Oocyte meiotic spindles are bipolar but assemble without centrosomes. Three Caenorhabditis elegans genes that contribute are that for the calponin homology domain protein, aspm-1; the katanin mei-1; and the kinesin-12 family member klp-18. The results indicate that both microtubule severing and ASPM-1 promote pole assembly, whereas KLP-18 promotes bipolarity.
Abstract
In many animals, including vertebrates, oocyte meiotic spindles are bipolar but assemble in the absence of centrosomes. Although meiotic spindle positioning in oocytes has been investigated extensively, much less is known about their assembly. In Caenorhabditis elegans, three genes previously shown to contribute to oocyte meiotic spindle assembly are the calponin homology domain protein encoded by aspm-1, the katanin family member mei-1, and the kinesin-12 family member klp-18. We isolated temperature-sensitive alleles of all three and investigated their requirements using live-cell imaging to reveal previously undocumented requirements for aspm-1 and mei-1. Our results indicate that bipolar but abnormal oocyte meiotic spindles assemble in aspm-1(-) embryos, whereas klp-18(-) and mei-1(-) mutants assemble monopolar and apolar spindles, respectively. Furthermore, two MEI-1 functions—ASPM-1 recruitment to the spindle and microtubule severing—both contribute to monopolar spindle assembly in klp-18(-) mutants. We conclude that microtubule severing and ASPM-1 both promote meiotic spindle pole assembly in C. elegans oocytes, whereas the kinesin 12 family member KLP-18 promotes spindle bipolarity.
INTRODUCTION
Oocyte meiosis includes two rounds of cell division—meiosis I and II—which produce a haploid oocyte pronucleus. In many animal species, including vertebrates and nematodes, these two meiotic cell divisions require bipolar spindles that, in contrast to mitotic spindles, assemble in the absence of centrosomes. These spindles are small and closely associated with the cell cortex, and they ultimately extrude two sets of chromosomes into polar bodies during highly asymmetric cell divisions (Fabritius et al., 2011). Although oocyte meiotic spindle assembly has been investigated in both vertebrate and invertebrate model systems, the genetic requirements for this process are poorly understood compared with mitotic spindle assembly.
Mitotic spindle assembly occurs through two parallel pathways: chromosome search and capture by growing microtubules initiated at the two centrosomes that dominate bipolar mitotic spindle structure, and chromosome-initiated microtubule assembly and the subsequent bipolar organization of antiparallel microtubules (Heald et al., 1997; Khodjakov et al., 2000; Meunier and Vernos, 2012). Oocyte meiotic spindles in many animals use only one of these pathways, relying entirely on acentrosomal assembly (Yamamoto et al., 2006; Muller-Reichert et al., 2010; Fabritius et al., 2011). Thus oocyte meiotic spindle assembly provides a simplified system for studying the shared pathway of acentrosomal spindle assembly.
With its powerful genetics and transparent anatomy, Caenorhabditis elegans provides an appealing model system for the investigation of acentrosomal oocyte meiotic spindle assembly dynamics (Yamamoto et al., 2006; Muller-Reichert et al., 2010; Fabritius et al., 2011). The oocyte meiosis I spindle begins to assemble late in oocyte maturation, when the nuclear envelope breaks down. Subsequently, by prometaphase and after fertilization, microtubule polymerization and organization results in the formation of a compact but bipolar spindle oriented roughly parallel to the oocyte cell cortex. By metaphase, the spindle shortens to form a tight barrel shape. The spindle then rotates such that its bipolar axis becomes orthogonal to the overlying plasma membrane. Subsequently, the paired homologous chromosomes separate and move toward each pole during anaphase, with one haploid set of paired sister chromatids extruded into a small polar body. In meiosis II, a small bipolar spindle again assembles, the two sister chromatids for each remaining homologue separate and move to opposite poles, and again one set is extruded into a second polar body, with the other set constituting the haploid oocyte contribution to the zygote genome (Albertson and Thomson, 1993; McNally et al., 2006).
Although positioning of the C. elegans oocyte meiotic spindle has been investigated extensively (Yang et al., 2003, 2005; Ellefson and McNally, 2009, 2011; van der Voet et al., 2009), less is known about its assembly. Three C. elegans genes known to contribute to oocyte meiotic spindle assembly are aspm-1, which encodes a protein with a single calponin homology domain and two IQ repeats (van der Voet et al., 2009); mei-1, which encodes the catalytic subunit of the microtubule-severing complex katanin (Clark-Maguire and Mains, 1994); and klp-18, which encodes a kinesin 12 (Segbert et al., 2003).
Both C. elegans aspm-1 and the Drosophila orthologue, Asp, are essential and required for proper execution of oocyte meiotic cell divisions (Riparbelli et al., 2002; van der Voet et al., 2009). In C. elegans oocytes, ASPM-1 is required for the meiotic spindles to align orthogonally and be in close proximity to the overlying plasma membrane (van der Voet et al., 2009; Ellefson and McNally, 2011). In mouse, ASPM-1 is expressed specifically in the primary sites of prenatal cortical neurogenesis and is localized to mitotic spindle poles (Bond et al., 2002; Fish et al., 2006). RNA interference (RNAi) knockdown of mouse ASPM-1 results in abnormal orientation of neural stem cell divisions and associated loss of cortical neurons (Fish et al., 2006). Moreover, mutations in ASPM-1 are the most common cause of microcephaly in humans (Bond et al., 2002). However, the molecular mechanisms underlying these ASPM-1 requirements are largely unknown.
RNAi knockdown of C. elegans klp-18/kinesin 12 results in chromosome misalignment and segregation defects during oocyte meiotic cell division (Segbert et al., 2003) and in assembly of a monopolar meiotic spindle (Wignall and Villeneuve, 2009). In vertebrates, the kinesin 12 family members hKlp2 and Kif15 are partially redundant with kinesin 5 for bipolar mitotic spindle assembly (Tanenbaum et al., 2009; Vanneste et al., 2009), although C. elegans klp-18 does not appear to be required for mitotic spindle assembly (Segbert et al., 2003). Vertebrate kinesin 12 family members form homodimers and may promote mitotic spindle bipolarity by cross-linking antiparallel microtubules through an interaction with the microtubule-binding protein TPX-2 (Tanenbaum et al., 2009; Vanneste et al., 2009; Sturgill and Ohi, 2013).
The C. elegans gene mei-1 encodes the p60 catalytic subunit of the widely conserved microtubule-severing complex called katanin (McNally and Vale, 1993; Hartman et al., 1998), previously shown in C. elegans to be required for proper assembly and orientation of oocyte meiotic spindles (Mains et al., 1990; Clandinin and Mains, 1993; Clark-Maguire and Mains, 1994; Srayko et al., 2000, 2006; Yang et al., 2003; McNally et al., 2006; McNally and McNally, 2011). The C-terminal region of MEI-1 contains the AAA ATPase microtubule-severing activity, whereas the N-terminal region binds to the p80 subunit MEI-2 (Mains et al., 1990; Clandinin and Mains, 1993; Clark-Maguire and Mains, 1994; Srayko et al., 2000, 2006; Yang et al., 2003; McNally et al., 2006; McNally and McNally, 2011). A partial reduction of mei-2 and hence microtubule severing was shown to prevent the reduction in oocyte meiotic spindle length that normally occurs during wild-type development (McNally et al., 2006). In addition, MEI-1 is required for recruitment of ASPM-1 to meiotic spindle poles, and a homozygous viable mei-1 allele with compromised microtubule-severing activity—ct103—can still recruit ASPM-1 to meiotic spindle poles (McNally and McNally, 2011; Gomes et al., 2013). This microtubule-severing-defective ct103 allele mediates the assembly of bipolar spindles that, although longer than normal, can still shorten but are often mispositioned. Another microtubule-severing-defective mutant with a similar phenotype was initially described as an allele of mei-1 but was later reported in a correction to be an allele of mei-2 (McNally and McNally, 2011; Gomes et al., 2013). Other alleles that more completely reduce mei-1 function have more severe spindle defects (Mains et al., 1990; Clandinin and Mains, 1993; Clark-Maguire and Mains, 1994; Srayko et al., 2000, 2006; Yang et al., 2003; McNally et al., 2006; McNally and McNally, 2011). Finally, the N-terminus of MEI-1 also can bind microtubules, and this may contribute to polar organization (McNally and McNally, 2011). In sum, these results led to the conclusion that MEI-1 has two distinct functions—microtubule severing and spindle pole organization (McNally and McNally, 2011)—although the role of microtubule severing is unclear.
After isolating conditional (heat-sensitive) alleles of C. elegans aspm-1, mei-1, and klp-18, we used live-cell imaging with transgenic fluorescent protein fusions to investigate the requirements for these loci in oocyte meiotic spindle assembly. Our results indicate that KLP-18 promotes spindle bipolarity, whereas both the MEI-1–dependent recruitment of ASPM-1 to the spindle and the microtubule-severing activity of MEI-1 contribute to spindle pole assembly.
RESULTS
Temperature-sensitive mutations with abnormal numbers of oocyte pronuclei map to the conserved genes bmk-1, aspm-1, klp-18, and mei-1
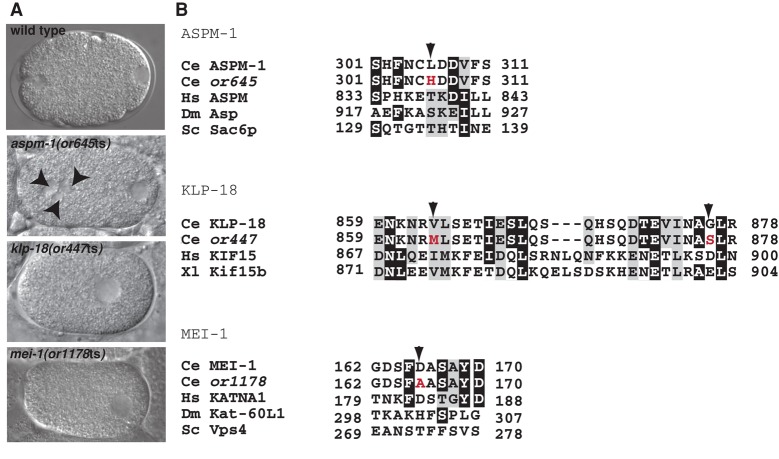
After completion of oocyte meiosis I and II in wild-type C. elegans zygotes, two spherical and haploid pronuclei appear, one from the egg and one from the sperm (Figure 1A; Albertson, 1984; Albertson and Thomson, 1993). To identify essential genes that mediate meiotic spindle assembly, we used Nomarski optics to examine live one-cell-stage embryos made by a collection of temperature-sensitive (ts) embryonic-lethal mutants at the restrictive temperature (Materials and Methods). We looked for mutants with either no or more than one oocyte pronucleus as indicator of defects in chromosome segregation during meiosis I or II and hence possibly defects in oocyte meiotic spindle assembly. Here we report our isolation of four such recessive ts mutants: or447ts, or627ts, or645ts, and or1178ts (Figure 1, Supplemental Figure S1, and 4).
FIGURE 1:
The ts mutant alleles of aspm-1, klp-18, and mei-1 have abnormal numbers of maternal pronuclei and encode missense mutations. (A) Nomarski images of one-cell-stage wild-type and mutant embryos. Embryos are positioned with the anterior (maternal) and posterior (paternal) pronuclei to the left and right, respectively; genotypes are indicated. Note the presence of extra maternal pronuclei in aspm-1(or645ts) mutant embryos (arrowheads) and the absence of maternal pronuclei in klp-18(or447ts) and mei-1(or1178ts) mutants. (B) Partial sequence alignments of orthologues from C. elegans, Homo sapiens (Hs), Xenopus laevis (Xl), Drosophila melanogaster (Dm), and Saccharomyces cerevisiae (Sc) with the wild-type and mutant C. elegans (Ce) proteins. Arrowheads indicate altered residues, with wild-type amino acids in black and mutant amino acids in red. Note the two mutations in klp-18. The alignment was performed using Boxshade. If the residue is identical to the column consensus, there is a black background; if the residue is similar to the column consensus, there is a gray background.
TABLE 1:
Embryonic viability of oocyte meiotic spindle–defective mutants.
| Allele | Homozygote embryonic viability (%) at | Heterozygote embryonic viability (%) at 26°C | |
|---|---|---|---|
| 15°C | 26°C | ||
| Wild type | 99 (n = 358) | 97 (n = 197) | – |
| aspm-1(or645) | 99 (n = 216) | 38 (n = 271) | 99 (n = 555) |
| klp-18(or447) | 91 (n = 308) | 4 (n = 504) | 99 (n = 260) |
| mei-1(or1178) | 98 (n = 127) | 0 (n = 262) | 99 (n = 256) |
| bmk-1(or627) | 79 (n = 175) | 1.4 (n = 561) | 99 (n = 403) |
| mei-1(ct46ct103) | 65 (n = 194) | ||
| bmk-1 deletion alleles | |||
| bmk-1(ok391) | 99 (n = 176) | ||
| bmk-1(tm969) | 99 (n = 154) | ||
| bmk-1 complementation test | |||
| bmk-1(or627)/bmk-1(ok391) | 40 (n = 341) | ||
| bmk-1(or627)/bmk-1(tm969) | 27 (n = 617) | ||
| bmk-1 recessive gain-of-function test | |||
| bmk-1(or627) + bmk-1(RNAi) | 77 (n = 731) | ||
Embryonic viability (percentage hatching) was scored for wild type and each ts mutant at permissive (15°C) and restrictive temperature (26°C) temperatures and for bmk-1 deletion alleles and mei-1(ct46ct103) mutants at 26°C (Materials and Methods). Embryonic viability at the restrictive temperature from heterozygous ts mutants was examined to determine whether the mutations are recessive or dominant.
After mapping the mutations and performing complementation tests, we sequenced amplified genomic DNA fragments from candidate genes for each mutation to identify the causal mutations (Figure 1B and Supplemental Figure S1A; Materials and Methods). First, we found that or627ts is an allele of bmk-1, the C. elegans orthologue of kinesin 5/Eg5 (Supplemental Figure S1A; Materials and Methods). In vertebrates, this kinesin is required for bipolar mitotic spindle assembly, but C. elegans bmk-1/kinesin 5 is not essential, is not required for meiotic spindle assembly, and has only a minor role in mitotic spindle assembly dynamics (Bishop et al., 2005; Saunders et al., 2007). Consistent with these previous studies, we found that or627ts is a recessive gain-of-function bmk-1 mutation (Materials and Methods). Although this mutation results in fully penetrant meiotic spindle defects (Table 1 and Supplemental Figure S1, B and C), we chose to focus further analysis on the three recessive, loss-of-function mutations we isolated in aspm-1, mei-1, and klp-18.
In genomic DNA from or645ts mutants, which failed to complement the deletion allele aspm-1(ok1208) for maternal-effect embryonic lethality, we found a single leucine-to-histidine change at codon 306 in the aspm-1 open reading frame, 5′ of sequences that encode the conserved calponin homology domain (Figure 1B). Similarly, or447ts failed to complement the deletion allele klp-18(ok2519) and contained two missense mutations that both affect the C-terminal coiled-coiled region of KLP-18: a valine-to-methionine change at codon 854, and a glycine to serine change at codon 876 (Figure 1B). Finally, or1178ts failed to complement the conditional allele mei-1(or646ts) and resulted in an aspartate-to-alanine change at codon 166 that affects sequences N-terminal to the conserved AAA ATPase domain (Figure 1B). The nonconditional deletion alleles aspm-1(ok1208) and klp-18(ok2519) both result in adult sterility, and studies of these gene requirements during early embryogenesis in C. elegans thus far have used RNAi to reduce gene function. To our knowledge, aspm-1(or645ts) and klp-18(or447ts) are the first conditional alleles identified for these loci, and mei-1(or1178ts) is the strongest ts allele isolated for this locus (O'Rourke et al., 2011); thus all three should prove useful for further studies of their requirements throughout development (Table 2).
TABLE 2:
Postembryonic phenotypes of oocyte meiotic spindle–defective mutants.
| Allele | Fertile adultsa (%) | Larval lethal (%) | Male (%) | Sterile (%) | n |
|---|---|---|---|---|---|
| aspm-1(or645) | 56 | 44 | 118 | ||
| klp-18(or447) | 67 | 4 | 29 | 117 | |
| mei-1(or642) | 98 | 2 | 120 | ||
| mei-1(or646) | 94 | 3 | 3 | 167 | |
| mei-1(or1178) | 66 | 33 | 129 | ||
| mei-2(sb39) | 85 | 15 | 107 |
Larval lethality, sterility, and gender were scored after L1 larvae were matured to adulthood at restrictive temperature (26°C).
aAll adults laid dead embryos.
To compare oocyte meiotic spindle assembly in live wild-type and mutant embryos, we used spinning-disk confocal microscopy and transgenic strains expressing translational fusions of GFP to β-tubulin and mCherry to Histone2B to mark microtubules and chromosomes during meiosis I spindle assembly (Materials and Methods). We used both our ts mutant alleles and RNAi knockdown, which result in similar phenotypes, to reduce gene functions (Figures 2 and 3A). Because the klp-18 gene is tightly linked to the mCherry:Histone2B transgene integration site (unpublished data), we used RNAi to reduce klp-18 function in most experiments.
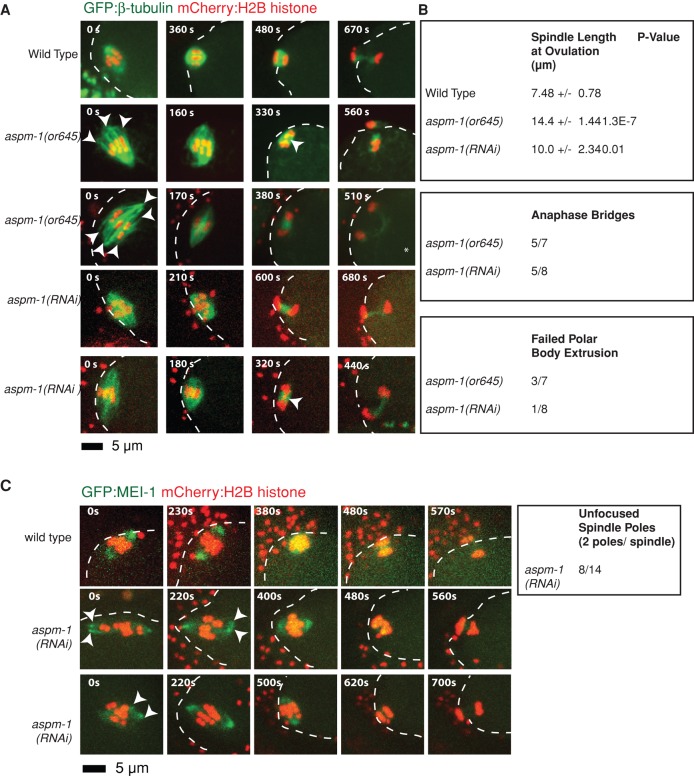
FIGURE 2:
aspm-1(-) mutants assemble long, bipolar oocyte meiotic spindles with unfocused pole ends and aberrantly organized chromosomes. (A) Spinning-disk confocal images were recorded over time during meiosis I in live wild-type (Supplemental Movie S1) and aspm-1(-) mutant (Supplemental Movies S2 and S3) embryos expressing mCherry:Histone2B and GFP:β-tubulin translational fusions to mark chromosomes and microtubules, respectively. Indicated time points begin at ovulation. A white dashed line marks the edge of the plasma membrane. In the first column, white arrowheads mark unfocused pole ends, and in the third column, arrowheads mark the lagging chromosomes during anaphase, as quantified in B. The asterisk indicates an embryo in which polar body extrusion failed. Bottom rows, examples of mutant embryos with more-focused poles. (B) Quantification of spindle defects in aspm-1(-) mutants. Meiotic spindles were measured directly after ovulation from one end of the pole to the other using spinning-disk confocal images. (C) MEI-1 marks unfocused pole ends in aspm-1(RNAi). Spinning-disk confocal images taken over time during meiosis I in live wild-type (Supplemental Movie S4) and aspm-1(-) mutant (Supplemental Movie S5) embryos expressing GFP:MEI-1 and mCherry:Histone2B translational fusions to mark spindle poles and chromosomes, respectively. Times indicated begin at ovulation. Scale bar as shown. Arrowheads indicate mutant spindle poles that initially appear fragmented but later coalesce into more-focused poles resembling those observed in wild-type embryos.
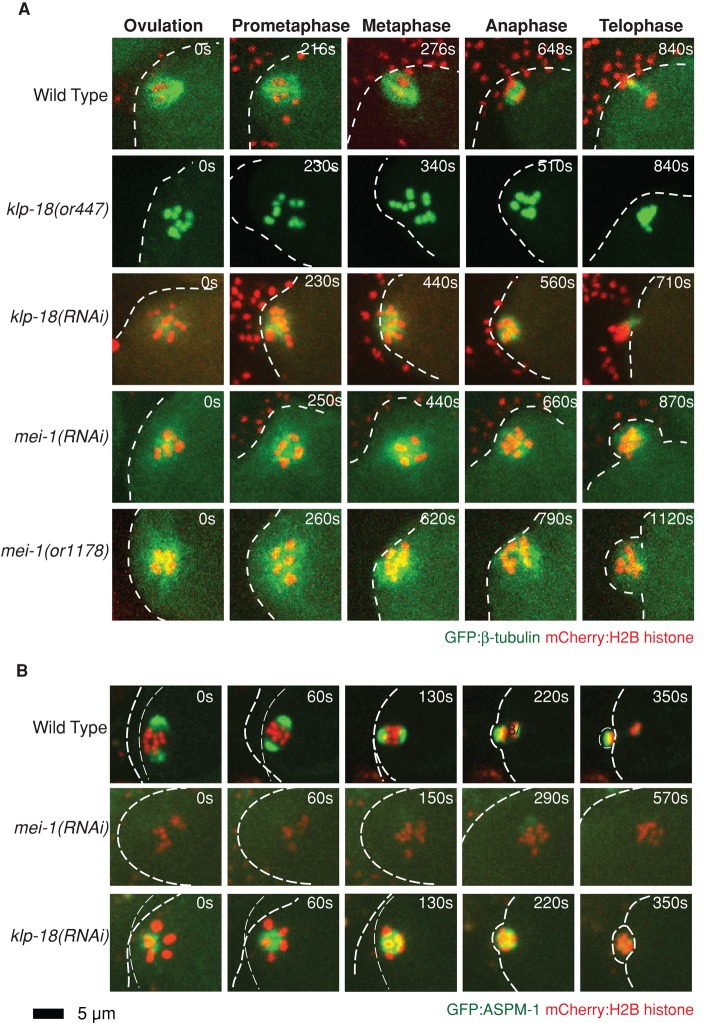
FIGURE 3:
Monopolar and apolar oocyte meiotic spindles assemble in klp-18(-) and mei-1(-) mutants, respectively. (A) Time-lapse spinning-disk confocal images from immobilized worms were recorded during meiosis I in wild-type (Supplemental Movie S1) and mutant zygotes (Supplemental Movies S6–S9) expressing mCherry:Histone2B and GFP:β-tubulin to mark chromosomes and microtubules, respectively, from ovulation to polar body extrusion, but only GFP:Histone2B marks the chromosomes in klp-18(or447) oocytes. Anterior is to the left, times indicated are relative to ovulation, and a white dashed line marks the edge of the zygote plasma membrane. In this and subsequent figures, each image shown is a projection of six consecutive frames taken at 1.5-μm intervals in a Z-stack for each time point. (B) ASPM-1 marks a single pole in klp-18(-) mutants and no pole in mei-1(-) mutants. Spinning-disk confocal images taken over time during meiosis I in live wild-type (Supplemental Movie S10) and mutant embryos (Supplemental Movies S11 and S12) expressing GFP:ASPM-1 and mCherry:Histone2B translational fusions to mark spindle poles and chromosomes, respectively. Times indicated begin at ovulation. Scale bar as shown.
aspm-1(or645ts) meiotic spindles are bipolar but assemble abnormally and are misoriented
Of the three mutants we isolated, we saw the least severe spindle defects in aspm-1(or645ts) mutants (Figures 2A). In previous studies that used RNAi to reduce its function, aspm-1 was shown to act with dynein to rotate the meiosis I spindle such that it orients orthogonally to the plane of the plasma membrane (Ellefson and McNally, 2009; van der Voet et al., 2009; Wignall and Villeneuve, 2009). We similarly found that bipolar meiosis I spindles assembled in aspm-1(or645ts) mutant zygotes and failed to rotate to an orthogonal orientation or to stay in close proximity to the plasma membrane (Figure 2A), followed in 4 of 15 cases by failed attempts to extrude chromosomes into a polar body (Figure 2B).
In addition to the previously reported meiotic spindle-positioning defects, we also observed earlier defects in spindle morphology in aspm-1(or645ts) mutants. After oocyte pronuclear envelope breakdown, ovulation, and fertilization, aspm-1(or645ts) mutants assembled meiosis I spindles that were large compared with wild type (Figure 2B). Before metaphase, the mutant spindles also appeared to have unfocused poles, often with more than a single focus of microtubule ends visible at one or both poles (Figure 2A and Supplemental Figure S2A), similar to a previous observation for a single fixed embryo (Wignall and Villeneuve, 2009). To further examine the integrity of the oocyte meiotic spindle poles, we reduced aspm-1 function in a transgenic strain that expresses a GFP fusion to MEI-1, which, as shown previously, localizes to the two poles in wild-type spindles (McNally et al., 2006). We again observed unfocused but bipolar spindles early in meiosis I, with more than a single focus of GFP:MEI-1 detected at one or both poles in most mutant embryos (Figure 2C).
Finally, the meiotic chromosomes in aspm-1(or645ts) mutants appeared disorganized. During prometaphase of meiosis I, chromosomes in some cases appeared more dispersed within the spindle relative to wild type (Figure 2A). Nevertheless, as the meiotic cell cycle progressed, the mutant spindles shortened and maintained bipolarity, and chromosomes congressed to a metaphase plate, more nearly resembling wild-type spindle morphology. However, we then observed lagging chromosomes during anaphase and telophase (Figure 2A). Defects in chromosome segregation and failed polar body extrusion presumably account for the penetrant phenotype of multiple maternal pronuclei observed using differential interference contrast microscopy, with nuclear membranes assembling around multiple chromosome masses present in oocytes upon the completion of meiosis I and II (Figure 1 and Table 1). We conclude that ASPM-1 has important roles in meiotic spindle assembly, positioning, and function but is not required for spindle bipolarity.
Bipolar oocyte meiotic spindles fail to assemble in both mei-1(-) and klp-18(-) mutants
In contrast to aspm-1(-) mutants, klp-18(-) and mei-1(-) mutants assembled oocyte meiotic spindles that appear to lack bipolarity (Figure 3A). Indeed, both klp-18(-) and mei-1(-) mutants frequently extruded all oocyte chromosomes into the first polar body, leaving zygotes with no maternal genome contribution (Table 1 and Figures 1A and 3A).
Although both mutants lacked bipolarity, the meiotic spindle assembly defects we observed in mei-1(-) and klp-18(-) mutants were distinct. In mei-1(-) mutants, the oocyte meiosis I spindle was highly disorganized: the chromosomes were variably dispersed without ever congressing to a metaphase plate, and microtubules appeared loosely organized. Ultimately, an interspersed assembly of microtubules and chromosomes was often extruded into a single, abnormally large polar body (Figure 3A). By contrast, during oocyte meiotic spindle assembly in klp-18(-) mutants, chromosomes also were initially scattered among loosely organized microtubules, but chromosomes and microtubules progressively congressed into a smaller area, with chromosomes at the periphery of a single bright focus of microtubule density (Figure 3A).
klp-18(-) mutant oocyte meiotic spindles are monopolar, whereas mei-1(-) mutant spindles are apolar
On the basis of their distinct morphologies, we hypothesized that mei-1(-) meiotic spindles are apolar and that klp-18(-) mutants assemble monopolar spindles. To further investigate whether klp-18(-) and mei-1(-) spindles are monopolar or apolar, we used ASPM-1 as a spindle pole marker. From previous studies of fixed embryos, ASPM-1 is known to localize to oocyte meiotic spindle poles (Wignall and Villeneuve, 2009; McNally and McNally, 2011; Gomes et al., 2013). To examine ASPM-1 localization dynamics in live oocytes, we used recombineering to construct a functional N-terminal translational fusion of GFP to ASPM-1 driven by the endogenous promoter (Materials and Methods). In wild-type transgenic oocytes, GFP:ASPM-1 marked both spindle poles before metaphase and persisted through anaphase (Figure 3B).
We next used transgenic strains expressing the GFP:ASPM-1 fusion to further compare mei-1(-) and klp-18(-) mutant oocytes (Figure 3B). Consistent with previous studies using fixed embryos (McNally and McNally, 2011; Gomes et al., 2013), GFP:ASPM-1 was nearly absent from the disorganized meiotic spindles observed in mei-1(-) mutant oocytes. By contrast, ASPM-1 brightly marked a single focus in klp-18(-) mutant spindles, consistent with a previous analysis of fixed specimens (Wignall and Villeneuve, 2009). These results provide the first analysis of C. elegans ASPM-1 localization in live embryos and support the conclusions that mei-1(-) oocytes lack meiotic spindle polarity and klp-18(-) mutants assemble monopolar spindles (Discussion).
mei-1 is required for monopolar spindle assembly in klp-18(-) mutants
We next wanted to identify genetic requirements for oocyte meiotic spindle pole assembly, using the monopolar spindles in klp-18(-) mutants as a simplified model for wild-type spindle pole assembly. Wild-type and mutant embryos with bipolar oocyte meiotic spindles capture and segregate chromosomes into a single metaphase plate that then transitions during anaphase into two nearly identical masses, sometimes connected by anaphase bridges in mutants. These dynamics made it difficult to directly and quantitatively compare bipolar spindle dynamics to the apolar and monopolar mei-1(-) and klp-18(-) spindles. We therefore limited our quantitative comparisons to mutants with apolar or monopolar oocyte meiotic spindles.
To use klp-18(-) mutants as a model for analyzing pole assembly, we first developed a simple quantitative measurement to compare differences in spindle size and organization. The measurement entails tracking the two-dimensional area occupied by oocyte chromosomes over time in projected Z-stacks. We then compared the area at ovulation, immediately after the egg has been fully engulfed by the spermathecae, with the smallest area occupied by the chromosomes during meiosis I (Figure 4; Materials and Methods).
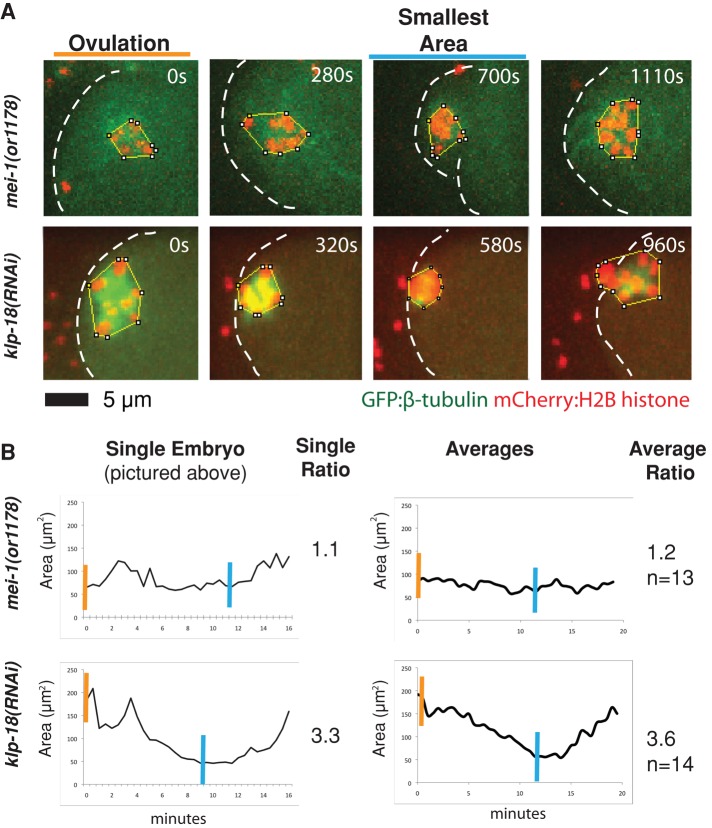
FIGURE 4:
Measuring the area occupied by chromosomes over time quantitatively distinguishes oocyte meiotic spindle-defective mutant phenotypes. (A) Spinning-disk confocal images were recorded over time during meiosis I in live wild-type and mutant embryos expressing mCherry:Histone2B and GFP:β-tubulin translational fusions to mark chromosomes and microtubules, respectively. (B) Left, the ImageJ polygon tool was used to measure the area occupied by chromosomes at each time point for the embryos shown in A. Examples of how the area was traced are shown in each image (A). Right, measurements beginning at ovulation and taken every 30 s, ending when chromosomes were extruded into a polar body. If chromosomes did not extrude into a polar body, the movie was ended after a failed attempt to extrude chromosomes. Averages for the indicated number of embryos are shown. Right, compaction ratios of the areas occupied at ovulation (orange lines) divided by the smallest area occupied during meiosis I (blue lines).
We found that klp-18(-) oocyte chromosomes were distributed over a relatively large area at ovulation but then congressed into a much smaller area (Figure 4). By contrast, chromosomes in mei-1(-) oocytes occupied a smaller area initially but generally failed to congress (Figure 4). We then calculated the ratio of the area occupied by oocyte chromosomes at ovulation over the smallest area occupied during meiosis I (Figure 4). In klp-18(-) mutants, this “area occupied by chromosomes” (AOC) ratio was on average 3.6, whereas in mei-1(-) mutants the AOC ratio was on average 1.2. As an independent measure of spindle pole assembly, we also compared the smallest areas occupied by chromosomes in mei-1(-) and klp-18(-) mutants and found that chromosomes came to occupy a smaller area in klp-18(-) than in mei-1(-) mutants (Figure 5). We conclude that oocytes fail both to assemble spindle poles and to organize chromosomes in mei-1(-) mutants, whereas klp-18(-) mutants assemble organized and monopolar oocyte meiotic spindles that appear to capture and bring chromosomes toward the monopole.
FIGURE 5:
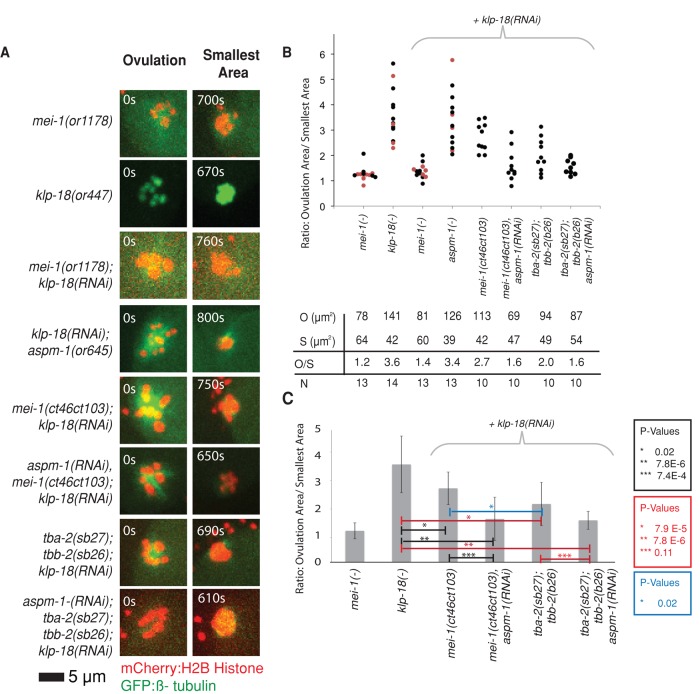
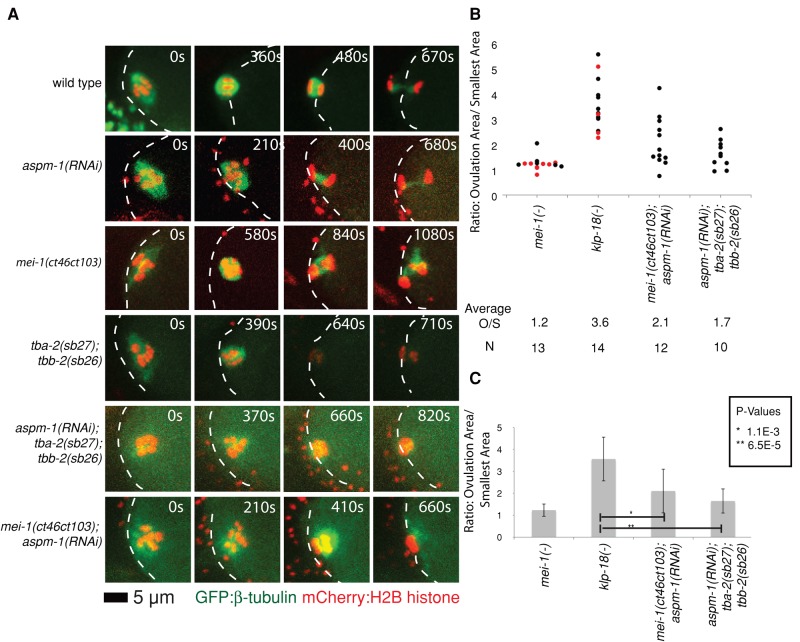
Assembly of monopolar oocyte meiotic spindles in klp-18 mutant requires both the katanin activity of MEI-1 and ASPM-1. (A) Representative spinning-disk confocal images were recorded over time during meiosis I in live mutant embryos (Supplemental Movies S6–S9 and S13–S18) expressing mCherry:Histone2B and GFP:β-tubulin translational fusions to mark chromosomes and microtubules, respectively, with the exception of klp-18(or447ts) oocytes, which expressed GFP:Histone2B only. The images shown are from ovulation and from the time during meiosis I at which the chromosomes occupied the smallest area. (B) Scatter plot showing the ratios of the areas occupied by chromosomes at ovulation over the smallest areas occupied during meiosis I. For mei-1(-) and klp-18(-), measurements were taken using RNAi (black dots) and ts mutations (red dots) to reduce gene function. Average areas for combined genotypes at ovulation (O), average smallest areas (S), average ratios (O/S), and number of embryos analyzed for each genotype (N) are shown below the graph. (C) Bar graph showing the average ratio of the areas occupied at ovulation over the smallest area occupied during meiosis I for selected genotypes. The p values from Student's test are indicated in black for observations made in the mei-1(ct46ct103) background, in red for tba-2(sb27);tbb-2(sb26) background, and in blue when comparing these two when klp-18 is knocked down. p < 0.05 indicates a statistically significant difference.
We next asked whether the reduction in meiotic spindle size that occurs over time in klp-18(-) mutants requires mei-1 function. We reasoned that a loss of any obvious spindle pole assembly might prevent the assembly of a monopolar spindle in klp-18(-) mutants. To this end, we examined oocyte meiotic spindle assembly dynamics in mutants lacking both mei-1 and klp-18 function (Figure 5 and Supplemental Figure S2). As expected, spindle assembly dynamics in double mutants resembled those observed in mei-1(-) single mutants: the initially dispersed oocyte chromosomes failed to congress and thus did not occupy a smaller area later in meiosis I. We conclude that mei-1 is required for assembly of monopolar meiotic spindles in klp-18(-) mutants, consistent with previous observations (McNally and McNally, 2011).
The microtubule-severing activity of MEI-1 is required for monopolar oocyte meiotic spindle assembly in klp-18(-) mutants
The MEI-1 protein could promote oocyte meiotic spindle assembly through two distinct functions: the microtubule severing mediated by its C-terminal AAA ATPase domain, and recruitment of ASPM-1 to the meiotic spindle (McNally and McNally, 2011). Although the partially microtubule severing defective mei-1(ct46ct103) allele is homozygous viable (Table 1) and assembles abnormally long but bipolar meiosis I spindles (McNally and McNally, 2011), we found that the spindle poles initially appeared loosely organized compared with wild type (Figures 6 and 7), although the microtubule signal was reduced in these mutants and we have not quantified this defect. The loosely organized poles could result from a role for microtubule severing in pole assembly or be an indirect consequence of the initially larger size of the mutant spindles relative to wild type. To distinguish between these possibilities, we asked whether the microtubule-severing activity of mei-1 is required for monopolar spindle assembly in klp-18(-) mutants. Intriguingly, the spindle dynamics in mei-1(ct46ct103); klp-18(-) double mutants was intermediate compared with that in mei-1(or1178ts) and klp-18(-) single mutants: the double mutant AOC ratio of 2.7 was lower than the 3.6 ratio observed in klp-18(-) mutants but greater than the 1.4 ratio observed after reducing all mei-1 function in klp-18(-) mutants (Figure 5). Similarly the smallest area occupied by chromosomes in the double mutant was less than that observed in mei-1(-) but greater than that observed in klp-18(-) single mutants (Figure 5). We conclude that the microtubule-severing activity of mei-1 plays a role in the assembly of monopolar oocyte meiotic spindles in klp-18(-) mutants.
FIGURE 6:
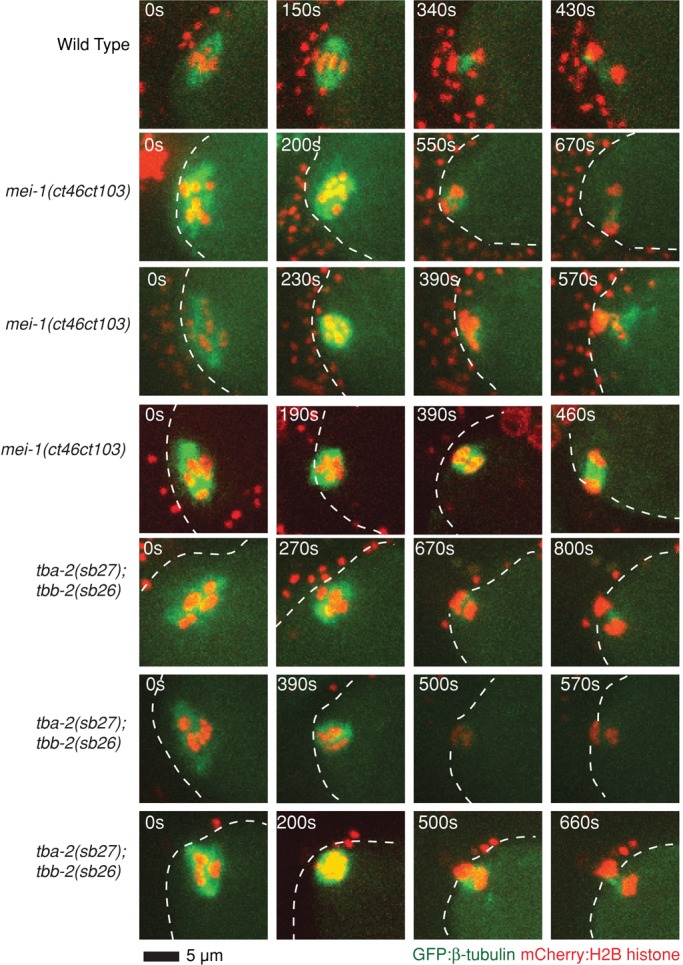
Microtubule-severing-defective mei-1(ct46ct103) and tba-2(sb27);tbb-2(sb26) mutant oocytes assemble bipolar oocyte meiotic spindles. Spinning-disk confocal images were recorded over time during meiosis I in live wild-type (Supplemental Movie S1) and mutant embryos (Supplemental Movies S19 and S20) expressing mCherry:Histone2B and GFP:β-tubulin translational fusions to mark chromosomes and microtubules, respectively. Indicated time points begin at ovulation. A white dashed line marks the edge of the plasma membrane. Scale bar as shown.
FIGURE 7:
Mutants lacking either mei-1–mediated microtubule severing or ASPM-1 are bipolar, whereas mutants lacking both are apolar. (A) Spinning-disk confocal images were recorded over time during meiosis I in live wild-type (Supplemental Movie S1) and mutant embryos (Supplemental Movies S2, S3, and S19–S22) expressing mCherry:Histone2B and GFP:β-tubulin translational fusions to mark chromosomes and microtubules, respectively. Indicated time points begin at ovulation. A white dashed line marks the edge of the plasma membrane. Scale bar as shown. (B) For mei-1(-) and klp-18(-), measurements were taken using RNAi (black dots) and ts mutations (red dots) to reduce gene function. Average ratios for combined genotypes (O/S) and the number of embryos analyzed for each genotype (N) are shown below the graph. (C) Bar graph showing the average ratio of the areas occupied at ovulation over the smallest area occupied during meiosis I for selected genotypes. The p values from Student's test are indicated; p < 0.05 indicates a statistically significant difference.
Although the mei-1(ct46ct103) allele has been shown to have substantially reduced microtubule-severing activity (McNally and McNally, 2011), the mutation also could have other effects on MEI-1 function that are responsible for the loss of spindle pole focusing observed in klp-18(-) mutants. To test independently whether microtubule severing plays a role in oocyte meiotic spindle pole focusing, we used mutations in the α- and β-tubulin genes tba-2 and tbb-2 (tba-2(sb27) and tbb-2(sb26)) that were isolated as suppressors of a dominant mei-1 allele with constitutive microtubule-severing activity; these tubulin mutants apparently are resistant to microtubule severing (Lu et al., 2004; Lu and Mains, 2005). We found that the AOC ratio and the smallest area occupied by chromosomes in tba-2(sb27); tbb-2(sb26); klp-18(-) triple mutants also were intermediate between the values observed for klp-18(-) single mutants and klp-18(-) mutants lacking all mei-1 function (Figure 5), further supporting a role for microtubule severing in oocyte meiotic spindle pole assembly.
aspm-1 is required for monopolar oocyte meiotic spindle assembly in klp-18(-) mutants that lack mei-1 microtubule-severing activity
We next asked whether aspm-1 is required for the compact monopolar appearance of the meiotic spindle in klp-18(-) oocytes as they approach completion of meiosis I. Two observations led us to suspect a role for aspm-1. First, as in mei-1(ct46ct103) mutants, the meiotic spindle poles in aspm-1(-) oocytes initially appeared loosely organized (Figures 2 and 6), consistent with a transient role in focusing spindle poles during prometaphase. Second, the ASPM-1 protein localized to a single monopolar focus in klp-18(RNAi) oocytes but was largely absent from apolar mei-1(-) spindles (Figure 3B). We therefore examined spindle assembly in live mutant oocytes lacking both aspm-1 and klp-18 function. We found that spindle dynamics in the double mutant were nearly identical to those in klp-18(-) single mutants, with AOC ratios of ∼3.4 and ∼3.6, respectively (Figure 5 and Supplemental Figure S2). However, we then considered the possibility that whereas a role for aspm-1 might not be apparent in the aspm-1(-); klp-18(-) double mutant, a contribution by aspm-1 might be detected in the already compromised context of the mei-1(ct46ct103); klp-18(-) double mutant. We therefore used RNAi to knock down both klp-18 and aspm-1 in mei-1(ct46ct103) mutants. We found that reducing both aspm-1 and microtubule-severing activity in a klp-18(-) mutant background did result in a more severe loss of spindle pole focusing compared with mei-1(ct46ct103); klp-18(RNAi) double mutant, with AOC ratios of 1.6 and 2.8, respectively (Figure 5). Similarly, reducing aspm-1 function in the tba-2(sb27); tbb-2(sb26); klp-18(-) mutant further reduced the AOC ratio compared with tba-2(sb27); tbb-2(sb26); klp-18(-), although in this case the reduction was not statistically significant, due in part to the more severe loss of compaction observed in the tubulin mutant background relative to the microtubule-severing mutant background (Figure 5). We conclude that mei-1 contributes to oocyte meiotic spindle pole focusing through both its microtubule severing activity and its recruitment of ASPM-1 to the spindle.
We relied on AOC ratios to quantitatively compare different mutant phenotypes with respect to monopolar spindle assembly, as we believe this measurement best represents the movement of chromosomes into a smaller volume over time. However, this measurement includes the initial area occupied by chromosomes, and one significant difference between mei-1(-) and klp-18(-) single mutants is that chromosomes initially occupy a significantly larger area in klp-18(-) mutants (Figures 4 and 5B). We suspect that this difference may be due to the reduced number of microtubules present in mei-1(-) mutants relative to wild-type zygotes (Srayko et al., 2006), although we have not quantified this phenotype in our analysis. Because this difference between mei-1(-) and klp-18(-) mutants may confound our reliance on AOC ratios, we also compared the smallest volume occupied by chromosomes during meiosis I in different mutant backgrounds (Figure 5B). When using this measurement, we also observed a significant difference between mei-1(-) and klp-18(-) single mutants (p = 2.9 × 10−5) and between klp-18(-) and mei-1(-); klp-18(-) double mutants (p = 0.003). Although we did not observe a significant difference when comparing this value between klp-18(-) mutants and double and triple mutants specifically lacking microtubule-severing activity, we did observe a significant difference when comparing klp-18(-) and klp-18(-); tba-2(sb27); tbb-2(sb26); aspm-1(RNAi) mutants (p = 0.02); this quadruple mutant exhibited the most-reduced AOC ratio of all mutants specifically lacking microtubule-severing but not all mei-1 function (Figure 5).
As a final test of whether both aspm-1 and microtubule severing contribute to spindle pole focusing, we used RNAi to reduce aspm-1 function in mei-1(ct46ct103) and tba-2(sb27); tbb-2(sb26) mutants. Whereas aspm-1(-) and mei-1(ct46ct103) single mutants and the tba-2(sb27); tbb-2(sb26) double mutant all assembled abnormal but bipolar oocyte meiosis I spindles, we found that the mei-1(ct46ct103); aspm-1(RNAi) double mutant and the tba-2(sb27); tbb-2(sb26); aspm-1(RNAi) triple mutant both were unable to assemble a bipolar spindle and instead assembled disorganized and apparently apolar spindles that resembled those observed in mutants lacking all mei-1 function (Figure 7), although the AOC ratios were more variable (Figures 5 and 7). These results support our conclusion that both aspm-1 and microtubule-severing activity contribute to meiotic spindle pole assembly. Finally, the AOC ratio in klp-18(-) mutants that lacked both microtubule severing and ASPM-1 was not as low as the AOC ratio in klp-18(-); mei-1(or1178ts) double mutants with a more complete loss of mei-1 function. Although this difference was not statistically significant, we cannot rule out the possibility that mei-1 acts through additional factors to promote bipolar oocyte meiotic spindle assembly.
DISCUSSION
By using live-cell imaging to quantify microtubule and chromosome dynamics during C. elegans oocyte meiotic spindle assembly, we confirmed previously identified requirements for aspm-1, mei-1, and klp-18, three essential genes known to be required for oocyte meiotic spindle positioning or assembly. Our results further indicate that the calponin homology domain protein ASPM-1, in addition to its previously documented role in spindle positioning, also contributes to assembly of the acentrosomal oocyte meiotic spindle poles and is required for chromosome segregation during anaphase. Furthermore, our results provide the first evidence that microtubule severing by MEI-1 also contributes to spindle pole focusing. Finally, our results confirm in live embryos the previous conclusion that klp-18/kinesin 12 is not required for spindle pole assembly but instead promotes oocyte meiotic spindle bipolarity.
klp-18 promotes oocyte meiotic spindle bipolarity
The conclusion that klp-18(-) mutants assemble monopolar oocyte meiotic spindles is based on three observations. First, during meiosis I in klp-18(-) mutants, the chromosomes are initially dispersed, but over time the chromosomes move toward each other to ultimately occupy a small volume. The chromosomes surrounded a central core of microtubule density as they congressed, consistent with the capture and movement of chromosomes by microtubules emanating from a monopole. Second, the oocyte meiotic spindle pole marker ASPM-1, found at both poles in wild type, localizes to a single focus in klp-18(-) mutants, and the dispersed chromosomes moved toward this ASPM-1 focus. Third, consistent with our live-cell analysis, immunofluorescence images of fixed embryos were consistent with monopolar oocyte meiotic spindles assembling in klp-18(-) mutants (Wignall and Villeneuve, 2009).
The human and mouse kinesin 12 family members hKlp2 and Kif15 are partially redundant with the kinesin 5 family member Eg5 in promoting mitotic spindle bipolarity (Tanenbaum et al., 2009; Vanneste et al., 2009). Chemical inhibition of Eg5 early in mitosis results in a loss of bipolarity, but chemical inhibition beginning at metaphase has no effect (Kapoor et al., 2000). Although RNAi knockdown of either hKlp2 or Kif15 alone does not affect bipolarity, knocking them down after chemical inhibition of Eg5 beginning at metaphase does lead to loss of mitotic spindle bipolarity (Tanenbaum et al., 2009; Vanneste et al., 2009). In C. elegans, neither klp-18/kinesin 12 nor bmk-1/kinesin 5 is required for mitotic spindle bipolarity (Tanenbaum et al., 2009; Vanneste et al., 2009). Although it is possible that they are more fully redundant during mitosis than the vertebrate orthologues, we did not detect any loss of mitotic spindle bipolarity in early embryos lacking both klp-18 and bmk-1 function (unpublished data).
Both kinesin 5 and 12 family members are believed to mediate bipolar spindle assembly by cross-linking and pushing in opposite directions antiparallel microtubules to promote separation of the two spindle poles (Tanenbaum et al., 2009; Vanneste et al., 2009; Sturgill and Ohi, 2013). For Eg5 the cross-linking results from the formation of homotetramers of the kinesin heavy chain, with the oppositely projecting motor domain dimers binding antiparallel microtubules (Weinger et al., 2011). In contrast, kinesin 12 family members form homodimers that can bind one microtubule, with cross-linking possibly achieved by binding Tpx2, which also binds microtubules. In C. elegans, RNAi knockdown of Tpx2 compromises mitotic spindle assembly, but no requirement for oocyte meiotic spindle assembly was reported (Ozlu et al., 2005). Thus it remains unclear whether and how klp-18 cross-links microtubules to promote meiotic spindle bipolarity in oocytes.
The temporal pattern of chromosome distribution in klp-18(-) mutants is distinct from that observed in C. elegans mutants with monopolar mitotic spindles in early embryonic cells (Kitagawa et al., 2009; Song et al., 2011). In these mutants, chromosomes are captured by astral microtubules emanating from a single centrosome and over time congress to form an arc peripheral to the monopole. We conclude that the acentrosomal meiotic spindle monopole in klp-18(-) mutants is more compact than with a centrosomal mitotic monopole, accounting for the movement of oocyte meiotic chromosomes into a much smaller volume.
mei-1 and aspm-1 promote oocyte meiotic spindle pole assembly
The microtubule-severing complex katanin comprises two subunits, p60 and p80, encoded in C. elegans by the genes mei-1 and mei-2 (Clark-Maguire and Mains, 1994; Hartman et al., 1998; Srayko et al., 2000). Inactivation of either gene results in similar meiotic spindle defects and embryonic lethality. Partial loss-of-function mutations have been used to identify requirements for mei-1 and mei-2 in oocyte meiotic spindle positioning (Yang et al., 2003; McNally et al., 2006) and for the decrease in spindle length that occurs around metaphase (McNally et al., 2006). More recently, an analysis of a mei-1 allele, ct103, isolated as an intragenic suppressor of a gain-of-function mei-1 allele, ct46 (Mains et al., 1990; Clandinin and Mains, 1993), showed that this homozygous viable katanin-defective mei-1 allele can recruit ASPM-1 to bipolar meiotic spindles and mediate a partial decrease in spindle length (McNally and McNally, 2011; Gomes et al., 2013). The mei-1(ct46ct103) allele was shown to have a substantial reduction in microtubule-severing activity compared with wild-type mei-1 when expressed in tissue culture cells and assayed by quantifying the immunofluorescence signal from microtubules (McNally and McNally, 2011; Gomes et al., 2013). These results suggest that the microtubule-severing activity of mei-1 is not required for oocyte meiotic spindle pole assembly. However, by quantifying spindle morphology dynamics over time in klp-18(-) mutants, which assemble monopolar spindles, we found that the microtubule-severing activity of katanin does contribute to meiotic spindle pole assembly. This conclusion is based on several observations. First, the partially katanin-defective allele mei-1(ct46ct103) assembles bipolar oocyte meiotic spindles, but the poles appear only loosely focused early in meiosis I. Second, decreasing microtubule-severing activity with mei-1(ct46ct103) or increasing resistance to microtubule severing with tba-2(sb27); tbb-2(sb26) leads to partial loss of monopolar spindle assembly in klp-18(-) mutants, as judged by the failure of oocyte chromosomes to move into as small a volume as they do in mutants lacking only klp-18 function. Third, whereas reducing aspm-1 function in klp-18(-) mutants does not significantly affect the movement of chromosomes into a smaller volume, reducing aspm-1 function in mutants that lack both klp-18 function and have reduced microtubule-severing activity further limited the ability of chromosomes to move into a smaller volume. This again supports a role for microtubule severing in pole assembly. Finally, double mutants that lack both aspm-1 function and microtubule severing fail to assemble bipolar spindles, even though the single mutants assemble bipolar, if initially loosely organized, spindles. These latter two observations support our conclusion that ASPM-1 recruitment by MEI-1 and MEI-1 microtubule-severing activity play independent roles in oocyte meiotic spindle pole assembly. However, microtubule severing appears to play a more significant role in pole assembly than does ASPM-1, as reducing ASPM-1 function alone in a klp-18(-) background did not significantly alter the movement of chromosomes into a more compact volume. Finally, whereas mei-1 is an essential gene, we do not know whether microtubule severing and pole assembly are each individually required for viability.
Our analysis does not provide mechanistic insight into how ASPM-1 and microtubule severing each promote pole assembly. Nevertheless, as illustrated in Figure 8, we speculate that severing of microtubules nucleated by chromatin during acentrosomal spindle assembly (Heald et al., 1996; Khodjakov et al., 2000) could liberate microtubules that contribute to spindle assembly (Burbank et al., 2006). Alternatively, it is possible that the mei-1(ct46ct103) and tba-2(sb27); tbb-2(sb26) mutants influence pole assembly not through microtubule severing itself, but perhaps through related interactions with microtubules that do not necessarily lead to severing. ASPM-1, whose polar recruitment depends on MEI-1 (McNally and McNally, 2011), in turn might bring minus ends into close proximity at spindle poles through its interactions with the NuMA-related protein LIN-5 and cytoplasmic dynein (van der Voet et al., 2009; Ellefson and McNally, 2011). Consistent with such a model, a GFP fusion to the C. elegans dynein heavy chain (GFP:DHC-1) was largely absent from mei-1(-) mutant spindles (Supplemental Figure S3).
FIGURE 8:
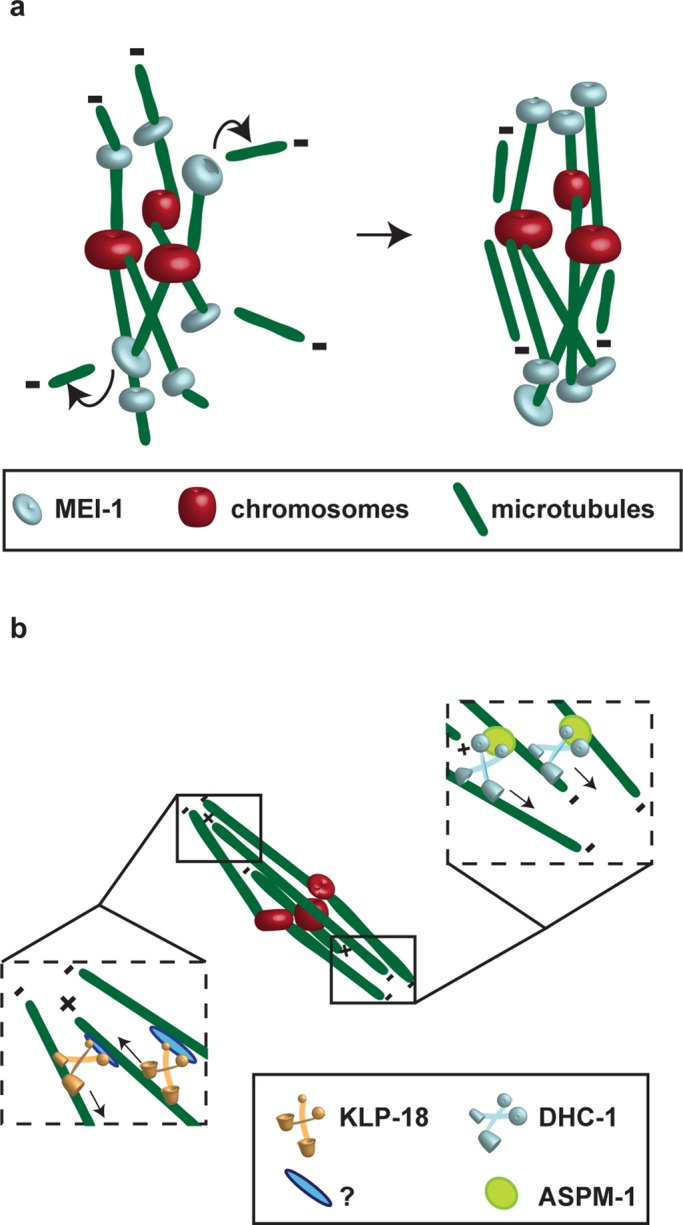
Models for how MEI-1 contributes to pole focusing by severing microtubules, whereas ASPM-1 cross-links parallel microtubules and KLP-18 promotes bipolarity by cross-linking antiparallel microtubules. (A) MEI-1 may mediate spindle pole assembly by severing chromatin-nucleated microtubules. (B) KLP-18 may promote bipolarity by cross-linking antiparallel microtubules with the help of an unknown factor. ASPM-1 may focus spindle poles by cross-linking the minus ends of parallel microtubules through a dynein-dependent mechanism; consistent with this model, GFP:DHC-1 is nearly absent from meiotic spindles in mei-1(-) oocyte meiotic spindles (Supplemental Figure S3). The recruitment of ASPM-1 to oocyte meiotic spindle poles requires MEI-1; however, it is not known whether this recruitment involves a direct or indirect interaction, and hence this relationship is not depicted.
In sum, our analysis identifies possible roles for two mei-1 functions—ASPM-1 recruitment and microtubule severing—in the assembly of oocyte meiotic spindle poles. However, we cannot rule out other roles for mei-1 in oocyte meiotic spindle assembly, involving, for example, the ability of the MEI-1 N-terminus to bind microtubules (McNally and McNally, 2011).
Quantitative analysis of oocyte meiotic spindle morphology–defective mutants can define spindle assembly pathways
Our understanding of oocyte meiotic spindle assembly has been limited in part by difficulties in observing oocyte meiotic cell divisions using live-cell imaging methods and by the very small size of the meiotic spindle. Our use of live imaging of oocyte meiotic spindle assembly in multiple mutant backgrounds, our quantification of spindle morphology dynamics, and our use of the spindle pole marker GFP:ASPM-1 together enabled us to identify previously unappreciated roles for genes known to be required for oocyte meiotic spindle assembly.
Our analysis of oocyte meiotic spindle assembly in addition benefited from the identification of conditional mutations in essential, maternally expressed genes. Many essential genes that are required for early embryogenesis also have earlier requirements during either gonadogenesis or germline development, with null alleles and RNAi often resulting in sterility and thus precluding or complicating the analysis of requirements during meiosis I and II. Conditional alleles also facilitate the construction and analysis of mutants lacking the function of multiple genes without having to rely entirely on the less efficient and less reproducible approach of using RNAi, especially when reducing multiple gene functions. Moreover, conditional mutations can be used to sensitize genetic backgrounds for modifier screens designed to identify additional factors (Dorfman et al., 2009; O'Rourke et al., 2011).
Further investigation of the molecular genetic pathways that control oocyte meiotic spindle assembly will benefit from continued efforts to identify conditional alleles of essential genes required for this process, the further application of live-cell imaging using fluorescent protein fusions to spindle-associated proteins, and the continued application of quantitative approaches to the analysis of spindle morphology and dynamics.
MATERIALS AND METHODS
C. elegans strains
N2 Bristol was used as the wild-type strain, with standard nematode protocols used as described (Brenner, 1974). The ts alleles were isolated in a screen for ts-embryonic lethal mutant using methods previously described (Kemphues et al., 1988; O'Connell et al., 1998; Encalada et al., 2000). The ts mutations were isolated from a lin-2(e1309) background and outcrossed at least three times with the N2 strain as described (Encalada et al., 2000). The ts strains were maintained at the permissive temperature of 15°C, and L4s were shifted to the restrictive temperature, 26°C, for 2–5 h before young adult worms were cut open to isolate mutant embryos for phenotypic analysis, bypassing larval lethality and sterility phenotypes that result from earlier upshifts (Table 2). The ts mutations and deletion alleles are listed by chromosomes: mei-1(or646ts) I; mei-1(or1178ts) I; mei-1(ct46ct103) I; aspm-1(or645ts) I; aspm-1(ok1208) I; mei-2(sb39ts) I; klp-18(or447ts) IV; klp-18(ok2519) IV; bmk-1(or627ts) IV; bmk-1(tm696) V; bmk-1(ok391) V. Transgenic GFP:β-tubulin and mCherry:Histone2B strains were derived from AZ244 (GFP:β-tubulin) and OD56 (mCherry:Histone2B), respectively. A GFP:ASPM-1 construct was made by recombineering using the fosmid WRM0624aG02 as previously described (O'Rourke et al., 2007). Transgenic strains were generated by microparticle bombardment as previously described (Praitis et al., 2001). The GFP:ASPM-1 fusion was functional, in that it rescued embryonic lethality when crossed into the aspm-1(or645ts) mutant background (unpublished data). Transgenic strains were crossed into a him-5(e1490) background, and males from the him-5 strains were used to introduce the transgenes into ts mutant strains.
Genetic analysis
Viability counts of embryos (percentage hatching) were determined by singling at least seven L4s onto individual plates, growing them at the permissive or restrictive temperature until eggs were laid, and then removing the mother and counting the eggs. We allowed the embryos to develop for 18–24 h and then counted the unhatched embryos. With all four alleles, we found that mutant/+ worms produced roughly one-fourth homozygous self-progeny (unpublished data), indicating that each recessive mutant phenotype is caused by mutation(s) at single or tightly linked loci. Complementation tests were performed by first generating him-8 or him-5 strains that were homozygous for a ts mutant allele; males from these strains were then crossed into heterozygous strains carrying the deletion alleles. Embryonic viability in the broods of F1s was scored at the restrictive temperature as described. Recessive gain-of-function test of bmk-1(or627ts) allele (using RNAi to knock down the mutant gene product) was performed after using hypochlorite to isolate embryos from homozygous mutant worms and leaving them in M9 overnight at 15°C. The synchronized hatched and starved homozygous mutant L1s were then put onto agar plates seeded with Escherichia coli expressing double-stranded bmk-1 RNA (Fraser et al., 2000; see later description) and matured to adulthood at 26°C. Young adults were then removed, and the eggs laid were scored for percentage hatching as described.
Positional cloning of TS mutant loci
Using visible markers, we found that or645ts resides within a 1.4-cM interval between unc-29 and lin-11 on linkage group 1 (unpublished data). Data from genome-wide RNAi knockdown screens (www.wormbase.org) indicate that aspm-1 is the only gene in this region that results in the production of one-cell zygotes with multiple maternal pronuclei, as observed in or645ts mutants. Furthermore, or645ts failed to complement the aspm-1 deletion allele ok1208 (unpublished data). Homozygous or645ts genomic DNA was purified using a DNeasy Blood and Tissue kit (Qiagen, Valencia, CA). We amplified overlapping DNA fragments that span the aspm-1 locus by PCR and purified the product using QIAquick PCR Purification kit (Qiagen). Overlapping DNA fragments were sequenced by Sequetech (Mountain View, CA).
To identify the causal mutation in or447ts mutants, we again used visible markers and mapped the mutation to a 0.11-cM interval between dpy-20 and bli-6 on linkage group 4. The only C. elegans gene within this interval that, when reduced in function using RNAi, results in oocyte meiotic cell division defects is the kinesin 12 family member klp-18. Moreover, in genetic crosses, or447ts failed to complement a nonconditional and likely null klp-18 deletion allele, ok2519 (unpublished data), consistent with or447ts being a klp-18 allele. From or447ts genomic DNA, we PCR amplified and sequenced overlapping DNA fragments that span the klp-18 locus.
Using a genome-wide single nucleotide polymorphism (SNP) mapping and whole-genome sequencing approach (Doitsidou et al., 2010), we mapped or1178ts to an ∼9-Mb region on Linkage group 1. The mei-1 gene was the most promising candidate in this region, based on both phenotypic similarity to or1178ts as reported in earlier studies of mei-1 mutants (Srayko et al., 2000) and the results from multiple genome-wide RNAi screens (www.wormbase.org). The allele or1178ts failed to complement the previously identified mei-1(or646ts) allele while complementing both aspm-1(or645ts) and mei-2(sb39ts) mutants, consistent with or1178ts being a mei-1 allele. From or1178ts genomic DNA, we PCR amplified and sequenced overlapping fragments spanning the mei-1 locus.
We used PCR-targeted SNP mapping to place or627ts within a small interval from 6.24 to 6.54 Mb on LG V (unpublished data). There were no annotated genes in this region that when knocked down by RNAi resulted in an or627ts-like mutant phenotype. We therefore used a genome interval pull-down method, followed by Illumina-based DNA sequencing, to identify any mutations within this interval that might be responsible for the embryonic lethality (O'Rourke et al., 2011). The DNA sequence data revealed a single missense mutation in one annotated gene within this interval (O'Rourke et al., 2011; Supplemental Figure S1B). In vertebrates, this kinesin is required for bipolar mitotic spindle assembly, but, surprisingly, analysis of RNAi knockdown defects and results from analysis of two bmk-1–deletion alleles indicate that C. elegans bmk-1/kinesin 5 is not essential, is not required for meiotic spindle assembly, and has only a minor role in mitotic spindle assembly dynamics (Bishop et al., 2005; Saunders et al., 2007). We therefore asked whether or627ts might be a recessive gain-of-function mutation. Consistent with this possibility, or627ts partially failed to complement the bmk-1 deletions alleles tm969 and ok391 (Table 1). More conclusively, when we used RNAi to knock down mutant bmk-1 function in homozygous or627ts worms, embryonic viability at the restrictive temperature was strongly rescued (Table 1).
RNA interference
RNAi to reduce the function of mei-1, klp-18 or aspm-1 was performed by three methods: feeding, injection, and soaking (Fire et al., 1998). RNAi by feeding was performed by placing L1 larvae synchronized by hypochlorite as described previously (Kamath et al., 2001). For codepletions, we used the same process, but we seeded plates with an equal mixture of the double-strand (ds) RNA–expressing E. coli strains. We maintained the affected worms at room temperature and examined their phenotypes in zygotes within whole-mounted young adults. The ts strains were treated in the same way but maintained at 15°C and shifted to 26°C as L4 larvae. RNAi experiments by injection were done as described previously (Zipperlen et al., 2001) 24 h before imaging. To create the double-stranded RNA, a PCR product was generated from the aspm-1 and klp-18 clones available from the Ahringer library (Fraser et al., 2000). RNAi was produced by in vitro transcription using RiboMAX kits (Promega, Madison, WI), and the sample was diluted to 0.8 μg/μl. Finally, RNAi by soaking was performed by purifying dsRNA and diluting in a soaking buffer according to WormBook (www.wormbook.org). L4 worms were soaked for 24 h at room temperature and imaged as young adults.
Microscopy
Live imaging of fluorescent fusion proteins during meiosis was accomplished by mounting embryos on 8% agar pads with 1 μl each of 1-μm polystyrene beads and M9 on microscope slides covered with a coverslip. Zygotes were analyzed on a DMI 4000B microscope (Leica, Wetzlar, Germany) fitted with a Leica 63×/1.40–0.60 HCX Plan Apo oil objective lens in a room maintained at 25°C. Time-lapse videos were obtained with a Hamamatsu (Hamamatsu, Japan) electron-multiplying charge-coupled device digital camera using Volocity software (PerkinElmer, Waltham, MA). Six stacks of 1.5-μm thickness were used to collect images every 10 s. After recording, the videos were cropped and adjusted for contrast in ImageJ (National Institutes of Health, Bethesda, MD). Measurements of chromosome compaction were taken using the ImageJ polygon selection tool to trace around the two-dimensional perimeter of chromosomes to obtain an area. The area of pixels was then multiplied by 0.214 μm2/pixel, a value specific to our Leica DMI 4000B.
Supplementary Material
Acknowledgments
We thank the C. elegans Genetic Center (funded by the National Institutes of Health National Center for Research Resources) and the Mitani lab for strains. This work was supported by National Institutes of Health Grant GM049869 (B.B.), the National Institutes of Health (S.L.), the Canadian Institute of Health Research (P.E.M.), and an American Heart Pre-doctoral Fellowship (A.C.). We are grateful to Chris Doe for comments on the manuscript and to members of the Bowerman lab for productive discussions.
Abbreviations used:
- AAA
ATPases associated with diverse cellular activities
- Asp
Abnormal Spindle
- aspm-1
Abnormal Spindle Morphology-1
- bmk-1
BiMC-related kinesin-1
- GFP
green fluorescent protein
- klp-18
kinesin-like protein-18
- mei-1
defective meiosis-1
- TPX-2
target protein for Xenopus kinesin-like protein-2
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-11-0687) on February 19, 2014.
*Present address: Department of Developmental Biology, Stanford University School of Medicine, Stanford, CA 94305.
REFERENCES
- Albertson DG. Formation of the first cleavage spindle in nematode embryos. Dev Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- Albertson DG, Thomson JN. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1993;1:15–26. doi: 10.1007/BF00710603. [DOI] [PubMed] [Google Scholar]
- Bishop JD, Han Z, Schumacher JM. The Caenorhabditis elegans Aurora B kinase AIR-2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles. Mol Biol Cell. 2005;16:742–756. doi: 10.1091/mbc.E04-08-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbank KS, Groen AC, Perlman ZE, Fisher DS, Mitchison TJ. A new method reveals microtubule minus ends throughout the meiotic spindle. J Cell Biol. 2006;175:369–375. doi: 10.1083/jcb.200511112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin TR, Mains PE. Genetic studies of mei-1 gene activity during the transition from meiosis to mitosis in Caenorhabditis elegans. Genetics. 1993;134:199–210. doi: 10.1093/genetics/134.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire S, Mains PE. mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics. 1994;136:533–546. doi: 10.1093/genetics/136.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M, Poole RJ, Sarin S, Bigelow H, Hobert O. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS One. 2010;5:e15435. doi: 10.1371/journal.pone.0015435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman M, Gomes JE, O'Rourke S, Bowerman B. Using RNA interference to identify specific modifiers of a temperature-sensitive, embryonic-lethal mutation in the Caenorhabditis elegans ubiquitin-like Nedd8 protein modification pathway E1-activating gene rfl-1. Genetics. 2009;182:1035–1049. doi: 10.1534/genetics.109.104885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefson ML, McNally FJ. Kinesin-1 and cytoplasmic dynein act sequentially to move the meiotic spindle to the oocyte cortex in Caenorhabditis elegans. Mol Biol Cell. 2009;20:2722–2730. doi: 10.1091/mbc.E08-12-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefson ML, McNally FJ. CDK-1 inhibits meiotic spindle shortening and dynein-dependent spindle rotation in C. elegans. J Cell Biol. 2011;193:1229–1244. doi: 10.1083/jcb.201104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encalada SE, Martin PR, Phillips JB, Lyczak R, Hamill DR, Swan KA, Bowerman B. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev Biol. 2000;228:225–238. doi: 10.1006/dbio.2000.9965. [DOI] [PubMed] [Google Scholar]
- Fabritius AS, Ellefson ML, McNally FJ. Nuclear and spindle positioning during oocyte meiosis. Curr Opin Cell Biol. 2011;23:78–84. doi: 10.1016/j.ceb.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Gomes JE, Tavernier N, Richaudeau B, Formstecher E, Boulin T, Mains PE, Dumont J, Pintard L. Microtubule severing by the katanin complex is activated by PPFR-1-dependent MEI-1 dephosphorylation. J Cell Biol. 2013;202:431–439. doi: 10.1083/jcb.201304174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman JJ, Mahr J, McNally K, Okawa K, Iwamatsu A, Thomas S, Cheesman S, Heuser J, Vale RD, McNally FJ. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 1998;93:277–287. doi: 10.1016/s0092-8674(00)81578-0. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J Cell Biol. 1997;138:615–628. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Busso C, Fluckiger I, Gonczy P. Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryos. Dev Cell. 2009;17:900–907. doi: 10.1016/j.devcel.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Lu C, Mains PE. Mutations of a redundant alpha-tubulin gene affect Caenorhabditis elegans early embryonic cleavage via MEI-1/katanin-dependent and -independent pathways. Genetics. 2005;170:115–126. doi: 10.1534/genetics.104.030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Srayko M, Mains PE. The Caenorhabditis elegans microtubule-severing complex MEI-1/MEI-2 katanin interacts differently with two superficially redundant beta-tubulin isotypes. Mol Biol Cell. 2004;15:142–150. doi: 10.1091/mbc.E03-06-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains PE, Kemphues KJ, Sprunger SA, Sulston IA, Wood WB. Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics. 1990;126:593–605. doi: 10.1093/genetics/126.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KP, McNally FJ. The spindle assembly function of Caenorhabditis elegans katanin does not require microtubule-severing activity. Mol Biol Cell. 2011;22:1550–1560. doi: 10.1091/mbc.E10-12-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier S, Vernos I. Microtubule assembly during mitosis—from distinct origins to distinct functions. J Cell Sci. 2012;125:2805–2814. doi: 10.1242/jcs.092429. [DOI] [PubMed] [Google Scholar]
- Muller-Reichert T, Greenan G, O'Toole E, Srayko M. The elegans of spindle assembly. Cell Mol Life Sci. 2010;67:2195–2213. doi: 10.1007/s00018-010-0324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell KF, Leys CM, White JG. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics. 1998;149:1303–1321. doi: 10.1093/genetics/149.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, et al. A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PLoS One. 2011;6:e16644. doi: 10.1371/journal.pone.0016644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Dorfman MD, Carter JC, Bowerman B. Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genet. 2007;3:e128. doi: 10.1371/journal.pgen.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozlu N, Srayko M, Kinoshita K, Habermann B, O'Toole ET, Muller-Reichert T, Schmalz N, Desai A, Hyman AA. An essential function of the C. elegans ortholog of TPX2 is to localize activated aurora A kinase to mitotic spindles. Dev Cell. 2005;9:237–248. doi: 10.1016/j.devcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli MG, Callaini G, Glover DM, Avides Mdo C. A requirement for the Abnormal Spindle protein to organise microtubules of the central spindle for cytokinesis in Drosophila. J Cell Sci. 2002;115:913–922. doi: 10.1242/jcs.115.5.913. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Powers J, Strome S, Saxton WM. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol. 2007;17:R453–R454. doi: 10.1016/j.cub.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segbert C, Barkus R, Powers J, Strome S, Saxton WM, Bossinger O. KLP-18, a Klp2 kinesin, is required for assembly of acentrosomal meiotic spindles in Caenorhabditis elegans. Mol Biol Cell. 2003;14:4458–4469. doi: 10.1091/mbc.E03-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MH, Liu Y, Anderson DE, Jahng WJ, O'Connell KF. Protein phosphatase 2A-SUR-6/B55 regulates centriole duplication in C. elegans by controlling the levels of centriole assembly factors. Dev Cell. 2011;20:563–571. doi: 10.1016/j.devcel.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko M, Buster DW, Bazirgan OA, McNally FJ, Mains PE. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 2000;14:1072–1084. [PMC free article] [PubMed] [Google Scholar]
- Srayko M, O'Toole ET, Hyman AA, Muller-Reichert T. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr Biol. 2006;16:1944–1949. doi: 10.1016/j.cub.2006.08.029. [DOI] [PubMed] [Google Scholar]
- Sturgill EG, Ohi R. Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr Biol. 2013;23:1280–1290. doi: 10.1016/j.cub.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol. 2009;19:1703–1711. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- van der Voet M, Berends CW, Perreault A, Nguyen-Ngoc T, Gonczy P, Vidal M, Boxem M, van den Heuvel S. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat Cell Biol. 2009;11:269–277. doi: 10.1038/ncb1834. [DOI] [PubMed] [Google Scholar]
- Vanneste D, Takagi M, Imamoto N, Vernos I. The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr Biol. 2009;19:1712–1717. doi: 10.1016/j.cub.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Weinger JS, Qiu M, Yang G, Kapoor TM. A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr Biol. 2011;21:154–160. doi: 10.1016/j.cub.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wignall SM, Villeneuve AM. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat Cell Biol. 2009;11:839–844. doi: 10.1038/ncb1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto I, Kosinski ME, Greenstein D. Start me up: cell signaling and the journey from oocyte to embryo in C. elegans. Dev Dyn. 2006;235:571–585. doi: 10.1002/dvdy.20662. [DOI] [PubMed] [Google Scholar]
- Yang HY, Mains PE, McNally FJ. Kinesin-1 mediates translocation of the meiotic spindle to the oocyte cortex through KCA-1, a novel cargo adapter. J Cell Biol. 2005;169:447–457. doi: 10.1083/jcb.200411132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HY, McNally K, McNally FJ. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C elegans. Dev Biol. 2003;260:245–259. doi: 10.1016/s0012-1606(03)00216-1. [DOI] [PubMed] [Google Scholar]
- Zipperlen P, Fraser AG, Kamath RS, Martinez-Campos M, Ahringer J. Roles for 147 embryonic lethal genes on C.elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 2001;20:3984–3992. doi: 10.1093/emboj/20.15.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.