Mouse cells deleted of all or different combinations of lamins are used to show that, when expressed at sufficiently high levels, lamin-A, -B1, and -B2 can each assemble into an evenly organized nuclear lamina and ensure the even distribution of nuclear pore complexes. By contrast, only lamin-A ensures the localization of emerin.
Abstract
The nuclear lamina (NL) consists of lamin polymers and proteins that bind to the polymers. Disruption of NL proteins such as lamin and emerin leads to developmental defects and human diseases. However, the expression of multiple lamins, including lamin-A/C, lamin-B1, and lamin-B2, in mammals has made it difficult to study the assembly and function of the NL. Consequently, it has been unclear whether different lamins depend on one another for proper NL assembly and which NL functions are shared by all lamins or are specific to one lamin. Using mouse cells deleted of all or different combinations of lamins, we demonstrate that the assembly of each lamin into the NL depends primarily on the lamin concentration present in the nucleus. When expressed at sufficiently high levels, each lamin alone can assemble into an evenly organized NL, which is in turn sufficient to ensure the even distribution of the nuclear pore complexes. By contrast, only lamin-A can ensure the localization of emerin within the NL. Thus, when investigating the role of the NL in development and disease, it is critical to determine the protein levels of relevant lamins and the intricate shared or specific lamin functions in the tissue of interest.
INTRODUCTION
As a major component of the nuclear lamina (NL), the type V intermediate filament proteins called lamins are implicated in a large number of functions (Dechat et al., 2008; Burke and Stewart, 2012; Butin-Israeli et al., 2012; Schreiber and Kennedy, 2013). Lamins are required for maintaining nuclear shape, and by forming a protein meshwork underneath the nuclear envelope, lamins are also believed to tether gene-poor regions of the chromatin to the nuclear periphery, which facilitates the formation of constitutive heterochromatin along with other proteins, such as HP1 (Worman et al., 1988; Ye and Worman, 1996; Ye et al., 1997; Solovei et al., 2013). In addition, lamins bind to other nuclear proteins, especially other NL proteins, to facilitate their nuclear retention and even distribution. Although most lamins assemble into the NL at the nuclear periphery, a small fraction of lamins can be found in the interior of the nucleus, where they may perform additional functions. Studies using tissue culture cells and in vitro assays suggest that lamins regulate basic cellular processes, including repressing gene expression (Reddy et al., 2008; Zullo et al., 2012), facilitating DNA replication (Meier et al., 1991; Spann et al., 1997; Shimi et al., 2008), and even promoting spindle morphogenesis (Tsai et al., 2006; Ma et al., 2009; Goodman et al., 2010). More recent studies using model organisms further show that lamins are required for proper building of at least some organs (Vergnes et al., 2004; Coffinier et al., 2010, 2011; Kim et al., 2011; Chen et al., 2013; Jung et al., 2013).
Despite the importance of lamins, understanding their molecular functions in a given cellular process is difficult due to the polymerized state of the proteins, the multitude of interactions with other NL proteins and chromatin, and the presence of more than one lamin gene in many organisms. Caenorhabditis elegans expresses only one lamin isoform, which is a B-type lamin. Thus studies in C. elegans have helped us to understand the role of the NL. However, the substantial amount of maternal supply in C. elegans has limited the use of null mutations to decipher definitively whether lamin-B is essential for various NL functions during embryogenesis (Liu et al., 2000; Haithcock et al., 2005).
The interest in understanding the NL and lamin's functions has increased dramatically since the discovery of various human diseases, referred to as laminopathies, associated with NL proteins (Butin-Israeli et al., 2012; Schreiber and Kennedy, 2013). Unfortunately, it has been difficult to understand the disease mechanism of laminopathies because all mammals have three lamin genes that express three major lamin proteins and several splicing variants in many cell types. For example, the three mouse genes Lmna, Lmnb1, and Lmnb2, express lamin-A or -C (often referred to as lamin-A/C and several other splicing variants), lamin-B1, and lamin-B2 (and a splicing variant lamin-B3), respectively. Lamin-A, -B1, and -B2 undergo differential modifications of their C-termini, which give rise to mature proteins that either lose (lamin-A) or retain a farnesyl group (B1 and B2; Corrigan et al., 2005; Adam et al., 2013; Jung et al., 2013). In addition, although all three lamins share the same tripartite domain structures of intermediate filament proteins, they are only ∼50% identical at the primary amino acid sequence level by pairwise comparisons. Thus the sequence conservation and variations in modification imply that lamins have both shared and distinct functions in mammals.
To understand how lamins and the NL perform various functions, it is critical to decipher how different lamins assemble into the NL and how lamins regulate the distribution of other NL proteins. Unfortunately, studies of the assembly of lamins into NL have yielded confusing and controversial results. The single B-type lamin in C. elegans is able to assemble into an evenly organized NL to regulate the distribution of other NL proteins (Liu et al., 2000). Similarly, in mammals, lamin-B1 and -B2 are able to properly assemble into a NL in many cells expressing no or very few A-type lamins (Vergnes et al., 2004; Coffinier et al., 2010, 2011; Kim et al., 2011; Eckersley-Maslin et al., 2013). Although these observations indicate that B-type lamins can assemble into a properly organized NL independent of A-type lamins, studies in HeLa cells suggest a hierarchical assembly (Shimi et al., 2008). Indeed, reduction of lamin-B1 causes dramatic disorganization of lamin-A/C and lamin-B2, which appears as loose lamin networks by immunofluorescence microscopy. By contrast, reducing either lamin-B2 or lamin-A/C has no obvious effect on the proper assembly of NL by the remaining two lamins (Shimi et al., 2008). This apparent dependence of lamin-A/C and lamin-B2 assembly on lamin-B1 does not, however, occur in several mouse cell types. For example, an even distribution of lamin-A/C is found around the nuclei of mouse keratinocytes deleted of both lamin-B1 and -B2 genes (Kim et al., 2011; Yang et al., 2011). Moreover, in a fraction of mouse embryonic fibroblasts (MEFs) expressing a truncated form of lamin-A/C, a mild lamin-B1 defect, manifested as small gaps of lamin-B1 immunostaining at the nuclear periphery, has been reported (Sullivan et al., 1999; Kubben et al., 2011). Thus, despite much effort, whether different lamins depend on one another to assemble into the NL and how such assembly regulates the localization and organization of other NL proteins has been unclear.
Recently we derived mouse embryonic stem cells (mESCs) deleted of all or different combinations of lamins. The mESCs harboring single, double, or triple lamin deletions exhibit euploidy and normal proliferation and can differentiate into fibroblast-like cells, beating cardiomyocytes, and neural progenitor cells in vitro (Kim et al., 2011, 2013). By using these mESCs and their differentiated cells, we report some general rules of lamin assembly and function. We discuss the implications of our findings in the context of organismal development and disease.
RESULTS
Lamin-B1 is required for organization of lamin-A/C and lamin-B2 in mESCs
Because mESCs deleted of different combinations of lamins can undergo self-renewal and differentiation (Kim et al., 2011, 2013), they offer an unprecedented opportunity to study whether lamins depend on one another for their assembly into the NL. In wild-type cells, A- and B-type lamins assemble into a dense meshwork, which appears as an evenly distributed layer underneath the nuclear envelope as revealed by conventional immunofluorescence microscopy. Previous studies showed that large-scale changes of lamin structures can be detected by immunofluorescence (Shimi et al., 2008; Jung et al., 2013). Thus we used immunostaining to analyze lamins and NL organization.
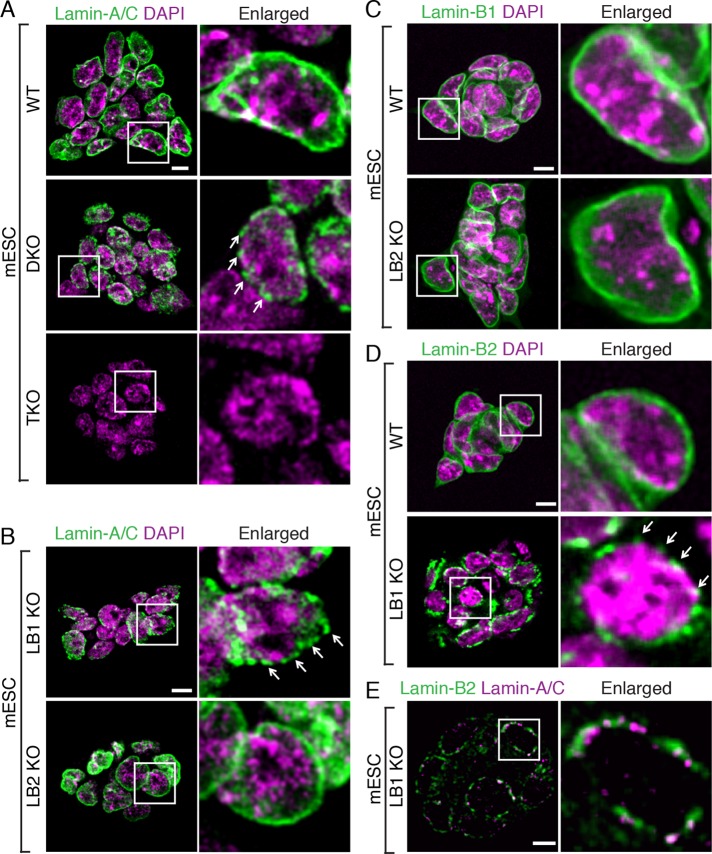
We first tested whether lamin-A/C could assemble into the NL in the absence of B-type lamins in mESCs. In wild-type mESCs, lamin-A/C was evenly distributed at the nuclear periphery, as expected (Figure 1A). In contrast, most (>95%) of Lmnb1−/−;Lmnb2−/− (lamin-B1 and -B2 double knockout [DKO]) mESCs had a discontinuous lamin-A/C distribution (Figure 1A, arrows). The lamin-A/C antibody was specific because no lamin-A/C could be detected by this antibody in the lamin-A/C, -B1, and -B2 triple-knockout (TKO; Lmna−/−;Lmnb1−/−;Lmnb2−/−) mESCs (Figure 1A; Kim et al., 2013). Of interest, lamin-A/C in >90% of the Lmnb1−/− (lamin-B1 KO) mESCs also appeared discontinuous (Figure 1B), whereas Lmnb2−/− (lamin-B2 KO) mESCs exhibited a normal-appearing lamin-A/C distribution (Figure 1B). Thus lamin-B1, but not lamin-B2, is sufficient for proper targeting of lamin-A/C into the NL of mESCs.
FIGURE 1:
Effect of lamin-B1 on assembly of lamin-A/C and lamin-B2 in mESCs. (A, B) Immunofluorescence images of lamin-A/C in wild-type (WT), Lmnb1−/−; Lmnb2−/− (DKO), Lmna−/−;Lmnb1−/−;Lmnb2−/− (TKO), Lmnb1−/− (LB1 KO), and Lmnb2−/− (LB2 KO) mESCs. Arrows indicate the discontinuous lamin-A/C staining along the nuclear periphery. (C) Lamin-B1 staining in WT and LB2 KO mESCs. (D) Lamin-B2 staining in WT and LB1 KO mESCs. Arrows indicate the discontinuous lamin-B2 staining along the nuclear periphery. (E) Double labeling of lamin-A/C and lamin-B2 in LB1 KO mESCs. Right, outlined nuclei enlarged. Scale bars, 5 μm.
We next tested whether lamin-B1 and -B2 regulate each other's distribution. In wild-type mESCs, both lamin-B1 and -B2 were similarly distributed at the nuclear rim (Figure 1, C and D). In lamin-B2 KO mESCs, lamin-B1 distribution was also normal (Figure 1C). However, in the nuclei of lamin-B1 KO mESCs, lamin-B2 appeared discontinuous (Figure 1D, arrows), similar to the distribution of lamin-A/C in the lamin-B DKO or lamin-B1 KO mESCs. Thus, in mESCs, lamin-B1 is not only able to assemble into the NL in the absence of lamin-B2 but is required for the proper association of lamin-A/C and lamin-B2 with the NL. Dual-color labeling shows that lamin-B2 and lamin-A/C exhibit limited colocalization in the absence of lamin-B1 (Figure 1E), suggesting that the two types of lamins assemble into separate but interacting structures, which is consistent with previous reports (Izumi et al., 2000; Shimi et al., 2008).
It is possible that lamin-B1 could influence the expression of lamin-B2 and lamin-A/C, thereby indirectly regulating their assembly and distribution. To test this, we examined the protein levels of lamin-A/C and lamin-B2 in mESCs by Western blotting analyses. We found that the amount of lamin-A/C was similar in wild-type, lamin-B single KO, and DKO mESCs (Supplemental Figure S1A). The lamin-B2 levels in wild-type and lamin-B1 KO mESCs were also similar (Supplemental Figure S1B). Thus lamin-B1 does not regulate the amount of lamin-A/C and lamin-B2 in mESCs.
Lamin-B1 is required for organization of lamin-A/C and lamin-B2 in differentiated cells
Differentiated cells such as fibroblasts are well spread, and as a result the nuclei are sufficiently flattened to observe the overall localization of lamins within the lamina by focusing on the nuclear surface. In contrast, mESCs are not well spread, making it difficult to visualize the lamina. Thus we treated the mESCs with retinoic acid for 7 d (Supplemental Figure S2A) following a published protocol (Suraneni et al., 2012) to differentiate the mESCs into embryonic stem cell–derived fibroblast-like cells (EDFCs) with flat and elongated morphologies (Supplemental Figure S2B). Consistent with our previous findings (Kim et al., 2013), wild-type, lamin-B DKO, and lamin TKO mESCs are all able to differentiate into EDFCs (Supplemental Figure S2B).
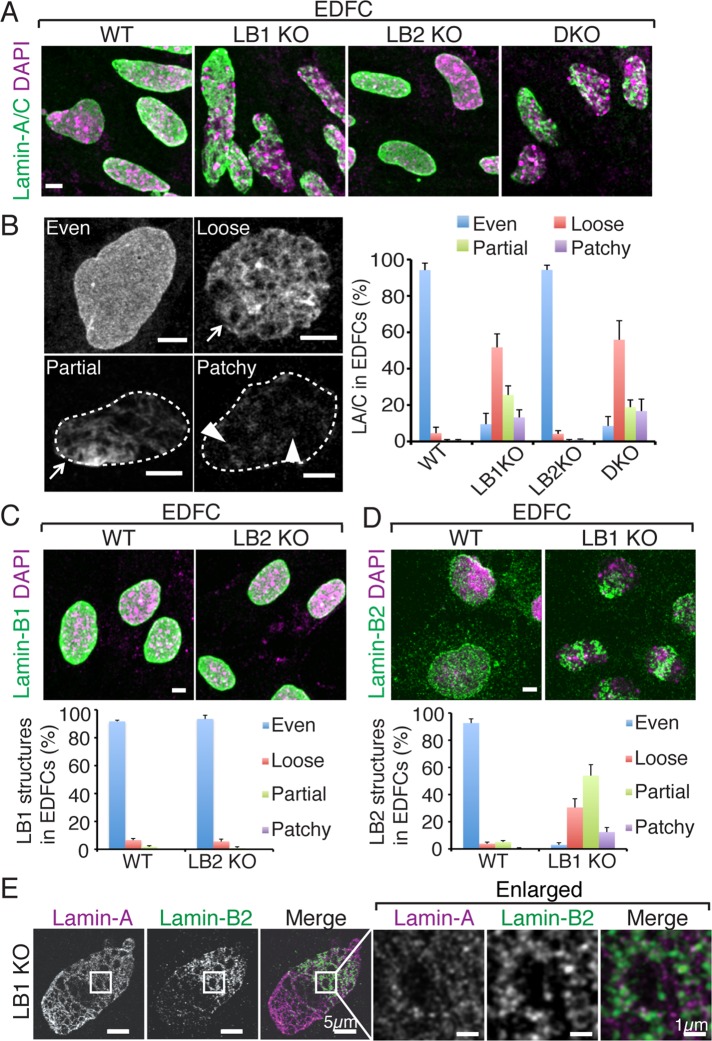
We found that most (>90%) of the wild-type or lamin-B2 KO EDFCs exhibited an even distribution of lamin-A/C on the surface of the nuclei (Figure 2, A and B). By contrast, most (>90%) of the lamin-B1 KO or lamin-B DKO EDFCs exhibited a disorganized lamin-A/C structure (Figure 2, A and B). These disorganized structures can be categorized as a loose meshwork throughout the NL, a partial network covering only a fraction of the nuclear surface, or small, dim patches over the nucleus (Figure 2B). The loose meshwork of lamin-A/C appeared as disorganized filamentous structures with large openings (Figure 2B, arrows), whereas the lamin-A/C patches appeared as short fibers or small aggregates (Figure 2B, arrowheads). These analyses suggest that lamin-A/C could still assemble into filamentous structures in the absence of lamin-B1, but the filaments failed to form a dense and even meshwork. The deletion of lamin-B1 did not affect the protein level of lamin-A/C in EDFCs (Supplemental Figure S3A). This suggests that lamin-B1 is required for proper assembly of lamin-A/C into the NL in EDFCs.
FIGURE 2:
Lamin-B1 regulates the assembly of lamin-A/C and lamin-B2 in EDFCs. (A) Immunofluorescence images of lamin-A/C in WT, LB1 KO, LB2 KO, and DKO EDFCs. (B) Examples of different lamin-A/C staining patterns in EDFCs categorized as evenly organized meshwork, loose meshwork, partial network, or patchy structures. Arrows indicate loose meshes of filamentous structures. Arrowheads indicate aggregates or short fibers. Dashed lines delineate the borders of the nuclei with partial and patchy lamin-A/C patterns. Right, different quantified and plotted lamin-A/C patterns. Error bars, SD (n = 3 experiments, with >200 cells counted in each experiment). (C, D) Lamin-B1 (C) or lamin-B2 (D) staining in wild-type or lamin-B single KO EDFCs, along with quantifications (plots at bottom), using the same methods as in B. (E) Double labeling of lamin-A/C and lamin-B2 in a LB1 KO EDFC. Scale bars, 5 μm, or as specified.
We next examined the organization of lamin-B1 and lamin-B2 in EDFCs. Lamin-B1 organization appeared normal in most (>90%) of the wild-type or lamin-B2 KO EDFCs (Figure 2C). Lamin-B2 was also distributed throughout the NL in the wild-type EDFCs, although the staining appeared punctate (Figure 2D). In lamin-B1 KO EDFCs, lamin-B2 in most nuclei (>90%) appeared as loose meshwork, partial network, or patches, similar to lamin-A/C in lamin-B1 KO EDFCs (Figure 2D). Previous studies showed that lamin-B2 has weaker interactions with itself and with other lamins (Schirmer, 2004) as compared with lamin-B1 and lamin-A/C. The punctate appearance of lamin-B2 throughout the NL in wild-type cells suggests that this lamin is expressed at a low level and is distributed by interactions with the other lamins (discussed further later). On removal of the abundant lamin-B1, lamin-B2 would have an increased chance to interact with itself and with lamin-A/C. If lamin-A/C is also expressed at low levels in these cells (discussed further later), this might explain why lamin-B2 appears as uneven filamentous networks/patches in lamin-B1 KO EDFCs as opposed to the punctate and even distribution in the NL of wild-type EDFCs. In addition, double labeling revealed that lamin-B2 and lamin-A/C filaments or patches partially coaligned in lamin-B1 KO EDFCs (Figure 2E). This suggests that lamin-B2 and lamin-A/C structures interact with each other, which is consistent with previous biochemical (Schirmer, 2004) and imaging analyses (Delbarre et al., 2006; Shimi et al., 2008). Similar to lamin-A/C, the lamin-B2 protein level was not affected upon lamin-B1 deletion in EDFCs (Supplemental Figure S3B). Taken together, our findings suggest that lamin-B1 is required for proper assembly of lamin-A/C and lamin-B2 in the NL in EDFCs.
Lamin-A/C can assemble into the NL in mouse embryonic fibroblasts deleted of B-type lamins
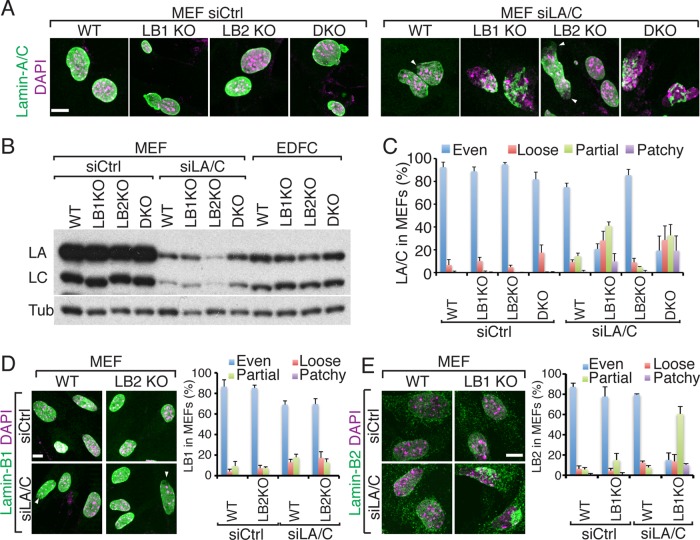
Although the foregoing studies suggest that lamin-B1 is required for organizing lamin-A/C in the NL in mESCs and EDFCs, lamin-A/C appears normal in MEFs whether they are derived from wild-type, lamin-B single KO, or DKO littermates (Figure 3A). Of interest, these MEFs expressed more lamin-A/C than did EDFCs (Figure 3B). This suggests that the assembly of lamin-A/C is dependent on its protein level. Consistent with this idea, lamin-B DKO EDFCs containing the highest amount of lamin-A/C as measured by immunofluorescence intensity (see Materials and Methods) formed mostly a loose meshwork, whereas those containing intermediate or low levels of lamin-A/C formed mainly partial networks or patches, respectively (Supplemental Figure S4A). We note that with use of structured illumination microscopy (SIM; Schermelleh et al., 2008), which increases the resolution of the conventional confocal microscope, the evenly distributed lamin-A/C meshwork visualized by confocal microscopy appeared less dense in lamin-B1 KO MEFs than in wild-type cells (Supplemental Figure S4B). This suggests that lamin-B1 may affect the packing of lamin-A/C filaments.
FIGURE 3:
Concentration-dependent organization of lamin-A/C in MEFs deleted of B-type lamins. (A) Immunofluorescence images of lamin-A/C in MEFs treated with control (siCtrl) or lamin-A/C (siLA/C) siRNAs. Arrowheads indicate gaps in the lamin-A/C staining. (B) Western blotting analyses of lamin-A (LA) and lamin-C (LC) in MEFs and EDFCs. Tubulin (Tub), loading control. (C) Quantification of lamin-A/C morphology in MEFs. Error bars, SD (n = 3 experiments, with >200 cells counted in each experiment). (D, E) Images of lamin-B1 (D) and lamin-B2 (E) localization in MEFs. Right, quantifications of lamin morphologies. Arrowheads in D indicate small lamin-B1–staining gaps. Scale bars, 10 μm.
If the assembly and distribution of lamin-A/C throughout the NL in lamin-B DKO MEFs is due to the high level of A-type lamins, reduction of the proteins by RNA interference (RNAi) in these cells should dramatically disrupt lamin-A/C organization. We found that when the amount of lamin-A/C in lamin-B1 KO and lamin-B DKO MEFs was reduced below the level found in EDFCs by small interfering RNA (siRNA; Figure 3B), loose/partial meshworks or patchy lamin-A/C structures appeared (Figure 3, A–C). However, reduction of lamin-A/C in wild-type or lamin-B2 KO MEFs did not affect lamin-A/C distribution in most (∼75%) of the cells analyzed (Figure 3A). In the remaining ∼25% of the cells, the lamin-A/C structures appeared mostly normal, with only small areas of the nuclei covered by defective lamin-A/C patches (Figure 3A, arrowheads).
We also examined lamin-B1 and lamin-B2 organization in MEFs. Most (>85%) of the wild-type or lamin-B2 KO MEFs contained evenly distributed lamin-B1 staining, as expected (Figure 3D). When the amount of lamin-A/C was reduced by RNAi in these cells, however, ∼70% contained normal-appearing lamin-B1 (Figure 3D), whereas the remaining ∼30% of the cells contained nuclei with small areas (<30%) of the NL containing abnormal lamin-B1 structures (Figure 3D, arrowheads). In contrast to the defective lamin-B2 structures formed in lamin-B1 KO EDFCs (Figure 2D), most (>80%) of lamin-B1 KO MEFs contained nuclear lamin-B2 distributed throughout the NL compared with controls (Figure 3E). However, reduction of lamin-A/C by RNAi in lamin-B1 KO MEFs resulted in the formation of loose/partial or patchy lamin-B2 structures in the NL of most nuclei (Figure 3E), whereas a similar reduction of lamin-A/C in wild-type MEFs did not disrupt lamin-B2 organization (Figure 3E). The lamin-B2 protein level was not affected by lamin-B1 deletion or lamin-A/C knockdown (Supplemental Figure S4C). Taken together, these analyses suggest that the distribution of a given lamin within the NL depends on the total lamin level present in the nucleus.
A single lamin can assemble throughout the NL when expressed at a high level
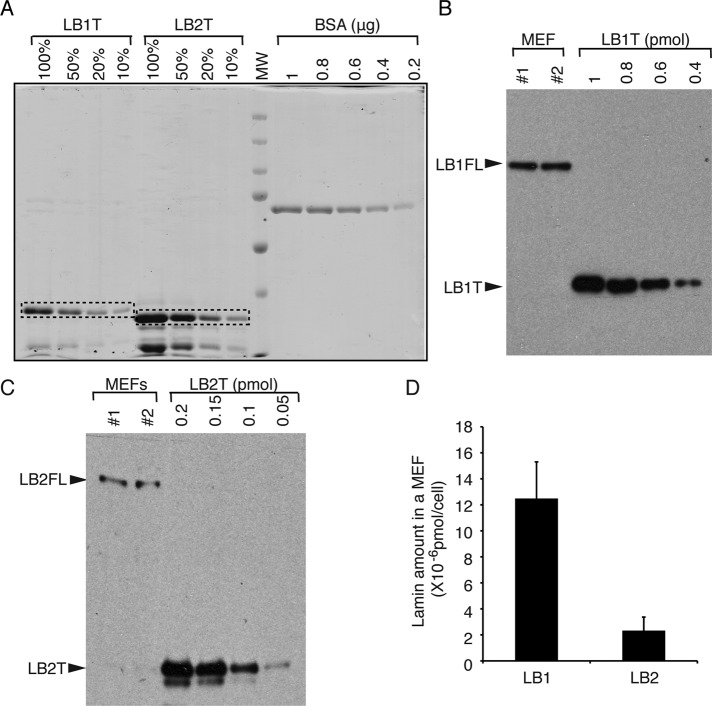
If the distribution of an individual lamin within the NL depends on the total lamin level, our prior observations would suggest that lamin-B1 is present at higher levels than lamin-B2 in MEFs. To measure the amount of lamin-B1 and lamin-B2 in cells, we used purified C-terminal fragments of lamin-B1 (amino acids [aa] 388–588) and lamin-B2 (aa 383–596) as standards (Figure 4A) to quantify each protein in cells using antibodies recognizing sequences within the C-terminal regions of each lamin (Figure 4, B and C). We found that one wild-type MEF contained (1.2 ± 0.3) × 10−5 pmol of lamin-B1 and (2 ± 1) × 10−6 pmol of lamin-B2 (mean ± SD; N = 3; Figure 4D). The approximately sixfold higher amount of lamin-B1 than lamin-B2 could explain why proper distribution of lamin-B2 depends on both lamin-B1 and lamin-A/C in MEFs. We found lamin-B2 levels to be similar in EDFCs and MEFs (Supplemental Figure S4D). Thus the low lamin-B2 concentration may explain why it appears punctate in wild-type EDFCs and MEFs (Figures 2D and 3E).
FIGURE 4:
Lamin-B1 is expressed at a higher level than lamin-B2 in MEFs. (A) Coomassie blue staining of the purified LB1T, LB2T, and BSA. LB1T and LB2T (dashed boxes) were quantified using BSA as the standard. (B, C) Lamin-B1 (B) and lamin-B2 (C) in the whole-cell lysates of 5 × 104 MEFs derived from two wild-type embryos (1 and 2) were quantified by Western blotting analyses using LB1T and LB2T as standards, respectively. Arrowheads indicate full-length (FL) LB, LB1T, or LB2T. (D) Amount of lamin-B1 and lamin-B2 in one MEF. Error bars, SD (n = 3 experiments).
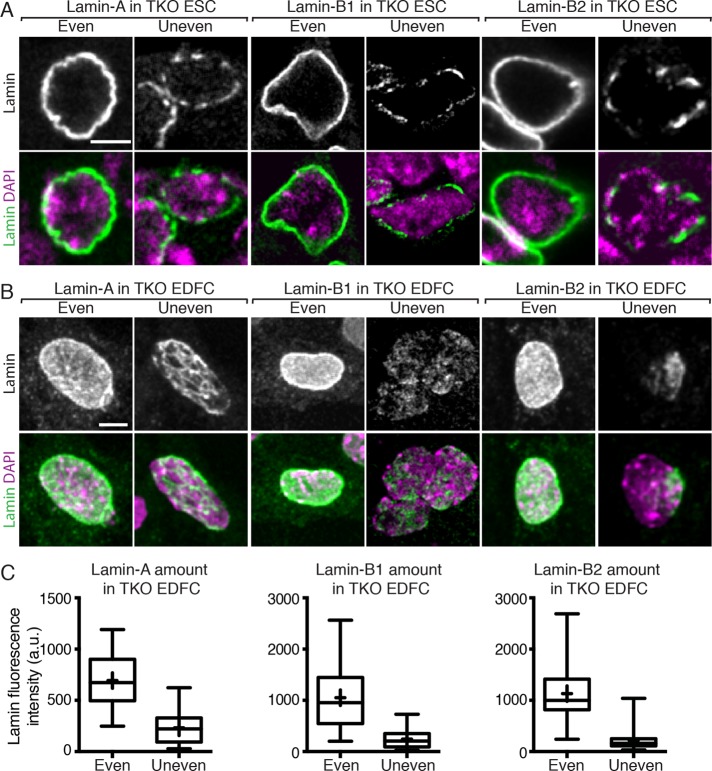
To directly test the idea that each lamin can assemble into an evenly organized NL when present at high levels, we transiently expressed untagged lamin-A, -B1, or -B2 in lamin TKO mESCs or EDFCs. The expression level of each exogenous lamin varied among individual cells after transfection followed by fixation and staining, which allowed us to correlate the distribution patterns of each lamin with its relative expression levels as measured by fluorescence intensity. We found that lamin TKO cells expressing high or low amounts of lamin-A, -B1, or -B2 exhibited an even or an uneven NL distribution, respectively (Figure 5, A–C). Lamin-B2, when expressed at high levels in TKO EDFCs, appeared as an even and continuous layer around the nucleus similar to that of lamin-B1 and lamin-A. This is consistent with our idea that the punctate lamin-B2 staining in wild-type EDFCs and MEFs is caused by the low protein concentration in these cells. Thus lamin-A, -B1, or -B2 can each assemble into an evenly organized NL when it is expressed at sufficient levels. This result also suggests that in cells expressing multiple lamins, the most highly expressed lamin facilitates the organization of the lamins that are expressed at low levels.
FIGURE 5:
Lamin-A, -B1, or -B2 alone can assemble into an even NL when expressed at a high level. Immunofluorescence images of untagged lamin-A, -B1, or -B2 expressed in TKO mESCs (A) or EDFCs (B). To visualize lamin distribution in cells expressing a low level of the protein, the brightness and contrast of the images were adjusted, which revealed the uneven structures. (C) The immunofluorescence intensity of each lamin in the nuclei of TKO EDFCs with even or uneven staining was quantified and is shown by box-and-whisker plots (a.u., arbitrary units). The upper and lower whiskers represent maximal and minimal values, respectively. The upper and lower borders of the box indicate the 75th and 25th percentiles, respectively. The line within the box represents the median. The cross indicates the mean value. More than 80 cells were analyzed in each condition. Scale bars, 5 μm.
Lamins function redundantly to ensure the even distribution of the nuclear pore complexes in differentiated cells
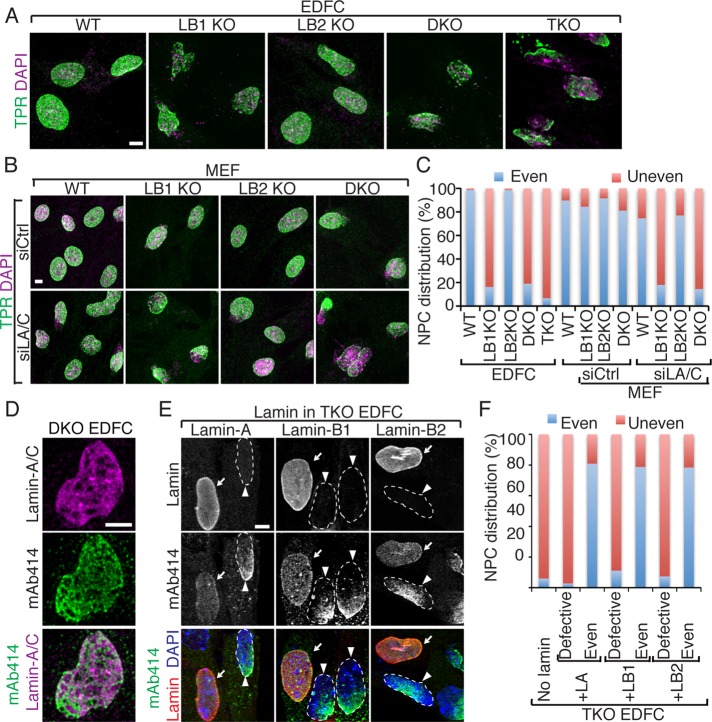
Both A- and B-type lamins can bind to nucleoporin (Nup) 153 (Al-Haboubi et al., 2011) and regulate the distribution of the nuclear pore complexes (NPCs; Sullivan et al., 1999; Kubben et al., 2011; Chen et al., 2013; Jung et al., 2013). Using antibodies recognizing TPR, a Nup found in the nuclear basket of the NPC (Frosst et al., 2002), we found that the distribution of NPCs appeared similar in wild-type, lamin-B single KO and DKO, and lamin TKO mESCs (Supplemental Figure S5A). We obtained similar results by using the monoclonal antibody 414 (mAb414) known to recognize four nucleoporins, Nup62, Nup153, Nup214, and Nup358 (Supplemental Figure S5B). This shows that lamins are not required for NPC distribution in mESCs. In contrast, most of the nuclei in lamin-B1 KO or lamin-B DKO EDFCs exhibited an uneven distribution of NPCs throughout the nuclear envelope, whereas the NPC distribution in lamin-B2 KO EDFCs appeared similar to the wild-type cells (Figure 6, A and C). Reduction of lamin-A/C by RNAi in wild-type, lamin-B single KO, or DKO MEFs revealed that both lamin-A/C and lamin-B1 could maintain the even NPC distribution in MEFs (Figure 6, B and C), whereas lamin-B2 was dispensable in all cells (Figure 6, A–C). Consistent with these results, the uneven distribution of NPCs appeared more severe in lamin TKO EDFCs than in lamin-B1 KO or lamin-B DKO EDFCs (Figure 6A). Dual-color labeling of NPCs and lamin-A/C in lamin-B DKO EDFCs showed that the defective lamin-A/C network coaligned with NPCs (Figure 6D).
FIGURE 6:
Lamins have a shared function in regulating NPC distribution in differentiated cells. (A, B) Distribution of NPCs in EDFCs (A) and MEFs (B) as revealed by immunofluorescence staining of TPR, a nucleoporin in the nuclear basket of NPCs. (C) Even and uneven NPC distribution were quantified and plotted, and >200 cells were analyzed in each of two experiments. (D) Double labeling of NPC (by mAb414) and lamin-A/C in the lamin-B DKO EDFC. (E) Double labeling of NPC (mAb414) and the indicated lamin expressed in TKO EDFCs. Arrows and arrowheads indicate cells with even or uneven distribution of NPCs, respectively. Dashed lines delineate the borders of nuclei in nontransfected cells. (F) Quantification of NPC distribution in TKO EDFCs that express either no lamin or the indicated lamin that exhibited defective or even distribution in the nucleus (>40 cells were quantified in each of two experiments). Scale bars, 5 μm.
Because EDFCs and MEFs containing abnormal lamin structures also exhibited altered distributions of NPCs, we asked whether each lamin, when expressed at high levels and forming an even NL, could ensure the even distribution of NPCs in differentiated cells. We found that the uneven distribution of NPCs in lamin TKO EDFCs was rescued when a single A- or B-type lamin was transiently expressed and assembled into a normal-appearing NL, whereas EDFCs expressing a given lamin at low levels and exhibiting a relatively disorganized NL still contained unevenly distributed NPCs (Figure 6F). Taken together, our findings suggest that lamin-A, -B1, or -B2 alone, when expressed at sufficient levels, can ensure the even distribution of NPCs in differentiated cells.
A specific requirement for lamin-A/C in retention and distribution of emerin in the nuclear envelope
To further study how different lamins function in organizing other nuclear peripheral proteins, we examined emerin, an inner nuclear membrane protein in the NL known to interact with lamin-A/C and lamin-B1 (Vaughan et al., 2001). Previous studies showed that lamin-A/C is required for the tethering of emerin to the nuclear envelope in various somatic tissue culture cell lines (Vaughan et al., 2001; Chen et al., 2012). Because our findings show that a single lamin, when expressed at sufficient levels to form an evenly organized NL, can support the proper distribution of NPC in lamin TKO cells, the seemingly lamin-A/C–specific function in emerin localization could be due to the insufficient amount of B-type lamins in cells previously examined. The various lamin-knockout cells that we generated allow us to test properly the role of each lamin in emerin localization.
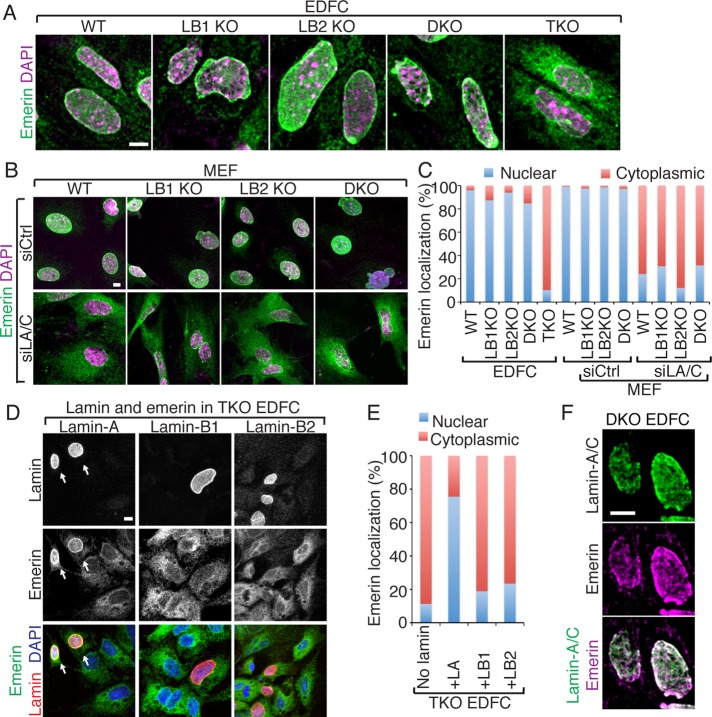
We found that emerin localization was similar in wild-type, lamin-B single KO and DKO, and lamin TKO mESCs (Supplemental Figure S5C). However, emerin was mislocalized into the cytoplasm in most (>90%) lamin TKO EDFCs (Figure 7, A and C), whereas most nuclei retained emerin in wild-type, lamin-B1 KO, lamin-B2 KO, and lamin-B DKO EDFCs (Figure 7, A and C). Because emerin expression levels were similar in all EDFCs (Supplemental Figure S3A), the mislocalization of emerin in the lamin TKO EDFCs was not due to the reduction of the protein. This suggests that lamin-A/C directly tethers emerin to the NL.
FIGURE 7:
A specific requirement for lamin-A in the nuclear retention and distribution of emerin. (A, B) Emerin distribution in EDFCs (A) and MEFs (B) as revealed by immunofluorescence microscopy. (C) Quantification of emerin localization based on whether emerin is solely nuclear or both nuclear and cytoplasmic in indicated cells and treatments. More than 200 cells were analyzed in each of two experiments. (D) Double labeling of emerin and the indicated lamin expressed in TKO EDFCs. Arrows indicate the nuclear retention of emerin in cells with even lamin-A distribution. (E) Quantification of emerin distribution in TKO EDFCs that expressed the indicated lamin (>50 cells were analyzed in each of two experiments). (F) Double labeling of emerin and lamin-A/C in the DKO EDFCs. The colocalized lamin-A/C (green) and emerin (magenta) appears white. Scale bars, 5 μm.
Next we studied emerin localization in wild-type and mutant MEFs. Although emerin exhibited normal nuclear localization in wild-type and lamin-B single or double KO MEFs, reduction of lamin-A/C by RNAi in these MEFs resulted in similar levels of emerin mislocalization into the cytoplasm (Figure 7, B and C). To demonstrate directly the dependence of emerin's NL-retention on lamin-A, we transiently expressed lamin-A, -B1, or lamin-B2 in the lamin TKO EDFCs and examined emerin localization. We found that only the expression of a sufficient amount of lamin-A, which supported the formation of a smooth NL, could relocate emerin from the cytoplasm to the NL in the TKO EDFCs (Figure 7, D and E). Consistent with the idea that lamin-A/C regulates emerin nuclear retention and organization, in lamin-B1 KO or lamin-B DKO EDFCs in which lamin-A/C was disorganized in the NL, emerin colocalized with the disorganized lamin-A/C structures (Figure 7F). These analyses demonstrate a distinct role of lamin-A in tethering emerin in the NL in differentiated cells.
DISCUSSION
With the creation and study of mESCs lacking all lamins, it has become clear that lamins are not essential for the basic proliferation and initial lineage differentiation of mESCs (Kim et al., 2011, 2013). Studies of lamins in the context of development and disease, on the other hand, have shown that these proteins are required for proper building and maintenance of at least some tissues and organs. How a given lamin is required for organ development and maintenance has, however, remained unclear. For example, lamin-B1 and -B2 single KO or DKO mice exhibit pronounced defects in the development of the forebrain (Vergnes et al., 2004; Coffinier et al., 2010, 2011; Kim et al., 2011). On the other hand, lamin-B DKO mice appear to have normal skin development and function (Kim et al., 2011; Yang et al., 2011). Further studies show that deletion of individual or both B-type lamins causes defects in tissues such as lung, bone, and diaphragm (Vergnes et al., 2004; Kim et al., 2011). In addition, recent studies in Drosophila, which expresses one lamin-B and one lamin-C (the Drosophila A-type lamin), show that lamin-B is specifically required in cyst stem cells to ensure proper organogenesis of the testis (Chen et al., 2013). Thus, to further dissect the developmental functions of lamins, it is critical to understand why different tissues exhibit different requirements for a specific lamin protein. Our findings reported here offer critical insights regarding the study of lamins in the context of development and disease and its interpretation.
We show that when expressed at a high level, a single A- or B-type lamin alone can assemble throughout the NL, which in turn is sufficient to ensure even distribution of NPCs throughout the nuclear envelope. Therefore, when interpreting lamin mutant phenotypes, it is critical to consider both the expression level of each lamin in the affected cells and the shared functions of lamins. For example, deletion of lamin-B1 causes a more severe brain defect than that of lamin-B2 in mice (Vergnes et al., 2004; Coffinier et al., 2010; Kim et al., 2011), which might suggest that lamin-B1 has more important functions in the brain than lamin-B2. However, our studies reported here would caution against such an interpretation, because if lamin-B1 is expressed at higher levels than lamin-B2 in the brain, one would expect to see phenotypic differences between the two lamin-B deletion mice. In addition, expression of A-type lamins during mouse embryogenesis is quite low in a number of tissues compared with B-type lamins (Houliston et al., 1988; Röber et al., 1989; Swift et al., 2013). This could explain why lamin-A/C KO mice die days after birth, whereas KO of each B-type lamin leads to lethality at birth (Sullivan et al., 1999; Vergnes et al., 2004; Coffinier et al., 2010; Kim et al., 2011; Kubben et al., 2011).
Lamin-B is essential in the Drosophila cyst stem cell lineage for testis development by regulating the localization of Nup153, which in turn ensures nuclear EGF signaling in these cells (Chen et al., 2013). Because our findings here show that when expressed at high levels individual A- or B-type lamins have shared functions in regulating NPCs, the seemingly tissue-specific function of lamin-B in the cyst stem cell lineage could be due simply to expression of lamin-C (the A-type lamin in Drosophila) at too low level to perform the function. Consistent with this interpretation, forced expression of lamin-C in the lamin-B–depleted cyst stem cell lineage results in a significant rescue of testis development (Chen et al., 2013).
Our findings also demonstrate that lamin-A, but not the B-type lamins, is specifically required for the proper distribution of emerin. Therefore, in efforts to dissect tissue-specific functions of lamins, it is critical to understand whether a particular lamin has unique functions in the affected tissue that is not shared by other lamins. By using the lamin KO combinations in ESCs and differentiated cells described in this study, it should be possible to define systematically the shared and specific functions of each lamin in a cell type–specific manner. This effort should greatly facilitate the study of how the lamins and the NL function in development and disease.
Lamins ensure the proper distribution of emerin and NPCs in differentiated cells but not in ESCs. The lamin-A–independent localization of emerin in ESCs may be due to the small amount of cytoplasm in ESCs compared with differentiated cells. Consequently, the small amount of cytoplasmic emerin in lamin-A–null ESCs may not cause an obvious change of nuclear emerin concentration. Alternatively, emerin may be retained in the ESC nucleus via other nuclear proteins. The reason why lamins are not required for proper distribution of NPCs may be more complicated. One factor that could influence the distribution of NPCs is the cytoskeletal force applied on nuclear pores. Lamins may play a role to counteract this force in light of their roles in nuclear mechanics (Lammerding et al., 2006; Swift et al., 2013; Zwerger et al., 2013). The limited cytoplasm in ESCs may lead to limited cytoskeleton forces acting on the nuclear membranes and NPCs, which would make lamins nonessential in NPC distribution. Alternatively, ESCs may use nuclear proteins other than lamins to counteract cytoskeletal forces exerted on the membranes and NPCs.
Because lamins are relatively insoluble and it has been very difficult to visualize lamin polymer structures in nuclei other than those derived from Xenopus oocytes (Aebi et al., 1986), how lamins assemble into the NL has been poorly understood. Our derivation of the lamin TKO ESCs and our finding that different amounts of lamin expression lead to different organization of filamentous structures should enable detailed studies of lamin assembly and their structure within the NL. For example, by developing inducible lamin expression in TKO cells, it will be possible to control the concentrations of lamins in cells and use live imaging to visualize the nucleation and growth of lamin filaments at different lamin concentrations. Using this tool, we will also be able to study the interaction of different lamins with one another during assembly and how mutant lamins disrupt the assembly and proper targeting of wild-type lamins and other NL-associated proteins. Note that our findings are based on epifluorescence microscopy and confocal microscopy, which can visualize large-scale structural changes of lamin assembly (Shimi et al., 2008; Jung et al., 2013). At this level of resolution, we show that each lamin, when expressed at sufficiently high levels, can form an evenly distributed NL. However, it is likely, given the limits of resolution of conventional light microscopy, that individual or different combinations of lamin(s) form distinct ultrastructures, which have different biological functions. Therefore it will be important to extend the use of superresolution microscopy and ultimately electron microscopy to further dissect lamin structure, using the cell types used in this study.
The lack of understanding of lamin assembly has limited our knowledge about how the NL participates in both the formation and regulation of the organization of constitutive heterochromatin. By combining the ability to induce the expression of different amounts of a specific lamin in TKO cells with the mapping of lamin–chromatin binding sites, it should be possible to study whether different lamins prefer different regions of chromatin and whether there are preferred genomic sites for lamin binding. These studies should open the door to further interrogate how other chromatin regulators or NL proteins work with lamins to promote the formation and maintenance of the heterochromatin.
MATERIALS AND METHODS
Cell derivation, culture, and differentiation
The derivation and culture of mESCs were described previously (Kim et al., 2011, 2013). EDFCs were derived from mESCs using a published protocol (Suraneni et al., 2012) with modifications. Briefly, mESCs were separated from feeder cells as described (Suraneni et al., 2012) and cultured in 2i medium (Ying et al., 2008) for one or two passages. Then mESCs were plated at a density of 125–250 cells/mm2 in gelatin-coated plates with differentiation medium (Knockout DMEM; Invitrogen, Carlsbad, CA), 15% fetal bovine serum (Invitrogen), 100 μM β-mercaptoethanol (Invitrogen), 2 mM l-glutamine (Glutamax; Invitrogen), 0.1 mM nonessential amino acids (Invitrogen), 50 U/ml penicillin/streptomycin (Invitrogen), and 0.4 μM retinoic acid (Sigma–Aldrich, St. Louis, MO). Cells were then allowed to differentiate for 7 d to generate EDFCs.
Primary MEFs were derived from embryonic day 13.5 mouse embryos as previously described (Okita et al., 2010) with minor modifications. Briefly, the internal organs were first removed from dissected embryos. The remaining bodies were minced and dissociated using 0.25% trypsin (Invitrogen). Then MEFs were collected and cultured in the MEF medium (the differentiation medium lacking retinoic acid). MEFs at passage 1 were frozen as stocks. MEFs at passages 2 and 3 were used in all experiments.
RNA interference
The sequence of Silencer Select lamin-A/C–specific siRNA (Invitrogen) is 5′-GGCUUGUGGAGAUCGAUAAtt-3’. For RNAi, MEFs were transfected using the RNAiMAX transfection reagent (Invitrogen) following the manufacturer's instructions. At 72 h after transfection, MEFs were collected for immunofluorescence and Western blotting analyses.
Immunofluorescence microscopy
Primary antibodies used for immunofluorescence microscopy are monoclonal mouse anti–lamin-A/C (1:1000; Active Motif, Carlsbad, CA), polyclonal rabbit anti–lamin-A (1:200; H-102; Santa Cruz Biotechnology, Dallas, TX), polyclonal rabbit anti–lamin B1 (1:1000; ab16048; Abcam, Cambridge, UK), mouse monoclonal anti–lamin-B2 (1:200; E3; Invitrogen), affinity-purified polyclonal chicken anti–lamin-B2 (1:1000; generated in-house; Kim et al., 2011), polyclonal rabbit anti-TPR (1:250; ab84516; Abcam), mouse monoclonal antibody 414 (mAb414 at 1:1000; Abcam), and polyclonal rabbit anti-emerin (1:200; FL-254; Santa Cruz Biotechnology) antibodies. Secondary antibodies used are Alexa 488 goat anti-mouse and Alexa 568 goat anti-rabbit antibodies (Invitrogen).
For all immunofluorescence experiments, cells were grown on gelatin-coated coverslips and then fixed by 4% paraformaldehyde (Sigma-Aldrich) for 10 min, followed by permeabilization using 0.1% Triton-100 (Sigma-Aldrich) in phosphate-buffered saline (PBS) for 10 min and then blocking by 4% bovine serum albumin (BSA) in PBS for 1 h. The incubation of cells with primary antibodies was done overnight at 4°C. Then cells were washed with 4% BSA in PBS, followed by incubation with secondary antibodies for 1 h at room temperature. Cells were washed with PBS, stained with 4′,6-diamidino-2-phenylindole, and mounted with Immu-Mount (Thermo Scientific, Waltham, MA).
Confocal images were acquired using a scanning confocal microscope (Leica SP5) with a 63×/1.4 objective and an electron-multiplying charge-coupled device camera. Images were processed using ImageJ (National Institutes of Health, Bethesda, MD). For quantification, a Nikon Eclipse E800 epifluorescence microscope with a PL APO 63×/1.4 oil objective was used to take images.
To measure lamin amounts in terms of immunofluorescence intensity, images were taken with the same exposure time. The border of each nucleus was manually drawn, and the average lamin fluorescence intensity of the nuclear region was measured by MetaMorph's Integrated Morphometry Analysis. The fluorescence intensity of an empty space near each cell was measured as background and subtracted from the lamin signal. Box-and-whisker plots were drawn using Prism software. Other quantifications were plotted using Excel (Microsoft, Redmond, WA).
Structured illumination microscopy
To prepare samples for superresolution structured illumination microscopy, cells were fixed in 3% paraformaldehyde for 10 min and permeabilized in 0.1% Triton X-100 in PBS 3 × 3 min on ice. The cells were then incubated in rabbit anti–lamin-A antibody (Dechat et al., 2007) diluted 1:3000 in PBS containing 5% normal goat serum in a humidified atmosphere at room temperature for 30 min. Then the cells were incubated in goat anti-rabbit Alexa 488 secondary antibody (Molecular Probes) diluted 1:400 in PBS for 30 min at room temperature. Images were taken using a Nikon SIM.
Western blotting analyses
Whole-cell lysates were generated using RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% deoxycholic acid, 10 μg/ml DNase I) and diluted in SDS–PAGE sample buffer. Cell lysates were separated in 10% SDS–PAGE and transferred onto nitrocellulose membranes. The membranes were blocked with 5% milk and probed with the following antibodies: polyclonal rabbit anti–lamin-B1 (1:5000; ab16048; Abcam), polyclonal chicken anti–lamin-B2 (1:10,000; Kim et al., 2011), monoclonal mouse anti–lamin-A/C (1:5000; Active Motif), polyclonal rabbit anti-emerin (1:2000; FL-254; Santa Cruz Biotechnology), or mouse monoclonal anti-tubulin (1:5000; DM1a; Sigma-Aldrich). Antibodies were detected with horseradish peroxidase–conjugated anti-mouse (1:5000) or anti-rabbit (1:5000) antibodies (Thermo Scientific) and West Pico Chemiluminescent Substrate (Thermo Scientific).
Expression of individual lamins in TKO mESCs and EDFCs
To express lamin-A, -B1, and -B2 in mESCs and EDFCs, the coding sequences for mouse Lmnb1, Lmnb2, and Lmna were PCR amplified from IMAGE clones 6816118, 5696459, and 4240057, respectively. The PCR products were cloned into pPyCAGIP (Chambers et al., 2003). The resulting constructs were confirmed by DNA sequencing (the full DNA sequence information is available upon request). The plasmids were transfected using Lipofectamine 2000 (Invitrogen) into lamin TKO mESCs grown in the 2i medium (Ying et al., 2008) or the TKO EDFCs after 3 d of differentiation. At 48–72 h after transfection, cells were collected for further analyses.
Quantification of lamin protein amounts in MEFs
Because the lamin-B1 and -B2 antibodies used in this study recognize epitopes in the C-terminus of each protein, we expressed the lamin-B C-termini in bacteria as hexahistidine fusion proteins and purified these proteins for quantitative Western blotting analyses. The coding sequences corresponding to aa 388–588 of mouse lamin-B1 (LB1T) and aa 383–596 of mouse lamin-B2 (LB2T) were PCR amplified and cloned into the EcoRI and SalI sites of the pET28a vector. The resulting constructs, pET28a-LB1T and pET28a-LB2T, were confirmed by DNA sequencing (the full DNA sequence information is available upon request). Histidine-LB1T and -LB2T proteins were expressed in BL21-CodonPlus bacteria and purified using nickel–nitrilotriacetic acid agarose beads (Qiagen) following the manufacturer's instructions. To determine the concentration of the lamin-B protein standards, different amounts of LB1T, LB2T, and BSA (Sigma-Aldrich) were separated by 10% SDS–PAGE and stained with Coomassie blue (Figure 4A). The intensity of BSA bands was quantified using Photoshop to generate a standard curve, which was then used to determine the concentration of LB1T and LB2T. To quantify lamin proteins in MEFs, cell lysates were generated using RIPA buffer at a density of 5 × 104 cells/μl and diluted in SDS sample buffer. Different amounts of cell lysates and standard proteins were analyzed in the same Western blot. Standard curves were generated using LB1T or LB2T, which were then used to quantify the respective lamin-B amounts from cell lysates on the same blot. Lamin-B1 and -B2 were detected using polyclonal rabbit anti–lamin-B1 (1:5000; ab16048; Abcam) and polyclonal chicken anti-lamin-B2 (1:10,000; Kim et al., 2011) antibodies, respectively.
Supplementary Material
Acknowledgments
We thank Ona Martin for technical assistance, Anne Goldman for helping with SIM, all members of the Zheng lab, and colleagues in the Embryology Department of the Carnegie Institution and the Biology Department of Johns Hopkins University for discussions. This work was supported by a Senior Scholar Award to Y.Z. from the Ellison Medical Foundation and National Institutes of Health Grants GM056312 (Y.Z.), GM106023 (a shared grant to Y.Z. and R.D.G.), and CA031760 (R.D.G).
Abbreviations used:
- DKO
lamin-B1 and -B2 double knockout
- EDFC
embryonic stem cell–derived fibroblast-like cell
- mESC
mouse embryonic stem cell
- TKO
lamin-A/C, -B1, and -B2 triple knockout
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-11-0644) on February 12, 2014.
REFERENCES
- Adam SA, Butin-Israeli V, Cleland MM, Shimi T, Goldman RD. Disruption of lamin B1 and lamin B2 processing and localization by farnesyltransferase inhibitors. Nucleus. 2013;4:142–150. doi: 10.4161/nucl.24089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi U, Cohn J, Buhle L, Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- Al-Haboubi T, Shumaker DK, Köser J, Wehnert M, Fahrenkrog B. Distinct association of the nuclear pore protein Nup153 with A- and B-type lamins. Nucleus. 2011;2:500–509. doi: 10.4161/nucl.2.5.17913. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol. 2012;14:13–24. doi: 10.1038/nrm3488. [DOI] [PubMed] [Google Scholar]
- Butin-Israeli V, Adam SA, Goldman AE, Goldman RD. Nuclear lamin functions and disease. Trends Genet. 2012;28:464–471. doi: 10.1016/j.tig.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chen C-Y, Chi Y-H, Mutalif RA, Starost MF, Myers TG, Anderson SA, Stewart CL, Jeang K-T. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell. 2012;149:565–577. doi: 10.1016/j.cell.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chen X, Zheng Y. The nuclear lamina regulates germline stem cell niche organization via modulation of EGFR signaling. Cell Stem Cell. 2013;13:73–86. doi: 10.1016/j.stem.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, Chang SY, Nobumori C, Tu Y, Farber EA, Toth JI, Fong LG, Young SG. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc Natl Acad Sci USA. 2010;107:5076–5081. doi: 10.1073/pnas.0908790107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, et al. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol Biol Cell. 2011;22:4683–4693. doi: 10.1091/mbc.E11-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan DP, Kuszczak D, Rusiñol AE, Thewke DP, Hrycyna CA, Michaelis S, Sinensky MS. Prelamin A endoproteolytic processing in vitro by recombinant Zmpste24. Biochem J. 2005;387:129–138. doi: 10.1042/BJ20041359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Shimi T, Adam SA, Rusiñol AE, Andres DA, Spielmann HP, Sinensky MS, Goldman RD. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proc Natl Acad Sci USA. 2007;104:4955–4960. doi: 10.1073/pnas.0700854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre E, Tramier M, Coppey-Moisan M, Gaillard C, Courvalin J-C, Buendia B. The truncated prelamin A in Hutchinson-Gilford progeria syndrome alters segregation of A-type and B-type lamin homopolymers. Hum Mol Genet. 2006;15:1113–1122. doi: 10.1093/hmg/ddl026. [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Bergmann JH, Lazar Z, Spector DL. Lamin A/C is expressed in pluripotent mouse embryonic stem cells. Nucleus. 2013;4:53–60. doi: 10.4161/nucl.23384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst P, Guan T, Subauste C, Hahn K, Gerace L. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J Cell Biol. 2002;156:617–630. doi: 10.1083/jcb.200106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman B, Channels W, Qiu M, Iglesias P, Yang G, Zheng Y. Lamin B counteracts the kinesin Eg5 to restrain spindle pole separation during spindle assembly. J Biol Chem. 2010;285:35238–35244. doi: 10.1074/jbc.M110.140749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, Mattout A, Gruenbaum Y, Liu J. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:16690–16695. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houliston E, Guilly MN, Courvalin JC, Maro B. Expression of nuclear lamins during mouse preimplantation development. Development. 1988;102:271–278. doi: 10.1242/dev.102.2.271. [DOI] [PubMed] [Google Scholar]
- Izumi M, Vaughan OA, Hutchison CJ, Gilbert DM. Head and/or CaaX domain deletions of lamin proteins disrupt preformed lamin A and C but not lamin B structure in mammalian cells. Mol Biol Cell. 2000;11:4323–4337. doi: 10.1091/mbc.11.12.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H-J, et al. Farnesylation of lamin B1 is important for retention of nuclear chromatin during neuronal migration. Proc Natl Acad Sci USA. 2013;110:E1923–E1932. doi: 10.1073/pnas.1303916110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan C-M, Gaiano N, Ko MSH, Zheng Y. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011;334:1706–1710. doi: 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Zheng X, Zheng Y. Proliferation and differentiation of mouse embryonic stem cells lacking all lamins. Cell Res. 2013;23:1420–1423. doi: 10.1038/cr.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubben N, et al. Post-natal myogenic and adipogenic developmental: defects and metabolic impairment upon loss of A-type lamins. Nucleus. 2011;2:195–207. doi: 10.4161/nucl.2.3.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- Liu J, Rolef Ben-Shahar T, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Tsai M-Y, Wang S, Lu B, Chen R, Iii JRY, Zhu X, Zheng Y. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat Cell Biol. 2009;11:247–256. doi: 10.1038/ncb1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J, Campbell KH, Ford CC, Stick R, Hutchison CJ. The role of lamin LIII in nuclear assembly and DNA replication, in cell-free extracts of Xenopus eggs. J Cell Sci. 1991;98:271–279. doi: 10.1242/jcs.98.3.271. [DOI] [PubMed] [Google Scholar]
- Okita K, Hong H, Takahashi K, Yamanaka S. Generation of mouse-induced pluripotent stem cells with plasmid vectors. Nat Protoc. 2010;5:418–428. doi: 10.1038/nprot.2009.231. [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Röber RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- Schermelleh L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC. The stability of the nuclear lamina polymer changes with the composition of lamin subtypes according to their individual binding strengths. J Biol Chem. 2004;279:42811–42817. doi: 10.1074/jbc.M407705200. [DOI] [PubMed] [Google Scholar]
- Schreiber KH, Kennedy BK. When lamins go bad: nuclear structure and disease. Cell. 2013;152:1365–1375. doi: 10.1016/j.cell.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J Cell Biol. 2012;197:239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M-Y, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- Vaughan A, Alvarez-Reyes M, Bridger JM, Broers JL, Ramaekers FC, Wehnert M, Morris GE, Whitfield WGF, Hutchison CJ. Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. J Cell Sci. 2001;114:2577–2590. doi: 10.1242/jcs.114.14.2577. [DOI] [PubMed] [Google Scholar]
- Vergnes L, Péterfy M, Bergo MO, Young SG, Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc Natl Acad Sci USA. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Yuan J, Blobel G, Georgatos SD. A lamin B receptor in the nuclear envelope. Proc Natl Acad Sci USA. 1988;85:8531–8534. doi: 10.1073/pnas.85.22.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Chang SY, Yin L, Tu Y, Hu Y, Yoshinaga Y, de Jong PJ, Fong LG, Young SG. An absence of both lamin B1 and lamin B2 in keratinocytes has no effect on cell proliferation or the development of skin and hair. Hum Mol Genet. 2011;20:3537–3544. doi: 10.1093/hmg/ddr266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Callebaut I, Pezhman A, Courvalin JC, Worman HJ. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J Biol Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- Ying Q-L, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- Zwerger M, Jaalouk DE, Lombardi ML, Isermann P, Mauermann M, Dialynas G, Herrmann H, Wallrath LL, Lammerding J. Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Hum Mol Genet. 2013;22:2335–2349. doi: 10.1093/hmg/ddt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.