the chromatin to facilitate repair (Yu et al. 2011). Furthermore, DDB1–DDB2 can interact with mammalian histone acetyl-transferases such as CBP/p300 and STAGA (a SAGA-like complex containing GCN5L), implying that DDB1–DDB2 complexes might recruit histone acetyltransferase activities to UV-induced damage sites (Datta et al. 2001; Martinez et al. 2001). Indeed, p53-dependent recruitment of CBP/p300 to UV sites via its interaction with CSB has been observed, accompanied by an increase in H3 acetylation (Fousteri et al. 2006; Rubbi and Milner 2003). Increased histone acetylation at the NER site is likely to contribute to the p53-induced chromatin relaxation that is induced by DNA damage. However, exactly how p53 influences the events occurring at NER sites is not clear, given that p53 is not recruited to sites of NER (Rubbi and Milner 2003).
Interestingly, in Drosophila, a decrease of H3K9 trimethylation in heterochromatin regions is observed in response to UV irradiation, and this depends on the lysine demethylase dKDM4B (Palomera-Sanchez et al. 2010). As for the histone acetylation, this histone demethylation is also dependent on the fly homolog of p53. In the absence of dKDM4B, CPDs are not efficiently removed. Demethylation of H3 K9me may be required to allow the subsequent acetylation of H3 K9 that occurs at sites of NER. Somewhat counterintuitive to the chromatin relaxation that occurs in response to UV irradiation, all three HP1 isoforms, alpha, beta, and gamma, are recruited to sites of UV damage (Luijsterburg et al. 2009). Reminiscent of the situation at DSBs, this recruitment of HP1 to sites of UV damage is not dependent on its binding to methylated H3 K9. Recruitment of HP1 appears to be important for NER because loss of HP1 in worms results in sensitivity to UV light (Luijsterburg et al. 2009).
UV-induced damage also activates the DDR. ATR is activated by the processing of UV-induced lesions into ssDNA repair intermediates (Giannattasio et al. 2010; Sertic et al. 2011; Vrouwe et al. 2011). H2AX becomes phosphorylated after UV irradiation (Marti et al. 2006). Furthermore, ATM, NBS1, and Rad51 colocalize with CPDs after UV irradiation (Oh et al. 2011). Given that the very high experimental UV doses also result in DSBs, it is possible that activation of the DDR by UV irradiation may not always be in response to the UV-induced lesions themselves.
The chromatin structure must be restored after NER. The histone H3/H4 chaperone CAF-1 is required for chromatin assembly after NER in vivo (Green and Almouzni 2003), specifically incorporating newly synthesized canonical histone H3.1 (Polo et al. 2006). Akin to the role of CAF-1 in assembling newly synthesized histones carrying H3 K56Ac following DSB repair, UV repair is also accompanied by a local increase in H3 K56Ac (Battu et al. 2011). Again reminiscent of the situation during DSB repair, H3 K56Ac is not required for NER, but it is required for inactivation of the DNA damage checkpoint after NER (Battu et al. 2011). In summary, the mechanisms of reestablishing the chromatin structure after DSB repair and NER appear to be very closely related, if not identical.
Unique to NER, however, is post-DNA repair histone ubiquitylation. Monoubiquitylated H2A appears at sites of NER (Zhu et al. 2009). Noteworthy, this does not happen in cells lacking XPA, XPG, or XPF, i.e., late-acting NER proteins, suggesting that the appearance of monoubiquitylated H2A is dependent on successful DNA repair. Monoubiquitylation of H2A is also dependent on DNA processing, as it is impaired in cells depleted of human EXO1 (Sertic et al. 2011). The E3 ligase Ring2 is required for this H2A monoubiquitylation, and it is also dependent on ATR and MDC1 but does not require H2AX (Bergink et al. 2006; Marteijn et al. 2009). Furthermore, knockdown of CAF-1 prevents the H2A monoubiquitylation after NER, suggesting that it occurs after chromatin reassembly at the site of NER. Functionally, the reason for this post-NER monoubiquitylation is currently unknown.
Conclusions and future challenges
It is now clear that DNA repair processes, the DNA damage response, and chromatin structural changes are tightly mechanistically linked. Furthermore, they are acting cooperatively to organize the efficient removal of DNA damage from the genome. Changes in the chromatin structure in response to DNA damage allow the cell to sense the presence and find the location of DNA damage within the genome, enable amplification of the damage signal, coordinate recruitment and regulation of the repair factors, as well as provide the molecular signal that tells the DNA damage checkpoint that DNA repair is complete. Clearly, chromatin is not playing a passive role in DNA repair; rather, chromatin is actively regulating and promoting nearly every aspect of DNA repair and the maintenance of genomic stability.
The wealth of knowledge on the mechanism of the DDR response in mammalian cells that is briefly covered in this review is impressive, given that much of it came from the study of repair foci and/or eventual cell death in response to DNA damage. The order of events during the DDR has been established by failure to observe recruitment of proteins to foci upon mutation or knockdown of other factors. However, the study of foci alone does not allow one to know the status of repair of the DNA lesion per se nor the stage of repair during which the factors are recruited. Kinetic analyses of DDR/repair factors locating to foci in wild-type cells lack the temporal resolution to yield accurate insight into events that occur in a coordinated manner or in very rapid succession in our cells. Foci analyses also fail to inform us as to how close or how far from the DNA lesion are the DDR factors located that are being recruited to the break. ChIP analysis of factor recruitment to defined sites of repair can yield this kind of information. Accordingly, several mammalian cell lines have been engineered to generate defined DSBs (i.e., DSBs created enzymatically versus damage resulting from genotoxic stress). For example, rare-cutting endonucleases I-SceI and I-PpoI offer predictable cutting patterns but suffer from problems such as persistent cutting/repair cycles and suboptimal DSB location within specialized chromatin structures within the rDNA (for I-PpoI) (Berkovich et al. 2007; Rodrigue et al. 2006; Rouet et al. 1994). Induction of the endonuclease AsiSI generates more than 100 DSBs (Iacovoni et al. 2010). However, at best, only around 10% of the cells carry the DSB at any particular location (Iacovoni et al. 2010) making it difficult to accurately correlate the timing of factor recruitment with respect to repair of the DNA lesion per se.
The inducible HO endonuclease system has been transformatory for the understanding of many different DSB repair pathways in yeast (Haber 2006) because the extent of DSB induction is so high that recruitment of proteins can be accurately correlated to the stage of DNA repair. Furthermore, yeast HO systems have been devised such that there is a single round of cutting and repair, enabling synchronization of DSB repair in the population. The field of mammalian DSB repair would be greatly aided by DSB repair systems equivalent to the yeast HO system. Ideally, such mammalian repair systems would allow the synchronous induction of an efficient DSB at a known location in the genome so that the intermediates of DNA repair can be detected in most cells of the population. Indeed, an extremely efficient HO endonuclease system was recently developed in mouse cells for the study of NHEJ repair by Ricky De Benedetti’s group (Sunavala-Dossabhoy and De Benedetti 2009). In the ideal mammalian DSB repair system, the repair of this DSB should be experimentally controllable in the absence of subsequent rounds of cutting and repair and produce a repaired product that can be distinguished from the uncut parental DNA. Such a system would enable extensive ChIP analyses of defined factors, as well as proteomic analyses to find new factors involved in DNA repair, in response to DNA damage during and after DNA repair. Site-specific inducible break systems will also allow the field to unambiguously determine whether the mechanisms of repair and DNA damage response are the same or different for breaks within heterochromatin versus euchromatin.
The development of systems that provide an immediate and direct read out of failure to repair DNA lesions, rather than waiting for cell death or fixing cells to look at repair foci, would also be invaluable in both the laboratory and the clinic. Coupled with methods to isolate specifically those cells that have failed to undergo repair, such approaches would enable siRNA and chemical library screens in order to isolate new players in the DDR, as well as isolate compounds that could be used for sensitization to radiation therapy.
Now that chromatin dynamics has an undeniable foothold in the repair field and with dozens of damage-induced histone modifications with no currently known function, there is no doubt that our understanding of the epigenetic regulation of genomic stability will increase dramatically in the coming years. Already, one can recognize a shift in the direction of research in the DNA repair field, as the years of data that have outlined the role of chromatin in DNA repair have provided a backdrop for increasingly elegant questions. Now, research is being conducted to further define not just the players in repair and the DDR but the control mechanisms that optimize the integrity of the process on the physiologically relevant chromatin template. It is becoming clear that chromatin changes coordinate many of the steps in the DDR, with specific transient histone modifications acting to recruit particular proteins at precise times. It is also clear that ATP-dependent nucleosome remodelers play a key role in promoting specific molecular events during the DDR. Furthermore, the multiple recent reports of context-specific repair responses, wherein the cell senses and evaluates the chromatin environment in which the DNA lesion lies and adjusts the molecular response accordingly, underline not only how tightly controlled DNA repair is but also how critical it is to develop improved assay systems to evaluate the complex role of chromatin in mammalian DNA repair processes.
Acknowledgments
We apologize to those in the field whose work we were not able to discuss due to space limitations.
Footnotes
Communicated by Erich Nigg
References
- Ahel D, Hořejší Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-[bgr] mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- Ayoub N, Jeyasekharan AD, Venkitaraman AR. Mobilization and recruitment of HP1β: a bimodal response to DNA breakage. Cell Cycle. 2009;8:2946–2951. [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Baldeyron C, Soria G, Roche D, Cook AJL, Almouzni G. HP1α recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battu A, Ray A, Wani AA. ASF1A and ATM regulate H3K56-mediated cell-cycle checkpoint recovery in response to UV irradiation. Nucleic Acids Res. 2011;39:7931–7945. doi: 10.1093/nar/gkr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Salomons FA, Hoogstraten D, Groothuis TAM, de Waard H, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20:1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- Bewersdorf J, Bennett BT, Knight KL. H2AX chromatin structures and their response to DNA damage revealed by 4Pi microscopy. Proc Natl Acad Sci. 2006;103:18137–18142. doi: 10.1073/pnas.0608709103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- Borde V, Robine N, Lin W, Bonfils S, Geli V, Nicolas A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen H-T, Bosque D, Callen E, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim J-E, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer AL, Sharma GG, Huang C-Y, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- Buard J, Barthes P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callén E, Wong N, Chen H-T, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. Around the world of DNA damage INO80 days. Cell. 2004;119:733–735. doi: 10.1016/j.cell.2004.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung WL, Turner FB, Krishnamoorthy T, Wolner B, Ahn S-H, Foley M, Dorsey JA, Peterson CL, Berger SL, Allis CD. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr Biol. 2005;15:656–660. doi: 10.1016/j.cub.2005.02.049. [DOI] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiácovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Xu X, Zhong X, Ahmed F, Zhong J, Liao J, Dykxhoorn DM, Weinstock DM, Pfeifer GP, Lieberman J. A PP4-phosphatase complex dephosphorylates [gamma]-H2AX generated during DNA replication. Mol Cell. 2008;31:33–46. doi: 10.1016/j.molcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RMA, Chiganças V, da Silva Galhardo R, Carvalho H, Menck CFM. The eukaryotic nucleotide excision repair pathway. Biochimie. 2003;85:1083–1099. doi: 10.1016/j.biochi.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ. γH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS One. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Bagchi S, Nag A, Shiyanov P, Adami GR, Yoon T, Raychaudhuri P. The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat Res/DNA Repair. 2001;486:89–97. doi: 10.1016/s0921-8777(01)00082-9. [DOI] [PubMed] [Google Scholar]
- Dellaire G, Kepkay R, Bazett-Jones DP. High resolution imaging of changes in the structure and spatial organization of chromatin, γ-H2A.X and the MRN complex within etoposide-induced DNA repair foci. Cell Cycle. 2009;8:3750–3769. doi: 10.4161/cc.8.22.10065. [DOI] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang C-Y, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen Y-CM, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Côté J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Duan M-R, Smerdon MJ. UV damage in DNA promotes nucleosome unwrapping. J Biol Chem. 2010;285:26295–26303. doi: 10.1074/jbc.M110.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsässer SJ, D’Arcy S. Towards a mechanism for histone chaperones. Biochima et Biophysica Acta (BBA)—Gene Regulatory Mechanisms. 2012 doi: 10.1016/j.bbagrm.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Schubert T, Marth C, Längst G, Rippe K. Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proc Natl Acad Sci. 2010;107:19873–19878. doi: 10.1073/pnas.1003438107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk M, Lukasova E, Gabrielova B, Ondrej V, Kozubek S. Chromatin dynamics during DSB repair. Biochimica et Biophysica Acta (BBA)—Molecular. Cell Res. 2007;1773:1534–1545. doi: 10.1016/j.bbamcr.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Chen H-T, Celeste A, Ward I, Romanienko PJ, Morales JC, Naka K, Xia Z, Camerini-Otero RD, Motoyama N, et al. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol. 2002;4:993–997. doi: 10.1038/ncb884. [DOI] [PubMed] [Google Scholar]
- FitzGerald JE, Grenon M, Lowndes NF. 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans. 2009;37:897–904. doi: 10.1042/BST0370897. [DOI] [PubMed] [Google Scholar]
- Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee S-H, Nickoloff JA, Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LHF. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Friedberg ECW, Graham C, Siede W, Wood RD, Schultz RA, Ellengberger T. DNA repair and mutagenesis. 2. ASM; Washington: 2006. [Google Scholar]
- Gale JM, Nissen KA, Smerdon MJ. UV-induced formation of pyrimidine dimers in nucleosome core DNA is strongly modulated with a period of 10.3 bases. Proc Natl Acad Sci. 1987;84:6644–6648. doi: 10.1073/pnas.84.19.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Follonier C, Tourrière H, Puddu F, Lazzaro F, Pasero P, Lopes M, Plevani P, Muzi-Falconi M. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol Cell. 2010;40:50–62. doi: 10.1016/j.molcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- Ginjala V, Nacerddine K, Kulkarni A, Oza J, Hill SJ, Yao M, Citterio E, van Lohuizen M, Ganesan S. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol. 2011;31:1972–1982. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Fahy D, Smerdon MJ. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol. 2006;13:902–907. doi: 10.1038/nsmb1152. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol. 2011;18:831–839. doi: 10.1038/nsmb.2077. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Green CM, Almouzni G. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 2003;22:5163–5174. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy GJ, Yang W, Gellert M. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc Natl Acad Sci. 2010;107:22487–22492. doi: 10.1073/pnas.1014958107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Santoro J, Kapetanaki MG, Hsieh CL, Gorbachinsky I, Levine AS, Rapić-Otrin V. The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 2008;68:5014–5022. doi: 10.1158/0008-5472.CAN-07-6162. [DOI] [PubMed] [Google Scholar]
- Guo R, Chen J, Mitchell DL, Johnson DG. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 2011;39:1390–1397. doi: 10.1093/nar/gkq983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair. 2006;5:998–1009. doi: 10.1016/j.dnarep.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Hara R, Sancar A. Effect of damage type on stimulation of human excision nuclease by SWI/SNF chromatin remodeling factor. Mol Cell Biol. 2003;23:4121–4125. doi: 10.1128/MCB.23.12.4121-4125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink BA, Tubbs AT, Dorsett Y, Bednarski JJ, Walker LM, Feng Z, Sharma GG, McKinnon PJ, Zhang J, Bassing CH, et al. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 2011;469:245–249. doi: 10.1038/nature09585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- Hiom K. Coping with DNA double strand breaks. DNA Repair. 2010;9:1256–1263. doi: 10.1016/j.dnarep.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Holthausen JT, Wyman C, Kanaar R. Regulation of DNA strand exchange in homologous recombination. DNA Repair. 2010;9:1264–1272. doi: 10.1016/j.dnarep.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Huber A, Bai P, de Murcia JM, de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair. 2004;3:1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Huen MSY, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MSY, Huang J, Leung JWC, Sy SMH, Leung KM, Ching Y-P, Tsao SW, Chen J. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol Cell. 2010;37:854–864. doi: 10.1016/j.molcel.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, DiTullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of [gamma]H2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol. 2010;191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Conrad S, Voss K-O, Zink D, Durante M, Löbrich M, Taucher-Scholz G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011;39:6489–6499. doi: 10.1093/nar/gkr230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wang X, Bao S, Guo R, Johnson DG, Shen X, Li L. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc Natl Acad Sci. 2010;107:17274–17279. doi: 10.1073/pnas.1008388107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S, Stucki M. MDC1: the art of keeping things in focus. Chromosoma. 2010;119:337–349. doi: 10.1007/s00412-010-0266-9. [DOI] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Keogh M-C, Kim J-A, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. A phosphatase complex that dephosphorylates [gamma]H2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007a;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- Kim J-A, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J Cell Biol. 2007b;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-K, Patel D, Choi B-S. Contrasting structural impacts induced by cis–syn cyclobutane dimer and (6–4) adduct in DNA duplex decamers: implication in mutagenesis and repair activity. Photochem Photobiol. 1995;62:44–50. doi: 10.1111/j.1751-1097.1995.tb05236.x. [DOI] [PubMed] [Google Scholar]
- Kim Y-C, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh K-S, Kraemer KH, Shiloh Y, Bustin M. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat Cell Biol. 2009;11:92–96. doi: 10.1038/ncb1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Müller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Florens L, MacDonald WH, Swanson SK, Glaser RL, Yates JR, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, Janicki SM, Ogiwara H, Kohno T, Kanno S-i, et al. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell. 2010;40:976–987. doi: 10.1016/j.molcel.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, Stucki M, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190:731–740. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro F, Giannattasio M, Puddu F, Granata M, Pellicioli A, Plevani P, Muzi-Falconi M. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair. 2009;8:1055–1067. doi: 10.1016/j.dnarep.2009.04.022. [DOI] [PubMed] [Google Scholar]
- Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M. Histone methyltrans-ferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-S, Park J-H, Kim S-J, Kwon S-J, Kwon J. A cooperative activation loop among SWI/SNF, [gamma]-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010a;29:1434–1445. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Goodarzi AA, Jeggo PA, Paull TT. 53BP1 promotes ATM activity through direct interactions with the MRN complex. EMBO J. 2010b;29:574–585. doi: 10.1038/emboj.2009.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Corsa CAS, Pan PW, Wu L, Ferguson D, Yu X, Min J, Dou Y. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 2010;30:5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Qiu J, Ratnakumar K, Laurent BC. RSC functions as an early double-strand-break sensor in the cell’s response to DNA damage. Curr Biol. 2007;17:1432–1437. doi: 10.1016/j.cub.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Gu J, Lu H, Shimazaki N, Tsai AG. Nonhomologous DNA end joining (NHEJ) and chromosomal translocations in humans. In: Nasheuer H-P, editor. Genome stability and human diseases. Springer; the Netherlands: 2010. pp. 279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-Y, Rai R, Li K, Xu Z-X, Elledge SJ. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1–Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc Natl Acad Sci U S A. 2005;102:15105–15109. doi: 10.1073/pnas.0507722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. MDC1 Maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Lowndes NF. The interplay between BRCA1 and 53BP1 influences death, aging, senescence and cancer. DNA Repair. 2010;9:1112–1116. doi: 10.1016/j.dnarep.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8[thinsp]A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, van Attikum H. Chromatin and the DNA damage response: the cancer connection. Mol Onc. 2011;5:349–267. doi: 10.1016/j.molonc.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, Dantuma NP, Lukas J, Vermeulen W. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J Cell Biol. 2009;186:835–847. doi: 10.1083/jcb.200902150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AGW, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra PV, Ahel D, Ryan DP, Weston R, Wiechens N, Kraehenbuehl R, Owen-Hughes T, Ahel I. DNA repair factor APLF is a histone chaperone. Mol Cell. 2011;41:46–55. doi: 10.1016/j.molcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochan TA, Venere M, DiTullio RA, Halazonetis TD. 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer Res. 2003;63:8586–8591. [PubMed] [Google Scholar]
- Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. INO80 and [gamma]-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Shen X. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol. 2009;10:373–384. doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang S-Y, Eppink B, Chung YM, Shalev G, Shema E, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang Y-G, Cuenin C, Li H, Wang Z-Q, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DVNP, Shimada M, Tauchi H, Suzuki H, Tashiro S, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S, Yasui A, Yokota J, Kohno T. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 2011;30:2135–2146. doi: 10.1038/onc.2010.592. [DOI] [PubMed] [Google Scholar]
- Oh K-S, Bustin M, Mazur SJ, Appella E, Kraemer KH. UV-induced histone H2AX phosphorylation and DNA damage related proteins accumulate and persist in nucleotide excision repair-deficient XP-B cells. DNA Repair. 2011;10:5–15. doi: 10.1016/j.dnarep.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomera-Sanchez Z, Bucio-Mendez A, Valadez-Graham V, Reynaud E, Zurita M. Drosophila p53 is required to increase the levels of the dKDM4B demethylase after UV-induced DNA damage to demethylate histone H3 lysine 9. J Biol Chem. 2010;285:31370–31379. doi: 10.1074/jbc.M110.128462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-H, Park E-J, Lee H-S, Kim SJ, Hur S-K, Imbalzano AN, Kwon J. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting [gamma]-H2AX induction. EMBO J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng A, Chen P-L. NFBD1, like 53BP1, is an early and redundant transducer mediating Chk2 phosphorylation in response to DNA damage. J Biol Chem. 2003;278:8873–8876. doi: 10.1074/jbc.C300001200. [DOI] [PubMed] [Google Scholar]
- Peng G, Lin S-Y. BRIT1/MCPH1 is a multifunctional DNA damage responsive protein mediating DNA repair-associated chromatin remodeling. Cell Cycle. 2009;8:3071–3072. doi: 10.4161/cc.8.19.9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto DMS, Flaus A. Structure and function of histone H2AX. In: Nasheuer H-P, editor. Genome stability and human diseases. Springer; the Netherlands: 2010. pp. 55–78. [Google Scholar]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. Ku80 removal from DNA through double strand break-induced ubiquitylation. J Cell Biol. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing Y, Yamazoe M, Hirota K, Dejsuphong D, Sakai W, Yamamoto KN, Bishop DK, Wu X, Takeda S. The epistatic relationship between BRCA2 and the other RAD51 mediators in homologous recombination. PLoS Genet. 2011;7:e1002148. doi: 10.1371/journal.pgen.1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Dai H, Multani AS, Li K, Chin K, Gray J, Lahad JP, Liang J, Mills GB, Meric-Bernstam F, et al. BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell. 2006;10:145–157. doi: 10.1016/j.ccr.2006.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan B, Smerdon MJ. Enhanced DNA repair synthesis in hyperacetylated nucleosomes. J Biol Chem. 1989;264:11026–11034. [PubMed] [Google Scholar]
- Rodrigue A, Lafrance M, Gauthier M-C, McDonald D, Hendzel M, West SC, Jasin M, Masson J-Y. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J. 2006;25:222–231. doi: 10.1038/sj.emboj.7600914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi CP, Milner J. p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. EMBO J. 2003;22:975–986. doi: 10.1093/emboj/cdg082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Fisher AEO, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Sánchez-Molina S, Mortusewicz O, Bieber B, Auer S, Eckey M, Leonhardt H, Friedl AA, Becker PB. Role for hACF1 in the G2/M damage checkpoint. Nucleic Acids Res. 2011;39:8445–8456. doi: 10.1093/nar/gkr435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bähler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Sandhu SK, Yap TA, de Bono JS. The emerging role of poly (ADP-ribose) polymerase inhibitors in cancer treatment. Curr Drug Targets. 2011;12:2034–2044. doi: 10.2174/138945011798829438. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Kiely R, McHugh PJ. The Ino80 chromatin-remodeling complex restores chromatin structure during UV DNA damage repair. J Cell Biol. 2010;191:1061–1068. doi: 10.1083/jcb.201006178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame J-C, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Sertic S, Pizzi S, Cloney R, Lehmann AR, Marini F, Plevani P, Muzi-Falconi M. Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc Natl Acad Sci. 2011;108:13647–13652. doi: 10.1073/pnas.1108547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30:3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Ma J-L, Oum J-H, Yanez Y, Lee SE. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol. 2005;25:3934–3944. doi: 10.1128/MCB.25.10.3934-3944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V (D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol Cell. 2009;34:535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun J-M, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–749. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Mun Tho L, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. In: George FVW, George K, editors. Advances in cancer research. Academic; New York: 2010. pp. 73–112. [DOI] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11–RAD50–NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- Sunavala-Dossabhoy G, De Benedetti A. Tousled homolog, TLK1, binds and phosphorylates Rad9; TLK1 acts as a molecular chaperone in DNA repair. DNA Repair. 2009;8:87–102. doi: 10.1016/j.dnarep.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Suquet C, Smerdon MJ. UV damage to DNA strongly influences its rotational setting on the histone surface of reconstituted nucleosomes. J Biol Chem. 1993;268:23755–23757. [PubMed] [Google Scholar]
- Sy SMH, Huen MSY. The 53BP1-EXPAND connection in chromatin structure regulation. Nucleus. 2010;1:3. doi: 10.4161/nucl.1.6.13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takedachi A, Saijo M, Tanaka K. DDB2 complex-mediated ubiquitylation around DNA damage is oppositely regulated by XPC and Ku and contributes to the recruitment of XPA. Mol Cell Biol. 2010;30:2708–2723. doi: 10.1128/MCB.01460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford DJ, Stewart BW. Micrococcal nuclease: its specificity and use for chromatin analysis. Int J Biochem. 1989;21:127–138. doi: 10.1016/0020-711x(89)90100-6. [DOI] [PubMed] [Google Scholar]
- Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley RT, Lacoste N, Jobin-Robitaille O, Allard S, Cote J. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol Cell Biol. 2005;25:8179–8190. doi: 10.1128/MCB.25.18.8179-8190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- Vempati RK, Jayani RS, Notani D, Sengupta A, Galande S, Haldar D. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J Biol Chem. 2010;285:28553–28564. doi: 10.1074/jbc.M110.149393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrouwe MG, Pines A, Overmeer RM, Hanada K, Mullenders LHF. UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J Cell Sci. 2011;124:435–446. doi: 10.1242/jcs.075325. [DOI] [PubMed] [Google Scholar]
- Wagner CR, Kuervers L, Baillie DL, Yanowitz JL. xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature. 2010;467:839–843. doi: 10.1038/nature09429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 Form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BSCPES. 53BP1, a medicator of the DNA damage checkpoint. Science. 2002;298:1435. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhai L, Xu J, Joo H-Y, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Ward IM, Minn K, van Deursen J, Chen J. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmerdam DO, Kanaar R. Dealing with DNA damage: relationships between checkpoint and repair pathways. Mutat Res Rev Mutat Res. 2010;704:2–11. doi: 10.1016/j.mrrev.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Wilson KA, Stern DF. NFBD1/MDC1, 53BP1 and BRCA1 have both redundant and unique roles in the ATM pathway. Cell Cycle. 2008;7:3584–3594. doi: 10.4161/cc.7.22.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JL, Singh N, Mer G, Chen J. MCPH1 functions in an H2AX-dependent but MDC1-independent pathway in response to DNA damage. J Biol Chem. 2007;282:35416–35423. doi: 10.1074/jbc.M705245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Shi Y, Mulligan P, Gay F, Landry J, Liu H, Lu J, Qi HH, Wang W, Nickoloff JA, et al. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat Struct Mol Biol. 2007;14:1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol. 2010;191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kim Y-S, Yang X-P, Li L-P, Liao G, Xia F, Jetten AM. The ubiquitin-interacting motif-containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007;67:6647–6656. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Dutt S, Xu R, Graves K, Juszczynski P, Manis JP, Shipp MA. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol Cell. 2009;36:110–120. doi: 10.1016/j.molcel.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Bailis JM. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010;20:402–409. doi: 10.1016/j.tcb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Teng Y, Waters R, Reed SH. How chromatin is remodelled during DNA repair of UV-induced DNA damage in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002124. doi: 10.1371/journal.pgen.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CCS, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- Yu Y, Teng Y, Liu H, Reed SH, Waters R. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc Natl Acad Sci U S A. 2005;102:8650–8655. doi: 10.1073/pnas.0501458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G, Zhu B. Histone variants and epigenetic inheritance. Biochimica et Biophysica Acta (BBA)—Gene Regulatory Mechanisms. 2011 doi: 10.1016/j.bbagrm.2011.06.007. (in press, corrected proof) [DOI] [PubMed] [Google Scholar]
- Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: to be or not to be. FEBS Lett. 2010;584:3717–3724. doi: 10.1016/j.febslet.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarebski M, Wiernasz E, Dobrucki JW. Recruitment of hetero-chromatin protein 1 to DNA repair sites. Cytometry Part A. 2009;75A:619–625. doi: 10.1002/cyto.a.20734. [DOI] [PubMed] [Google Scholar]
- Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang Q-E, Ray A, Wani G, Han C, Milum K, Wani AA. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J Biol Chem. 2009;284:30424–30432. doi: 10.1074/jbc.M109.044982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Wani G, Arab HH, El-Mahdy MA, Ray A, Wani AA. Chromatin restoration following nucleotide excision repair involves the incorporation of ubiquitinated H2A at damaged genomic sites. DNA Repair. 2009;8:262–273. doi: 10.1016/j.dnarep.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]