Abstract
The genomic era has enabled research projects that use approaches including genome-scale screens, microarray analysis, next-generation sequencing, and mass spectrometry–based proteomics to discover genes and proteins involved in biological processes. Such methods generate data sets of gene, transcript, or protein hits that researchers wish to explore to understand their properties and functions and thus their possible roles in biological systems of interest. Recent years have seen a profusion of Internet-based resources to aid this process. This review takes the viewpoint of the curious biologist wishing to explore the properties of protein-coding genes and their products, identified using genome-based technologies. Ten key questions are asked about each hit, addressing functions, phenotypes, expression, evolutionary conservation, disease association, protein structure, interactors, posttranslational modifications, and inhibitors. Answers are provided by presenting the latest publicly available resources, together with methods for hit-specific and data set–wide information retrieval, suited to any genome-based analytical technique and experimental species. The utility of these resources is demonstrated for 20 factors regulating cell proliferation. Results obtained using some of these are discussed in more depth using the p53 tumor suppressor as an example. This flexible and universally applicable approach for characterizing experimental hits helps researchers to maximize the potential of their projects for biological discovery.
INTRODUCTION
The past decade has witnessed huge advances in the power and scope of analytical technologies based on genomic data. These methods, which include the functional identification of genes using traditional genetic and RNA interference (RNAi)-based knockdown screens (Forsburg, 2001; Boutros and Ahringer, 2008), the identification of DNA and RNA populations by microarray analysis or next-generation sequencing (Capaldi, 2010; Niedringhaus et al., 2011; Ozsolak and Milos, 2011), and the identification of proteins, complexes, and their modifications using mass spectrometry–based proteomics (Walther and Mann, 2010), have transformed biological research. Many researchers can now turn to these techniques to address specific biological questions or, by performing larger-scale or high-throughput experiments, discover genes, transcripts, and proteins involved in their system of interest.
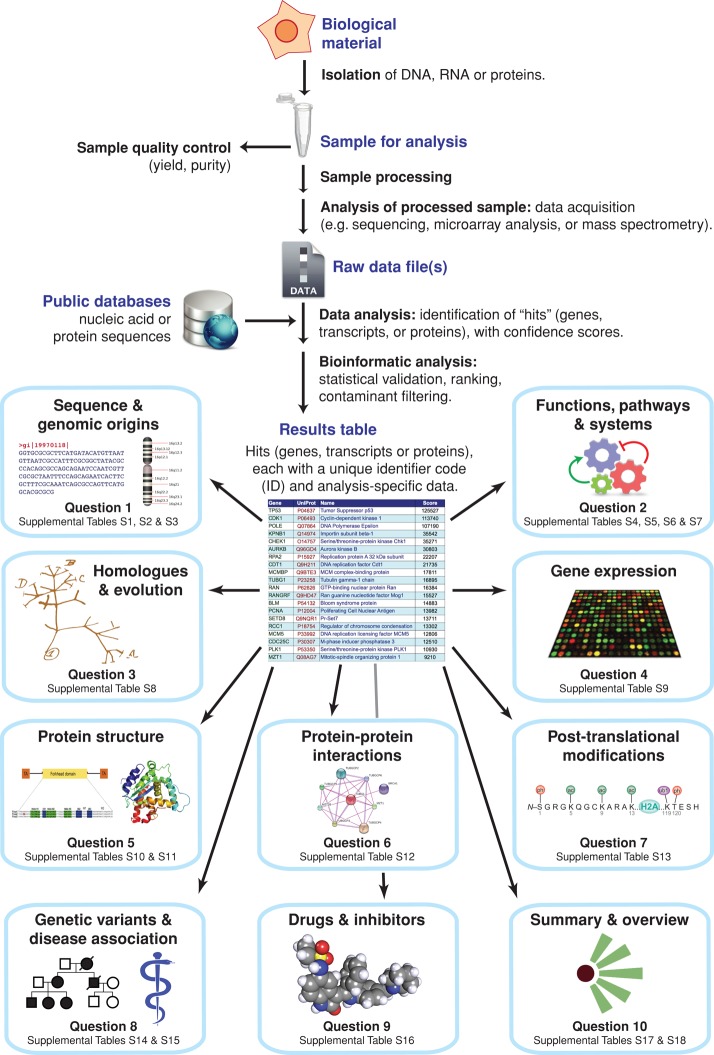
Although these technologies differ greatly in their principles and mechanistics, their general approaches share a similar form. A typical workflow (Figure 1) involves first the careful isolation of DNA, RNA, or proteins from the biological sample of interest, followed by a quality control step. The subsequent analysis increasingly depends on highly specialized instrumentation and technical expertise and is usually performed not by biologists themselves but by analytical platforms within core facilities of institutes or outsourced to external companies. Raw data from these analyses are analyzed computationally, resulting in the identification of multiple gene, transcript, or protein hits, that is, entries from public sequence databases, each described using a unique identifier code (ID; in some cases known as an accession code), plus other data pertinent to the experiment in question, such as a confidence score or intensity measurement. The resulting hits table may typically undergo further bioinformatic analyses, including statistical validation, ranking, and, in some cases, identification and removal of known contaminant entries.
FIGURE 1:
Generalized workflow for the analysis of DNA, RNA, or protein samples and questions about the hits identified. Nucleic acid or protein samples isolated from the biological material of interest are processed, then analyzed by various methods. Raw analytical data are then matched to entries in public databases, generating a results table listing the genes, transcripts, or proteins (hits) identified. For each of these hits, 10 questions relating to their features, functions, and other properties are shown (blue boxes). Each question is addressed by a section in the text, plus one or more supplemental tables containing examples of hyperlinks to entries in online resources.
Unfortunately, this hits table is often where platform-driven analysis stops, leaving the research biologist with a list of often unfamiliar gene, transcript, or protein names, abbreviations, and IDs, of which he or she has the task of making sense. Researchers faced with such a list will naturally be curious to find out more about each of the hits, to determine whether they are interesting and worthy of investing time and resources for follow-up studies. They may wish to know whether the hit was previously reported to have an involvement in their biological system of interest or whether it is novel and what is known about its functions, structure, interactions, and so on.
Fortunately, help is at hand. The past decade has also seen the emergence of a plethora of high-quality database resources providing information about the functions of genes, transcripts, and proteins for many organisms. These provide multiple gateways for the biologist, allowing access to information relating to nucleotide or amino acid sequences, genomic origins, evolutionary conservation, expression in cells and tissues, and association with disease processes. Further protein-related information centers on enzymatic or other functions, biological processes in which they are involved, domain and three-dimensional structures, interaction partners, posttranslational modifications, and the possibility of modulating their activities using small-molecule inhibitors.
Database resources providing such information are necessarily based on gene, (less commonly) transcript, or protein IDs. Analytical outputs generate hit lists of usually one ID type. However, types of ID can be interconverted (albeit not always perfectly), meaning that the resources available for consultation are not restricted by the ID type generated by the analysis, allowing the data-mining net to be cast as widely as possible. Thus a gene function database should be considered of equal relevance for exploring the roles of protein hits as a protein-based resource. Conversely, a protein domain structure database should be considered of equal interest for investigating potential functions of the products of gene- or transcript-based hits as a gene-based resource.
This review aims to contribute toward satisfying the desires of research biologists to explore the functions of protein-encoding genes and their transcripts and products. Ten questions are posed to guide the characterization of a given hit, each question being answered by the presentation of one or more Internet-based resources that provide reliable and relevant information, are freely accessible, and are described in peer-reviewed publications. Resources are included on the basis of quality, comprehensiveness, and usability. Another important parameter is that hit-specific entries in these resources should be directly accessible via standard gene, transcript, or protein IDs.
Each answer is accompanied by Web links to specific entries (“deep links”) in relevant databases, presented in the accompanying supplemental tables. The utility of these resources is illustrated using a hypothetical data set of 20 factors that regulate aspects of cell proliferation. Of these, particular focus is drawn to DNA polymerase (Kornberg, 1990), the cyclin-dependent kinase Cdk1 (Dorée and Hunt, 2002), and the tumor suppressor p53 (Lane et al., 2010), at least one being present in all organisms, enabling the outputs of various resources to be compared.
Straightforward methods are also described for biologists to access these resources by generating one-click links from results spreadsheets directly to database entries and by supplementing results tables with information to annotate each hit. These approaches provide an efficient and flexible means for biologists working with any genome-based technology and experimental species to retrieve reliable information to enhance biological discovery without the need for bioinformatic training, programming experience, or specialist software.
QUESTION 1: WHAT IS THE SEQUENCE OF THE HIT, AND WHAT ARE ITS GENOMIC ORIGINS?
Hits from screens and analytical experiments may be in the form of genes (identified by unique codes, names, or symbols) or nucleotide or amino acid sequences (identified by unique IDs). In either case, it is often worthwhile to visit the relevant page of the home database, that is, the primary or official repository of information about that hit, before embarking on visits to online resources of more specific functional information.
For gene-based experimental hits from model organisms, the home database would be the corresponding species-specific gene database (Supplemental Table S1). Here gene pages can be accessed using species-specific gene nomenclature and codes, which may differ from those used by major sequence databases, with the majority also being directly accessible using sequence IDs of standard types. Species-specific gene databases allow rapid access to information about genotypes, phenotypes, and the availability of mutant strains and related resources, making these preferred first ports of call for gene-based studies.
For sequence-based hits, the relevant home database would be the primary repository for the sequence (Supplemental Table S2). Visiting such a resource allows the biologist to touch base with the experimental origins of the sequence and identify the research team or project from which the data originate. These resources also allow rapid retrieval of nucleotide or amino acid sequences in FASTA and other standard formats, which, although unlikely to shed light on questions of gene or protein function, is useful for bioinformatic procedures for which direct linking is not possible. Which site is considered the home database depends on the source of the sequence information used in the analysis. However, this logic also works in reverse: for several analytical techniques, the research biologist (or the analyst, at the biologist's direction) has a choice of sequence database from which hits can be identified on the basis of experimental data. As sequence databases become more comprehensive, genome (or proteome) coverage alone will no longer become the principal criterion for making such choices; the quality of annotation and user experience may also contribute to decisions regarding which sequence databases are preferred.
Nucleotide sequence–based hits
The longest-established repositories of nucleotide sequences are GenBank (Benson et al., 2014), the European Molecular Biology Laboratory Nucleotide Sequence Database (EMBL-Bank), which is part of the European Nucleotide Archive (ENA; Pakseresht et al., 2014), and the DNA Data Bank of Japan (DDBJ) (Kosuge et al., 2014). These resources collaborate to share sequence information, so that all GenBank/EMBL/DDBJ entries can be retrieved from each of the host websites. The Reference Sequence (RefSeq) database (Pruitt et al., 2014) aims to provide a single entry for each nucleic acid or protein molecule, making explicit the relationships between genes, transcripts, and proteins. GenBank/EMBL/DDBJ and RefSeq entries are stored in the Nucleotide database of the National Center for Biotechnology Information (NCBI; NCBI Resource Coordinators, 2014), making this an ideal home database for accessing nucleotide entries of these types.
The Ensembl resource (Flicek et al., 2014) comprises sequences of genomic DNA, transcripts, and predicted polypeptide products, with data originating from genome sequencing projects, mostly from vertebrates. Updated versions are released on an approximately quarterly basis to incorporate genome reassemblies and the integration of new sequence and annotation data; this ensures that the database continues to improve in reliability, although one downside is that Ensembl IDs periodically become outdated and replaced. The main Ensembl resource is complemented by the Ensembl Genomes database (Kersey et al., 2014), comprising entries from sequenced bacteria, fungi, plants, protists, and invertebrate Metazoa (Supplemental Table S3).
For species whose genomes have not been completely sequenced (e.g., Xenopus laevis), Gene Indices (Lee et al., 2005) are a useful source of sequence information. Here multiple transcripts are assembled into tentative consensus sequences that can be used as references for gene expression or (following in silico translation) proteomics studies.
Protein hits
For protein sequences, the UniProt resource (UniProt Consortium, 2014) comprises entries from the curated Swiss-Prot and the noncurated TrEMBL (translated EMBL nucleotide) databases. Entries from both use UniProt IDs (known as accession codes), which present a standard nomenclature used by the majority of protein-oriented programs and resources. Swiss-Prot, sometimes referred to as the gold-standard protein database, combines stable IDs with rich, expert-curated annotation relating to the protein's composition and biological functions. TrEMBL entries derive from automatically in silico–translated nucleotide entries from the ENA and Ensembl databases but lack functional annotation. With new versions being released approximately monthly, the UniProt resource manages to combine the best of both worlds: rich annotation together with regularly updated entries. An additional advantage of UniProt is that for most entries, official gene symbols are included in the protein entry headings, providing a means for gene- as well as protein-oriented database resources to be directly accessed.
Another protein resource of note is the NCBI's nonredundant (nr) database, compiled from entries originating from GenBank/EMBL/DDBJ, RefSeq, Swiss-Prot, and protein-structure databases. Entries, which are accessible from the NCBI protein portal (NCBI Resource Coordinators, 2014), all have two IDs: one from the database of origin, plus a GenInfo Identifier (GI) number. Combining entries in this way, nr has the advantage of being very comprehensive in its coverage, but it has disadvantages such as the rapid turnover of GI numbers, inconsistency in nomenclature between source databases, and poor functional annotation.
A protein database that achieved popularity among proteomics researchers was the International Protein Index (IPI; Kersey et al., 2004); however, in 2011 this was discontinued and entries integrated into UniProt (Griss et al., 2011). The removal in 2014 of IPI cross-references in UniProt entries finally rendered the IPI obsolete.
Genomic context
For hits of any type, users may wish to access information about the genomic contexts of the relevant genes, including chromosomal location, gene length and orientation, proximal genes, and relevant genomic features in the vicinity. Many species-specific gene databases contain an embedded genome viewer presenting concise genomic information, and often this is sufficient. However, when more detailed information is required, including multiple alignments with other sequences and features, direct access to the relevant locus within a specialized genome browser is desirable. Probably the most comprehensive cross-species tool for visualizing and aligning sequences in their genomic contexts is the UCSC Genome Browser (Karolchik et al., 2014), which can be queried using all major gene, transcript, or protein IDs. Here a diagram shows the gene's location within the relevant chromosome, and, below, a panel presents a graphical view of the relevant genomic region, with a multiple alignment of various sequences, including splice-variant mRNAs and expressed sequence tags, features such as single-nucleotide polymorphisms (SNPs) and repeats, and features derived from ENCODE project data (Rosenbloom et al., 2013). Also shown are multiple alignments between the gene of interest and orthologues from related species. A complementary functionality is provided by the genome browser within Ensembl, with flexible options for the export of genomic sequence data.
Because navigation and exploration of genomes are not the themes of this review, for further information readers are directed to the Nature Genetics free online series of tutorials, “A user's guide to the human genome” (Wolfsberg et al., 2002). Although these guides may be a little outdated and human-centric, the principles remain unchanged and are applicable to the navigation of genomes of many organisms.
QUESTION 2: WHAT ARE THE KNOWN FUNCTIONS OF THE GENE AND ITS PRODUCTS?
This question is probably the most significant of them all: what is the gene of interest for, what does its product do, and in what processes is it involved? Given the complexity of biological systems, these straightforward questions often yield diverse and incomplete sets of answers.
One of the first paradigms in molecular biology was the “one gene, one enzyme” hypothesis (Horowitz, 1995), and although this proved to be a great oversimplification, many gene-product hits from screens may correspond to proteins with characterized enzymatic activities. Many other proteins may have structural functions or play roles in signaling pathways or regulating gene expression. In each of these cases researchers will be interested to retrieve essential information plus contextual information relating to the functions of the hits identified in their experiment. Essential information would include the following. For enzymes: substrates and products and molecules that modulate their activity, such as allosteric activators and cofactors; for structural proteins: the relevant cellular structures and partner molecules with which they collaborate to maintain the structure; for signaling molecules: upstream regulators and downstream targets, plus other components, such as scaffold proteins; and for gene expression modulators: members of protein complexes that modulate chromatin and affect transcription. Contextual information could include the following. For enzymes: metabolic pathways in which they are involved; for structural proteins: the role of the structure in the life of the cell; for signaling pathways: an overview of relevant pathways, from initial stimuli to ultimate responses; and for gene expression regulators: their roles during differentiation and development. For all proteins, information about the location(s) within or outside of the cells in which they function is of great interest and relevance. Online resources may provide such information in a variety of means, ranging from a single word, a line of text, a paragraph, a summary diagram, or a full-length review article.
Literature searching and functional summaries
One obvious possible starting point for the retrieval of known functions of a gene or protein of interest is a search of the biomedical literature, using tools such as PubMed, Google Scholar, and others (Lu, 2011; Supplemental Table S4). However, this can result in the retrieval of hundreds of titles, linked to abstracts but not necessarily full-text articles. A more efficient strategy is to extract pertinent sentences from publications using “smart” literature-mining tools such as Textpresso for model organisms (Müller et al., 2004) and iHOP (Fernandez et al., 2007). Although a helpful step forward, these tools still leave the user with typically dozens (or hundreds) of disconnected sentences to sift through and interpret. More convenient still would be a concise executive summary of the known properties of the gene or protein. Entries in the NCBI Gene database (NCBI Resource Coordinators, 2014) contain, for better-characterized genes, a single-paragraph summary of the functions of the gene product in its physiological and (if relevant) pathological context. Similarly, the curated (Swiss-Prot) entries in UniProt have a General annotation (Comments) section in which functions, activity, subunit structure, and other properties are listed, broken down into categories, and well referenced. For human genes, more extensive expert-curated information is provided by the Online Mendelian Inheritance in Man (OMIM) resource (Amberger et al., 2011). Entries here contain well-referenced descriptions of the identification of the gene and its functions, allelic variants and association with disease, and the biochemical properties of the product. OMIM entries can also be retrieved by searching with gene identifiers for nonhuman model organisms.
Ontology resources
One widely used approach to the functional characterization of gene products is the use of controlled-vocabulary ontology terms (Supplemental Table S5). This allows hits to be compared, sorted, and grouped on the basis of their properties. The Gene Ontology (GO) project (Blake, 2013) uses three categories of ontology term—molecular function, biological process, and cellular component—based on data from gene or protein resources and published literature. GO classification has a hierarchical structure, terms being applicable at different levels. For a given hit, an interactive hierarchical GO graph is viewable at Ensembl (within the transcript-based display). A detailed listing of all applicable GO terms for a factor of interest is rapidly accessible from QuickGO (Huntley et al., 2009), although this comprehensive output often contains considerable redundancy. It is often more desirable to represent each GO category by one or very few concise terms, so-called GO slims, but these are curated and accessed independently of the main GO project. PANTHER (Mi et al., 2013) is a resource that classifies proteins from 82 organisms based on evolutionary relationships. Curated functional information is provided for genes, transcripts, and proteins in the form of “slim” terms for the three GO categories plus two functional categories of its own: Protein class and Pathway.
Although undoubtedly helpful, there are notable drawbacks to using ontology terms to characterize gene products. First, the high rate of research output makes it difficult for assignment of terms to keep pace. Second, in several cases, terms are inferred from those of orthologous proteins, introducing assumptions that may not always hold true. Third, ontology labels largely fail to capture the dynamic nature of proteins during the life of a cell or organism. For example, a protein may be cytoplasmic in interphase, nuclear in prophase, associate with the mitotic spindle in metaphase, and be rapidly degraded in anaphase; recording such dynamic behavior is crucial for understanding this protein's function but would not be reflected by the corresponding ontology terms.
Enzymes, signaling pathways, and systems
Where it is clear from summary information or ontology annotation that the gene product of interest has enzymatic activity, researchers may wish to dig deeper to find out more about the enzyme: its known substrates, products, and means of regulation. For this, specialized enzyme information resources can be rapidly accessed and are worth visiting (Supplemental Table S6). The IntEnz resource (Fleischmann et al., 2004) is home to the official Enzyme Commission nomenclature; its website provides a clear overview of reactions catalyzed by each enzyme and lists other relevant molecules such as cofactors. For more detailed enzymatic information, BRENDA (Schomburg et al., 2013) presents an extremely comprehensive resource, each enzyme record having subsections relating to structure, enzyme–ligand interactions, inhibitors and activating molecules, catalytic parameters, and reaction conditions. Also recorded is information relating to the cloning, expression, purification, and engineering of the enzyme, plus connections with disease.
For proteins with roles in signaling pathways, a pathway diagram showing at a glance the role of the protein of interest can be a helpful starting point. One of the original resources providing information in this format is BioCarta Pathways (Nishimura, 2001), with its colorful, cartoon-like (but expert-curated) pathway diagrams. Complementing these are the more electrical circuit–like diagrams of the Kyoto Encyclopedia of Genes and Genomes (KEGG) PATHWAY resource (Kanehisa et al., 2014). On both of these sites, clicking a protein component within the diagram links to gene- or protein-specific information.
The most comprehensive resource encompassing enzymes and signaling molecules in their cellular contexts is Reactome (Croft et al., 2014). Here molecules small and large are recorded together with characterized “events,” which include enzymatic reactions, intermolecular binding, and intracellular transport. Querying Reactome lists reactions and pathways involving the molecule in question; clicking one of these opens an interactive, zoomable diagram of the reaction or pathway, in the context of cellular compartments and membranes, with steps involving the queried gene product highlighted.
Accompanying the rise of systems biology approaches (Kirschner, 2005), a systems-based means to retrieving information on gene and protein function can also be useful. The NCBI's BioSystems resource (Geer et al., 2010) presents a single portal for accessing information on the involvement of molecules in biological systems, with data originating from resources including KEGG, Reactome, and others. A BioSystems search first generates a list of systems in which the gene product plays a role, with headings that may range from the extremely general (e.g., “intracellular”) to the very specific (e.g., “CDT1 association with the CDC6:ORC:origin complex”). Selection of a heading opens a page containing a one-paragraph description of the system, a pathway diagram (where appropriate), and a multitabbed section providing links to relevant genes and proteins and to related biological systems.
Project-specific databases
Functional information more relevant to a particular biological process can in some cases be obtained from Web-based databases created to disseminate data generated by specific research projects (Supplemental Table S7). Data from genome-scale knockout or knockdown projects are particularly relevant, as users can be almost certain to obtain some functional information relating to their genes of interest: at least whether they are essential for viability, plus obvious and more subtle loss-of-function phenotypes. A pioneering example of this is PhenoBank, which describes and shows movies of phenotypes obtained from a genome-wide RNAi-knockdown screen in Caenorhabditis elegans early embryos (Sönnichsen et al., 2005). Another such resource is the Schizosaccharomyces pombe gene database, PomBase (Wood et al., 2012), which records phenotypes obtained from a genome-wide deletion screen (Kim et al., 2010; Hayles et al., 2013).
The MitoCheck database, based on human genes, shows movies of time-lapse fluorescence microscopy experiments, as well as inferred phenotypes, from genome-scale RNAi screens. Initially created to record the effects of human gene knockdowns on chromosome behavior during the cell cycle (Neumann et al., 2010), this resource is being complemented by data sets from subsequent RNAi screens investigating additional cellular processes. Also included are data on the subcellular localization and protein interactions of gene products required for cell division (Hutchins et al., 2010). The MitoCheck database can be searched using human gene symbols, synonyms, or UniProt IDs, plus gene terms for orthologous nonhuman genes, making this a unique and valuable cross-species functional resource.
QUESTION 3: WHAT HOMOLOGUES OF THIS GENE (OR PROTEIN) ARE KNOWN? HOW WELL HAS IT BEEN CONSERVED THROUGH EVOLUTION?
There are several reasons for wanting to identify genes or proteins of closely related sequence to the one of interest. First, for a poorly annotated transcript or protein (e.g., one with a sequence ID but no gene information) the issue may simply be one of identification: an identical (or virtually identical) sequence from the same species may provide sufficient information to allow gene identification and further exploration. Second, for a hit originating from a less well-annotated database, orthologues from closely related species may provide richer functional annotation and greater availability of research resources (including mutant strains, recombinant proteins, and antibodies) for follow-up studies. Third, knowing the extent of conservation of the gene through evolution indicates how fundamental its role is: genes well conserved throughout the kingdom of life likely play central roles in vital cell processes, whereas those with more limited conservation likely have more specialized roles in certain classes of organism.
When considering homologous genes or proteins, the distinction between orthologues (in which sequence divergence follows speciation) and paralogues (in which divergence follows gene duplication) should be borne in mind (Fitch, 2000). For many genes or proteins, homologues of both types have been identified by automated methods, together with inferences about their evolutionary history. However, for the many gene products whose sequence conservation is low or patchy, formal identification of orthologues and paralogues is a highly skilled task, the domain of specialist bioinformaticians.
From a gene or protein of interest, one can quickly identify known orthologues and paralogues via Ensembl (Supplemental Table S8). Each Ensembl gene page contains links to an orthologue page and a paralogue page, and then to pairwise alignments of cDNA and protein sequences. Although these orthologues and paralogues are based on Ensembl IDs, the pages themselves can be accessed directly via a variety of gene, nucleotide, and protein ID types. The MitoCheck database, searchable using human and nonhuman gene names, lists Ensembl-predicted paralogues and orthologues in a concise format, the latter being linked to species-specific gene databases where appropriate.
HomoloGene (NCBI Resource Coordinators, 2014) is the NCBI's resource for automated retrieval of gene and protein homologues from 21 completely sequenced genomes. Querying by gene symbol or NCBI-based nucleotide or protein ID generates a list of gene orthologues, alongside each being a corresponding orthologous protein (all based on RefSeq entries), with a graphical representation of conserved domains. Links from this page include those to a multiple sequence alignment, a table of pairwise alignment scores, and literature references. An additional useful feature of HomoloGene is its statement on evolutionary conservation—for example, “gene conserved in fungi/metazoa group.”
For those wanting to perform de novo searches for entries with very similar nucleotide or protein sequences to hits of interest, probably the best-known method is the BLAST algorithm (Altschul et al., 1990). BLAST searches can be launched via direct Web links from the NCBI and UniProt websites for nucleotide or protein hits, respectively, with matches typically listed in decreasing order of quality. Because searches such as BLAST are relatively processor intensive and can take a few minutes to complete, a more efficient approach is to retrieve identified lists of proteins that share a minimum sequence identity. When one has a protein or nucleotide entry and wants to identify a set of closely related proteins from any species, a very fast and direct method is the UniProt Reference Clusters (UniRef) facility (Suzek et al., 2007). UniRef can display a list (“cluster”) of UniProt entries, from all species, whose sequences share at least 50%, at least 90%, or 100% identity to the query. In cases in which the name of the hit is obscure or a gene identifier is absent, accessing UniRef (e.g., at the 90% identity level) quickly displays the set of highly similar proteins, some of which may be better annotated than the query. However, UniRef listings lack information about which entries within the cluster have the closest identity to the protein of interest, making this utility unsuitable to judge which protein is the closest homologue in the same or other species.
One graphical approach to the identification of gene or protein homologues in their evolutionary context is the phylogenetic tree. TreeFam (Schreiber et al., 2014) allows rapid access to phylogenetic trees representing families of related genes from genome-sequenced animals (plus budding and fission yeasts, two flagellates, and Arabidopsis as an outgroup set of reference species). Querying by gene or protein ID selects the appropriate gene family, for which information is presented in two views. The Summary view displays a highly compact tree showing the extent of conservation of genes from the family within various taxonomic ranks. The Gene Tree view displays a rooted, scaled phylogenetic tree in which each of the nodes (Ensembl genes) is labeled with gene name and species, together with a domain diagram of the corresponding protein.
QUESTION 4: HOW IS THIS GENE EXPRESSED IN CELLS OR TISSUES, AND HOW DOES THIS CHANGE UNDER EXPERIMENTAL CONDITIONS?
Valuable indications as to the potential involvement of a gene in a biological process of interest can be obtained from data relating to its pattern of expression within the organism and how this changes during development or in response to cell stress or drug treatment conditions. For experimental model organisms, summaries of gene expression in physiological contexts relevant to that species are most often included in the species-specific gene databases; those listed in Supplemental Table S1 all have “expression” data sections, except for SGD (budding yeast), for which expression data are hosted by the SPELL database (Hibbs et al., 2007; Supplemental Table S9).
Because the major high-throughput gene expression technologies (microarray analysis and next-generation sequencing) are nucleic acid based, the majority of expression data are at the transcriptional level. The two largest repositories of gene expression data obtained with such technologies are the NCBI's Gene Expression Omnibus (GEO; Barrett et al., 2013) and the European Bioinformatics Institute (EBI)–hosted ArrayExpress (Rustici et al., 2013), which also imports GEO entries. These resources provide public access to billions of expression data entries, together with corresponding experimental details. However, with so much data available, instead of accessing potentially hundreds of experimental records corresponding to a particular gene, it is almost always preferable to obtain an overview of the relevant data first. This facility is admirably provided by the EBI's Gene Expression Atlas (Petryszak et al., 2014), a gene-oriented database containing a curated subset of ArrayExpress data, accessible using virtually all gene, nucleotide, or protein IDs. Subsections summarize expression of the selected gene in tissues (for some species accompanied by a diagram of the organism with expressing tissues highlighted), by cell types, cell lines, and disease states, and in response to drug and other experimental treatments. Results are provided with links to the original data at ArrayExpress.
For expression detected at the protein level, The Human Protein Atlas (Asplund et al., 2012) contains image-based data from immunohistochemical and immunocytochemical analyses for thousands of human gene products. Each gene page summarizes the subcellular location of its product and provides relative quantifications of antibody staining (from negative to strong) across a range of normal tissues and organs, cancer tissues, and cell lines. Clicking a summary result leads to a detailed page of staining data, and then to high-resolution micrograph images. In each case, full information about the antibody used is provided.
QUESTION 5: WHAT IS THE COMPOSITION OF THE PROTEIN, IN TERMS OF DOMAINS, SEQUENCE MOTIFS, OR THREE-DIMENSIONAL STRUCTURE?
Domains and sequence motifs
For a gene or protein with which one is unfamiliar, a visual overview of conserved domains and functional sites in the protein can give useful insights, for example, into catalytic activities, binding of cofactors and interaction partners, and subcellular localization. Some of these properties are conferred by larger protein domains, well conserved in terms of sequence and structure, whereas other functions depend on short linear motifs (SLiMs) of just a few amino acids in length (Hunt, 1990). Domains and motifs within many proteins have been identified and reported in published studies, and many of them are included in curated UniProt entries. However, for a more comprehensive coverage of such features, one should turn to specialist resources, which use automated sequence or structure-based classification algorithms, in some cases complemented by manual curation.
Several resources providing for the identification of protein domains have been developed; these differ in approaches, definitions, and algorithms, thus generating complementary sets of classifications. Major protein-domain resources driven primarily by sequence data include Pfam (Finn et al., 2014), SMART (Letunic et al., 2012), PANTHER (Mi et al., 2013), and InterPro (Hunter et al., 2012; Supplemental Table S10). In contrast, domain classification by CATH-Gene3D (Sillitoe et al., 2013; Lees et al., 2014) is driven mainly by protein three-dimensional (3D) structure data, including the overall architecture, subdomain folding, and secondary structural elements. Programs for identifying SLiMs within a protein of interest include PROSITE (Sigrist et al., 2013), the Eukaryotic Linear Motif (ELM) database (Dinkel et al., 2014), Minimotif Miner (Mi et al., 2012), and Scansite (Obenauer et al., 2003).
Each of these domain or motif identification resources can be accessed independently. However, a more efficient approach is to employ a program that uses several methods and generates a graphical output integrating all identified features in a single display. Typically with these programs, “mousing over” a feature triggers a pop-up box providing further information. The ideal program would use all of the foregoing domain and motif identification methods, generating a single clear and concise diagram; because no single program includes all of these methods, the prominent resources are described separately.
The NCBI's Conserved Domain Database (CDD; Marchler-Bauer et al., 2013) generates a very compact diagram representing superfamilies, domains, and functional sites of the protein, together with annotation of specific residues required for activities such as enzymatic catalysis or binding to DNA. Parameters used to define matches can be refined to allow identification of features at different sensitivity thresholds. Mousing over a feature opens a box providing a functional description and (where available) a 3D structure image. Proteins harboring similar domain architectures can be displayed via a single-click link to the CDART website (Geer et al., 2002). The CDD is thus an excellent starting point for identifying key features of a protein, before using other programs to perform deeper exploration.
The InterPro resource (Hunter et al., 2012) identifies protein features in four categories: families, domains, repeats, and sites, based on signatures defined by multiple partner databases. The outputs are displayed in a clear graphical multialignment, each feature hit being a Web link to the relevant entry in the home resource. DASty (Villaveces et al., 2011) uses the Distributed Annotation System to delegate protein feature annotation to different servers in parallel. The protein's complete amino acid sequence is shown, and below, a graphical multialignment shows features returned from various sources. These include InterPro domains, structural elements, and functional sites manually curated by UniProt, plus a selection of predicted SLiMs. The ELM functional site prediction tool (Dinkel et al., 2014) displays Pfam and SMART domains, globular and ordered (or disordered) regions, and secondary structural elements, plus a large battery of SLiMs, both from sequence-based predictions and curated from the literature, in a graphical multialignment format. Below the protein feature diagram, a table provides detailed information on the sequence segments corresponding to features in the display.
The ANNIE protein sequence annotation and interpretation environment (Ooi et al., 2009) runs >20 search algorithms in parallel on an input sequence to identify compositional and secondary structural features and matches to various SLiMs and other sequence motifs. Protein domains are identified by real-time searches using the HMMER, IMPALA, and RPS-BLAST algorithms. A unique feature of this resource is its Interactive View display environment: within the graphical multialignment, mousing over any identified feature reveals more information about that match and its quality, whereas “dragging over” a segment allows the user to zoom in on a region of interest—if desired, all the way to the amino acid sequence.
Three-dimensional structures
Because a protein's structure provides the key to understanding the mechanism of its function, insights can often be gained by exploring structures, especially for proteins complexed with physiologically relevant molecules such as interacting proteins or peptides, nucleic acids, substrates or cofactors, or small-molecule inhibitors. The definitive resource for protein 3D structures is the Protein Data Bank (PDB), for which records can be readily retrieved from the Research Collaboratory for Structural Bioinformatics (RCSB) website (Rose et al., 2013), its partner site PDB in Europe (PDBe; Gutmanas et al., 2014), and the NCBI's Molecular Modeling Database (MMDB; Madej et al., 2012; Supplemental Table S11). These resources offer complementary search and display options, but all allow the inspection of structures online using interactive Web-based viewers and the download of structure data files for offline viewing using the latest 3D-structure exploration software.
In many cases there are multiple PDB records for a given protein; these often correspond to protein constructs of different lengths and proteins complexed with different molecules. Searching the foregoing resources by gene or protein ID yields the set of relevant records, each title being a brief description of the protein plus any complexed molecules. Alternatively, a graphical overview of PDB records corresponding to a given gene product is provided by PDBsum (de Beer et al., 2014). Here a domain diagram of the full-length protein is shown, and immediately below, a graphical alignment of constructs whose structures have been solved, with secondary structural elements depicted schematically. For a given construct, clicking its schematic diagram opens a sequence alignment with the full-length protein, whereas clicking its PDB code opens a page displaying a wealth of structural, biochemical, and functional information, with links to structure viewers and to downloading the structure file.
Returning to DASty, this program cleverly integrates sequence-based domain and feature prediction with 3D modeling. When a protein's 3D structure is available, this appears above the graphical multialignment in an interactive Jmol viewer (Herraez, 2006), allowing zooming in and out, rotation, and display in different formats. Clicking a domain or feature within the alignment highlights corresponding residues within the primary sequence and on the 3D structure, allowing researchers to identify 3D juxtapositions of domains and features within the protein.
QUESTION 6: WHICH PROTEIN INTERACTION PARTNERS HAVE BEEN REPORTED FOR THIS GENE PRODUCT?
Following the maxim “by your friends shall you be known,” further understanding of a protein's functions may be gained by identifying other proteins with which it interacts. It is increasingly recognized that most cellular processes are controlled by proteins acting in the context of complexes or “molecular machines” (Alberts, 1998) and that the specificity and coordination of intracellular signaling pathways is due to a large degree to interactions between signaling molecules and scaffold proteins (Good et al., 2011). Thus information about interactions of gene products can provide useful physiological context to understanding their biological functions.
Numerous protein–protein interaction databases have been developed, and thus a plethora of information is accessible, with a variety of options for retrieval and display (Supplemental Table S12). Some standardization between these resources is being achieved, as 12 prominent interaction databases have joined the International Molecular Exchange (IMEx) consortium to share curation and annotation of interaction data (Orchard et al., 2012). IntAct (Orchard et al., 2014) and BioGRID (Chatr-Aryamontri et al., 2013) are both IMEx-member protein–protein interaction resources that display interactions initially in a table format, with options for these to be displayed graphically. In the IntAct results table, the protein of interest (molecule A) appears next to information about each interacting protein (molecule B), including methods by which interaction was established (e.g., tandem affinity purification or two-hybrid assay), with links to literature references. The Graph tab generates a simple interactive interaction diagram, centered on molecule A. In BioGRID, gene products interacting with the protein of interest are tabulated in two formats: an uncluttered Summary, listing interactors by gene symbol, with synonyms and one-line descriptions; and a Sortable Table, listing the species, types of experiment used to establish interaction, and links to literature. The Graphical Viewer button opens a radial interaction diagram centered on the molecule of interest, with multiple options including filtering out interactions from low- or high-throughput studies or those discovered by different experimental approaches.
The STRING resource (Franceschini et al., 2013) generates by default a colorful interactive network diagram centered on the protein of interest, surrounded by its interacting partners. Each interacting protein is represented by a colored ball, labeled by gene symbol. Clicking each ball opens an information box containing a description, domain, and (where available) 3D structure; clicking an edge (connecting line) opens a box detailing the evidence for that interaction. Because STRING defines “interaction” based on diverse criteria, including “experiments” (such as coprecipitation or yeast two-hybrid assays), “coexpression,” and “textmining,” care should be exercised in interpreting the network diagram to ensure that interactions shown are of a type relevant to the issue in question. This can be achieved via the color coding of the network edges by interaction type, and a filter can be applied to restrict displayed interactions to those of a certain type—for example, “experiments.”
Interaction databases are invaluable resources for providing information about known partners and networks in which the gene products are involved. However, protein–protein interactions are certainly not always constitutive, and cell contexts—for example, cell-cycle and developmental stages and responses to stresses—count for a great deal. Currently, as with ontology terms, these dynamic aspects are rarely conveyed by interaction databases, leaving room for future developments to these resources.
QUESTION 7: WHAT POSTTRANSLATIONAL MODIFICATIONS HAVE BEEN REPORTED FOR THIS PROTEIN?
The covalent addition of chemical moieties to the side chains of particular amino acid residues is a highly prevalent and versatile mechanism by which proteins are regulated in both eukaryotes and prokaryotes (Walsh, 2006; Deribe et al., 2010) and likely plays important regulatory roles in virtually all cellular processes. The number of different types of posttranslational modification (PTM) known to exist runs into the hundreds (UniProt lists >450) and continues to increase. Because several residues within a protein may potentially be modified, with different PTMs present in various combinations, the potential repertoire of distinct species of modified protein in a cell is astronomical.
The modification status of a protein is highly dynamic, in many cases depending on the activities of enzymes that catalyze the addition and removal of the PTMs, those of proteins that bind depending on modification status, and the relative colocalization of all these players within the cell—sometimes to different parts of an organelle. These properties often in turn depend on cellular contexts such as cell type, cell-cycle phase, and cellular stresses, including drug treatments. Thus researchers wanting to know whether their gene product of interest is modified in vivo need to take into consideration this biological and experimental contextual information; the storage and retrieval of these metadata poses a particular challenge for PTM databases.
Hundreds of proteins have been the subject of focused PTM-related publications, and UniProt makes a major effort to incorporate these findings into its reviewed (Swiss-Prot) entries. For better-characterized proteins, the Post-translational modification subsection of UniProt's General annotation (Comments) reports which residues are known to be modified, which enzymes catalyze these modifications (where identified), and their functional consequences, with literature links. The power of modern mass spectrometry–based proteomics to identify, with high confidence and on a fairly large scale, several (but certainly not all) PTMs in proteins isolated from cells or tissues (Young et al., 2010) means that modifications identified from more-focused studies can be complemented by high-quality PTM data sets from larger-scale studies. However, whereas the resultant data explosion can be readily accommodated by specialist PTM databases, those protein resources that rely on manual curation to ensure quality are likely to lag behind in terms of up-to-dateness.
Arguably the most extensively studied protein PTM is the phosphorylation of tyrosine, serine, and threonine residues, and protein phosphorylation resources have taken the lead regarding the curation of PTM data and their retrieval with the necessary contextual information (Supplemental Table S13). The most comprehensive of these is PhosphoSitePlus (Hornbeck et al., 2012), in which each protein record includes a functional summary, and then modification sites (phosphorylation, plus several other types) are shown graphically in the context of the protein's domains. An accompanying table lists PTM sites with their surrounding amino acid sequences, comparing those from orthologous proteins in related species. Each modified residue has its own record page, displaying experimental and contextual information and literature references. Complementary information on protein phosphorylation is provided by the Phospho.ELM (Dinkel et al., 2011) and PHOSIDA (Gnad et al., 2011) resources.
Several databases include information about specific PTM types, but trawling though each individually would be an unnecessarily tedious exercise. A more efficient approach is to search a meta-database, one bringing together data originating from several separate databases. For PTM-related information the most comprehensive resource is dbPTM (Lu et al., 2013), whose entries cover 96 types of modification and originate from UniProt, with specialist databases of PTMs including protein phosphorylation, glycosylation, S-nitrosylation, ubiquitylation, and methylation, plus their own literature text-mining efforts. Here the location of each experimentally determined PTM is shown in a protein-domain diagram; beneath is a table listing the PTMs with some sequence context, Web links to the databases of origin, and literature references.
Another approach to determining whether a protein may be posttranslationally modified is the use of PTM predictors. These programs scan a protein's sequence, scoring each residue as a candidate for modification based on quantitative evaluation of the match between the sequence immediately surrounding the residue and patterns of amino acid preferences of modifying enzymes for substrate targeting. Such analyses can provide helpful indications as to which residues of a protein might be modified, together with suggestions of enzymes capable of catalyzing the addition. Nevertheless, this approach is ultimately limited, as the sequence preferences of many modifying enzymes are unknown, and these programs rarely consider additional crucial determinants such as longer-range enzyme–substrate contacts and the combination of spatial and substrate exclusivity (Alexander et al., 2011). PTM predictors have been discussed and evaluated (Eisenhaber and Eisenhaber, 2010; Que et al., 2010).
QUESTION 8: HAVE GENETIC VARIATIONS TO THIS HIT BEEN REPORTED, AND ARE THEY ASSOCIATED WITH HUMAN DISEASE PROCESSES?
Mutations and structural variations
The identification of genetic variations giving rise to distinct phenotypes is of course a cornerstone of the genetic approach to understanding biological processes. Genetic variations range in scale from one-base-pair SNPs, to genomic structural variations (GSVs) of tens to millions of base pairs, to complete gene deletions or knockouts. Observed phenotypes resulting from such variations depend on the biological conditions used by researchers to assay and characterize the variant cells and organisms and can range from the loosely descriptive to the highly quantitative. Such phenotypic data can be presented in a variety of formats, including text, tables, multidimensional images, and videos, all of which can be incorporated into modern Web-based gene resources.
For well-studied organisms, much relevant information on genetic variations and corresponding phenotypes is present in species-specific gene databases (Supplemental Table S1), and so for exploring known variations in hits from model experimental organisms and their biological consequences, the relevant gene page from such a resource is often the best place to start. More comprehensive, multispecies repositories of genetic variations are stored in specialist resources such as dbSNP for SNPs (Bhagwat, 2010) and DGVa and dbVAR for GSVs (Lappalainen et al., 2013; Supplemental Table S14). Information from these and other sources is available via Ensembl, for which gene pages can be accessed from multiple ID types. In the relevant Ensembl gene page, under Gene-based displays, one option is the Variation table; this provides an overview of all genetic variations identified for that gene. Here initially types of variation are listed, each accompanied by a brief description and the number of times a variation of that type has been found in the gene of interest. Clicking Show for a variation type opens a table displaying full information about all occurrences for that gene.
At the protein level, reviewed UniProt entries list documented amino acid variants under the Sequence annotation (Features) section, within three categories: Alternative sequence, Natural variant, and Mutagenesis. Alongside each variant are links to literature and source databases (where appropriate) and a graphic illustrating the position of the variation within the protein. When the variant has functional or pathological consequences, these are briefly described.
Association with human diseases
A major motivation for studying many biological processes is to gain insight into causes of human disease, and thus it is often of interest to establish whether genes or proteins of interest are reported to be associated with pathological states. For information linking genes to human diseases, useful starting points are the manually curated summaries within NCBI Gene (Summary and Phenotypes sections), OMIM (Gene Function section), and UniProt (Involvement in disease section). Supplementing these are more specialized resources linking genetic variations and expression abnormalities to clinical conditions. Although efforts are underway to standardize such data and centralize them in a single portal such as the NCBI's ClinVar (Landrum et al., 2014), the current diversity of complementary disease resources means that these still warrant separate descriptions.
A comprehensive categorization of associations between human genes and diseases is provided by the Genetic Association Database (Becker et al., 2004; Supplemental Table S15). Querying by gene term generates a table of published instances in which the involvement of that gene in a disease has been tested. Each entry reports the disease name and class and associated terms, plus numerous links, including one to the corresponding publication, often with a one-line summary of the study's conclusions. For well-studied genes the full output may contain considerable redundancy, and the database also reports negative associations (i.e., when a gene–disease association was tested and not found to exist), although these can be filtered from the output. Another valuable resource linking genes to diseases is KEGG (Kanehisa et al., 2014), in which a list of diseases associated with a gene of interest is shown in the relevant entry in the KEGG GENES database. Clicking a disease identifier links to the relevant entry in KEGG DISEASE, providing a full description of that disorder, the nature of the genetic association, etiological factors, and molecular markers, plus pharmacological agents with which it can be treated. The Comparative Toxicogenomics Database (Davis et al., 2013) incorporates a complementary approach: in addition to recording associations between genes and disease from manual curation, this resource contains gene–disease associations inferred on the basis of reported interactions between gene products and compounds and between compounds and disease. Inferences are given a score that is used to rank the (often long) resultant list of gene–disease associations.
Two broad classes of genetic disease in which connections between variations and symptoms have been most closely studied are developmental disorders and cancer. For the former, searching the DECIPHER resource (Bragin et al., 2014) by gene name generates a table that lists documented occurrences of consented patients harboring variations in that gene, the relevant variations, and descriptions of associated phenotypic symptoms. Clicking a record reveals more information about the patient, plus a genome browser indicating the position of the relevant variation relative to genes and other features.
The most intensely studied class of disease at the molecular level is almost certainly cancer, and probably the most comprehensive resource of somatically acquired genetic variations linked to human neoplasms is COSMIC (Forbes et al., 2011). For a given gene, an overview page contains an embedded genome browser providing a graphical summary of cancer-linked variations. Further gene-related information includes a breakdown of variation types, their distribution in different tissues, tables of specific mutations, and histograms of mutation frequency within protein domains, as well as lists of relevant studies and literature references. Cancer cells harboring mutations in certain genes may exhibit altered sensitivity to particular pharmaceutical agents. When appropriate, such drugs are listed, linking to their relevant entries in the Genomics of Drug Sensitivity in Cancer (GDSC) resource (Yang et al., 2013), providing interactive graphical representations of a wealth of data relating to drug sensitivity and biomarkers.
QUESTION 9: ARE INHIBITORS OF THE GENE PRODUCT KNOWN, AND IS THE GENE “DRUGGABLE”?
The ability of a gene product to be specifically and potently inhibited by a small-molecule inhibitor provides great potential for its biochemical activities to be studied in vitro and for establishing its involvement in biological processes in cells and in vivo systems. Blocking the action of specific proteins involved in pathological processes is of course one principal means of treating disease.
For a drug or chemical database to be useful for biological data mining, it must be queryable on the basis of target IDs, using standard gene or protein nomenclature. One such resource is ChEMBL (Bento et al., 2014), a database of bioactive small molecules (Supplemental Table S16). Because dozens or hundreds of interacting molecules may be recorded for a given target, ChEMBL provides interactive graphs displaying distributions of their properties, allowing the user to narrow a set of compounds for examination. Selected compounds are presented in a sortable table, which includes structure diagrams, physicochemical properties, and results of relevant bioassays. Another such resource is DrugBank (Law et al., 2014), a comprehensive database of pharmaceuticals and inhibitors. When a protein is recorded as a target of such a molecule, a UniProt ID-based search reveals protein information, followed by a table of relevant interacting molecules, each being a link to the full record from the main database.
The Comparative Toxicogenomics Database records links between genes and interacting chemicals, including effects of compounds on the expression of genes, as well as the activities of their products. Each gene page displays the top 10 interacting chemicals (by number of literature references) in a bar chart. Clicking a chemical name opens a list of corresponding interactions; alternatively, clicking the Chemical Interactions tab opens a large sortable table of all interacting chemicals, with one-line descriptions of each interaction, plus literature references.
The canSAR resource (Bulusu et al., 2014) integrates gene, protein, functional, and chemical interaction information from numerous sources. Following a target search, its Screening & Chemistry output displays interactive pie charts allowing the user to filter interacting compounds on the basis of bioactivity type (binding, inhibition, etc.). Compounds can also be filtered by the number of their physicochemical properties that fit with the Rule of Five (Ro5), used as a rule of thumb to judge a molecule's likelihood of being a successful oral in vivo pharmaceutical agent (Lipinski et al., 1997). Clicking Inspect after applying a filter leads to a series of interactive graphs relating to this subset of chemicals, including scatter plots of physicochemical properties, plus a sortable table of chemical structures, properties, bioassay results, and literature references.
An alternative approach to retrieving such information is to search a database of assays involving compounds and targets, such as PubChem BioAssay (Wang et al., 2014). Querying this resource generates a list of titles of biological assays (but not compound names) in which the query gene or protein was a target, plus literature references. Results can be refined with a single click—for example, to select those in which compounds inhibited with submicromolar (or subnanomolar) IC50 values. Each title links to a full record of information about the assay protocol, compounds used, and results; each compound links to its entry in the PubChem Compound database (Wang et al., 2009), where full chemical data are displayed, and the molecule's structure can be visualized in a 2D or 3D viewer.
Returning to the gene product, the term “druggability” refers to “the likelihood of being able to modulate a target with a small-molecule drug” (Owens, 2007). It is estimated that only 1/10 of human genes encode products that are potentially druggable, with less than half of these being associated with disease (Hopkins and Groom, 2002). Despite this limitation, the potential of gene products to be inhibitable is relevant, as it could be a factor for deciding which hits from a screen are deemed interesting for follow-up studies. For gene products with characterized three-dimensional structures, the DrugEBIlity resource takes a domain-based approach, using algorithms to assess multiple PDB records for the likely presence of binding sites for Ro5-compliant molecules. Querying by UniProt ID reveals a protein page showing sequence and domain information, followed by Tractability and Druggability scores, combined into an overall Ensemble Druggability assessment. The canSAR resource offers a complementary functionality, providing for a given protein a table of druggable or tractable domains, linking to diagrams detailing the interactions between these and relevant ligand molecules.
The set of possible combinations between proteins and small-molecule compounds recorded in public databases presents billions of potential docking interactions, a large proportion of which are uncharacterized, many possibly harboring significant pharmaceutical potential. Massive in silico efforts are underway to assess these potential interactions, and the recently launched Drugable portal allows access to data relating to compounds predicted to dock with each target protein, correlated with tissue-expression profiles (Reardon, 2013).
QUESTION 10: I WOULD LIKE TO KNOW EVERYTHING ABOUT THIS GENE (OR PROTEIN)! HOW CAN I ACCESS AS MUCH RELEVANT INFORMATION AS POSSIBLE?
The primary bioinformatic databases described so far provide a wealth of data relevant to the gene or protein of interest, but for the information-hungry biologist, visiting each site in turn is not likely to be the most efficient way of accessing these resources. A more effective approach is to perform a cross-database search (which queries multiple resources in parallel, generating many outputs) or to access a summary website (which provides an overview of information gathered from primary database resources).
Both the NCBI's “GQuery” Global Cross-Database Search and the EBI Search (also known as EB-eye) allow the user to query all of their hosted databases in one operation; these multisearches can be initiated via a direct Web link (Supplemental Table S17). In both cases, for each database the number of hits is shown, alongside links to results lists for that database. The Bioinformatic Harvester (Liebel et al., 2005) is a utility that launches searches of multiple resources simultaneously, retrieving information about the gene or protein of interest from human, mouse, rat, zebrafish, or Arabidopsis. Queries of any type (names or IDs of genes or proteins, or any text term) are first used to generate a list of relevant proteins (IPI identifiers and one-line descriptions). On selection of a protein, Bioinformatic Harvester performs the searches, displaying results from each resource in a separate frame, helpfully organized in a multitabbed Web page. A subsequent project from the Harvester team demonstrated a proof-of-principle that all publicly accessible scientific data from literature, databases, and laboratory-hosted Web pages could be made accessible via a Google-style interface, using distributed search engine technologies (Lütjohann et al., 2011). Resources such as these, which would grow as more data sets are linked, may evolve into invaluable additions to the data-mining toolkit in the future.
Even with multisearch approaches such as these, exploring each resource to retrieve relevant information can involve much effort. More convenient are the overviews of genes and their products provided by summary websites, which compile relevant information for display in an easily readable manner (Supplemental Table S18). For EBI Search results, alongside the listing of the number of hits in each database is the EBI Gene & Protein Summary. This provides a useful overview of the properties of a gene and its products, within five tabs: gene, expression, protein, protein structure, and literature (for five species: human, mouse, budding yeast, Drosophila, and C. elegans). InterMine database technology integrates information about genes and proteins from multiple sources (Smith et al., 2012) and powers overview facilities for several experimental species via a series of interconnected Web portals: FlyMine, YeastMine, MouseMine, RatMine, ZebrafishMine, metabolicMine (including human genes), and modMine (flies and worms). These sites provide a fairly comprehensive distillation of functional information, with clear navigation via tabs and sections. For human genes, GeneCards (Stelzer et al., 2011) presents a compilation of properties and functions of the gene of interest (and products) from numerous primary bioinformatic sources in a long scroll-down format. This output includes functional summaries from NCBI Gene and UniProt, genomic and expression data, and links to commercial reagents such as recombinant proteins, antibodies, inhibitors, and oligonucleotides for RNAi. A complementary functionality for human proteins is performed by neXtProt (Gaudet et al., 2013), which collates and summarizes a wealth of properties in an information-packed multitabbed format.
QUICK LINKS FROM HITS TO INTERNET-BASED RESOURCES
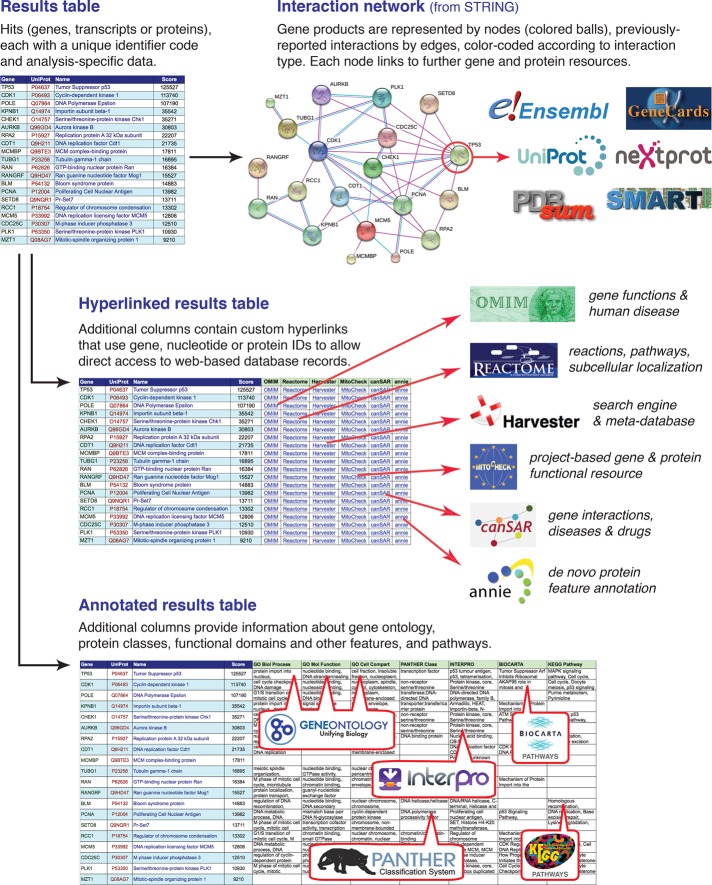
The answers to the foregoing 10 questions describe only a small selection of the dozens or hundreds of freely accessible bioinformatic databases available. However, free and user-friendly software specifically designed to allow quick access to relevant entries within these resources from results tables is not commonplace. One method allowing easy and direct access to appropriate entries within bioinformatic resources is the creation of a hyperlinked results table (Figure 2). Because all major spreadsheet programs allow the generation of hyperlinks incorporating contents of cells within the table, additional columns can be created containing custom hyperlinks that use hit-based gene, nucleotide, or protein identifiers, allowing users one-click direct access to relevant pages within Web-based resources. The creation of these hyperlinks is straightforward (Supplemental Method S1) and can be automated within most spreadsheet programs, thus negating the requirement for specialist data-mining software.
FIGURE 2:
Approaches for obtaining functional information about experimentally identified gene, transcript, or protein hits. Freely available software tools can be used to obtain information about features and functions of genes, transcripts, or proteins in a results table from multiple sources. Generation of an interaction network shows at a glance the nature of any previously reported interactions between members of a set of hits, each of which can be explored using the resources indicated. Making a hyperlinked results table allows one-click access from each hit directly to relevant pages from a wide range of resources. Creating an annotated results table containing controlled-vocabulary terms or keywords from a range of sources allows hits to be classified and sorted on the basis of these terms. Step-by-step protocols for performing these analyses are presented in the Supplemental Materials.
Online databases may be based around DNA, transcript, or protein IDs, with the majority using either gene symbols/codes or protein (UniProt) IDs, although some can be accessed using several ID types. To maximize the available options of resources directly accessible from results tables, a worthwhile exercise is to obtain gene symbols for transcripts or protein hits (Supplemental Method S2) or to obtain UniProt IDs for gene or transcript hits (Supplemental Method S3).
Clearly, with such a large number of online databases for which hyperlinks can be made, users should develop a familiarity with resources available for their given organism and make the hyperlinks targeted for that species and relevant to the biological questions being investigated.
Data set–wide information retrieval
Hyperlinked results tables allow one-at-a-time direct access to relevant information for each hit, but for larger sets of hits a more efficient approach for obtaining and managing such information is the annotated results table (Figure 2). Here additional columns are created containing pertinent information for each hit, such as ontology terms, protein domains and features, and pathways. Once in this format, hits can be easily sorted and categorized on the basis of these properties (e.g., using the AutoFilter facility within Excel [Microsoft, Redmond, WA]). It is also perfectly feasible to create a results table including multiple hyperlinks and annotation columns.
Several Web-based programs accept the input of multiple gene, nucleic acid, or protein IDs (with ID conversion if necessary), forming a table with additional columns containing feature annotations (Supplemental Table S19). These tables can be easily exported and integrated with the original results spreadsheet to form an annotated results table. PANTHER’s Gene List table provides annotation with GO-slim terms in the three categories, plus its own Protein class and Pathway terms. DAVID (Huang da et al., 2009b) generates a Functional Annotation Table containing full GO terms, OMIM diseases, InterPro and SMART features, and BioCarta and KEGG pathways. UniProt’s Results table can be customized with additional annotation columns including keywords, protein domains, disease associations, PubMed references, and even the full amino acid sequence. Step-by-step procedures for generating annotated results tables using these three resources are provided in Supplemental Methods S4–S6.
It is sometimes desirable to generate a visual representation of domains and other features for a set of proteins by inputting their IDs in batch rather than one at a time (as described for Question 5). The CDD provides for the submission and analysis of multiple proteins, allowing one-at-a-time display of domain structures, motifs, and key residues. In contrast, SMART allows input of many protein IDs, displaying their domains and features in a scroll-down multiple-protein view.
One useful approach to assessing known relationships between gene products in an experimental data set is the interaction network diagram (Figure 2). This can be created using STRING, in which, after the input of a hit list, a network diagram is generated with connections color coded by interaction type (Supplemental Method S7). This analysis provides an at-a-glance display of which subsets of genes or proteins share membership of complexes or systems. Each protein links to several database resources, and the page contains links to data set–wide overviews of occurrence (gene conservation), coexpression, and evidence for the mutual association of subsets of factors within the data set. Complementing this, input of gene, transcript, or protein hits to DAVID generates an Annotation Summary Results page with a Pathways subsection. Here the presence of one or more gene products from the input list within various pathways (from resources including BioCarta, KEGG, and Reactome) is indicated, and pathway diagrams can be displayed with the relevant proteins highlighted and flashing. More sophisticated analyses of the properties of a set of interacting gene products are possible using the Cytoscape network visualization and analysis software (Smoot et al., 2011). A large range of apps (formerly called plugins) is readily available, providing a huge repertoire of visualization and analysis options, including the integration of information from other data sets and annotation sources, and statistical analyses (Saito et al., 2012; Lotia et al., 2013).
After the retrieval of functional information about a set of experimental genes, transcripts, or proteins, a further analytical step could be to deduce which of their properties or ontology terms are enriched relative to a background data set, thus giving an indication of which classes of gene or protein or biological systems are overrepresented and thus predominate within the data set. These analyses are performed using bioinformatics enrichment tools. Discussion and comparison of such programs is beyond the scope of this article and is the subject of recent reviews (Hedegaard et al., 2009; Huang da et al., 2009a; Hung et al., 2012; Kouskoumvekaki et al., 2013).
Functional exploration of cell proliferation factors
The resources described here were applied to a hypothetical data set of 20 factors regulating aspects of cell proliferation: DNA replication, cell division, and genome stability. A hyperlinked table uses these factors to provide direct access to a selection of resources (Supplemental Table S20), which readers are invited to try out and compare to assess their suitability for their own research projects. Although no attempt is made to comprehensively report and evaluate the outputs of these resources, their application to the 20 cell proliferation factors (in particular p53) highlighted issues relating to resource utility for the following aspects of gene or protein function.
Ontology annotation.
The factors were analyzed for ontology terms in an attempt to retrieve straightforward descriptions of their functions. One-at-a-time searching using QuickGO returned GO terms for all 20 factors in all three categories. For p53, this yielded 97 distinct terms for biological process, 28 for molecular function, and 16 for cellular component. DAVID returned even more (213, 31, and 24, respectively), as its GO output covers a full range of hierarchical levels. The large volume of p53 annotations, albeit comprehensive, appeared in places contradictory (positive and negative regulation of apoptosis), redundant (ion binding, cation binding, zinc ion binding), and overwhelming (localization to seemingly every subcellular structure). Thus, valid although these assignments may be, they are meaningful only in their biological contexts. Aiming for a more efficient approach, annotation of the 20 factors with GO-slim terms was tried using PANTHER. This software recognized 17 of the proteins, provided GO-slim and protein-class annotation for 15, pathway terms for 5, and cell-component information for 1. This highlights issues encountered with ontology analysis: retrieval of comprehensive information may be useful for computational classification, but for human-readable summaries current outputs may appear both incomplete and too complete.
Protein–protein interactions.
Analysis of the 20 factors using STRING revealed that they form an interconnected interaction network (Figure 2). Choosing the “more” option expanded the network, introducing further factors recognizably involved in regulating cell proliferation. For p53, IntAct and BioGRID retrieved >500 interacting proteins (somewhat overwhelming network diagrams!), making the task of establishing which are biologically important a major challenge. Some indication of interaction significance was provided by table ranking (IntAct by confidence, BioGRID by reporting frequency); for both resources, p53’s top interactor was MDM2, its best-characterized regulator. However, which of the 500 are biologically validated, play roles in p53-mediated pathways, and regulate cell proliferation? Addressing such questions requires exploration beyond primary databases, integrating several data sources using more sophisticated software tools.
Protein feature identification.
The outputs of eight protein feature annotation programs were assessed using p53, a transcription factor with three functional domains (N-terminal transactivation, central DNA binding, C-terminal tetramerization), plus sites of interaction with ions, DNA, and regulatory proteins. All three domains are represented by Pfam and reflected in the outputs of Gene3D, InterPro, CDD, Dasty, and Annie, with only the central Pfam domain appearing in ELM. Gene3D identifies two domains based on structure, but SMART only a “low-complexity region.” In addition, manually curated annotations of experimentally determined functional regions and sites proved invaluable. CDD displays p53’s dimerization, DNA-binding, and zinc-binding sites, whereas ELM indicates docking sites for MDM2 and cyclins, plus verified nuclear localization and export sequences. Dasty in addition shows UniProt-curated regions of p53 required for binding physiological interactors. Thus information from each resource is different and complementary, and ideally all should be consulted to obtain the maximum information.
CONCLUSIONS AND PERSPECTIVES
Modern research biology increasingly relies on projects that use data derived from genome sequencing to make discoveries. The past decade saw a huge increase in the generation of sequence and experimental data, as well as in the number of databases relating to gene and protein sequence and function. A major challenge that has arisen is finding means of making optimal use of these resources to characterize and explore hits from larger-scale data sets in a way that makes sense to the research biologist and ultimately leads to discoveries of significance to the scientific community.
The enhancement of plain data spreadsheets to generate hyperlinked and annotated results tables is a flexible and universally applicable approach that facilitates the exploration and characterization of experimental hits from discovery projects. This procedure is relatively easily integrated as a last step in analytical workflows, using, for example, macros in Excel and online tools such as DAVID. For analytical service providers such as genomics, proteomics, and screening platforms, such enhancements give added value to the products they provide. For researchers, the presence of such links and annotations gives them the opportunity to easily categorize experimental hits on the basis of biological properties and allows them to pursue their curiosity as far as online resources allow.
The rise in recent years of several independent database resources covering the same territory has in many cases led to increasing data standardization and exchange. This period has also seen the emergence of meta-databases, cross-database search engines, and overview sites providing unified portals for information retrieval—resources particularly beneficial for researchers exploring hits from larger-scale studies. This being the case, support for expert-curated primary databases is still vital, as these remain the crucial points of contact with data-providing research teams and retain responsibility for data curation, quality control, and ensuring that connections are maintained between online data and peer-reviewed publications.
Cross-database searches such as those from the NCBI and the EBI and the Bioinformatic Harvester can query dozens of resources, identifying hundreds of relevant entries. Internet-wide search tools can query thousands of sources, potentially retrieving billions of documents and data files. Evidently, as the rapid expansion of data continues, the danger increasingly looms of encountering a “too much information” situation. The challenge for developers of information-retrieval software will inevitably shift from enabling access to ever-larger data quantities to ensuring that data are delivered in a meaningful way: organized, categorized, and presented such that they can be interpreted and evaluated by researchers worldwide.
Although various programs are available for exploring the properties of sets of genes, transcripts, and proteins, the ideal software tool, in my opinion, has yet to be created. Such software would be 1) free: publicly available, cross-platform, and open source, with database architecture and algorithms described in peer-reviewed publications; 2) comprehensive: able to draw on a wide variety of leading database resources; 3) updated: regularly, coordinated with releases of major sequence and functional databases; 4) smart: capable of automatically recognizing ID types and thus determining the relevant species and relationships between genes and their products; and 5) flexible: allowing a choice of analytical methods and output formats.
Continued increases in database comprehensiveness, usability, and integration can be expected in the future and are to be welcomed. So many genes and their products have undergone some degree of characterization, and so many biological processes have begun to be described in molecular terms, yet there remain a great many genes bearing the “uncharacterized” label. Thus, for researchers whose projects involve the discovery of genes, transcripts, or proteins and their functional characterization, with all the database resources available, paradoxically their most interesting hits may be the ones for which there is the least information to be found.
Supplementary Material
Acknowledgments
I extend sincerest thanks to the numerous curators, developers, and support staff of bioinformatic resources who provided valuable information during the researching and writing of this article, in several cases implementing suggested improvements to their software. Grateful thanks also go to the many colleagues past and present who provided helpful information, advice, and comments on the manuscript. During the preparation of this article I worked in the laboratories of J.-M. Peters and M. Méchali, both of whom I gratefully acknowledge for their support and guidance. I was funded by the European Commission (FP6 Integrated Project LSHG-CT-2004-503464 MitoCheck), the Austrian Science Fund (Special Research Programme Chromosome Dynamics), the European Research Council (FP7/2007-2013 Grant 233339 ORICODE), a postdoctoral fellowship from La Fondation pour la Recherche Médicale, and the Centre National de la Recherche Scientifique.
Abbreviations used:
- CDD
Conserved Domain Database
- CNRS
Centre National de la Recherche Scientifique
- DDBJ
DNA Data Bank of Japan
- EBI
European Bioinformatics Institute
- ELM
eukaryotic linear motif
- ENA
European Nucleotide Archive
- GDSC
Genomics of Drug Sensitivity in Cancer
- GEO
Gene Expression Omnibus
- GI
GenInfo Identifier
- GO
Gene Ontology
- GSV
genomic structural variation
- ID
identifier code
- IMEx
International Molecular Exchange
- IPI
International Protein Index
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NCBI
National Center for Biotechnology Information
- nr
nonredundant
- OMIM
Online Mendelian Inheritance in Man
- PDB
Protein Data Bank
- PTM
posttranslational modification
- RCSB
Research Collaboratory for Structural Bioinformatics
- Ro5
Rule of Five
- SLiM
short linear motif
- SNP
single-nucleotide polymorphism
Footnotes
Periodic updates to the Supplemental Materials for this article will be made available on the author's website: http://www.jrahutchins.net.
The author declares that no conflict of interest exists.
REFERENCES
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Alexander J, et al. Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci Signal. 2011;4:ra42. doi: 10.1126/scisignal.2001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM) Hum Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- Asplund A, Edqvist PH, Schwenk JM, Ponten F. Antibodies for profiling the human proteome—The Human Protein Atlas as a resource for cancer research. Proteomics. 2012;12:2067–2077. doi: 10.1002/pmic.201100504. [DOI] [PubMed] [Google Scholar]
- Barrett T, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2014;42:D32–D37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento AP, et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42:D1083–D1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat M. Searching NCBI's dbSNP database. Curr Protoc Bioinformat. 2010 doi: 10.1002/0471250953.bi0119s32. Chapter 1, Unit 1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA. Gene Ontology annotations and resources. Nucleic Acids Res. 2013;41:D530–D535. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- Bragin E, Chatzimichali EA, Wright CF, Hurles ME, Firth HV, Bevan AP, Swaminathan GJ. DECIPHER: database for the interpretation of phenotype-linked plausibly pathogenic sequence and copy-number variation. Nucleic Acids Res. 2014;42:D993–D1000. doi: 10.1093/nar/gkt937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulusu KC, Tym JE, Coker EA, Schierz AC, Al-Lazikani B. canSAR: updated cancer research and drug discovery knowledgebase. Nucleic Acids Res. 2014;42:D1040–D1047. doi: 10.1093/nar/gkt1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi AP. Analysis of gene function using DNA microarrays. Methods Enzymol. 2010;470:3–17. doi: 10.1016/S0076-6879(10)70001-X. [DOI] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, et al. The Comparative Toxicogenomics Database: update 2013. Nucleic Acids Res. 2013;41:D1104–D1114. doi: 10.1093/nar/gks994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer TA, Berka K, Thornton JM, Laskowski RA. PDBsum additions. Nucleic Acids Res. 2014;42:D292–D296. doi: 10.1093/nar/gkt940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- Dinkel H, Chica C, Via A, Gould CM, Jensen LJ, Gibson TJ, Diella F. Phospho.ELM: a database of phosphorylation sites—update 2011. Nucleic Acids Res. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel H, et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2014;42:D259–D266. doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorée M, Hunt T. From Cdc2 to Cdk1: when did the cell cycle kinase join its cyclin partner. J Cell Sci. 2002;115:2461–2464. doi: 10.1242/jcs.115.12.2461. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Eisenhaber F. Prediction of posttranslational modification of proteins from their amino acid sequence. Methods Mol Biol. 2010;609:365–384. doi: 10.1007/978-1-60327-241-4_21. [DOI] [PubMed] [Google Scholar]
- Fernandez JM, Hoffmann R, Valencia A. iHOP Web services. Nucleic Acids Res. 2007;35:W21–W26. doi: 10.1093/nar/gkm298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. Homology a personal view on some of the problems. Trends Genet. 2000;16:227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- Fleischmann A, Darsow M, Degtyarenko K, Fleischmann W, Boyce S, Axelsen KB, Bairoch A, Schomburg D, Tipton KF, Apweiler R. IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 2004;32:D434–D437. doi: 10.1093/nar/gkh119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. The art and design of genetic screens: yeast. Nat Rev Genet. 2001;2:659–668. doi: 10.1038/35088500. [DOI] [PubMed] [Google Scholar]
- Franceschini A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet P, et al. neXtProt: organizing protein knowledge in the context of human proteome projects. J Proteome Res. 2013;12:293–298. doi: 10.1021/pr300830v. [DOI] [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, Liu C, Shi W, Bryant SH. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 2011;39:D253–D260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griss J, Martin M, O'Donovan C, Apweiler R, Hermjakob H, Vizcaino JA. Consequences of the discontinuation of the International Protein Index (IPI) database and its substitution by the UniProtKB “complete proteome” sets. Proteomics. 2011;11:4434–4438. doi: 10.1002/pmic.201100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmanas A, et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2014;42:D285–D291. doi: 10.1093/nar/gkt1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J, Wood V, Jeffery L, Hoe KL, Kim DU, Park HO, Salas-Pino S, Heichinger C, Nurse P. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol. 2013;3:130053. doi: 10.1098/rsob.130053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedegaard J, et al. Methods for interpreting lists of affected genes obtained in a DNA microarray experiment. BMC Proc. 2009;3(Suppl 4):S5. doi: 10.1186/1753-6561-3-s4-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006;34:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]
- Hibbs MA, Hess DC, Myers CL, Huttenhower C, Li K, Troyanskaya OG. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz NH. One-gene-one-enzyme: remembering biochemical genetics. Protein Sci. 1995;4:1017–1019. doi: 10.1002/pro.5560040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hung JH, Yang TH, Hu Z, Weng Z, DeLisi C. Gene set enrichment analysis: performance evaluation and usage guidelines. Brief Bioinform. 2012;13:281–291. doi: 10.1093/bib/bbr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T. Protein sequence motifs involved in recognition and targeting: a new series. Trends Biochem Sci. 1990;15:305. [Google Scholar]
- Hunter S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley RP, Binns D, Dimmer E, Barrell D, O'Donovan C, Apweiler R. QuickGO: a user tutorial for the web-based Gene Ontology browser. Database (Oxford) 2009;2009:bap010. doi: 10.1093/database/bap010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JRA, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, et al. Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 2014;42:D546–D552. doi: 10.1093/nar/gkt979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- Kim DU, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MW. The meaning of systems biology. Cell. 2005;121:503–504. doi: 10.1016/j.cell.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Kornberg A. The private life of DNA polymerase I. Methods Enzymol. 1990;182:783–788. doi: 10.1016/0076-6879(90)82060-f. [DOI] [PubMed] [Google Scholar]
- Kosuge T, Mashima J, Kodama Y, Fujisawa T, Kaminuma E, Ogasawara O, Okubo K, Takagi T, Nakamura Y. DDBJ progress report: a new submission system for leading to a correct annotation. Nucleic Acids Res. 2014;42:D44–D49. doi: 10.1093/nar/gkt1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskoumvekaki I, Shublaq N, Brunak S. Facilitating the use of large-scale biological data and tools in the era of translational bioinformatics. 2013 doi: 10.1093/bib/bbt055. doi:10.1093/bib/bbt055. [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DP, Cheok CF, Lain S. p53-based cancer therapy. Cold Spring Harb Perspect Biol. 2010;2:a001222. doi: 10.1101/cshperspect.a001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen I, et al. DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res. 2013;41:D936–D941. doi: 10.1093/nar/gks1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law V, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Tsai J, Sunkara S, Karamycheva S, Pertea G, Sultana R, Antonescu V, Chan A, Cheung F, Quackenbush J. The TIGR Gene Indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res. 2005;33:D71–D74. doi: 10.1093/nar/gki064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees JG, Lee D, Studer RA, Dawson NL, Sillitoe I, Das S, Yeats C, Dessailly BH, Rentzsch R, Orengo CA. Gene3D: Multi-domain annotations for protein sequence and comparative genome analysis. Nucleic Acids Res. 2014;42:D240–D245. doi: 10.1093/nar/gkt1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebel U, Kindler B, Pepperkok R. Bioinformatic “Harvester”: a search engine for genome-wide human, mouse, and rat protein resources. Methods Enzymol. 2005;404:19–26. doi: 10.1016/S0076-6879(05)04003-6. [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Lotia S, Montojo J, Dong Y, Bader GD, Pico AR. Cytoscape app store. Bioinformatics. 2013;29:1350–1351. doi: 10.1093/bioinformatics/btt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CT, Huang KY, Su MG, Lee TY, Bretana NA, Chang WC, Chen YJ, Huang HD. dbPTM 3.0: an informative resource for investigating substrate site specificity and functional association of protein post-translational modifications. Nucleic Acids Res. 2013;41:D295–D305. doi: 10.1093/nar/gks1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. PubMed and beyond: a survey of Web tools for searching biomedical literature. Database (Oxford) 2011;2011:baq036. doi: 10.1093/database/baq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütjohann DS, Shah AH, Christen MP, Richter F, Knese K, Liebel U. “Sciencenet”—towards a global search and share engine for all scientific knowledge. Bioinformatics. 2011;27:1734–1735. doi: 10.1093/bioinformatics/btr181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madej T, et al. MMDB: 3D structures and macromolecular interactions. Nucleic Acids Res. 2012;40:D461–D464. doi: 10.1093/nar/gkr1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi T, et al. Minimotif Miner 3.0: database expansion and significantly improved reduction of false-positive predictions from consensus sequences. Nucleic Acids Res. 2012;40:D252–D260. doi: 10.1093/nar/gkr1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014;42:D7-D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedringhaus TP, Milanova D, Kerby MB, Snyder MP, Barron AE. Landscape of next-generation sequencing technologies. Anal Chem. 2011;83:4327–4341. doi: 10.1021/ac2010857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura D. A view from the Web: BioCarta. Biotech Software Internet Rep. 2001;2:117–120. [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi HS, Kwo CY, Wildpaner M, Sirota FL, Eisenhaber B, Maurer-Stroh S, Wong WC, Schleiffer A, Eisenhaber F, Schneider G. ANNIE: integrated de novo protein sequence annotation. Nucleic Acids Res. 2009;37:W435–W440. doi: 10.1093/nar/gkp254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard S, et al. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat Methods. 2012;9:345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard S, et al. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens J. Research highlight: determining druggability. Nat Rev Drug Discov. 2007;6:187. [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakseresht N, et al. Assembly information services in the European Nucleotide Archive. Nucleic Acids Res. 2014;42:D38–D43. doi: 10.1093/nar/gkt1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak R, et al. Expression Atlas update—a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–D932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que S, Wang Y, Chen P, Tang YR, Zhang Z, He H. Evaluation of protein phosphorylation site predictors. Protein Pept Lett. 2010;17:64–69. doi: 10.2174/092986610789909412. [DOI] [PubMed] [Google Scholar]
- Reardon S. Project ranks billions of drug interactions. Nature. 2013;503:449–450. doi: 10.1038/503449a. [DOI] [PubMed] [Google Scholar]
- Rose PW, et al. The RCSB Protein Data Bank: new resources for research and education. Nucleic Acids Res. 2013;41:D475–D482. doi: 10.1093/nar/gks1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, et al. ENCODE Data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustici G, et al. ArrayExpress update—trends in database growth and links to data analysis tools. Nucleic Acids Res. 2013;41:D987–D990. doi: 10.1093/nar/gks1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg I, et al. BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA. Nucleic Acids Res. 2013;41:D764–772. doi: 10.1093/nar/gks1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber F, Patricio M, Muffato M, Pignatelli M, Bateman A. TreeFam v9: a new website, more species and orthology-on-the-fly. Nucleic Acids Res. 2014;42:D922–D925. doi: 10.1093/nar/gkt1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillitoe I, et al. New functional families (FunFams) in CATH to improve the mapping of conserved functional sites to 3D structures. Nucleic Acids Res. 2013;41:D490–D498. doi: 10.1093/nar/gks1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RN, et al. InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics. 2012;28:3163–3165. doi: 10.1093/bioinformatics/bts577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Stelzer G, et al. In-silico human genomics with GeneCards. Hum Genomics. 2011;5:709–717. doi: 10.1186/1479-7364-5-6-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaveces JM, Jimenez RC, Garcia LJ, Salazar GA, Gel B, Mulder N, Martin M, Garcia A, Hermjakob H. Dasty3, a WEB framework for DAS. Bioinformatics. 2011;27:2616–2617. doi: 10.1093/bioinformatics/btr433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CT. Posttranslational Modification of Proteins: Expanding Nature's Inventory. Greenwood Village, CO: Roberts and Company; 2006. [Google Scholar]
- Walther TC, Mann M. Mass spectrometry-based proteomics in cell biology. J Cell Biol. 2010;190:491–500. doi: 10.1083/jcb.201004052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Suzek T, Zhang J, Wang J, He S, Cheng T, Shoemaker BA, Gindulyte A, Bryant SH. PubChem BioAssay: 2014 update. Nucleic Acids Res. 2014;42:D1075–D1082. doi: 10.1093/nar/gkt978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfsberg TG, Wetterstrand KA, Guyer MS, Collins FS, Baxevanis AD. A user's guide to the human genome. Nat Genet. 2002;32(Suppl):1–79. doi: 10.1038/ng961. [DOI] [PubMed] [Google Scholar]
- Wood V, et al. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 2012;40:D695–D699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NL, Plazas-Mayorca MD, Garcia BA. Systems-wide proteomic characterization of combinatorial post-translational modification patterns. Expert Rev Proteomics. 2010;7:79–92. doi: 10.1586/epr.09.100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.