Abstract
Iron homeostasis dysregulation has been regarded as an important mechanism in neurodegenerative diseases. The H63D and C282Y polymorphisms in the HFE gene may be involved in the development of sporadic amyotrophic lateral sclerosis (ALS) through the disruption of iron homeostasis. However, studies investigating the relationship between ALS and these two polymorphisms have yielded contradictory outcomes. We performed a meta-analysis to assess the roles of the H63D and C282Y polymorphisms of HFE in ALS susceptibility. PubMed, MEDLINE, EMBASE, and Cochrane Library databases were systematically searched to identify relevant studies. Strict selection criteria and exclusion criteria were applied. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the strength of associations. A fixed- or random-effect model was selected, depending on the results of the heterogeneity test. Fourteen studies were included in the meta-analysis (six studies with 1692 cases and 8359 controls for C282Y; 14 studies with 5849 cases and 13,710 controls for H63D). For the C282Y polymorphism, significant associations were observed in the allele model (Y vs C: OR=0.76, 95%CI=0.62-0.92, P=0.005) and the dominant model (YY+CY vs CC: OR=0.75, 95%CI=0.61-0.92, P=0.006). No associations were found for any genetic model for the H63D polymorphism. The C282Y polymorphism in HFE could be a potential protective factor for ALS in Caucasians. However, the H63D polymorphism does not appear to be associated with ALS.
Keywords: Amyotrophic lateral sclerosis, HFE, C282Y, H63D, Polymorphism, Meta-analysis
Introduction
The HFE gene, so named because of its dominant role in iron homeostasis and preventing hemochromatosis (i.e., iron overload in the blood) (1), is located on the short arm of chromosome 6 at location 6p21.3 (2). Mutations of HFE have been associated with iron overload. Loss of iron homeostasis in the brain may lead to oxidative stress and inflammation (3). HFE testing for the two main mutations (H63D and C282Y) should be performed in all patients with primary iron overload and unexplained increased transferrin saturation and/or serum ferritin values (4), which have been implicated in neurodegenerative diseases (5).
As the most common adult-onset neurodegenerative disease, amyotrophic lateral sclerosis (ALS) is characterized by degeneration of lower and upper motor neurons in the brain and spinal cord. ALS causes progressive muscle weakness and paralysis. Patient mortality generally occurs within 3 to 5 years after disease onset, due to respiratory insufficiency (6-8). ALS is nonfamilial (sporadic) in 90 to 95% of cases (9). The etiology of ALS remains unknown, and disease-modifying treatments are limited. However, two mutations of HFE, C282Y and H63D, have been associated with various clinical endpoints, including ALS, and iron overload (10), leading researchers to speculate whether iron accumulation might be a possible mechanism contributing to its pathophysiology (11,12).
Recent studies have analyzed the effect of the C282Y polymorphism on ALS, but the results have been conflicting (13-17). Some of these studies involved only a few hundred cases, too few for a reliable assessment. Three previous meta-analyses (18-20) regarding the H63D polymorphism and ALS yielded contradictory outcomes and missed some important studies. Although a few studies have sought to catalog the association between various HFE polymorphisms and ALS, most such publications lacked statistical power or did not present data for combined genotypes (21,22). Therefore, to address this literature gap, we studied the combined effect of the C282Y and H63D mutations on the risk of ALS, by conducting a meta-analysis of available studies with a case-control or cohort design.
Material and Methods
Selection of eligible studies
Two reviewers independently scrutinized studies on the associations between HFE polymorphisms and ALS. The PubMed, MEDLINE, EMBASE, and Cochrane Library databases, from their inception to September 26, 2012, were searched to identify potentially relevant studies. The following search strategy was used, which combined both the medical subject heading (MeSH) and text words (“hemochromatosis” or “HFE” or “C282Y” or “Cys282Tyr” or “H63D” or “His63Asp”) and (“amyotrophic lateral sclerosis” or “ALS”) and (“polymorphism*” or “variant*” or “genotype” or single nucleotide polymorphisms “SNPs”). The related articles of each included study generated by PubMed and relevant reviews were also screened, to avoid the omission of any potentially relevant study. When necessary, the authors of abstracts were contacted to obtain missing data. The search was done without restriction on language, but we only included published articles written in English.
Selection criteria
Two reviewers independently identified potentially relevant studies and evaluated each trial according to predetermined eligibility criteria. Studies were selected if they met the following criteria: 1) association study, using a case-control or cohort design, in sporadic ALS subjects diagnosed by El Escorial World Federation of Neurology guidelines; 2) available data for C282Y or H63D or HFE mutations with risk of ALS; and 3) the genotype distribution in the controls were in Hardy-Weinberg equilibrium (HWE).
The exclusion criteria were defined as: 1) abstracts, reviews, and animal studies; 2) useless data reported, genotype number, or frequency not included; and 3) genotype distribution in the control population was not consistent with HWE. If the same author published more than one study using the same case series, then only the most recent study or the study with the largest sample size was included in the meta-analysis.
Data extraction
After duplicate studies were removed and any additional studies were added, two investigators independently extracted data from each study in a standard fashion and entered the information into a common database. When discrepancies arose, all investigators assessed the data. The following information was collected: first author, year of publication, country, ethnicity, characteristics, study design, sample sizes of patients and controls, genotype numbers, minor allele frequency and numbers, and P value for HWE. The article by Praline et al. (13) used the review by Merryweather-Clarke et al. (21) to select five French series and pooled controls from those series. Therefore, we combined the data and analyzed the pooled controls from those series as one group and compared them with the cases.
Statistical analysis
Odds ratios (ORs) with 95% confidence intervals (CIs) for genotypes and alleles were used to assess the strength of association between HFE polymorphisms and ALS. The ORs were calculated for the allele contrast, additive genetic, recessive genetic, and dominant genetic models.
Heterogeneity, which was interpreted as the proportion of the total variation that was contributed by between-study variation, was examined with the I2 statistic. The effect of heterogeneity was measured with a quantitative measure, I2=100%×(Q−d.f.)/Q. If there was a statistical difference in terms of heterogeneity (P<0.10, I2>50%), then the random-effects model was used to estimate the pooled OR (23,24). Otherwise, the pooled OR was estimated by the fixed-effects model (25).
Because the frequencies of C282Y and H63D depend on ethnicity, and this effect is more pronounced in men than in women (26), subanalyses on ethnicity were performed when assessing clinical heterogeneity. The ethnic groups were defined as 1) people of continental European ancestry, including the United States, France, Germany, Ireland, Italy, or other; and 2) other ethnic groups (Asian, African, or other). Turks, Arabs, and Hispanics were not included in the European category. If heterogeneity still persisted after this stratification, the studies included in the analyses were inspected post hoc manually.
Sensitivity analysis was performed by deleting studies one by one to examine the influence of an individual data set on the pooled OR. An asymmetric plot suggested possible publication bias. The Egger test with a P value less than 0.05 was considered to indicate statistically significant publication bias (27). All statistical tests were performed with STATA version 11.0 (StataCorp., USA). All P values were two-sided. A P value less than 0.05 for any test was considered to be statistically significant.
Results
Study characteristics
The literature search of the PubMed, MEDLINE, EMBASE, and Cochrane Library databases resulted in 50 articles (Figure 1). After we excluded 17 duplicate articles, we screened the titles, abstracts, and full texts of the remaining 33 articles against the inclusion/exclusion criteria, which resulted in 10 articles. Three articles were excluded: one because the allele frequencies in control subjects deviated from HWE (2), another because it had no usable data (21), and the third because it was written in Chinese. Despite our best efforts, such as searching other related references and e-mailing/faxing all of the authors, we could not acquire usable data for the Merryweather-Clarke study (21). Of the remaining articles, we verified two primary articles with negative results by contacting the authors. Finally, seven articles fulfilling the inclusion criteria were identified and included in the meta-analysis (13-18,28).
Figure 1. Flow diagram of study.
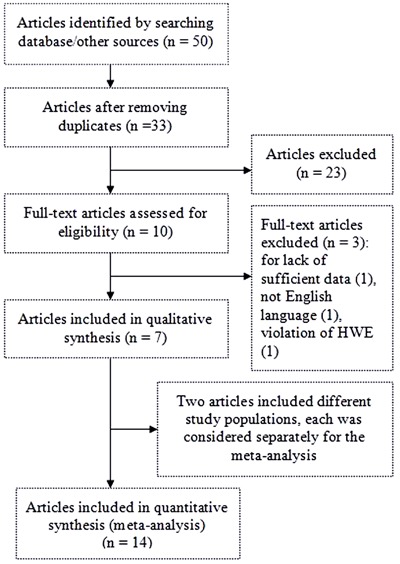
One article, by Goodall et al. (17), presented the genotype frequencies separately according to studies in Birmingham and Ireland. Therefore, each of these studies was considered separately for the meta-analysis. In another article, by van Rheenen et al. (18), cases were recruited from seven different countries (the Netherlands, Belgium, Germany, United Kingdom, Ireland, Sweden, and Switzerland). Each country was considered separately for the meta-analysis.
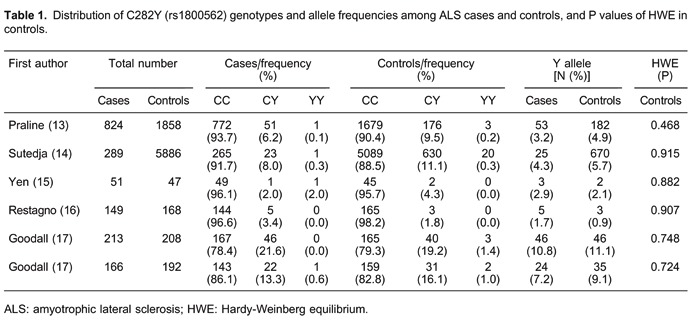
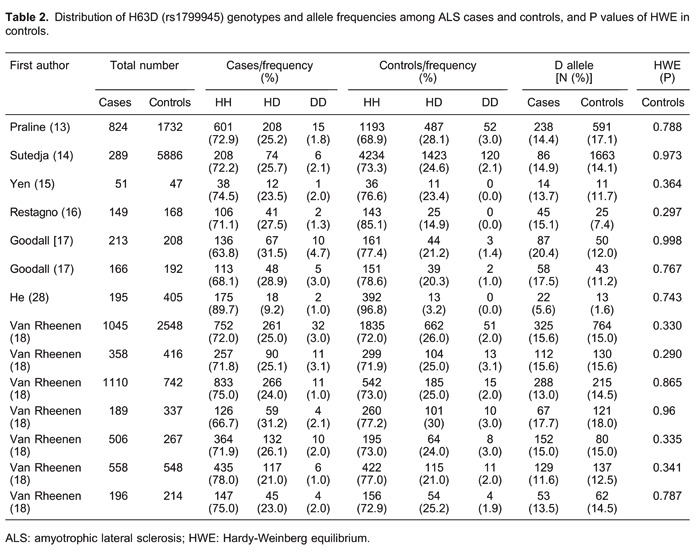
In total, 14 studies (six studies with 1692 cases and 8359 controls for the C282Y polymorphism, and 14 studies with 5849 cases and 1710 controls for the H63D polymorphism) were included in the meta-analysis. The main characteristics of the included studies are summarized in Supplementary Table S1 and Tables 1 and 2. In all the studies, minor allele frequencies were calculated for the controls (all >0.05).
Quantitative data synthesis
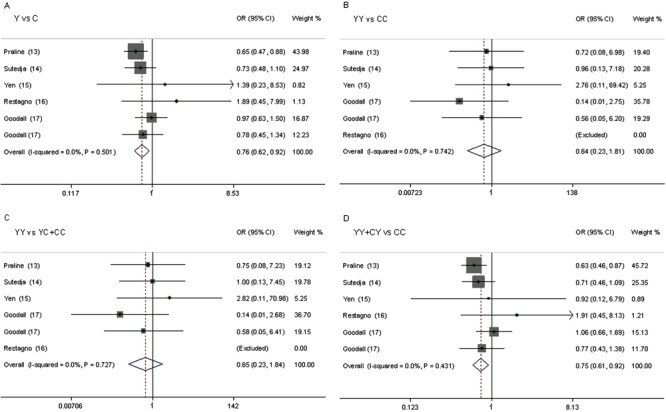
Table 3 shows the results for the associations between the risk of ALS and the C282Y and H63D polymorphisms in the different models. We obtained evidence of an association between a decreased risk of ALS and the C282Y polymorphism in the allele model (Y vs C: OR=0.76, 95%CI=0.62-0.92, P=0.005) and in the dominant model (YY+CY vs CC: OR=0.75, 95%CI=0.61-0.92, P=0.006). There was no significant difference between study heterogeneity (I2=0.0%; Figure 2).
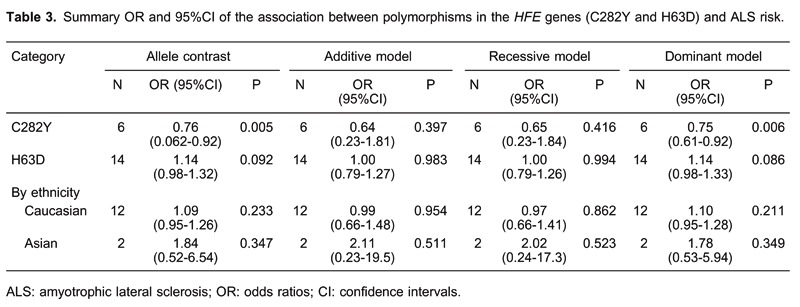
Figure 2. Forest plot of the assocition between ALS and the HFE C282Y mutation. A, Y vs C; B, YY vs CC; C, YY vs YC+CC; D, YY+CY vs CC.
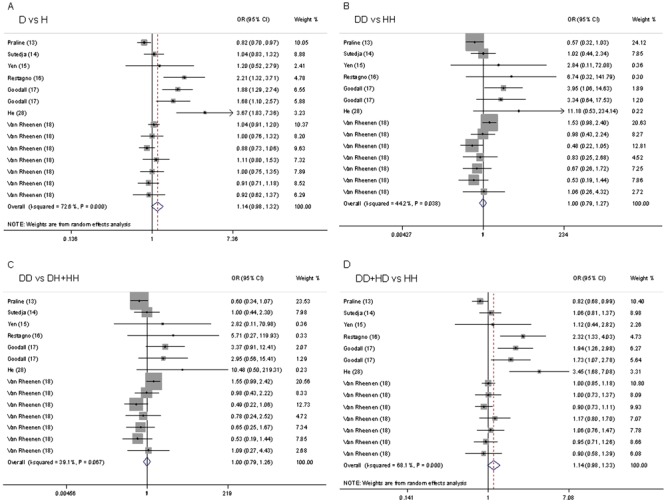
We did not observe evidence for an association of the H63D polymorphism with ALS risk in any of the models (D vs H: OR=1.14, 95%CI=0.98-1.31; DD vs HH: OR=1.00, 95%CI=0.79-1.27; recessive model: OR=1.00, 95%CI=0.79-1.26; dominant model: OR=1.14, 95%CI=0.98-1.33; Figure 3). In subgroup analysis by ethnicity, the differences between the allele, additive, recessive, and dominant models were not significant in the Caucasian or Asian subgroup.
Figure 3. Forest plot of the association between ALS and the HFE H63D mutation. A, D vs H; B, DD vs HH; C, DD vs DH+HH; D, DD+HD vs HH.
Sensitivity analysis
We examined the influence of each individual data set to the pooled OR in the meta-analysis. There was no substantial modification of our estimates after we excluded individual studies (data not shown), indicating that the results were stable.
Discussion
It is plausible that the HFE gene could be a risk factor for ALS (13,14,16,17). Many studies have explored the relationship of the HFE gene with ALS across populations worldwide. However, most of these studies have had small sample sizes and inconclusive findings. To overcome these shortcomings, meta-analysis can be used.
Meta-analysis is a powerful tool that can detect small effects in human genetic association studies (29). By increasing the sample size, this approach can reduce the incidence of false-positive or false-negative associations produced by chance. Previous meta-analyses have been performed to investigate the relationship of the H63D polymorphism of the HFE gene with ALS (18-20). However, the association of ALS with other genetic variants of the HFE gene awaits further analysis. Considering previous association studies of the HFE gene with ALS (13-18,28), five studies suggested that H63D mutations in HFE play a role in ALS pathogenesis in various populations (13,14,16,17,28). Two studies failed to find an association (15,18).
In the present meta-analysis, we used data from 14 observational studies. We found evidence for a significant association for the C282Y polymorphism, especially in the allele model, but no evidence for a relationship of the H63D polymorphism with ALS risk. These results are consistent with the findings of previous meta-analyses (14,18) and may confirm that the H63D polymorphism is not associated with ALS risk. One possible explanation is that the HFE C282Y homozygous genotype is particularly common in Northern European populations and, therefore, the HFE C282Y allele frequency was high (30).
Because all seven of the included studies were case-control studies, we did not perform a subgroup analysis by study design. We observed heterogeneity among the studies, which led us to employ a random-effects model. We performed a sensitivity analysis, removing each study one by one and rerunning the model to determine the effect on the overall estimate. The estimates changed very little when a study was removed. This finding suggests that the results were stable. Owing to the limited number of included studies, we did not perform a publication bias test (31-34).
Compared with previous meta-analyses, our meta-analysis has some specific strengths. First, the C282Y polymorphism was added, and a larger sample size was used to estimate its effect in the meta-analysis. To the best of our knowledge, no previous meta-analysis has explored the role of the C282Y polymorphism in the development and progression of ALS. To achieve a more reliable and comprehensive conclusion on the roles of both variants, we performed a meta-analysis to assess the association between H63D and C282Y polymorphisms and ALS risk. Second, we did not find any significant between-study heterogeneity in our meta-analysis. In contrast, significant heterogeneity among studies was a limitation in the meta-analysis by van Rheenen et al. (18). Moreover, we were able to perform a stratified subgroup analysis for the H63D polymorphism to explore the potential influence of ethnicity on the effects. The results of the subgroup analysis suggested that the H63D polymorphism was not a susceptibility factor for ALS in Caucasians or Asians. Because all five of the included studies of the C282Y polymorphism were from Europe (13-17), we did not perform a subgroup analysis by ethnicity. These results have not been found in previous meta-analyses. Third, we performed a sensitivity analysis to estimate the influence of each study on the overall estimate, the results of which suggested that the result was stable. In view of these findings, we are convinced that the results of our meta-analysis should be more reliable than the results of previous meta-analyses.
In this meta-analysis, all of the available results from case-control studies were pooled, which significantly increased the statistical power of the study. Nevertheless, to some extent, the results should be interpreted with caution, in light of some limitations. First, ALS is a multifactorial disease that involves complex interactions between environmental and genetic factors (35). We did not consider certain factors, such as living habits and employment, that could affect the significance of the independent role of HFE polymorphisms in ALS development. Second, the number of studies of C282Y was relatively small. Investigations involving large study samples of different ethnicities are necessary for a more reliable evaluation of its associations. Third, although we made every effort to find all appropriate publications, we may have missed some studies or retrieved some studies erroneously. Fourth, we did not consider studies published in languages other than English or data presented in abstracted form. This procedure may have caused a disproportionate exclusion of negative data, and may have biased our summary OR toward significance. Fifth, we cannot exclude the possibility that some of the positive results were biased, due to undetected population stratification in the original case-control samples, although the replication of these findings in clinic-based samples would argue against this possibility.
In spite of these limitations, for ALS candidate genes, we believe that the positive loci identified in our systematic meta-analysis represent particularly promising HFE mutations that warrant follow-up with high priority.
In conclusion, our data support the hypothesis that a change in iron metabolism may confer susceptibility to neurodegenerative diseases, such as ALS. The results of this meta-analysis suggest that the C282Y polymorphism in HFE could be a potentially protective factor for ALS in Caucasians. However, the H63D polymorphism does not appear to be associated with ALS risk. Studies showed that carriers of the DD+HD genotypes had higher serum ferritin values than those with the HH genotype for the HFE H63D mutation. The HFE C282Y and H63D mutations were associated with alterations in iron status in blood donors in a gender-dependent manner (36,37). To obtain a better understanding of the potential mechanism for ALS in humans, large well-designed epidemiological studies on the susceptibility to ALS are needed to confirm this association. It will also be necessary to consider genetic factors, gender, and environmental risk factors.
Supplementary material
Acknowledgments
The authors wish to thank all families for participating in this study. We also thank L.H. van den Berg for his kindness in providing materials.
Footnotes
First published online February 18, 2014.
References
- 1.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 2.Wang XS, Lee S, Simmons Z, Boyer P, Scott K, Liu W, et al. Increased incidence of the Hfe mutation in amyotrophic lateral sclerosis and related cellular consequences. J Neurol Sci. 2004;227:27–33. doi: 10.1016/j.jns.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell RM, Lee SY, Randazzo WT, Simmons Z, Connor JR. Influence of HFE variants and cellular iron on monocyte chemoattractant protein-1. J Neuroinflammation. 2009;6:6. doi: 10.1186/1742-2094-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos PC, Krieger JE, Pereira AC. Molecular diagnostic and pathogenesis of hereditary hemochromatosis. Int J Mol Sci. 2012;13:1497–1511. doi: 10.3390/ijms13021497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaur H, Halliwell B. Evidence for nitric oxide-mediated oxidative damage in chronic inflammation. Nitrotyrosine in serum and synovial fluid from rheumatoid patients. FEBS Lett. 1994;350:9–12. doi: 10.1016/0014-5793(94)00722-5. [DOI] [PubMed] [Google Scholar]
- 6.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N Engl J Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 7.Traynor BJ, Codd MB, Corr B, Forde C, Frost E, Hardiman OM. Clinical features of amyotrophic lateral sclerosis according to the El Escorial and Airlie House diagnostic criteria: A population-based study. Arch Neurol. 2000;57:1171–1176. doi: 10.1001/archneur.57.8.1171. [DOI] [PubMed] [Google Scholar]
- 8.Hardiman O, van den Berg LH, Kiernan MC. Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7:639–649. doi: 10.1038/nrneurol.2011.153. [DOI] [PubMed] [Google Scholar]
- 9.Cudkowicz ME, Kenna-Yasek D, Sapp PE, Chin W, Geller B, Hayden DL, et al. Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann Neurol. 1997;41:210–221. doi: 10.1002/ana.410410212. [DOI] [PubMed] [Google Scholar]
- 10.Njajou OT, Houwing-Duistermaat JJ, Osborne RH, Vaessen N, Vergeer J, Heeringa J, et al. A population-based study of the effect of the HFE C282Y and H63D mutations on iron metabolism. Eur J Hum Genet. 2003;11:225–231. doi: 10.1038/sj.ejhg.5200955. [DOI] [PubMed] [Google Scholar]
- 11.Kwan JY, Jeong SY, van Gelderen P, Deng HX, Quezado MM, Danielian LE, et al. Iron accumulation in deep cortical layers accounts for MRI signal abnormalities in ALS: correlating 7 tesla MRI and pathology. PLoS One. 2012;7: doi: 10.1371/journal.pone.0035241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothstein JD. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann Neurol. 2009;65(Suppl 1):S3–S9. doi: 10.1002/ana.21543. [DOI] [PubMed] [Google Scholar]
- 13.Praline J, Blasco H, Vourc'h P, Rat V, Gendrot C, Camu W, et al. Study of the HFE gene common polymorphisms in French patients with sporadic amyotrophic lateral sclerosis. J Neurol Sci. 2012;317:58–61. doi: 10.1016/j.jns.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 14.Sutedja NA, Sinke RJ, Van Vught PW, Van der Linden MW, Wokke JH, van Duijn CM, et al. The association between H63D mutations in HFE and amyotrophic lateral sclerosis in a Dutch population. Arch Neurol. 2007;64:63–67. doi: 10.1001/archneur.64.1.63. [DOI] [PubMed] [Google Scholar]
- 15.Yen AA, Simpson EP, Henkel JS, Beers DR, Appel SH. HFE mutations are not strongly associated with sporadic ALS. Neurology. 2004;62:1611–1612. doi: 10.1212/01.WNL.0000123114.04644.CC. [DOI] [PubMed] [Google Scholar]
- 16.Restagno G, Lombardo F, Ghiglione P, Calvo A, Cocco E, Sbaiz L, et al. HFE H63D polymorphism is increased in patients with amyotrophic lateral sclerosis of Italian origin. J Neurol Neurosurg Psychiatry. 2007;78:327–327. doi: 10.1136/jnnp.2006.092338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodall EF, Greenway MJ, van Marion I, Carroll CB, Hardiman O, Morrison KE. Association of the H63D polymorphism in the hemochromatosis gene with sporadic ALS. Neurology. 2005;65:934–937. doi: 10.1212/01.wnl.0000176032.94434.d4. [DOI] [PubMed] [Google Scholar]
- 18.van Rheenen W, Diekstra FP, van Doormaal PT, Seelen M, Kenna K, McLaughlin R, et al. H63D polymorphism in HFE is not associated with amyotrophic lateral sclerosis. Neurobiol Aging. 2013;34:1517–1517. doi: 10.1016/j.neurobiolaging.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 19.Ellervik C, Birgens H, Tybjaerg-Hansen A, Nordestgaard BG. Hemochromatosis genotypes and risk of 31 disease endpoints: meta-analyses including 66,000 cases and 226,000 controls. Hepatology. 2007;46:1071–1080. doi: 10.1002/hep.21885. [DOI] [PubMed] [Google Scholar]
- 20.Lill CM, Abel O, Bertram L, Al-Chalabi A. Keeping up with genetic discoveries in amyotrophic lateral sclerosis: the ALSoD and ALSGene databases. Amyotroph Lateral Scler. 2011;12:238–249. doi: 10.3109/17482968.2011.584629. [DOI] [PubMed] [Google Scholar]
- 21.Merryweather-Clarke AT, Pointon JJ, Jouanolle AM, Rochette J, Robson KJ. Geography of HFE C282Y and H63D mutations. Genet Test. 2000;4:183–198. doi: 10.1089/10906570050114902. [DOI] [PubMed] [Google Scholar]
- 22.Pointon JJ, Merryweather-Clarke AT, Carella M, Robson KJ. Detection of C282Y and H63D in the HFE gene. Genet Test. 2000;4:115–120. doi: 10.1089/10906570050114803. [DOI] [PubMed] [Google Scholar]
- 23.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 24.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 26.Pietrangelo A. Hereditary hemochromatosis - a new look at an old disease. N Engl J Med. 2004;350:2383–2397. doi: 10.1056/NEJMra031573. [DOI] [PubMed] [Google Scholar]
- 27.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He X, Lu X, Hu J, Xi J, Zhou D, Shang H, et al. H63D polymorphism in the hemochromatosis gene is associated with sporadic amyotrophic lateral sclerosis in China. Eur J Neurol. 2011;18:359–361. doi: 10.1111/j.1468-1331.2010.03158.x. [DOI] [PubMed] [Google Scholar]
- 29.Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20:439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 30.Santos PC, Cancado RD, Terada CT, Rostelato S, Gonzales I, Hirata RD, et al. HFE gene mutations and iron status of Brazilian blood donors. Braz J Med Biol Res. 2010;43:107–114. doi: 10.1590/S0100-879X2009007500031. [DOI] [PubMed] [Google Scholar]
- 31.Egger M, Smith GD. Bias in location and selection of studies. BMJ. 1998;316:61–66. doi: 10.1136/bmj.316.7124.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 33.Sutton AJ, Duval SJ, Tweedie RL, Abrams KR, Jones DR. Empirical assessment of effect of publication bias on meta-analyses. BMJ. 2000;320:1574–1577. doi: 10.1136/bmj.320.7249.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sterne JA, Gavaghan D, Egger M. Publication and related bias in meta-analysis: power of statistical tests and prevalence in the literature. J Clin Epidemiol. 2000;53:1119–1129. doi: 10.1016/S0895-4356(00)00242-0. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt S, Allen KD, Loiacono VT, Norman B, Stanwyck CL, Nord KM, et al. Genes and Environmental Exposures in Veterans with Amyotrophic Lateral Sclerosis: the GENEVA study. Rationale, study design and demographic characteristics. Neuroepidemiology. 2008;30:191–204. doi: 10.1159/000126911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos PC, Cancado RD, Pereira AC, Schettert IT, Soares RA, Pagliusi RA, et al. Hereditary hemochromatosis: mutations in genes involved in iron homeostasis in Brazilian patients. Blood Cells Mol Dis. 2011;46:302–307. doi: 10.1016/j.bcmd.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 37.Merryweather-Clarke AT, Pointon JJ, Shearman JD, Robson KJ. Global prevalence of putative haemochromatosis mutations. J Med Genet. 1997;34:275–278. doi: 10.1136/jmg.34.4.275. [DOI] [PMC free article] [PubMed] [Google Scholar]


