Chromatin remodeling enzymes can also have nonhistone roles, broadening their biological functions. It is shown that Kdm3a binding to cellular chaperones in the cytoplasm is relevant for morphogenetic events leading to infertility in enzymatically null mice. This provides evidence that Kdm3a is not just a histone modifier.
Abstract
The lysine demethylase Kdm3a (Jhdm2a, Jmjd1a) is required for male fertility, sex determination, and metabolic homeostasis through its nuclear role in chromatin remodeling. Many histone-modifying enzymes have additional nonhistone substrates, as well as nonenzymatic functions, contributing to the full spectrum of events underlying their biological roles. We present two Kdm3a mouse models that exhibit cytoplasmic defects that may account in part for the globozoospermia phenotype reported previously. Electron microscopy revealed abnormal acrosome and manchette and the absence of implantation fossa at the caudal end of the nucleus in mice without Kdm3a demethylase activity, which affected cytoplasmic structures required to elongate the sperm head. We describe an enzymatically active new Kdm3a isoform and show that subcellular distribution, protein levels, and lysine demethylation activity of Kdm3a depended on Hsp90. We show that Kdm3a localizes to cytoplasmic structures of maturing spermatids affected in Kdm3a mutant mice, which in turn display altered fractionation of β-actin and γ-tubulin. Kdm3a is therefore a multifunctional Hsp90 client protein that participates directly in the regulation of cytoskeletal components.
INTRODUCTION
Normal development requires precision and sufficient plasticity to adapt to environmental and genetic changes. The recent discovery of the reversible nature of histone methylation has generated interest into two gene families encoding demethylase enzymes, as they play fundamental roles by mediating timely expression of developmental genes. This is illustrated by the disease phenotypes observed in animal models and human patients (Kooistra and Helin, 2012) associated with mutation in some of these genes. Jumonji domain (JmjC)-containing proteins form a large family of oxoglutarate-dependent dioxygenases capable of removing methyl groups from arginine and lysines of histones (Klose and Zhang, 2007). Knockdown of JmjC proteins gives rise to a wide range of phenotypes from embryonic lethality to no discernible abnormality (Takeuchi et al., 1995; Catchpole et al., 2011; Iwamori et al., 2011; Ishimura et al., 2012). Despite the fact that many histone demethylases bind to a large number of genes, their silencing or knockdown alters the expression of relatively few genes with rather modest changes in expression levels. This suggests that histone demethylases may not have a major role in the regulation of gene expression but rather may be fine modulators of chromatin epigenetic states (Kooistra and Helin, 2012). Although normal development can be potentially disrupted by the synergistic effect of small changes in the expression of functionally related genes, the modest effect on transcription raises the interesting possibility that many histone demethylases may alter development through nonhistone functions as well (Webby et al., 2009; Lu et al., 2010).
Kdm3a (Jhdm2a, Jmjd1a, KIAA0742, TSGA) is a jumonji-domain protein able to remove mono- and dimethyl groups of histone H3 Lys-9 (H3K9me1 and H3K9me2) (Yamane et al., 2006). Kdm3a knockdown leads to immotile round-headed spermatozoa (globozoospermia) and metabolic syndrome associated with obesity and diabetes (Okada et al., 2007; Inagaki et al., 2009; Tateishi et al., 2009; Liu et al., 2010) through the regulation of genes involved in histone replacement during spermatogenesis and regulation of gene expression during adipogenesis. It has recently been shown that lack of Kdm3a leads to male-to-female sex reversal at non-Mendelian ratios, through misregulation of the sex-determining gene sry (Kuroki et al., 2013). Kdm3a is highly expressed in testis and at lower levels in fat tissue and can be readily purified from HeLa cells (Tsukada and Zhang, 2006). Kdm3a was shown to be a coactivator of estrogen and androgen receptors (Yamane et al., 2006) and sry (Kuroki et al., 2013); further studies showed that Kdm3a is also involved in many cellular processes, including progression through the cell cycle (Cho et al., 2011), embryonic (Loh et al., 2007) and adult (Ma et al., 2008) stem cell renewal, and differentiation of vascular smooth muscle (Lockman et al., 2007) and parietal endoderm (Herzog et al., 2012). Increased methylation of histone H3K9 in testis after Kdm3a knockdown has been found when spermatids are fractionated by stage (Liu et al., 2010) but not when whole testis extracts are analyzed (Okada et al., 2007), illustrating the stage-specific and modulatory nature of this demethylase. Nevertheless, global depletion of mono- and dimethylated histones is consistently observed following Kdm3a overexpression or in vitro incubation with purified Kdm3a protein (Yamane et al., 2006). Thus the function of Kdm3a as a histone lysine demethylase has been extensively demonstrated.
Kdm3a was shown to participate in the transcriptional control of transition proteins and protamines leading to postmeiotic chromatin condensation defects (Okada et al., 2007; Liu et al., 2010). It is thus postulated that the lack of DNA condensation underlies the abnormal sperm head shaping in Kdm3a animal models (Okada et al., 2010). The formation and shaping of the mammalian sperm head is a complex process that involves sequential changes in the nucleus and Golgi-derived acrosome in concert with the transient appearance of a microtubular manchette (Kierszenbaum and Tres, 2004). In mice, acrosome formation begins at stage 2 of round spermatids with the deposition of proacrosomal granules on the apical pole of the nuclear surface (Bowen, 1925). The acrosomal granules fuse, forming a vesicle surrounded by an acrosomal membrane anchored to the nucleus through an actin-rich plate called the acroplaxome (Hermo et al., 2010). The acrosomal sac starts to flatten and begins the gradual spreading over the nuclear surface. At steps 7 and 8, while the Golgi apparatus moves away from the apical pole, the flagellum engages within the implantation fossa at the caudal end of the nucleus forming the head-tail coupling apparatus (HTCA). At step 9, the nucleus starts to elongate, forming a dorsal and ventral surface in tight association with the acrosome and manchette microtubules forming the perinuclear ring that delineates its intersection with the acroplaxome (see diagram in Figure 2A later in this article). These tubulin- and actin-based structures play major roles in shaping the spermatid head by providing a structural platform for clutching forces during nuclear elongation (Kierszenbaum et al., 2003). The most common causes of round-headed spermatozoa are often associated with acrosomal and manchette defects (Dam et al., 2007). The intrinsic complexity of sperm head shaping renders difficult the assignment of a single molecular event to the causation of globozoospermia. Interestingly, some endogenous Kdm3a protein is present in the cytoplasm during spermatogenesis (Okada et al., 2007), but the potential contribution of non–histone-related roles for Kdm3a during spermatogenesis has not been investigated.
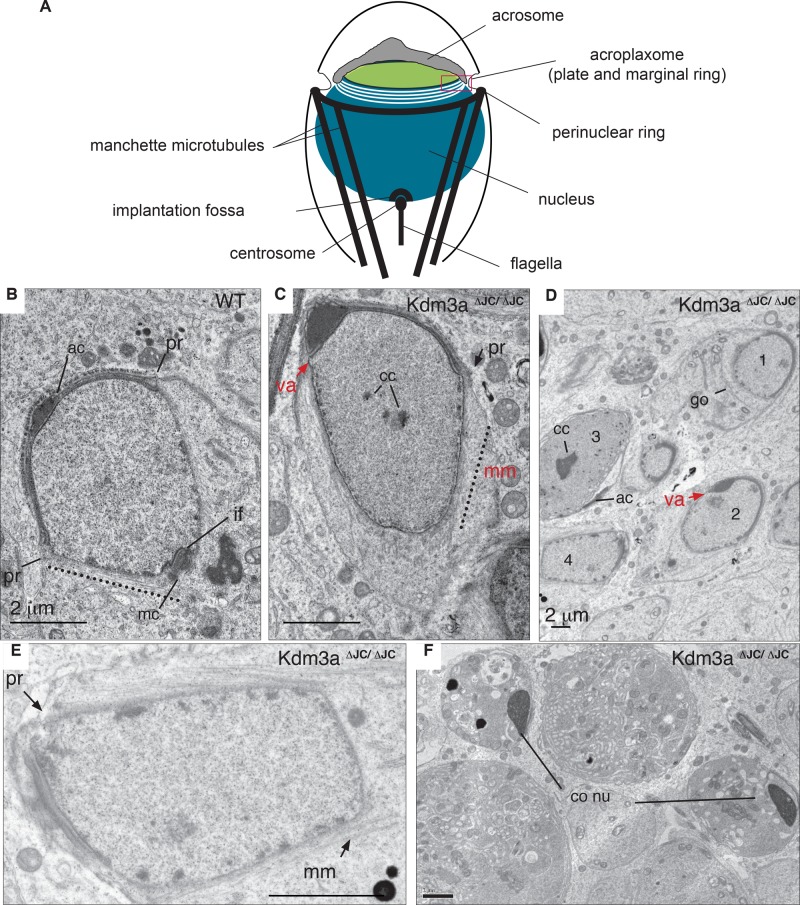
FIGURE 2:
Ultrastructural defects of Kdm3aΔJC/ΔJC precede and persist after chromatin condensation. (A) The diagram illustrates cellular structures observed by electron micrographs. (B) Representative electron micrograph of a wild-type (WT) spermatid in early step 9. The acrosome (ac) has flattened and spreads symmetrically over the nuclear envelope. Microtubules of the manchette (dotted line) emanate from the perinuclear ring (pr) parallel to the nuclear surface. The caudal end of the nucleus is indented by the implantation fossa (if), with the mother centriole closely apposed (mc). (C) Step 9 spermatid in homozygous testis shows absence of ventral acrosome (va) surface (red arrow) and microtubules of the manchette (mm, dotted line) extending beyond the perinuclear ring (pr). Caudal end of nucleus remains round without traces of implantation fossa (if) in this or other planes of this spermatid section. Electrodense portions in nucleus represent condensing chromatin (cc). (D) Low magnification of homozygous sections illustrating the extent of abnormal spermatids. Golgi has moved to caudal end (go; spermatid 1). One-sided acrosome without ventral acrosome (va; 2). Strongly condensed chromatin (cc; 3). Disrupted manchette (4). (E) Spermatid 4 shown in (C) presents disrupted perinuclear ring (pr) with only a few manchette microtubules (mm). Several cc domains are observed. (F) Condensed spermatid from homozygous animals displays dark nucleus, sign of chromatin compaction and if impacted by HTCA. Scale bars: 2 μm.
In this paper, we show that Kdm3a mutant mice have cytoplasmic defects preceding histone replacement and chromatin compaction that significantly contribute to arrest spermatid elongation and produce rounded sperm heads as previously reported for these models (Okada et al., 2007; Liu et al., 2010). Electron microscopy shows that elongating spermatids in homozygous animals lacking Kdm3a catalytic domain (Kdm3aΔJC) have deformed acrosomes, incomplete manchettes, and detached centrosomes. Similar but milder features are observed in a second mouse model with a gene-trap insertion within Kdm3a (Kdm3aGT), which presents normal electrodense (condensed) DNA but spermatid heads that are still rounded. We confirm the existence of a short Kdm3a isoform that is enzymatically active, expanding the complexity of Kdm3a regulation. This isoform is not interrupted by the gene-trap insertion, and although transcript levels are very low, this isoform may contribute toward the phenotype amelioration observed in the second model. We found Kdm3a to interact with the cellular chaperones: Hsp90, Cct/TriC, and Vcp. Multiple independent experimental designs addressed the specificity of Kdm3a interaction with Hsp90 and its requirement for Hsp90 chaperoning for its demethylase activity. Antibodies to an Hsp90 lysine residue known to be dynamically methylated (Abu-Farha et al., 2011; Donlin et al., 2012) indicate that this site is not modified by Kdm3a. Nevertheless, Hsp90 showed altered association with cytoplasmic components in somatic and germ cells of Kdm3a mutant mice, providing evidence that the cytoskeletal defects are a direct consequence of inactive Kdm3a. Our work provides molecular evidence for a previously unknown role of Kdm3a in the extensive cytoskeletal rearrangements required for spermatogenesis to proceed normally.
RESULTS
Kdm3a mouse models
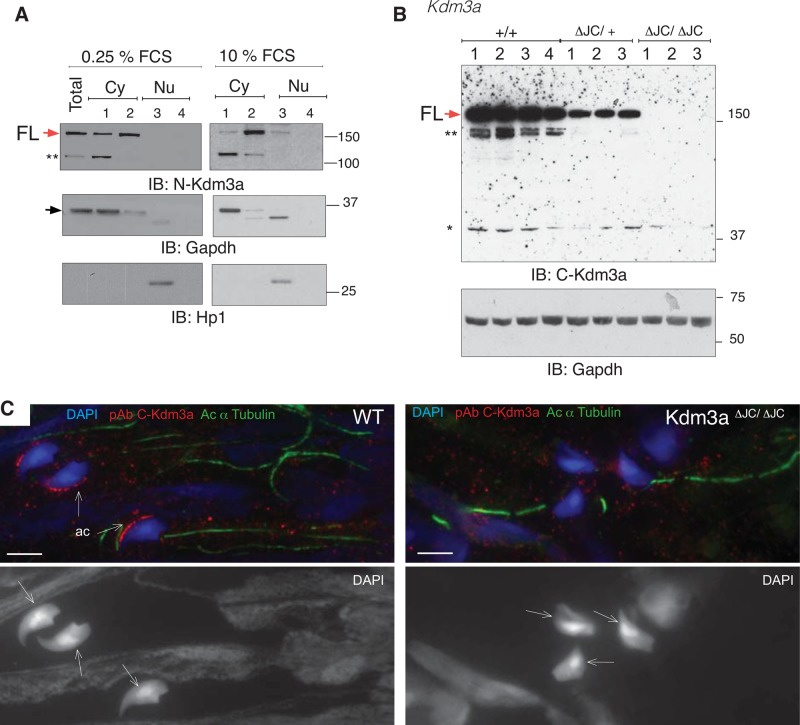
To expand the functional studies of Kdm3a, we generated two mouse models. First, we rederived a previously published targeted mutant allele that lacks the Kdm3a catalytic domain JmjC (Kdm3aΔJC; Tateishi et al., 2009; Figure 1A). In previous studies, these mice provided evidence for the role of Kdm3a in the regulation of metabolic gene expression and resistance to obesity (Tateishi et al., 2009). Homozygous males were shown to be infertile, with testes significantly smaller than those of wild-type littermates (Okada et al., 2007). Mice containing recombination Kdm3a alleles (ΔJC) were identified by PCR (Figure 1, A and B). Western blot analysis with a Kdm3a-specific polyclonal antibody directed to the N-terminal end of Kdm3a not contained within the deletion (Figure 1A, pAb N-Kdm3a) revealed the absence of full-length protein in homozygous cells and the appearance of low levels of deletion product in heterozygous and homozygous cells (Figure 1C). As shown previously, epididymis of Kdm3aΔJC/ΔJC animals shows a drastic decrease of spermatozoa and the few found have deformed heads (Figure 1D). Heterozygous animals were fertile (Table 1), but inspection of testis cross-sections revealed aberrant accumulation of large nuclei in the lumen of seminiferous tubules (Figure 1E and F).
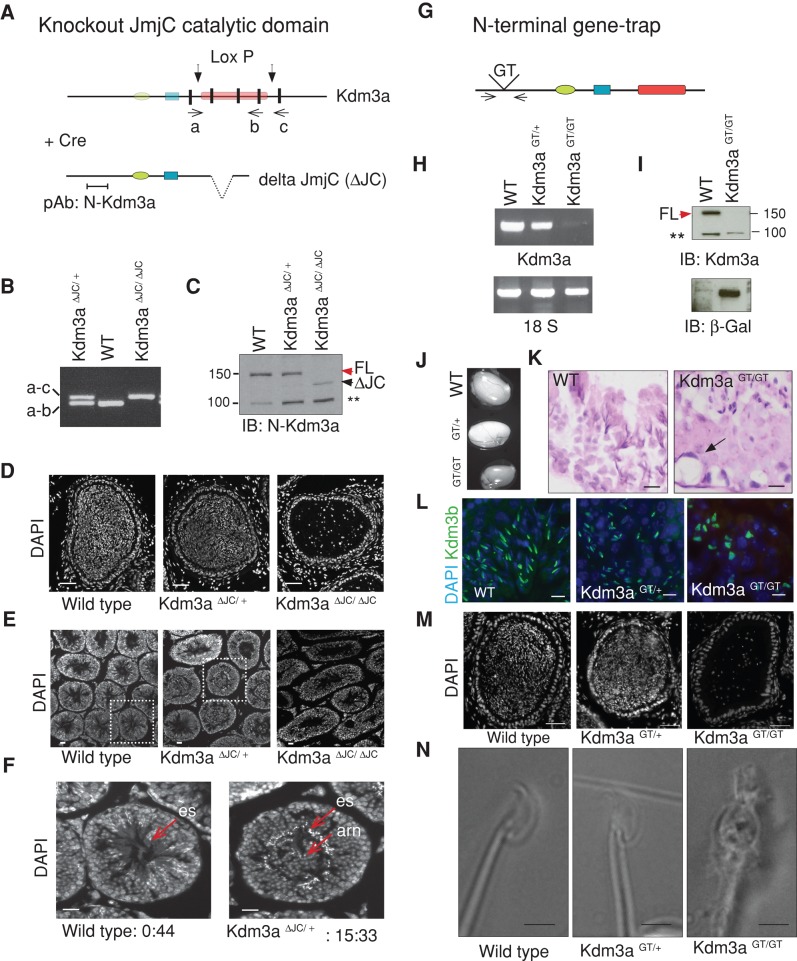
FIGURE 1:
Two Kdm3a mouse models present arrested spermatogenesis with globozoospermia. (A) Diagram illustrates Cre-mediated deletion of exons 22–24 containing JmjC catalytic domain of Kdm3a in Kdm3aΔJC mice (Tateishi et al., 2009). (a), (b), and (c) indicate the position of primers used to genotype mutant mice. Color code: green oval, C6-type zinc finger (ZF); blue, LXXLL nuclear receptor binding; red, JmjC domain; black, exons surrounding and affected by deletion. pAb, polyclonal antibody. (B) Genomic PCR identifying ΔJC deletion mutants. (C) Immunoblot of total MEF extracts with Kdm3a antibody (N-terminal) shows absence of full-length Kdm3a protein (FL, red arrow) in Kdm3aΔJC/ΔJC. A shorter Kdm3a protein product result of Cre-induced deletion is indicated (ΔJC, black arrow). A nonspecific band is indicated (*). (D) DAPI staining of Kdm3aΔJC/ΔJC epididymis reveals few mature sperm with rounded nuclei. (E) DAPI staining of testis cross-sections shows some abnormal tubules in Kdm3aΔJC/+ (boxed region). Kdm3aΔJC/ΔJC show arrested spermatogenesis. (F) Higher magnification of indicated regions from (E). es, elongated spermatids; arn, accumulation of round nuclei. Numbers represent the proportion of abnormal to normal tubules in each slide. (G) Diagram illustrates the position of the gene-trap insertion in Kdm3aGT/GT. Arrows indicate primers used for RT-PCR shown in (H). (H) Absence of Kdm3a transcript in Kdm3aGT/GT MEFs. Loading control used was 18S. (I) Immunoblot of total MEF extracts with a Kdm3a antibody directed to the N-terminus shows absence of full-length protein (FL, red arrow) in Kdm3aGT/GT. β-gal antibody detects the gene-trap product Kdm3aGT/GT only. (J) Kdm3aGT/GT testes are smaller than wild-type (WT) and heterozygous littermates. (K) Hematoxylin and eosin stain show disorganized tubules in testis sections of homozygous. (L) Immunohistochemistry of testis sections with a commercial antibody to Kdm3b (green) stained the acrosome of elongating spermatids. Note abnormal acrosome shape in Kdm3aGT/GT sections only. (M) DAPI panel shows absence of mature sperm in homozygous-GT epididymis. (N) Bright-field images show round-headed spermatozoon in Kdm3aGT/GT mice.
TABLE 1:
Heterozygous Kdm3a animals are fertile.
| Mouse model | Genotype (%)a | Total born | ||
|---|---|---|---|---|
| Wild-type | Heterozygous | Homozygous | ||
| ΔJC | 23 | 59 | 18 | 159 |
| GT | 31 | 45 | 24 | 127 |
aPercentage of genotypes in offspring from heterozygous crosses of knockout (ΔJC) and gene-trap (GT) mice. Each genotype is present in the expected Mendelian ratios.
The second mutant mouse allele contains a gene-trap insertion (Kdm3aGT) within intron 5–6 of the Kdm3a locus (Figure 1G). Heterozygous animals derived from germ line–transmitting male chimeras were backcrossed onto C57BL6J females. Reverse transcriptase followed by PCR and Western blot analysis revealed that Kdm3aGT/GT mice lack Kdm3a full-length transcript (Figure 1H) and protein (Figure 1I). Kdm3aGT/GT males are infertile, whereas heterozygous animals produce offspring (Table 1). Kdm3aGT/GT testes are also significantly smaller than those of wild-type and heterozygous littermates (Figure 1J). Cross-sections of Kdm3aGT/GT adult testes show disorganized stratification with many elongated spermatids at the edge of the lumen with abnormally shaped spermatid heads (Figure 1K) and acrosomes (Figure 1L). Cross-sections of adult epididymis showed absence of mature spermatozoa in Kdm3aGT/GT (Figure 1M).
Thus the use of two different mouse models for Kdm3a confirmed but also extended previously described phenotypes associated with a role of this gene in spermatogenesis. The detection of abnormally accumulated cells in the lumen of heterozygous testis suggests that Kdm3a function during spermatogenesis is dose dependent.
Ultrastructural characterization of spermatogenesis in Kdm3a models
We characterized the structural defects of these models by transmission electron microscopy (TEM). The earliest discernible phenotype of Kdm3aΔJC/ΔJC mice is the failure of the acrosome to spread bilaterally over the nucleus (Figure 2C), extending previous reports of failure of these mice to develop ventral surface, as determined by Alcian blue staining (Liu et al., 2010). Prominent cytoplasmic abnormalities in step 9 spermatids were abrupt termination of the acroplaxome (Figure 2D), ruptured perinuclear ring, and poorly developed manchette with discontinued microtubules that were often misoriented (Figure 2E). The perinuclear ring is the intersection of the acroplaxome with the manchette. Normally, a bundle of filamentous actin is found in the acroplaxome, bridging the acrosome sac with the nuclear lamina. In Kdm3aΔJC/ΔJC animals, this is evident on the dorsal side of the acrosome only (Figure 2D). Importantly, aggregates of condensed chromatin for fully condensed nuclei are observed in many elongating spermatids (Figure 2). Spermatogenesis arrests at step 9, as described previously, although a few Kdm3aΔJC/ΔJC condensed spermatids can be observed; these, in spite of a normally electrodense nucleus, show disarrayed cytoplasm (Figure 2F). Absence of an implantation fossa forming the HTCA in Kdm3aΔJC/ΔJC was frequent (unpublished data).
The Kdm3a gene-trap model has milder phenotypes. Kdm3aGT/GT spermatids display normal acrosome spreading as compared with control littermates but show asymmetric descent of the manchette and membranous folds not observed in wild type (Figure 3, A and B), as well as failure of the HTCA to implant in the nuclear fossa (Figure 3D). Most spermatids, found at the right plane, lacked the typical nuclear indentation demarcating the implantation site of the mother centriole; instead, a detached electrodense proteinaceous plaque was observed (Figure 3D). Many spermatids have normally condensed chromatin (Figure 3E), but those that reach the epididymis still have rounded heads and abnormal acrosome shape (Figure 3F).
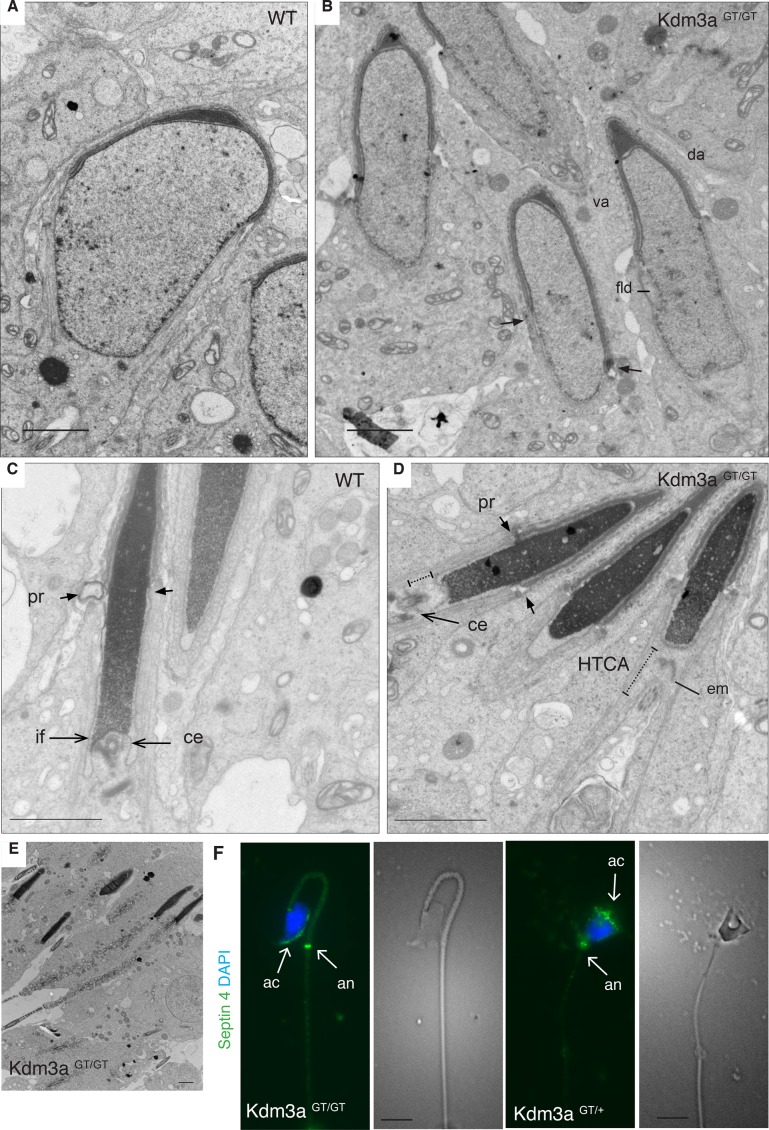
FIGURE 3:
Kdm3a gene-trap model presents milder structural defects. (A) Representative electron micrograph of a wild-type (WT) spermatid in early step 9. (B) Homozygous animals have ventral (va) and dorsal (da) acrosome surfaces, but manchette descends asymmetrically (arrows), and nuclear membrane has folds (fld). (C) The mother centriole (ce) is engaged in implantation fossa (if) in WT condensed spermatids. (D) Condensed spermatids from homozygous animals have detached mother centriole (ce) without clear implantation fossa (if) and detached electrodense material (em). (E) Low magnification of a homozygous-GT tubule illustrates the extent and frequency of condensed spermatids. (F) Epididymal spermatozoa from heterozygous animals stained with septin 4 (annulus and acrosomal marker, green) show acrosome expanding over a normal sperm head. Homozygous sperm shows cube-like head with horizontal acrosomes. Scale bars: A–E, 2 μm; F, 10 μm.
The presence of spermatids with condensed DNA in the gene-trap model dissociates chromatin from other structural defects contributing to the infertility phenotype of Kdm3a mutant mice. Both genetic models provide evidence for cytoplasmic defects that could account for or contribute to the globozoospermia phenotype of Kdm3a mutant mice. The lack of ventral acrosome surface and asynchronous descent of the manchette could be part of the same mechanism perturbed to a different extent. The difference in severity between Kdm3a mice may be due to the hypomorphic nature often observed in gene-trap models (Voss et al., 1998), although reverse transcription PCR (RT-PCR) and Western blots show negligible levels of full-length wild-type Kdm3a transcript and protein (Figure 1, H and I).
Cloning of a new murine Kdm3a isoform
There are several predicted isoforms for mouse and human Kdm3a (Ensembl). The milder severity of Kdm3a gene-trap mice prompted us to consider the existence of an alternative isoform not affected by the gene-trap insertion that could partially compensate for the lack of the full-length protein. We designed primers following a prediction from Ensembl in which a transcript with a unique exon contained within intron 11–12 would encode for a short Kdm3a isoform (Figure 4A). Using mRNA from Kdm3aGT/GT testis, we cloned a properly spliced transcript containing the three major Kdm3a domains and an alternative exon not found in the full-length transcript (Figure 4B, unique exon). Quantitative RT-PCR shows that transcript levels of this isoform, named isoform-2 (i2) is unaltered in adult testis from Kdm3aGT/GT mice (Figure 4C), indicating that the gene-trap insertion does not affect its expression in this tissue. Kdm3a-i2 is therefore likely transcribed from a promoter downstream from the gene-trap insertion. However, RNA expression of Kdm3a-i2 is much lower than full-length Kdm3a relative to glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Figure 4C). For testing whether Kdm3a-i2 encodes an enzymatically active histone demethylase, Kdm3a-i2 was cloned in-frame with a green fluorescent protein (GFP) tag and transfected into human retinal pigmented epithelium cell line RPE1 (Supplemental Figure S1A). The overexpressed protein localized to the nucleus and cytoplasm (Figures 4C and S1B) regardless of whether the tag was positioned at the N- or C-terminal end of the protein (unpublished data). Decreased immunostaining of transfected cells with antibodies to lysine-methylated histones indicates that this newly cloned Kdm3a isoform is also an active histone lysine demethylase (Figure 4, D and E).
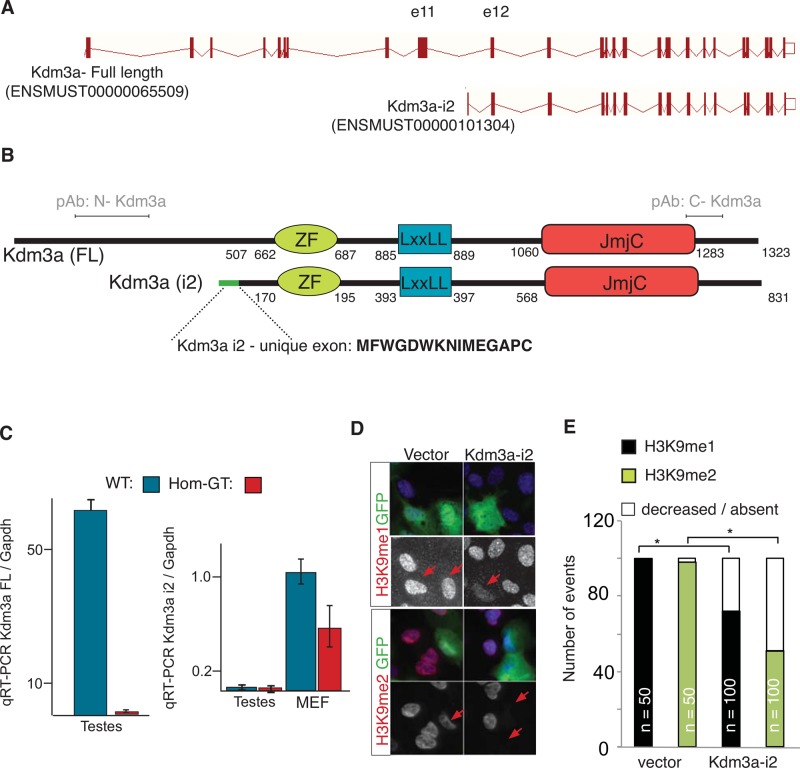
FIGURE 4:
Kdm3a encodes two protein isoforms. (A) Intron-exon diagram of Kdm3a. Short isoform i2 was cloned following ENSEMBL prediction (ID numbers between parentheses). First and unique Kdm3a-i2 exon is contained within intron 11. (B) Diagram illustrates the amino acid position of Kdm3a protein domains in full-length (FL) and isoform 2 (i2). Green line, i2-exclusive exon; amino acid sequence shown in bold. pAb indicates the relative position of amino terminal Kdm3a single polyclonal antibody used along this study. (C) qRT-PCR from testis and MEF RNA show a decrease of Kdm3a-FL and unaltered transcript levels of i2 in Kdm3aGT/GT testis. A slight reduction of Kdm3a-i2 is observed in Kdm3aGT/GT primary MEFs. (D) RPE1 cells transfected with GFP tag fused upstream of Kdm3a-i2. Monoclonal antibodies to mono- or dimethylated Lys-9 of histone H3 were used to determine the state of histone methylation (red) in transfected cells (green). Arrows point to transfected cells. (E) Quantitation of (D). n = number of cells counted for each condition. *p < 0.05, Fisher's exact test.
In summary, we show that Kdm3a encodes at least two enzymatically active isoforms. This newly identified protein could provide partial rescue, explaining the decreased severity of Kdm3aGT/GT mice.
Identification of Kdm3a-interacting proteins in testis
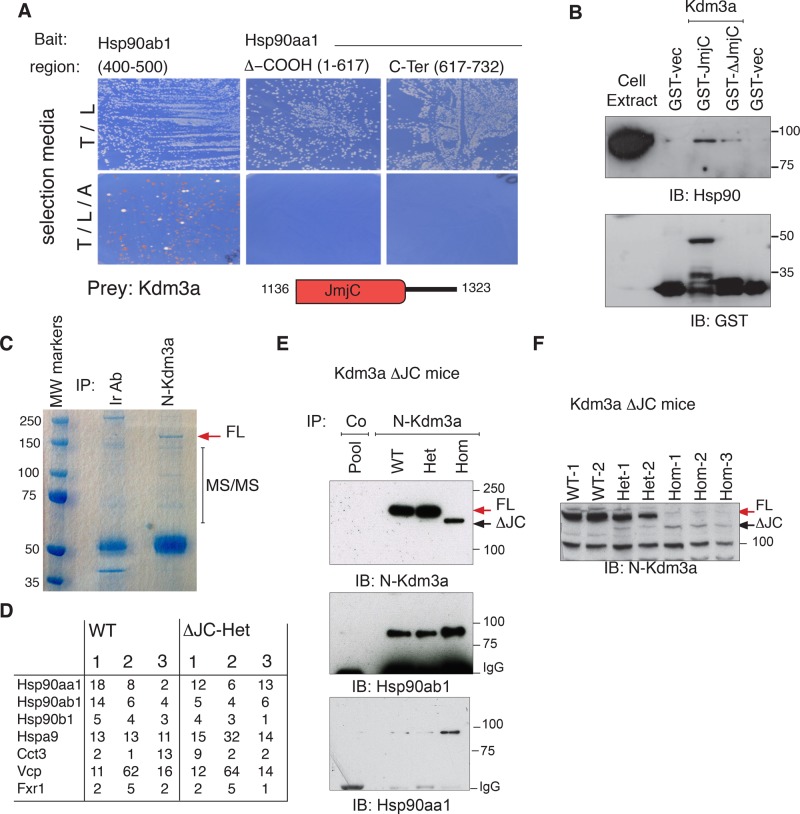
Originally, Kdm3a was isolated in a two-hybrid screen using different domains of Hsp90 as baits. Hsp90 chaperones, one of the most abundant cellular proteins, play multiple roles assisting client proteins that must undergo large structural changes for biological activity (Kim et al., 2013). We used different domains of Hsp90aa1 (Hsp90α) and Hsp90ab1 (Hsp90β) and found that the catalytic domain of Kdm3a interacts with the middle region of Hsp90ab1. The N-terminal and C-terminal dimerization domains of Hsp90aa1 failed to interact with Kdm3a under these experimental conditions (Figure 5A). Kdm3a interaction with Hsp90 was further validated through glutathione S-transferase (GST) pull downs, which showed that both the catalytic JmjC domain (GST-JmjC) and a C-terminal truncation without the catalytic domain (GST-ΔJmjC) interact with Hsp90, albeit with different efficiency as determined with pan-Hsp90 antibodies (Figure 5B).
FIGURE 5:
Kdm3a interacts with Hsp90 and other cellular chaperones in vitro and in vivo. (A) Positive clones identified from a large two-hybrid screen using Hsp90 as baits were purified, retransformed into yeast, and mated to reassess interactions. The catalytic domain of Kdm3a can rescue amino acid deficiency of the parental yeast strain only when Hsp90ab1 is the bait. Diagram illustrates JmjC region contained in two-hybrid and GST constructs. (B) Bacterially expressed C-terminal end of Kdm3a-GST: GST-JmjC (aa 1136–1323), GST-ΔJmjC (aa 1283–1323), and GST vectors were incubated with HeLa cell extracts. Proteins bound to glutathione beads were resolved and blotted with the indicated antibodies to determine interactions of GST constructs with Hsp90. (C) TrueBlue stain of whole adult testis extracts immunopurified with Kdm3a or an irrelevant antibody used as control (Ir Ab). The gel section indicated was cut for mass spectrometry (MS). (D) Table shows the number of unique peptides identified with >99% confidence for three independent immunopurifications resolved as shown in (C) for each genotype. Only proteins shared by the six runs and not found in control samples are included. (E) An antibody directed to the N-terminal end of Kdm3a pulls down endogenous Hsp90 from testis. Note that the in-frame deletion Kdm3a protein encoded by the floxed Kdm3a allele still binds Hsp90aa1 and Hsp90ab1 strongly. (F) Total extracts from adult testis immunoblotted with Kdm3a show absence of full-length (FL) and low levels of truncated (ΔJC) Kdm3a protein in Kdm3aΔJC/ΔJC animals. Kdm3a-FL (red arrow) in wild-type (WT) and heterozygous (Het) but not control pool is indicated. 1–3 identify an animal for each genotype.
To investigate the interaction profile of endogenous Kdm3a, we immunopurified testis extracts with antibodies to Kdm3a; this was followed by mass spectrometry (MS). We used extracts from wild-type and Kdm3aΔJC/+ animals. Three independent pull downs for each genotype from mice age 1.5–6 mo were resolved in polyacrylamide gels, and the indicated regions were cut and analyzed by MS (Figure 5C). In agreement with two-hybrid studies, we found multiple cellular chaperones from the Hsp90 family; we also found CCt chaperonins in Kdm3a pull downs absent in control samples (Figure 5D). There is also abundant representation of the chaperone valsolin-containing protein Vcp and the RNA-binding protein Fxr1. Kdm3a interaction with Hsp90 was further validated by immunoblotting with antibodies specific to Hsp90aa1 and Hsp90ab1. Pull-down extracts from wild-type and heterozygous mice specifically contain Hsp90ab1 and Hsp90aa1, indicating that Kdm3a interacts with both chaperones (Figure 5E). Interestingly, the truncated Kdm3a protein of Kdm3aΔJC/ΔJC mice is bound and immunopurified by an antibody directed to the N-terminal region of Kdm3a (Figure 5E and F). As determined by the relative abundance of Hsp90 to immunopurified Kdm3a proteins, the truncated form of Kdm3a binds Hsp90 chaperones more efficiently or in greater proportion than the full-length Kdm3a protein. Importantly, the catalytic domain of Kdm3a found in the two-hybrid screen to interact with Hsp90ab1 (Figure 5A) is deleted in Kdm3aΔJC/ΔJC mice (Figure 1A), but, as seen with the truncated GST-ΔJmjC construct, the C-terminal end of Kdm3a contributes to this interaction (Figure 5B). This observation suggests that multiple residues are responsible for Kdm3a interaction with Hsp90; alternatively, as Kdm3a may be rendered unstable through the deletion, it may have increased affinity for Hsp90.
In summary, these results show that Kdm3a interacts specifically with Hsp90 chaperones in vivo.
Substrate or client?
Kdm3a interaction and regulation of cytosolic chaperones would be consistent with the ultrastructural defects observed in Kdm3a mouse models. The chaperonin containing TCP1 (t-complex polypeptide 1/Cct/TriC) forms two heterogeneous eight-membered complexes (CCT1-8) (Dekker et al., 2011) that mediate the folding and assembly of a small subset of nascent polypeptides, preventing their aggregation within the crowded cytoplasmic environment (Kim et al., 2013) and playing a significant role in the cytoskeletal rearrangements required to produce mature spermatids (Willison et al., 1989; Sternlicht et al., 1993; Souès et al., 2003). Ccts chaperonins are essential for the de novo folding of actin and tubulin (Llorca et al., 2000), which form part of the acroplaxome and microtubular manchette affected in Kdm3a mouse models. Likewise, a critical requirement for Hsp90 in the extensive microtubular rearrangements during spermatogenesis has been documented (Yue et al., 1999), as has Hsp90 functional interplay with CCT chaperonins (Kim et al., 2013). Moreover the chaperone functions of Hsp90 are regulated by multiple posttranslational modifications (Li and Buchner, 2013), including lysine methylation by SmyD2 (Abu-Farha et al., 2011). This modification is in turn important for myocyte organization (Donlin et al., 2012). Misregulation of the posttranslational state of these cellular chaperones could thus contribute to the observed cytoplasmic defects of Kdm3a mutant mice. Conversely, Hsp90 could regulate Kdm3a functions instead, and the structural defects observed could be a consequence of the direct participation of Kdm3a on cytoplasmic components. For example, Hsp90 was found to modulate the stability of Kdm4b histone demethylase (Ipenberg et al., 2013) and to enhance the histone methyltransferase activity of SmyD2 (Abu-Farha et al., 2008).
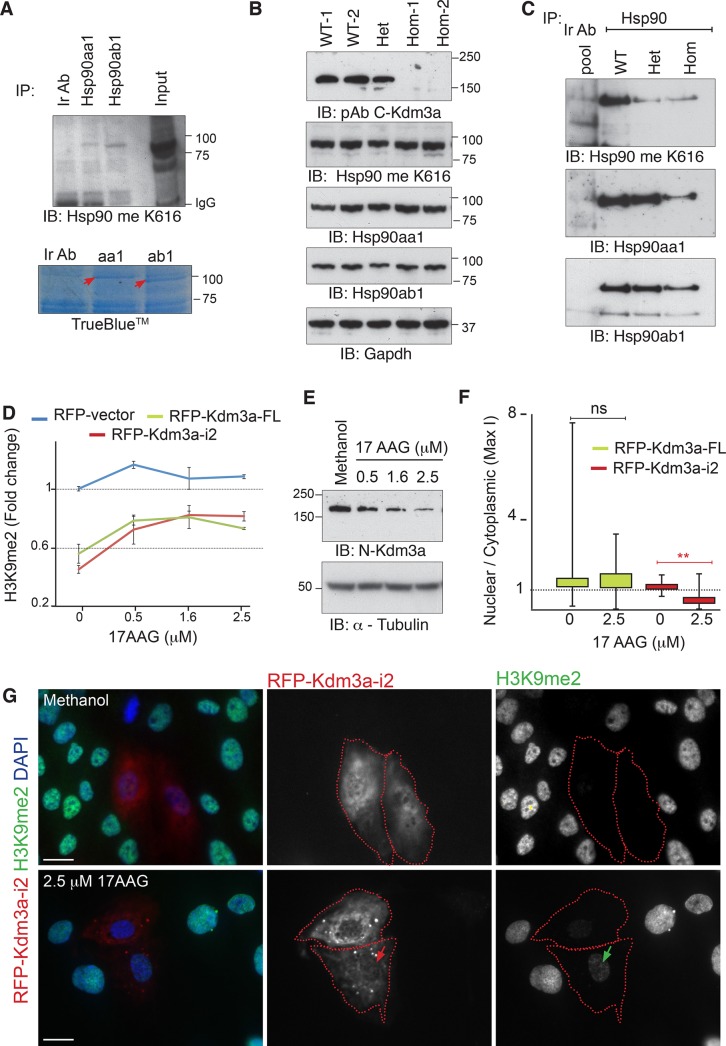
We first investigated the methylation state of Hsp90 in the absence of Kdm3a demethylase activity through the use of tailored antibodies to methylated K616 (Hsp90 meK616) of Hsp90 (Donlin et al., 2012). Individual pull downs of Hsp90aa1 and Hsp90ab1 indicate that both Hsp90 chaperones are methylated in vivo (Figure 6A). Neither total extracts (Figure 6B) nor Hsp90 pull downs (Figure 6C) from Kdm3aΔJC/ΔJC testis show significant changes in the methylation levels of Hsp90 as detected with meK616 antibody, indicating that Kdm3a does not regulate the methylation state of this lysine residue. It has been shown, however, that Hsp90 has multiple methylation sites (Abu-Farha et al., 2011), and it thus remains possible that other methylated lysines of Hsp90 could be regulated by Kdm3a.
FIGURE 6:
Kdm3a is an Hsp90 client protein. (A) Total extracts from ΔJC Kdm3a testis were immunopurified with the indicated Hsp90 antibodies and immunoblotted with an antibody to methylated Lys-616 (Me K616) of Hsp90. TrueBlue staining shows the gel fraction used to confirm these pull downs by MS. (B) Total extracts from testis immunoblotted with the indicated antibodies show comparable levels of methylated Hsp90 as detected with Me K616 antibody. (C) The same unaltered trend in methylation is observed when antibody-purified Hsp90 pull downs are blotted. A seemingly reduced methylation state of Hsp90 is observed in Kdm3aΔJC/+ pull downs relative to wild-type (WT) extracts. This difference is not detectable in shorter exposures of hsp90aa1 blots (unpublished data). (D) Fold change in the methylation levels of histones as detected with a monoclonal antibody to dimethylated H3K9. Each point represents the ratio between the mean intensity of H3K9 signal in transfected cells relative to their neighboring untransfected cells. The error bars represent the SEM between three independent experiments. More than 100 cells were counted for each condition. (E) Endogenous levels of Kdm3a protein are increasingly reduced as Hsp90 is inhibited with 17AAG in RPE1 cells. Tubulin is used as a loading control. (F) Box plots illustrate the subcellular distribution of RFP-Kdm3a isoforms in cells grown at the indicated 17AAG dose. Maximal pixel intensity of the nuclear and cytoplasmic compartments was measured as indicated in Figure S1C. * indicates statistical significance (p < 0.001, t test). (G) Change of subcellular distribution of Kdm3a-i2 after Hsp90 inhibition. Note nuclear depletion (red arrow) is often accompanied by increased H3K9 methylation (green arrow). Scale bars: 20 μM.
We then investigated whether Kdm3a requires Hsp90 for its demethylase activity, using histone methylation as a readout. Red fluorescent protein (RFP)- or GFP-tagged full-length or i2 Kdm3a (Supplemental Figure 1A) were transfected in parallel with their tag control. Six hours later, the cells were treated with the Hsp90 inhibitor 17AAG. The dimethylated state of histone H3 Lys-9 (H3K9me2) was assessed 48 h after transfection. Both tagged isoforms display nuclear and cytoplasmic distribution, albeit Kdm3a-FL nuclear presence is very strong (Supplemental Figure 1B). We observed a marked decrease in the intensity of tagged-Kdm3a upon treatment with Hsp90 inhibitors and a shift in the subcellular distribution, suggesting Hsp90 may regulate Kdm3a protein stability and localization. To dissociate the loss of Kdm3a demethylase activity from these observations, we measured H3K9me2 levels in cells with high levels of tagged-Kdm3a only (Figure 6D). These results indicate that the enzymatic activity of both Kdm3a isoforms depends on Hsp90, as their demethylation capacity decreases when Hsp90 is inhibited. The effect of Hsp90 on Kdm3a levels was confirmed by treating RPE1 cells with 17AAG followed by immunodetection of endogenous KDM3A protein (Figure 6E). The change in subcellular distribution was investigated by determining the ratio of nuclear to cytoplasmic fluorescence intensity of Kdm3a-transfected cells grown in solvent control or in the presence of 17AAG (Figure S1C). These measurements confirm a significant shift of RFP-Kdm3a-i2 but not RFP-Kdm3a-FL toward the cytoplasm upon Hsp90 inhibition (Figure 6F and G).
These results uncover a complex dependence of Kdm3a on Hsp90 to fulfill its biological functions. Interestingly, Hsp90 did not significantly affect the nuclear-cytoplasmic distribution of the full-length Kdm3a isoform but altered the distribution of Kdm3a-i2. This is perhaps because the short isoform (i2) does not contain the N-terminal region encoding a previously described nuclear localization signal (Brauchle et al., 2013). The dependence of both isoforms on Hsp90 for their enzymatic activity therefore suggests a specific requirement of the Hsp90 chaperone machinery for the demethylation reaction. Moreover, the absence of detectable changes in the methylation state of Hsp90 in Kdm3a mutant mice suggests the cytoskeletal defects uncovered in this study may reflect a direct role of Kdm3a in the cytoplasm rather than these being a consequence of Kdm3a-mediated dysregulation of the chaperone system in Kdm3a mutant mice.
Cytoplasmic localization of Kdm3a
Previous publications have observed cytoplasmic distribution of Kdm3a by means of immunofluorescence of testis sections (Okada et al., 2007) and somatic cells (Okada et al., 2007; Sar et al., 2009; Yang et al., 2009; Yamada et al., 2012). We extended these findings using a fractionation kit (Qproteome) that allows the sequential isolation of soluble and organelle-bound proteins from cytoplasm and nucleus, respectively. Subcellular fractionation of mouse embryonic fibroblasts (MEFs) followed by immunoblotting shows nuclear and cytoplasmic distribution of Kdm3a (Figure 7A). The endogenous Kdm3a full-length protein is nuclear and cytoplasmic in exponentially growing cultures (Figure 7A, 10% FCS), as observed for transfected RFP-KDM3A-FL in exponentially growing RPE1 cells, although the nuclear signal of the tagged protein is stronger (Figures 6F and S1B). In resting cells, however, Kdm3a is almost exclusively cytoplasmic (Figure 7A, 0.25% FCS). There is also a marked shift between soluble and membrane-bound Kdm3a depending on the growth conditions (Figure 7A, compare lanes 1 and 2 of 0.25 and 10% FCS). Finally, the use of an antibody directed to the C-terminal end of Kdm3a that does not recognize Kdm3aΔJC/ΔJC truncated protein (Figure 7B) shows Kdm3a presence in the cytoplasm with strong enrichment in the acrosome of wild-type but not Kdm3aΔJC/ΔJC mature spermatids (Figure 7C).
FIGURE 7:
Cytoplasmic distribution of endogenous Kdm3a. (A) Subcellular fractionation of serum-starved and exponentially growing MEFs immunoblotted with the indicated antibodies. Sequential disruption of cell and nuclear membranes gives rise to two cytoplasmic fractions: soluble (1) and membrane (2) and two nuclear fractions: soluble (3) and cytoskeletal (4). GAPDH and HP1 are controls for cytoplasmic and nuclear fractions, respectively. Red and black arrows indicate FL-Kdm3a and GAPDH proteins, respectively. (B) Total extracts from four wild-type (WT) and 6 ΔJC Kdm3a mice immunoblotted with pAb antibody directed to the C-terminal region of Kdm3a (see also Figures 6B and S2A). The immunizing peptide of this commercially available antibody is likely contained within ΔJC deletion, as no band is observed in any of the three Kdm3aΔJC/ΔJC mice analyzed. GAPDH is used as loading control. ** and * indicate potentially nonspecific bands. (C) Immunohistochemistry shows Kdm3a localization in acrosomes (ac) of wild-type (WT) mature spermatids but not Kdm3aΔJC/ΔJC as detected with C-Kdm3a antibody described in (B). Arrows indicate the dorsal surface of the spermatid on DAPI-stained nuclei.
These results indicate that Kdm3a distribution within cells is dynamic and influenced by growth conditions. These findings are consistent with the hypothesis that Kdm3a plays a direct role in the cytoplasm of maturing spermatids.
Investigation of the role of Kdm3a in the cytoplasm
To begin to explore the cytoplasmic role of Kdm3a, we used three independent approaches, bearing in mind that Kdm3a could interact with but might not necessarily demethylate nonhistone substrates. We reasoned that, if Kdm3a requires Hsp90 for its biological functions, differential associations of these chaperones between controls and homozygous animals evidenced by partition along sucrose gradients or IP/MS would provide information on the state of the cytoskeleton and potential Kdm3a-interacting proteins in the cytoplasm. Likewise, if Kdm3a has nonhistone substrates in the cytoplasm, these would be evidenced as differentially methylated proteins between Kdm3a homozygous and control animals.
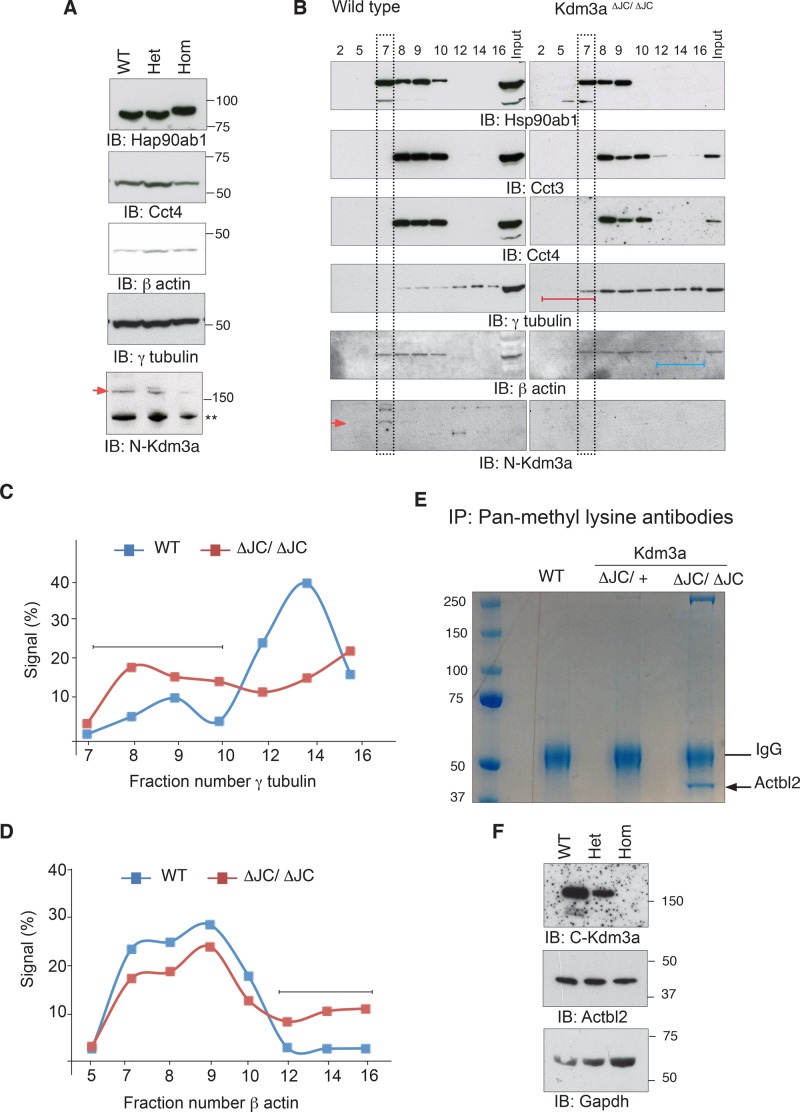
Actin and tubulins are among the most abundant components of the acrosome and manchette found in multiple polymerization states (Kierszenbaum et al., 2011). Their dependence on Cct and Hsp90 chaperones for maturation led us to explore their state in Kdm3aΔJC/ΔJC mutant testis. Total testis extracts show comparable levels of Hsp90ab1 and Cct4 used as indicators of Hsp90 and chaperonin II complex, respectively (Figure 8A). Levels of β-actin and γ-tubulin are also seemingly unaltered (Figure 8A). Sucrose gradients of cytoplasmic extracts from testis show instead slightly altered fractionation of chaperonins and Hsp90 in Kdm3aΔJC/ΔJC (Figure 8B). Hsp90ab1 migration along Kdm3aΔJC/ΔJC gradients is more compact, while that of Cct3 is more widespread compared with wild-type gradients (Figures 8B and S2). Most striking is the differential partition of γ-tubulin and β-actin. In homozygous testis, γ-tubulin is found in early fractions, suggesting that a significant amount of the cellular pool of γ-tubulin dissociates easily or is not part of large polymers (Figures 8B and S2, red lines). This is illustrated by measuring the relative distribution of γ-tubulin along each gradient (Figure 8C). On the contrary, β-actin shows increased presence in late fractions of the homozygous samples, indicating that in these mice actin is part of larger complexes, perhaps aggregates (Figure 8B, blue line, and D). These gradients also confirm the presence of Kdm3a in the cytoplasm, as detected with the two polyclonal antibodies validated in this study (Figures 8B and S3A). The altered state of cytoskeletal components relative to chaperone complexes in Kdm3a homozygous animals also extends to somatic cells (Figure S4).
FIGURE 8:
Altered state of cytoskeletal components in the cytoplasm of Kdm3aΔJC/ΔJC mice. (A) Total extracts from testis immunoblotted with the indicated antibodies show comparable levels between Kdm3a genotypes. (B) Cytoplasmic extracts from testis fractionated in discontinuous sucrose gradients (40–70%). The migration of each protein along the gradient was determined with the relevant antibodies. Lines represent the early soluble components (red) and late larger complexes (blue). Sedimentation of the bulk of cytoplasmic proteins is indicated (black rectangles). Red arrowhead indicates FL-Kdm3a, see also Figure S3A. (C) Percentage of signal intensity of each fraction relative to the sum of intensities along the fractions plotted in this graph illustrates the distribution profiles of γ-tubulin in wild-type (WT) and Kdm3aΔJC/ΔJC sucrose gradients. See Figure S2C for measurements. (D) Percentage intensity of each fraction relative to the sum of intensities along the fractions plotted in this graph illustrates the distribution profiles of β actin in wild-type (WT) and Kdm3aΔJC/ΔJC sucrose gradients. Black lines point to the most differing aspect of gradients between genotypes. (E) Total testis extracts were immunopurified with a pool of antibodies to mono-, di-, and trimethylated lysines. A single band is observed in Kdm3aΔJC/ΔJC pull downs only, subsequently identified by MS as Actbl2. (F) Total extracts from testis immunoblotted with the indicated antibodies show comparable levels of Actbl2 between Kdm3a genotypes.
Immunopurification of total testis extracts with antibodies to methylated lysines revealed a single band differentially present in Kdm3aΔJC/ΔJC samples that belongs to the actin family, β-actin–like protein 2 (κ-actin or Actbl2; Figure 8D). One of the identified peptides for this protein presents evidence of a potentially methylated lysine residue (Figure S5A). We then immunopurified Hsp90 chaperone complexes from age-matched wild-type, Kdm3aΔJC/+, and Kdm3aΔJC/ΔJC testis; this was followed by MS. Hsp90 interactions are usually transient; protein extracts were therefore incubated with antibodies for 1 h in the presence of sodium molybdate, thereby freezing the Hsp90 chaperone cycle and stabilizing interactions with client proteins (Taipale et al., 2012). This provides a snapshot of the state of Hsp90 complexes for each genotype. These experiments show largely overlapping hits for Hsp90 pull downs between the three Kdm3a genotypes absent in all the controls (WT, Kdm3aΔJC/+, and Kdm3aΔJC/ΔJC) performed with an irrelevant antibody (Supplemental Table S2). Actbl2 appears to be found exclusively in Kdm3aΔJC/ΔJC pull downs, but the previously identified methylated peptide was not found, and none of the remaining peptides found present evidence for methylation (Figure S5B). Total extracts immunoblotted with Actbl2 antibodies show that the overrepresentation of Actbl2 in Kdm3aΔJC/ΔJC pull downs is not due to increased expression levels (Figure 8F). Cct chaperonins also seem enriched in Hsp90 pull downs from Kdm3aΔJC/ΔJC extracts, particularly Cct7, which is found only in Kdm3aΔJC/ΔJC samples (Table S3). These findings are consistent with the increased overlap between Hsp90 and Cct chaperonins observed in Kdm3aΔJC/ΔJC gradients (Figure S2D) and the initial finding of Cct proteins in Kdm3a pull downs (Figure 5D). Furthermore, among all the methylated peptides identified in Hsp90 pull downs, 11 were found in Kdm3aΔJC/ΔJC samples only (Table S3). From these, Cct2 (Ttc chaperonin 1 beta; Figure S5C) and the cytoplasmic dyneins Dc1l1 and Dyhc1 could directly contribute to the cytoplasmic defects observed in Kdm3a mutant testis.
These three independent approaches provide consistent evidence on the altered state of cellular chaperones and on the components of microtubule and actin networks in animals lacking Kdm3a demethylase activity. The differential state of cellular chaperones in Kdm3aΔJC/ΔJC samples could reflect the state of actin or other folding targets altered in Kdm3a mutants.
DISCUSSION
In this study, we begin to unravel the functional complexity of Kdm3a. We present genetic and biochemical evidence that cytoskeletal abnormalities contribute to the male infertility phenotype previously reported for Kdm3a mouse models. The use of two Kdm3a models has allowed us to separate cytoplasmic from chromatin condensation defects. The most severe knockout model has acroplaxome, manchette, and HTCA abnormalities accompanying variable states of DNA condensation. In the milder gene-trap model, DNA condenses fairly normally, but manchette and centriolar defects persist, and mature spermatids still have rounded heads. The variable penetrance of these traits perhaps reflects the different dependence of multiple processes on Kdm3a activity. The Kdm3a isoform newly identified here may contribute to phenotype amelioration in the second mouse model. We further demonstrate that Kdm3a interacts with and requires Hsp90 for its demethylation activity and protein stability. We show that Kdm3a localizes not only to the nucleus but also to different cytoplasmic compartments depending on growth conditions and Hsp90 activity. We found Kdm3a to be enriched in the acrosome, which is in turn disrupted in Kdm3aΔJC/ΔJC mutants and provide evidence that the state of actin and tubulin components that form these structures is altered, although their protein levels remain unchanged. We therefore propose that Kdm3a is directly involved in the cytoskeletal rearrangements required to produce mature spermatids. Whether this is through Kdm3a interactions with actin, tubulin, or dynein components identified as candidates in this study or their posttranslational modification (PTM) remains to be investigated.
The original finding of the dynamic distribution of Kdm3a in the cytoplasm of spermatids (Okada et al., 2007) suggested that Kdm3a could have a cytoplasmic function. Hsp90 chaperones have also been reported among Kdm3a pulled-down proteins from somatic cells (Brauchle et al., 2013). Our ultrastructural analysis revealed cytoskeletal defects known to depend on actin, tubulin, and cellular chaperones (Kierszenbaum et al., 2011); biochemical evidence then uncovered their abnormal partition after Kdm3a loss of function. This could be a consequence of altered functional capacity of cellular chaperones, known to regulate protein folding and to be dynamically methylated (Abu-Farha et al., 2011), but the methylation state of Hsp90 was unchanged in Kdm3a homozygous mice. Instead, we found that Kdm3a protein stability and enzymatic activity is highly dependent on Hsp90, raising the possibility that the observed cytoskeletal defects could therefore be a direct consequence of Kdm3a activity on cytoplasmic components for which it would require Hsp90 chaperoning.
Actbl2 is differentially enriched in Kdm3aΔJC/ΔJC samples purified with antibodies to methyl lysines and Hsp90. Elongation of the spermatid head requires extensive restructuring of actin filaments in the leading edge of the acrosome and nuclear envelope (Kierszenbaum et al., 2003). Changes in the actin network are driven by a large variety of biochemical properties that involve nucleation of monomers and elongation of filaments to produce the many distinct actin architectures observed in living organisms (Blanchoin et al., 2014). Biochemical evidence suggests that the properties of actin filaments may vary according to the mix of actin isoforms in the filament (Perrin and Ervasti, 2010). If Actbl2 is posttranslationally altered in Kdm3a mutant spermatids, its association with other actin monomers may perturb filament formation and disrupt acrosome development, giving rise to the abnormal acrosome observed in Kdm3aΔJC/ΔJC mice. The importance of the correct spatiotemporal sequence in actin dynamics is illustrated by the many disease states associated with altered actin cytoskeleton (Blanchoin et al., 2014). Indeed, Actbl2 is associated with hepatocellular carcinomas (Chang et al., 2011; Junrong et al., 2011). Interestingly, Kdm3a may play a significant role during the malignant transformation of hepatocellular carcinomas (Yamada et al., 2012). Furthermore, this study found that in 80% of the samples expressing Kdm3a protein, this protein localized exclusively to the cytoplasm (Yamada et al., 2012). Actbl2 is therefore a very strong candidate to molecularly underlie Kdm3a mutant acrosomes, and this functional interaction may extend to somatic cells. Cytoplasmic dyneins are also associated with microtubules of the manchette for transport (Yoshida et al., 1994). These proteins found to be lysine methylated in human cells (Hornbeck et al., 2012; Cao et al., 2013) are thus also potential candidates via which Kdm3a could impact the cytoplasmic aspect of spermatogenesis found altered in Kdm3a mutant mice.
None of the proteins analyzed in this study shows altered levels in Kdm3aΔJC/ΔJC samples. Likewise, publicly available microarrays in which Kdm3a has been silenced (Herzog et al., 2012) show no significant change in the transcript levels of Actbl2, dyneins, or Ccts, indicating the impact of Kdm3a on these cytoplasmic components is not at the transcriptional level. The effect on these components could thus be through direct protein interaction and/or posttranslational modifications. The functional requirement of Hsp90 for Kdm3a demethylase activity raises the possibility that Kdm3a cytoplasmic targets could be posttranslationally modified as they are being folded, with the modifications contributing to their tertiary structure or polymerization state. The consistent lack of the ventral surface of the acrosome in Kdm3a mutant mice is intriguing. Could posttranslational modifications act to demarcate spatial boundaries between dorsal and ventral acrosome surfaces in otherwise continuous filaments of actin? Posttranslational modifications of ubiquitous and abundant proteins may thus confer spatial compartmentalization. Another possibility is that the ventral and dorsal surfaces of the acrosome could contain different ratios of actin isoforms.
Only a small fraction of cellular proteins are folded by Cct chaperonin, and from these only 5–10% will require further interaction with Hsp90 chaperones (Kim et al., 2013). It is thus likely that Kdm3a interacts with the Hsp90 chaperone system independently from chaperonins fulfilling other functions not directly related to protein folding. Histone and nonhistone substrates for Kdm3a are not mutually exclusive scenarios and warrant further investigation. We analyzed only one methylated residue of Hsp90, but it is known to have at least two methylatable residues (Abu-Farha et al., 2011); the effect of Kdm3a on these alternative sites remain to be investigated. The methylation state of Cct chaperonins also deserves further investigation.
Hsp90 and Cct chaperones are also known to play a role in chromatin remodeling by modulating binding and release of transcriptional regulatory machineries (Liu and DeFranco, 1999; Freeman and Yamamoto, 2001) and binding to heterochromatin in germ cells (Souès et al., 2003). We investigated the effect of Hsp90 on Kdm3a histone demethylase activity and found that it largely depends on these chaperones. Interestingly, the enzymatic activity, protein stability, and subcellular localization of Kdm3a depended on Hsp90. Further studies would be particularly relevant for androgen-responsive genes known to be regulated both by Hsp90 and Kdm3a (Freeman and Yamamoto, 2001; Yamane et al., 2006) and for the cellular response to hypoxia in which Kdm3a and HIF1-α levels are regulated by oxygen (Beyer et al., 2008; Pollard et al., 2008; Wellmann et al., 2008) and Hsp90 (Minet et al., 1999), respectively. Interestingly, we also found the JmjC domain of Kdm6a to interact with Hsp90aa1 (Figure S6), suggesting that interaction between JmjC-containing proteins and Hsp90 chaperone machinery is a common feature and may be relevant in the enzymatic activity of this family in general.
In summary, our work has only just begun to illustrate the complex regulatory and spatial landscape of Kdm3a, providing evidence for a wider functional role of this lysine demethylase beyond histone modification in the nucleus.
MATERIALS AND METHODS
Kdm3a mouse models
Kdm3aΔJC mice Kdm3aΔJC ES cells (a gift from Y. Zhang, Harvard Medical School) were used to produce chimeric animals upon microinjection into C57BL6J blastocysts and to establish a parental Kdm3a colony. Heterozygous males were then crossed to C57BL6 females expressing ubiquitous Cre, and the resulting mice containing recombination Kdm3a alleles (ΔJC) were identified by PCR. LacZ positive, F1 mice were crossed with a ubiquitously expressing Cre line to produce knockout offspring lacking the JmjC catalytic domain. Subsequent generations were backcrossed to the C57Bl6 strain and genotyped with primers A, B, and C (Tateishi et al., 2009) listed in Table S1.
Kdm3aGT mice were generated by microinjection of XR0062 ES cell lines (129/ola), obtained from the Sanger Institute Gene Trap Resource (www.genetrap.org), into C57BL6J blastocysts with transmission of the mutation on a C57 background. The cells used contained a gene-trap insertion (Kdm3aGT) within intron 5–6 of the mouse Kdm3a locus. Genotypes were identified by PCR of genomic DNA with primers GT1, GT2, and GT3 described in Table S1. Expression levels of the full-length transcript in gene-trapped mice were determined with the primer pair GT4.
Antibodies
The antibodies used along this study are to KDM3A (12835; Proteintech; and NB100-77282; Novus Biologicals, Littleton, CO); KDM3B (IHC 00189; Bethyl Laboratories, Montgomery, TX); Cct4 (ARP34271; Aviva); anti–mono- and dimethylated lysines (ab23366 and ab76118; Abcam, Cambridge, MA); anti-GFP (sc-8334), monoclonal to GAPDH (5019A-2; Imgenex, San Diego, CA), β-gal (A-11132; Molecular Probes), GST (C83271; LSBio); HP1a (clone 15.1952; Upstate); γ-tubulin (GTU-88; Sigma-Aldrich, St. Louis, MO), β-actin (ab8229; Abcam), Hsp90ab1 (MAB32861; R&D Systems), Hsp90aa1 (10713715; Pierce, Rockford, IL), Actbl2 (ab134977; Abcam). Secondary antibodies: anti-mouse and anti-rabbit F(ab′)2 immunoglobulin G Alexa Fluor 488 and 584 (Molecular Probes) for immunofluorescence or horseradish-conjugated peroxidase for immunoblots.
Cell lines, transfections, and cell fractionation
Telomerase-immortalized human retinal pigment epithelial cells (hTERT-RPE1) were obtained from Clontech (Mountain View, CA). Transfections were done in RPE1 cells at 80% confluency and were seeded the day before transfection without antibiotic. Lipofectamine 2000 was used following the manufacturer's guidelines for 5 h. MEFs were isolated by mincing E11.5- to E13.5-d embryos in DMEM supplemented with antibiotics and serum. MEFs were grown in 3% O2 incubators. Subcellular fractionation of MEFs was performed using Qproteome Cell Compartment kit according to the manufacturer's instructions (Qiagen, Valencia, CA).
Primers, RT-PCR, and qRT-PCR
All primers used below are listed in Table S1. First-strand cDNA was synthesized with random primers and Superscript III (Invitrogen, Carlsbad, CA). qPCRs were set up using the Quantitect SYBR green PCR kits (Qiagen). Individual reactions totaled 10 μl with a final primer concentration of 0.5 mM.
Cloning of Kdm3a-i2 isoform
Kdm3a-i2 was amplified from homozygous Kdm3a-GT MEF RNA or wild-type testis RNA and reverse transcribed with random primers; this was followed by PCR with primers designed from ENSEMBL prediction ENSMUST00000101304 as listed (Table S1). The PCR product was cloned into pGem-Teasy (Promega, Hayward, CA) vector and sequenced to verify correct splicing and open reading frame.
Tagged constructs
Tagged constructs were made using cDNA from homozygous Kdm3a-GT MEF (GFP) or wild-type testis (RFP) as template with primers listed in Table S1. Kdm3a-i2 was subcloned in-frame into pEGFP or RFP-vector. Kdm3a-FL was subcloned in-frame into RFP vector. Primers contained XhoI and BamHI restriction sites (Table S1).
TEM
Adult mice testes were fixed in 2% paraformaldehyde, 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer plus 0.04% CaCl2 overnight. Several cuts at the edges of the testes were done to allow penetration of fixative.
Immunofluorescence
Mouse testes utilized for tissue sections were fixed in ice-cold 4% methanol-free formaldehyde/phosphate-buffered saline (PBS) for at least 1 h (formaldehyde from TAAB Bioscience). Fixed material was dehydrated in methanol, wax embedded, and microtome sectioned. In all cases, cells and sections were incubated with primary antibodies overnight at 4°C. Secondary antibody was incubated for 2 h at room temperature or overnight at 4°C. Investigation of Kdm3a presence in the cytoplasm of spermatids was done on dissected tubules (Kotaja et al., 2004) squashed on glass slides and frozen on dry ice; this was followed by methanol and acetone fixation (Zhou et al., 2009).
Protein extracts and immunoprecipitations
Total cell extracts were obtained by lysing cells in 150 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% octylphenoxypolyethoxyethanol (IGEPAL), 1% Triton, 1 mM EDTA, 0.5% deoxycholate with cOmplete EDTA-free proteinase inhibitors (Roche, Indianapolis, IN), and Benzoase DNase (Novagen). For immunoprecipitations, cells were lysed in 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10% glycerol, 5 mM EDTA, and 0.5% IGEPAL with Benzoase DNase and protease inhibitors. Successive washes were carried out with reducing IGEPAL content to a final concentration of 0.01%, with final washes using only Tris-HCl, NaCl, EDTA, and 10% glycerol. Primary antibody was generally incubated overnight at 4°C, this was followed by 45 min with Dynabeads (Invitrogen) to concentrate immunoglobulin complexes. Hsp90 pull downs were done as previously described (Taipale et al., 2012). For the identification of methylated peptides, proteins extracts were prepared in the presence of 160 μM of the demethylase inhibitor 5-carboxy-8-hydroxyquinoline (SML0067; Sigma-Aldrich).
Sucrose gradients
Testis cytoplasmic extracts were prepared by hypotonic shock. Briefly, gently dislodged testes were taken to single-cell suspension in ice-cold PBS by passaging them 10 times through a 1-ml plunger. After being rinsed twice in PBS, cells underwent 5 min of centrifugation at 1000 × g. Cells were suspended in 0.1 × TBS (50 mM Tris, pH 7.5, 150 mM NaCl)–8% sucrose and left until they became swollen (for 30–60 min) on ice. Swollen cells were concentrated by 10 min of centrifugation at 2500 × g. All supernatant was removed, and cells were resuspended in 152 μl of 8% sucrose and 1.37 ml of lysis buffer (1 mM HEPES, pH 7.2, 0.5% IGEPAL, 0.5 mM MgCl2, 0.1% β-mercaptoethanol, cOmplete EDTA-free protease inhibitor); this was followed by 10 min of centrifugation at 2500 × g. An aliquot of this cleared cytoplasmic extract represented gradient input; the remaining extract (1 ml approximately) was layered on top of a discontinuous sucrose gradient formed by 1 ml of 40%, 1 ml of 50%, and 2 ml of 70% sucrose layers in gradient buffer (10 mM PIPES, pH 7.2, 0.1% Triton X-100, 0.1% β-mercaptoethanol). Gradients were run overnight at 40,000 rpm in a Beckman ultracentrifuge. Fractions of 250 μl were collected manually from the top of the gradients. Nocodazole (10 μg/ml) and cytochalasin B (5 μg/ml) were added to testis extracts during the 30–60 min incubation when indicated. Sucrose gradients of somatic cells were done with primary MEFs. In these cells, microtubules were destabilized for 90 min before cell lysis.
MS
For analysis of gel fractions, pulled-down proteins were fractionated on an SDS–PAGE gel and stained with TrueBlue (Egedon). The indicated gel chunks were excised and cut into 1-mm cubes; this was followed by in-gel digestion using a ProGest Investigator in-gel digestion robot (Genomic Solutions, Ann Arbor, MI) using standard protocols (Shevchenko et al., 1996). These gel cubes were destained by washing with acetonitrile and subjected to reduction and alkylation before digestion with trypsin at 37°C. Peptides were extracted with 10% formic acid and concentrated down to 20 μl using a SpeedVac (ThermoSavant, Thermo Fisher Scientific, Lafayette, CO). The peptides were then separated using a NanoLC Ultra 2D Plus loading pump and NanoLC AS-2 autosampler equipped with a nanoflex cHiPLC chip-based chromatography system (Eskigent, Redwood City, CA) and a ChromXP C18-CL trap and column (Eskigent), and eluted with a gradient of increasing acetonitrile containing 0.1% formic acid (5–35% acetonitrile in 45 min, 35–50% in a further 3 min, followed by 95% acetonitrile to clean the column, before reequilibration to 5% acetonitrile). The eluent was sprayed into a TripleTOF 5600 electrospray tandem mass spectrometer (ABSciex, Foster City, CA) and analyzed in information-dependent acquisition mode; we performed 250 ms of MS followed by 100-ms tandem mass spectrometry (MS/MS) analyses on the 20 most intense peaks seen by MS. The MS/MS data file generated was analyzed using the ProteinPilot (version 4.5 beta) Paragon algorithm (ABSciex) against the SwissProt database (www.ebi.ac.uk/uniprot, accessed February 2013) with both no species restriction and mouse-only, trypsin as the cleavage enzyme, and carbamidomethyl modification of cysteines. For bead analysis, pulled-down proteins were digested on beads at 37°C overnight using trypsin without prior reduction or alkylation. The peptides were then desalted using C18 Zip Tips (Millipore, Billerica, MA) and loaded onto a PepMap100 RSLC nanotrap column (Thermo Fisher Scientific) at a flow rate of 1 μl/min. Peptides were separated on a 15-cm PepMap100 RSLC analytical column (Thermo Fisher Scientific) using a Dionex Ultimate RSLC nano system and eluted with a gradient of increasing acetonitrile containing 0.1% formic acid (1.6–32% acetonitrile in 45 min, 32–90% in a further 0.1 min, followed by 4.6 min at 90% acetonitrile and back to 1.6% to re-equilibrate the column). Peptides were eluted into a Q-Exactive mass spectrometer (Thermo Fisher Scientific) using a flow rate of 300 nl/min and a spray voltage of 4 kV. Proteome Discoverer (version 1.4) was used to create peak lists from raw data and to search against the Swissprot Mus musculus database (accessed November 2013). Search parameters were set as follows: precursor mass tolerance of 10 ppm, fragment ion mass tolerance to 20 mmu, and enzyme as trypsin, allowing two missed cleavages. No static modifications were specified. However, the dynamic modification of single, di-, and trimethylation was allowed. Data were then further analyzed using Scaffold 4 (version 4.2.1).
Two-hybrid screens and GST subcloning
A mouse cDNA library was prepared by reverse transcribing mouse embryonic eye RNA with oligo-dT primers containing a BamHI site and ligated to EcoRI adaptors. cDNA products were digested with BamHI/EcoR1 and cloned onto BamHI/EcoR1 sites of pGKBT7 (Invitrogen) previously linearized with ClaI. The generated library was transformed by electroporation to a pJ69-a yeast strain. The indicated region of Hsp90ab1 was cloned onto a pGADT7 bait vector, and this transformed into the mating-compatible strain AH109-a. Strains were mated as described by the vector manufacturer (Invitrogen). Interacting proteins were identified by growing mated cultures in triple-selection media lacking adenine, leucine, and tryptophan. Larger colonies were assessed for X-gal staining, and positive clones were streaked onto new triple-selection media. cDNA from grown colonies was then amplified by PCR and sequenced with pGADT7 primers (Invitrogen). DNA was purified from selected clones and retransformed onto yeast for further validation. For GST constructs, isolated plasmids were transformed into bacteria, amplified, and digested with EcoRI/XhoI to clone onto pGEX-4-T1. Hsp90aa1-ΔC and Hsp90aa1-C terminus baits are in pAS2-1 vectors (Obermann et al., 1998).
Supplementary Material
Acknowledgments
We are indebted to Yi Zhang, Olena V. Taranova, W. Obermann, and A. Tarakhovsky for their generous gift of reagents. We thank the staffs at the Wellcome Trust Sanger Institute and the Institute of Genetics and Molecular Medicine microinjection facility for generation of transgenic mice and colony maintenance and Matthew Pearson for imaging scripts. We thank Nick Gilbert for expert biochemical advice and Pleasantine Mill and Javier Caceres for careful reading of the manuscript.
Abbreviations used:
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HTCA
head-tail coupling apparatus
- IP
immunopurified
- JmjC
jumonji domain
- MEF
mouse embryonic fibroblast
- MS
mass spectrometry
- PTM
posttranslational modification
- RFP
red fluorescent protein
- TEM
transmission electron microscopy
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0471) on February 19, 2014.
REFERENCES
- Abu-Farha M, Lambert J-P, Al-Madhoun AS, Elisma F, Skerjanc IS, Figeys D. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteomics. 2008;7:560–572. doi: 10.1074/mcp.M700271-MCP200. [DOI] [PubMed] [Google Scholar]
- Abu-Farha M, Lanouette S, Elisma F, Tremblay V, Butson J, Figeys D, Couture J-F. Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. J Mol Cell Biol. 2011;3:301–308. doi: 10.1093/jmcb/mjr025. [DOI] [PubMed] [Google Scholar]
- Beyer S, Kristensen MM, Jensen KS, Johansen JV, Staller P. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J Biol Chem. 2008;283:36542–36552. doi: 10.1074/jbc.M804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- Bowen RH. Notes on the topography of the Golgi apparatus in gland cells. Science. 1925;61:545–546. doi: 10.1126/science.61.1586.545. [DOI] [PubMed] [Google Scholar]
- Brauchle M, et al. Protein complex interactor analysis and differential activity of KDM3 subfamily members towards H3K9 methylation. PLoS One. 2013;8:e60549. doi: 10.1371/journal.pone.0060549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X-J, Arnaudo AM, Garcia BA. Large-scale global identification of protein lysine methylation in vivo. Epigenetics. 2013;8:477–485. doi: 10.4161/epi.24547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchpole S, Spencer-Dene B, Hall D, Santangelo S, Rosewell I, Guenatri M, Beatson R, Scibetta AG, Burchell JM, Taylor-Papadimitriou J. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int J Oncol. 2011;38:1267–1277. doi: 10.3892/ijo.2011.956. [DOI] [PubMed] [Google Scholar]
- Chang K-W, Chou A, Lee C-C, Yeh C, Lai M-W, Yeh T-S, Chen T-C, Liang K-H, Yeh C-T. Overexpression of κ-actin alters growth properties of hepatoma cells and predicts poor postoperative prognosis. Anticancer Res. 2011;31:2037–2044. [PubMed] [Google Scholar]
- Cho H-S, et al. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res. 2011;71:655–660. doi: 10.1158/0008-5472.CAN-10-2446. [DOI] [PubMed] [Google Scholar]
- Dam AHDM, Feenstra I, Westphal JR, Ramos L, van Golde RJT, Kremer JAM. Globozoospermia revisited. Hum Reprod Update. 2007;13:63–75. doi: 10.1093/humupd/dml047. [DOI] [PubMed] [Google Scholar]
- Dekker C, Roe SM, McCormack EA, Beuron F, Pearl LH, Willison KR. The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. EMBO J. 2011;30:3078–3090. doi: 10.1038/emboj.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin LT, et al. Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes Dev. 2012;26:114–119. doi: 10.1101/gad.177758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Yamamoto KR. Continuous recycling: a mechanism for modulatory signal transduction. Trends Biochem Sci. 2001;26:285–290. doi: 10.1016/s0968-0004(01)01834-5. [DOI] [PubMed] [Google Scholar]
- Hermo L, Pelletier R-M, Cyr DG, Smith CE. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 2: changes in spermatid organelles associated with development of spermatozoa. Microsc Res Tech. 2010;73:279–319. doi: 10.1002/jemt.20787. [DOI] [PubMed] [Google Scholar]
- Herzog M, Josseaux E, Dedeurwaerder S, Calonne E, Volkmar M, Fuks F. The histone demethylase Kdm3a is essential to progression through differentiation. Nucleic Acids Res. 2012;40:7219–7232. doi: 10.1093/nar/gks399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck P. V, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Tachibana M, Magoori K, Kudo H, Tanaka T, Okamura M, Naito M, Kodama T, Shinkai Y, Sakai J. Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells. 2009;14:991–1001. doi: 10.1111/j.1365-2443.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- Ipenberg I, Guttmann-Raviv N, Khoury HP, Kupershmit I, Ayoub N. Heat shock protein 90 (Hsp90) selectively regulates the stability of KDM4B/JMJD2B histone demethylase. J Biol Chem. 2013;288:14681–14687. doi: 10.1074/jbc.C113.462770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura A, Minehata K, Terashima M, Kondoh G, Hara T, Suzuki T. Jmjd5, an H3K36me2 histone demethylase, modulates embryonic cell proliferation through the regulation of Cdkn1a expression. Development. 2012;139:749–759. doi: 10.1242/dev.074138. [DOI] [PubMed] [Google Scholar]
- Iwamori N, Zhao M, Meistrich ML, Matzuk MM. The testis-enriched histone demethylase, KDM4D, regulates methylation of histone H3 lysine 9 during spermatogenesis in the mouse but is dispensable for fertility. Biol Reprod. 2011;84:1225–1234. doi: 10.1095/biolreprod.110.088955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junrong T, et al. Proteomic identification of CIB1 as a potential diagnostic factor in hepatocellular carcinoma. J Biosci. 2011;36:659–668. doi: 10.1007/s12038-011-9101-6. [DOI] [PubMed] [Google Scholar]
- Kierszenbaum AL, Rivkin E, Tres LL. Acroplaxome, an F-actin-keratin-containing plate, anchors the acrosome to the nucleus during shaping of the spermatid head. Mol Biol Cell. 2003;14:4628–4640. doi: 10.1091/mbc.E03-04-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum AL, Rivkin E, Tres LL. Cytoskeletal track selection during cargo transport in spermatids is relevant to male fertility. Spermatogenesis. 2011;1:221–230. doi: 10.4161/spmg.1.3.18018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum AL, Tres LL. The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Arch Histol Cytol. 2004;67:271–284. doi: 10.1679/aohc.67.271. [DOI] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Kotaja N, Kimmins S, Brancorsini S, Hentsch D, Vonesch J-L, Davidson I, Parvinen M, Sassone-Corsi P. Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat Methods. 2004;1:249–254. doi: 10.1038/nmeth1204-249. [DOI] [PubMed] [Google Scholar]
- Kuroki S, et al. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science. 2013;341:1106–1109. doi: 10.1126/science.1239864. [DOI] [PubMed] [Google Scholar]
- Li J, Buchner J. Structure, function and regulation of the Hsp90 machinery. Biomed J. 2013;36:106–117. doi: 10.4103/2319-4170.113230. [DOI] [PubMed] [Google Scholar]
- Liu J, DeFranco DB. Chromatin recycling of glucocorticoid receptors: implications for multiple roles of heat shock protein 90. Mol Endocrinol. 1999;13:355–365. doi: 10.1210/mend.13.3.0258. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhou S, Liao L, Chen X, Meistrich M, Xu J. Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J Biol Chem. 2010;285:2758–2770. doi: 10.1074/jbc.M109.066845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, Martín-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J. 2000;19:5971–5979. doi: 10.1093/emboj/19.22.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockman K, Taylor JM, Mack CP. The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ Res. 2007;101:e115–e123. doi: 10.1161/CIRCRESAHA.107.164178. [DOI] [PubMed] [Google Scholar]
- Loh Y-H, Zhang W, Chen X, George J, Ng H-H. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, Gudkov AV, Stark GR. Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Chiang C-HJ, Ponnusamy K, Ming G-L, Song H. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells. 2008;26:2131–2141. doi: 10.1634/stemcells.2008-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet E, Mottet D, Michel G, Roland I, Raes M, Remacle J, Michiels C. Hypoxia-induced activation of HIF-1: role of HIF-1α-Hsp90 interaction. FEBS Lett. 1999;460:251–256. doi: 10.1016/s0014-5793(99)01359-9. [DOI] [PubMed] [Google Scholar]
- Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- Okada Y, Tateishi K, Zhang Y. Histone demethylase JHDM2A is involved in male infertility and obesity. J Androl. 2010;31:75–78. doi: 10.2164/jandrol.109.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin BJ, Ervasti JM. The actin gene family: function follows isoform. Cytoskeleton (Hoboken) 2010;67:630–634. doi: 10.1002/cm.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard PJ, Loenarz C, Mole DR, McDonough Ma, Gleadle JM, Schofield CJ, Ratcliffe PJ. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1α. Biochem J. 2008;416:387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- Sar A, Ponjevic D, Nguyen M, Box AH, Demetrick DJ. Identification and characterization of demethylase JMJD1A as a gene upregulated in the human cellular response to hypoxia. Cell Tissue Res. 2009;337:223–234. doi: 10.1007/s00441-009-0805-y. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Souès S, Kann M-L, Fouquet J-P, Melki R. The cytosolic chaperonin CCT associates to cytoplasmic microtubular structures during mammalian spermiogenesis and to heterochromatin in germline and somatic cells. Exp Cell Res. 2003;288:363–373. doi: 10.1016/s0014-4827(03)00248-9. [DOI] [PubMed] [Google Scholar]
- Sternlicht H, Farr GW, Sternlicht ML, Driscoll JK, Willison K, Yaffe MB. The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc Natl Acad Sci USA. 1993;90:9422–9426. doi: 10.1073/pnas.90.20.9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9:1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Zhang Y. Purification of histone demethylases from HeLa cells. Methods. 2006;40:318–326. doi: 10.1016/j.ymeth.2006.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss AK, Thomas T, Gruss P. Efficiency assessment of the gene trap approach. Dev Dyn. 1998;212:171–180. doi: 10.1002/(SICI)1097-0177(199806)212:2<171::AID-AJA3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Webby CJ, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325:90–93. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- Wellmann S, Bettkober M, Zelmer A, Seeger K, Faigle M, Eltzschig HK, Bührer C. Hypoxia upregulates the histone demethylase JMJD1A via HIF-1. Biochem Biophys Res Commun. 2008;372:892–897. doi: 10.1016/j.bbrc.2008.05.150. [DOI] [PubMed] [Google Scholar]
- Willison K, Lewis V, Cordeli J, Dean C, Miller K, Lyon MF, Marsh M, Ord OX. The t complex polypeptide 1 (TCP-1) is associated with the cytoplasmic aspect of Golgi membranes. Cell. 1989;57:621–632. doi: 10.1016/0092-8674(89)90131-1. [DOI] [PubMed] [Google Scholar]
- Yamada D, et al. Role of the hypoxia-related gene, JMJD1A, in hepatocellular carcinoma: clinical impact on recurrence after hepatic resection. Ann Surg Oncol. 2012;19(Suppl 3):S355–S364. doi: 10.1245/s10434-011-1797-x. [DOI] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Yang J, Ledaki I, Turley H, Gatter KC, Montero J-CM, Li J-L, Harris AL. Role of hypoxia-inducible factors in epigenetic regulation via histone demethylases. Ann NY Acad Sci. 2009;1177:185–197. doi: 10.1111/j.1749-6632.2009.05027.x. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ioshii SO, Imanaka-Yoshida K, Izutsu K. Association of cytoplasmic dynein with manchette microtubules and spermatid nuclear envelope during spermiogenesis in rats. J Cell Sci. 1994;107:625–633. doi: 10.1242/jcs.107.3.625. [DOI] [PubMed] [Google Scholar]
- Yue L, Karr TL, Nathan DF, Swift H, Srinivasan S, Lindquist S. Genetic analysis of viable Hsp90 alleles reveals a critical role in Drosophila spermatogenesis. Genetics. 1999;151:1065–1079. doi: 10.1093/genetics/151.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Du Y-R, Qin W-H, Hu Y-G, Huang Y-N, Bao L, Han D, Mansouri A, Xu G-L. RIM-BP3 is a manchette-associated protein essential for spermiogenesis. Development. 2009;136:373–382. doi: 10.1242/dev.030858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.