Autophagosome–lysosome fusion requires the autophagosomal SNARE syntaxin 17. Syntaxin 17 interacts with the HOPS-tethering complex. HOPS is required for syntaxin 17–dependent autophagosome–lysosome fusion, besides its function in endolysosomal fusion.
Abstract
Membrane fusion is generally controlled by Rabs, soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs), and tethering complexes. Syntaxin 17 (STX17) was recently identified as the autophagosomal SNARE required for autophagosome–lysosome fusion in mammals and Drosophila. In this study, to better understand the mechanism of autophagosome–lysosome fusion, we searched for STX17-interacting proteins. Immunoprecipitation and mass spectrometry analysis identified vacuolar protein sorting 33A (VPS33A) and VPS16, which are components of the homotypic fusion and protein sorting (HOPS)–tethering complex. We further confirmed that all HOPS components were coprecipitated with STX17. Knockdown of VPS33A, VPS16, or VPS39 blocked autophagic flux and caused accumulation of STX17- and microtubule-associated protein light chain (LC3)–positive autophagosomes. The endocytic pathway was also affected by knockdown of VPS33A, as previously reported, but not by knockdown of STX17. By contrast, ultraviolet irradiation resistance–associated gene (UVRAG), a known HOPS-interacting protein, did not interact with the STX17–HOPS complex and may not be directly involved in autophagosome–lysosome fusion. Collectively these results suggest that, in addition to its well-established function in the endocytic pathway, HOPS promotes autophagosome–lysosome fusion through interaction with STX17.
INTRODUCTION
Macroautophagy (referred to as autophagy hereafter) is an intracellular degradation system that delivers cytoplasmic components such as large molecules and organelles to the lysosome. Autophagy consists of several sequential steps: autophagosome formation, autophagosome–lysosome fusion, degradation of autophagic materials, efflux of degradation products, and autophagic lysosome reformation (Tooze and Yoshimori, 2010; Mizushima et al., 2011; Chen and Yu, 2013). The molecular mechanism regulating the fusion of autophagosomes with lysosomes has not been fully established. It is likely that the canonical intracellular fusion machinery composed of Rabs, soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs), and tethers controls autophagosome–lysosome fusion.
The role of the Rab GTPase Rab7 in autophagosome–vacuole/lysosome fusion has been extensively studied. Autophagosomes accumulate in a Saccharomyces cerevisiae mutant that lacks the yeast Rab7 orthologue, Ypt7 (Kirisako et al., 1999). Mammalian Rab7 also regulates the maturation of late autophagic vacuoles dependent on its GTPase activity (Gutierrez et al., 2004; Jäger et al., 2004). Recently we reported that the Qa-SNARE syntaxin 17 (STX17) is inserted into completed autophagosomes via its unusual C-terminal hairpin-like structure and mediates autophagosome–lysosome fusion by binding to its partner SNAREs—synaptosomal-associated protein 29 (SNAP29; Qbc-SNARE) and vesicle-associated membrane protein 8 (VAMP8; R-SNARE)—in mammalian cells (Itakura et al., 2012b). These fusion factors are functionally conserved in Drosophila (Takáts et al., 2013). In yeast, although several SNAREs are involved in autophagosome–vacuolar fusion (Darsow et al., 1997; Sato et al., 1998; Fischer von Mollard and Stevens, 1999; Ishihara et al., 2001; Ohashi and Munro, 2010), there are no reports of an autophagosomal SNARE in yeast.
On the other hand, the involvement of tethering factors acting between the autophagosome and lysosome is less clear. Homotypic fusion and protein sorting (HOPS) is a conserved protein complex consisting of vacuolar protein sorting 11 (Vps11), Vps16, Vps18, Vps33, Vps39, and Vps41 (Rieder and Emr, 1997; Seals et al., 2000; Wurmser et al., 2000). It was originally identified in yeast as a tethering complex that is now known to be generally important for vacuolar/lysosomal fusion events from yeast to mammals (Balderhaar and Ungermann, 2013; Solinger and Spang, 2013). Metazoans have two Vps33 isoforms, Vps33A and Vps33B. It has been proposed that Vps33A functions in endolysosomal fusion in the HOPS complex, whereas Vps33B belongs to another hexameric tethering complex—class C core vacuole/endosome tethering (CORVET), which is mainly implicated in endosomal fusion (Akbar et al., 2009; Balderhaar and Ungermann, 2013). The requirement of the HOPS complex in the autophagy pathway is relatively clear in yeast. Yeast Vps18 is required for vacuolar biogenesis and autophagosome–vacuole fusion (Rieder and Emr, 1997). Recently it was reported that depletion of Vps33 impairs autophagosome–vacuole fusion in Aspergillus nidulans (Pinar et al., 2013). However, the involvement of the HOPS complex in autophagosome–lysosome fusion in mammals remains rather controversial. Liang et al. (2008) showed that knockdown of VPS16 reduces microtubule-associated protein light chain 3 (LC3)–lysosomal-associated membrane protein 1 (LAMP1) colocalization, even though LC3 puncta do not accumulate. Another HOPS component, VPS18, is also reported to be critical for autophagosome clearance in the mouse brain (Peng et al., 2012). However, Ganley et al. (2011) showed that autophagic flux is not affected when VPS16 is depleted in mouse cells. Therefore it is necessary to obtain additional mechanistic information on how HOPS functions in the autophagy pathway.
Furthermore, the role of ultraviolet irradiation resistance-associated gene (UVRAG), which is a subunit of the class III phosphatidylinositol 3-kinase (PtdIns 3-kinase) complex also interacting with HOPS, is controversial. UVRAG was originally reported to function in the autophagosome formation step together with Beclin 1 (Liang et al., 2006). Later it was proposed that UVRAG is a homologue of yeast Vps38 and is not required for autophagosome formation (Itakura et al., 2008; Farre et al., 2010; Knaevelsrud et al., 2010). In yeast, Vps38 is included in a class III PtdIns 3-kinase complex specific to the vacuolar protein sorting pathway (Kihara et al., 2001; Burda et al., 2002). In a separate study, UVRAG was shown to interact with HOPS and be required for autophagosome–lysosome fusion (Liang et al., 2008). UVRAG also plays other roles in antiapoptosis (Yin et al., 2011) and chromosomal stability (Zhao et al., 2012).
A potential problem underlying these controversies is that any factors that affect endolysosomes could secondarily inhibit autophagosome–lysosome fusion or autophagic degradation (Rusten and Simonsen, 2008; Lieberman et al., 2012), which makes it difficult to identify bona fide fusion factors. In the present study, we searched for STX17-interacting proteins to identify the factors directly involved in the autophagosome–lysosome fusion process. We found that STX17 interacts with HOPS and that HOPS promotes STX17-dependent autophagosome–lysosome fusion in addition to its well-known function in endolysosomal fusion. By contrast, UVRAG seems not to be directly involved in autophagosome–lysosome fusion.
RESULTS
STX17 interacts with the HOPS complex
To search for proteins that function in the autophagosome–lysosome fusion step together with STX17, we expressed human FLAG-tagged STX17 in HEK 293 cells, immunoprecipitated them with anti-FLAG antibody, and subjected them to highly sensitive direct nanoflow liquid chromatography/tandem mass spectrometry (Natsume et al., 2002). As a result, two HOPS components, VPS33A and VPS16, were identified as STX17-binding proteins. We confirmed that FLAG-STX17 interacts with Myc-tagged VPS33A (Figure 1A) and green fluorescent protein (GFP)–tagged VPS16 (Figure 1B). The interaction was not or only slightly enhanced under starvation conditions (Supplemental Figure S1A). By contrast, the interaction between VPS33A and STX18 (another endoplasmic reticulum [ER] SNARE) was not detected under either growing or starvation conditions (Supplemental Figure S1B). In addition, interactions between STX17 and all other HOPS components—VPS11, VPS18, VPS39, and VPS41—were detected (Figure 1, C and D). Furthermore, endogenous VPS33A and VPS16 were precipitated with GFP-STX17 (Figure 1E). However, STX17 did not interact with the other homologue of yeast Vps33, VPS33B (unpublished data). Because a previous report showed that the HOPS components also interact with UVRAG (Liang et al., 2008), we tested whether UVRAG was included in the same STX17 complex. However, UVRAG was not precipitated with GFP-STX17 (Figure 1E), whereas it was indeed precipitated with GFP-VPS33A (Figure 1F). These results suggest that STX17 associates with the HOPS complex but not with UVRAG.
FIGURE 1:
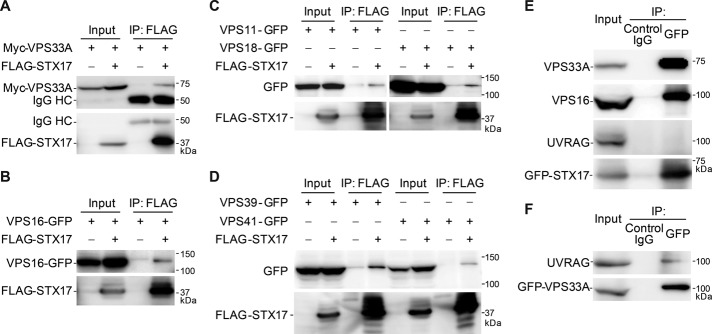
STX17 interacts with HOPS subunits. (A–D) STX17 interacts with VPS33A (A), VPS16 (B), and other components of the HOPS complex (C, D). HEK 293T cells were transfected with FLAG-STX17 and the indicated Myc- or GFP-tagged HOPS subunits. Three days later, cells were lysed and immunoprecipitated with anti-FLAG antibody (M2 affinity gel). Input corresponds to 6% of the cell lysate used for each immunoprecipitation. (E) STX17 interacts with endogenous VPS33A and VPS16 but not with UVRAG. HeLa cells stably expressing GFP-STX17 were cultured in starvation medium for 2 h, followed by immunoprecipitation using mouse IgG1 isotype control or anti-GFP antibody. The immunoprecipitates were then immunoblotted with anti-VPS33A, anti-VPS16, anti-UVRAG, and anti-STX17 antibodies. (F) Cell lysates of HeLa cells stably expressing GFP-VPS33A were subjected to immunoprecipitation using anti-GFP antibody and endogenous UVRAG was detected.
HOPS translocates to STX17-positive autophagosomes
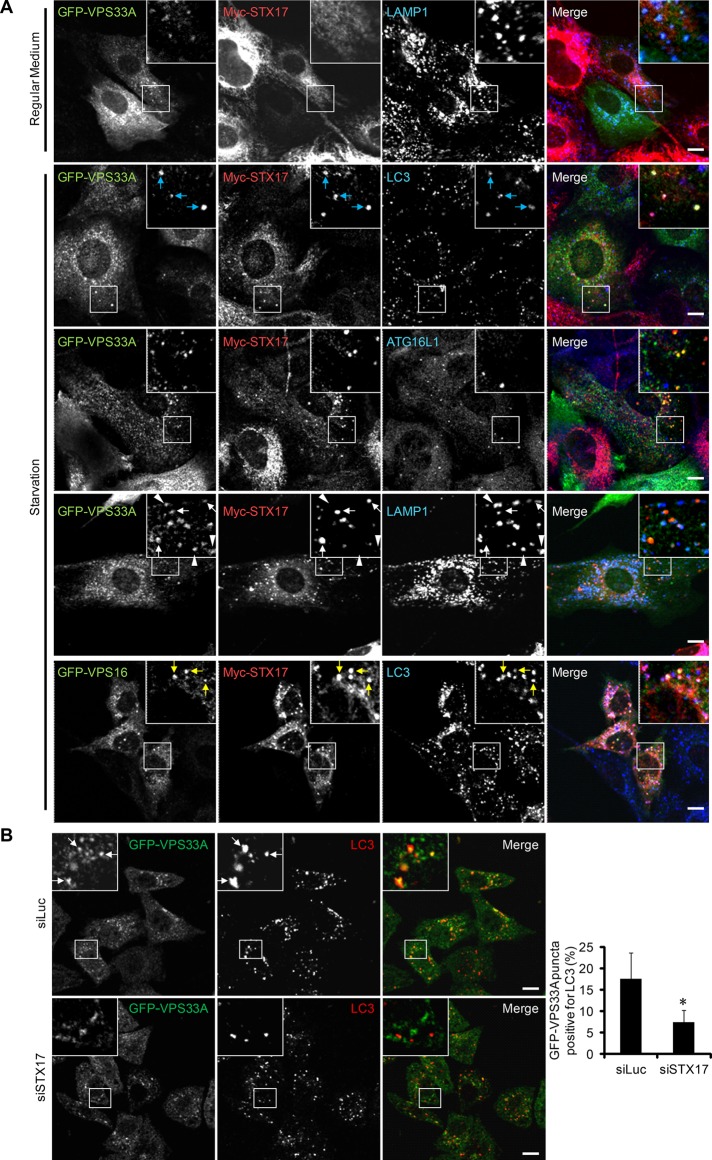
We further confirmed the interaction between STX17 and HOPS by immunocytochemistry. Under growing conditions, Myc-STX17 showed reticular and tubular patterns, which should represent the endoplasmic reticulum and mitochondria, as previously reported (Itakura et al., 2012b; Hamasaki et al., 2013; Figure 2A). GFP-VPS33A localized mainly to LAMP1-positive endolysosomes and did not colocalize with STX17 (Figure 2A). Under starvation conditions, Myc-STX17 formed punctate structures, and most of them colocalized with the autophagosomal marker LC3 (Figure 2A). Moreover, GFP-VPS33A translocated to these STX17- and LC3-positive structures, and these structures were frequently closely associated with LAMP1 signals but not completely integrated. Some GFP-VPS33A and LAMP1 double-positive structures were negative for Myc-STX17, suggesting that not all the GFP-VPS33A was translocated to autophagosomes. However, GFP-VPS33A did not colocalize to ATG16L1-positive isolation membranes (also known as phagophores; Figure 2A). VPS16 also colocalized with STX17 and LC3. These data suggest that the HOPS components are recruited to completed autophagosomes, where STX17 is also localized.
FIGURE 2:
VPS33A and VPS16 translocate to STX17-positive autophagosomes. (A) MEFs stably coexpressing Myc-STX17 and either GFP-VPS33A or GFP-VPS16 were cultured in regular medium or starvation medium for 2 h. Cells were costained with anti-Myc antibody and anti-LAMP1, anti-LC3, or anti-ATG16L1 antibody and analyzed by immunofluorescence microscopy. Structures positive for VPS33A, STX17, and LC3 are indicated by blue arrows, structures positive for VPS33A and STX17 but not for LAMP1 are indicated by white arrows, structures positive for VPS33A and LAMP1 but not for STX17 are indicated by white arrowheads, and structures positive for VPS16, STX17, and LC3 are indicated by yellow arrows. Signal color is indicated by the color of the text. Scale bars,10 μm. (B) HeLa cells stably expressing GFP-VPS33A were treated with siRNA against luciferase or STX17. After 72 h, cells were starved for an additional 2 h. Then cells were fixed, stained with anti-GFP and anti-LC3 antibodies, and analyzed by confocal microscopy. Structures positive for GFP-VPS33A and LC3 are indicated by white arrows. Scale bars, 10 μm. Quantification of the colocalization rate between endogenous LC3 and GFP-VPS33A is shown in the graph. Data represent mean ± SEM of at least 10 different cells. *p < 0.05 compared with the luciferase siRNA–treated group.
We also notified that some of the STX17- and LC3-positive structures were negative for VPS33A or VPS16 (Figure 2A), suggesting that recruitment of HOPS to autophagosomes is preceded by that of STX17. We then examined whether STX17 is required for the recruitment of the HOPS complex to autophagosomes. Knockdown of STX17 reduced the colocalization between GFP-VPS33A and LC3 (Figure 2B). These results suggest that STX17 promotes the autophagosomal association of HOPS.
The HOPS complex is required for autophagic activity
Although the role of the HOPS complex in the autophagy pathway is somewhat controversial (Liang et al., 2008; Ganley et al., 2011; Peng et al., 2012), our data clearly demonstrate that HOPS localizes on the autophagosomes and interacts with the autophagosomal SNARE STX17. This finding prompted us to further determine whether HOPS is required for autophagic activity.
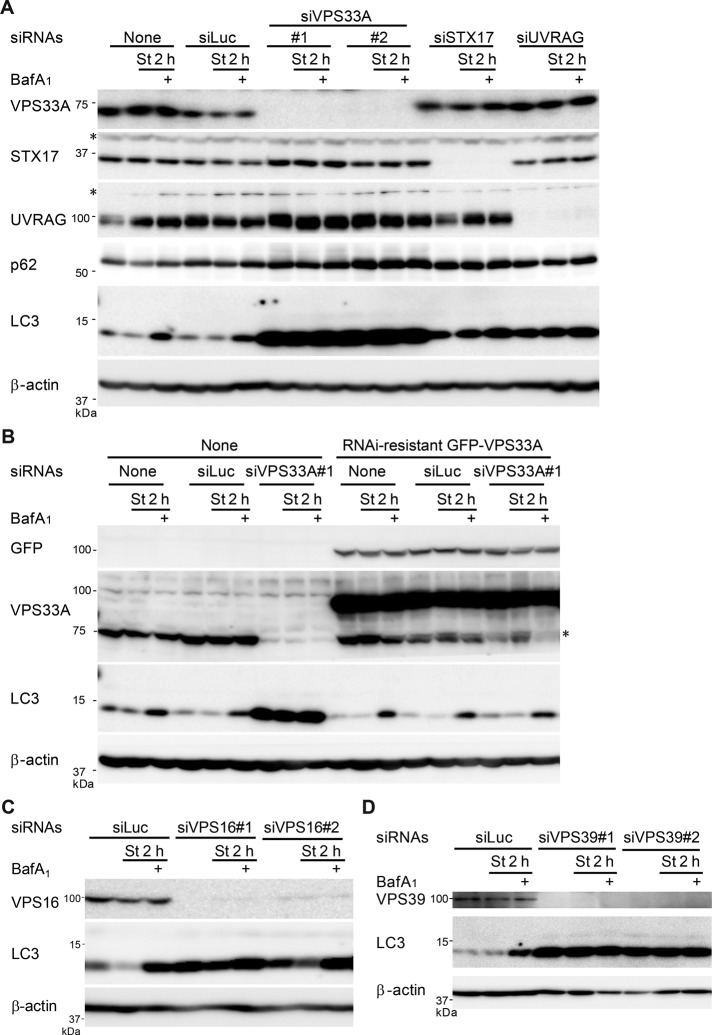
When VPS33A, STX17, or UVRAG was silenced with siRNAs, LC3-II accumulated, even under growing conditions (Figure 3A). LC3-II level was not further increased by treatment of the vacuolar ATPase inhibitor bafilomycin A1, suggesting impaired LC3-II turnover in these cells (Figure 3A). Moreover, starvation-induced degradation of p62, a selective autophagy substrate, was suppressed in these cells (Figure 3A), indicating that autophagic flux is blocked.
FIGURE 3:
Silencing of HOPS complexes blocks autophagic flux. (A) HeLa cells were treated with siRNA oligonucleotides against luciferase, VPS33A, STX17, or UVRAG. Then cells were cultured in regular or starvation medium for 2 h with or without 100 nM bafilomycin A1 and analyzed by immunoblotting using the indicated antibodies. An asterisk indicates a nonspecific band. (B) HeLa cells stably expressing siRNA-resistant GFP-VPS33A were treated with siRNA oligonucleotides against luciferase or VPS33A. Cells were cultured and analyzed as in A. An asterisk indicates a degradation product of GFP-VPS33A, which appears at a position similar to that of endogenous VPS33A. (C, D) HeLa cells were also treated with siRNA oligonucleotides against luciferase, VPS16 (C), or VPS39 (D) and analyzed as in A.
Expression of small interfering RNA (siRNA)–resistant VPS33A, which has silent mutations in the siRNA target sequence, restored the impaired autophagic flux caused by VPS33A knockdown (Figure 3B), confirming that the phenotype of VPS33A depletion is not due to an off-target effect. Furthermore, knockdown of other HOPS components, VPS16 (Figure 3C) and VPS39 (Figure 3D), phenocopied the effect of VPS33A depletion.
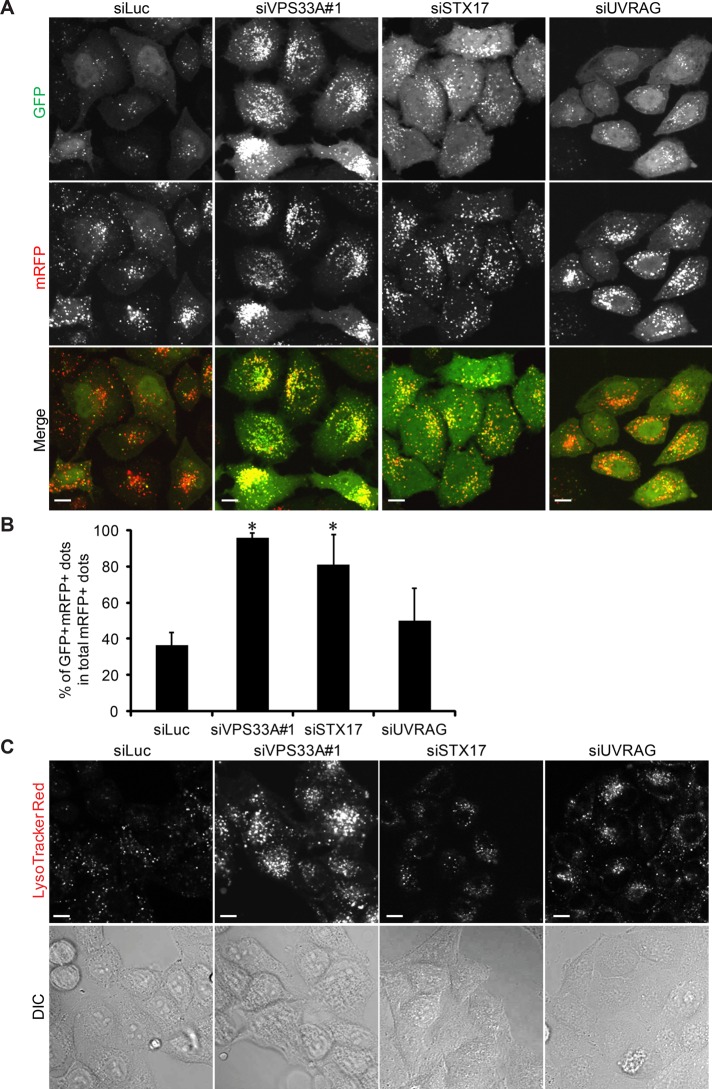
To further investigate the mechanisms by which HOPS knockdown impaired autophagic degradation, we used HeLa cells stably expressing the monomeric red fluorescent protein (mRFP)–GFP-LC3 tandem reporter (Kimura et al., 2007). GFP fluorescence is rapidly quenched in the lysosomal acidic environment, whereas mRFP fluorescence remains (Kimura et al., 2007). In control siRNA–treated cells, starvation treatment caused accumulation of many red puncta (which indicate autolysosomes) with a smaller number of yellow puncta (which indicate autophagosomes; Figure 4A). However, knockdown of VPS33A or STX17 led to massive accumulation of yellow puncta, suggesting that transition of double GFP- and mRFP-positive autophagosomes to mRFP-positive, GFP-negative autolysosomes is impaired (Figure 4, A and B). This could be due to decrease in autophagosome–lysosome fusion or increase in lysosomal pH. To discriminate between these two possibilities, we performed LysoTracker Red staining in HeLa cells treated with control, VPS33A, or STX17 siRNA. As a result, compared with control siRNA–treated cells, the LysoTracker signal was not decreased in VPS33A or STX17 siRNA–treated cells and in fact was rather increased in VPS33A siRNA–treated cells (Figure 4C). These results suggest that HOPS is required for autophagic activity and probably functions in a late step of the autophagy pathway.
FIGURE 4:
Knockdown of VPS33A and STX17 impairs autophagosome maturation. (A) HeLa cells stably expressing the mRFP-GFP-LC3 construct were transfected with siRNA against luciferase, VPS33A, STX17, or UVRAG. After 72 h, cells were starved for an additional 2 h, then fixed and analyzed by confocal microscopy. Scale bars, 10 μm. (B) Quantification of the results in A. Ratio (%) of double GFP- and mRFP-positive LC3 puncta (autophagosomes) per total mRFP-positive LC3 puncta (autophagosomes and autolysosomes). Data are mean ± SEM for 10 randomly selected images. *p < 0.05 compared with the luciferase siRNA–treated group. (C) Confocal images of live siLuciferase-, siVPS33A-, or siUVRAG-transfected HeLa cells stained with LysoTracker Red dye (top). Bottom, differential interference contrast images. Scale bars, 10 μm.
The HOPS complex is required for autophagosome–lysosome fusion
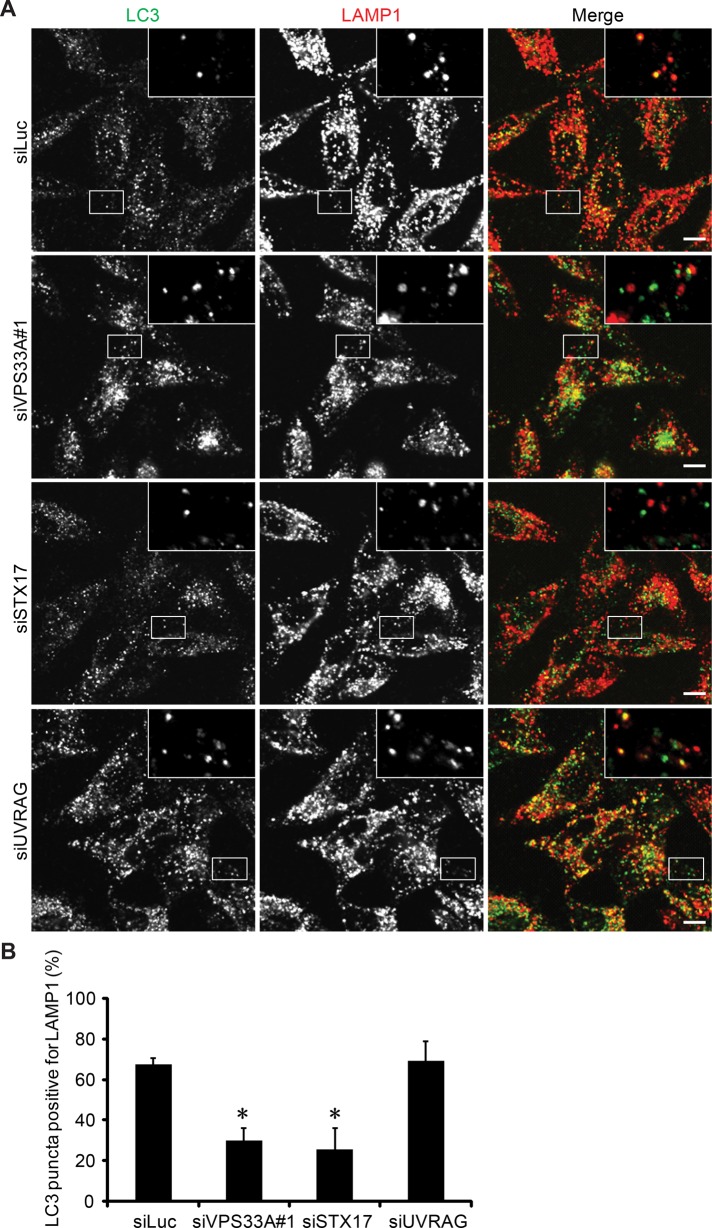
We then directly tested whether the HOPS complex is required for the fusion of autophagosomes and lysosomes like its binding partner STX17. Lysosomal hydrolase inhibitors E-64-d, pepstatin A, and leupeptin were used in HeLa cells to stabilize LC3 in autolysosomes. In control siRNA–treated cells, a high rate of LC3–LAMP1 colocalization was observed after 24-h serum starvation in the presence of the lysosomal inhibitors (Figure 5). However, the LC3–LAMP1 colocalization rate was notably lower in VPS33A and STX17 siRNA–treated cells (Figure 5). Moreover, knockdown of VPS33A or another HOPS component, VPS39, caused accumulation of double STX17- and LC3-positive, LAMP1-negative structures, which should represent complete autophagosomes, under either growing (Figure 6) or starvation (Supplemental Figure S2) conditions, suggesting a defect in both basal and starvation-induced autophagy. Consistent with the immunofluorescence data, electron microscopy showed accumulation of autophagosomes in VPS33A- or STX17-knockdown cells (Figure 7). Of importance, most of these autophagosomes were not degraded. In addition, consistent with its role as a stabilizer of the trans-SNARE complex (Xu et al., 2010), the amount of STX17-VAMP8 complex was reduced in VPS33A-knockdown cells (Supplemental Figure S3). Taken together, these data suggest that the HOPS complex is a bona fide fusion factor in the autophagy pathway.
FIGURE 5:
Knockdown of VPS33A and STX17 reduces colocalization of endogenous LC3 and LAMP1. (A) HeLa cells were transfected with siRNA against luciferase, VPS33A, STX17, or UVRAG. After 48 h, cells were serum starved in the presence of 10 μM E-64-d, 10 μg/ml pepstatin A, and 1 μg/ml leupeptin for an additional 24 h. Then cells were fixed, stained with anti-LC3 and anti-LAMP1 antibodies, and analyzed by confocal microscopy. Scale bars, 10 μm. (B) Quantification of colocalization rate between endogenous LC3 and LAMP1 as shown in A. The ratio of double LC3- and LAMP1-positive puncta per total LC3-positive puncta is expressed as mean ± SEM values of 10 randomly selected areas at the cell periphery. Analysis was performed in MetaMorph software. *p < 0.05 compared with luciferase siRNA–treated group.
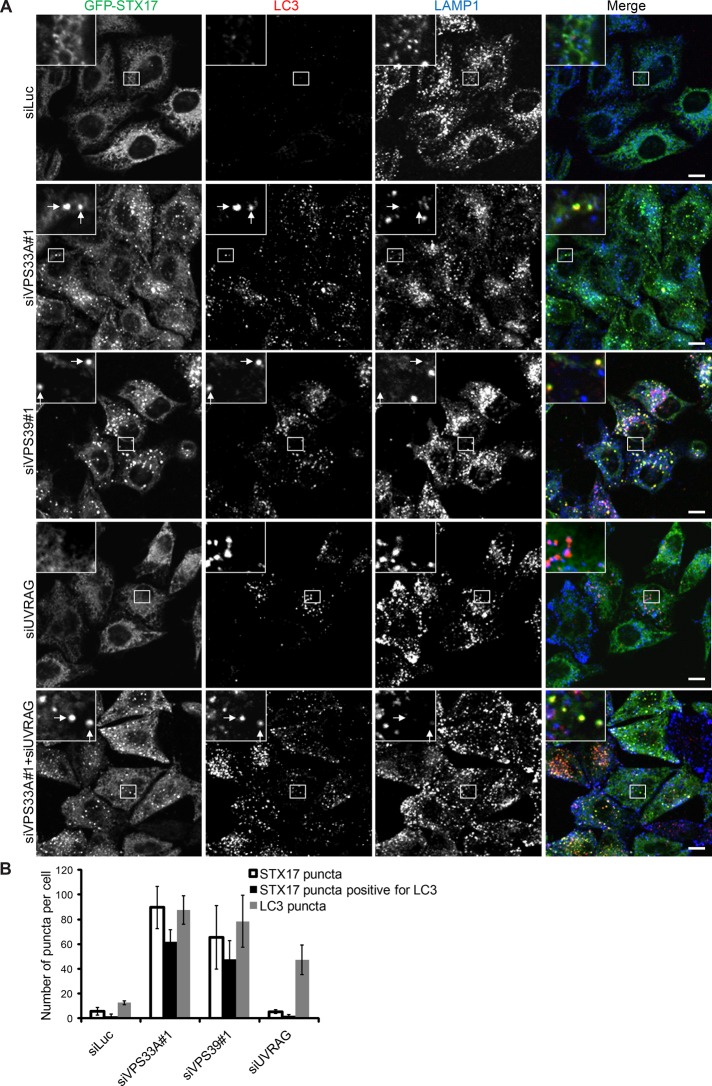
FIGURE 6:
Knockdown of VPS33A and VPS39 causes accumulation of double STX17- and LC3-positive structures. (A) HeLa cells stably expressing GFP-STX17 were treated with siRNA against luciferase, VPS33A, VPS39, UVRAG, or VPS33A + UVRAG. Then cells cultured in regular growth medium were analyzed by immunofluorescence microscopy using anti-LC3 and anti-LAMP1 antibodies. Structures positive for STX17 and LC3 but not for LAMP1 are indicated by arrows. Scale bars, 10 μm. (B) Quantification of the number of puncta positive for STX17, LC3, or both STX17 and LC3 per cell in each siRNA-treated group. Data are presented as mean ± SEM value of 10 different cells.
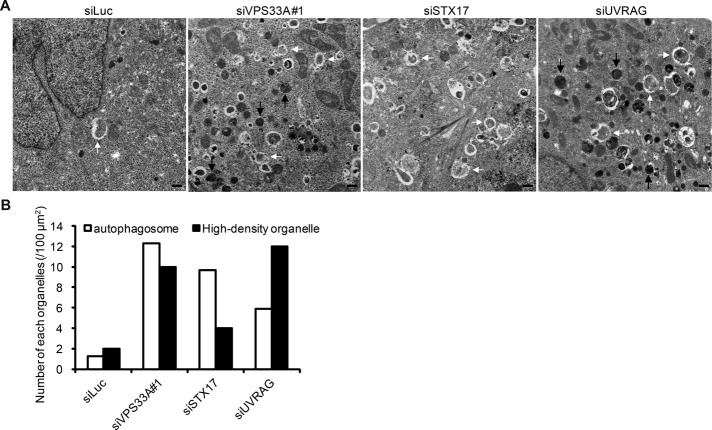
FIGURE 7:
Knockdown of VPS33A and UVRAG causes accumulation of autophagosomes and endolysosome-like structures. (A) HeLa cells treated with siRNA against luciferase, VPS33A, STX17, or UVRAG were cultured in starvation medium for 1 h and subjected to conventional electron microscopy analysis. White arrows indicate autophagosomes; black arrows indicate electron-dense endolysosome-like structures. Scale bars, 500 nm. (B) Quantification of the number of autophagosomes and high-density organelles from at least 50 randomly selected areas.
Endocytic pathway is affected in VPS33A-knockdown cells but not in STX17-knockdown cells
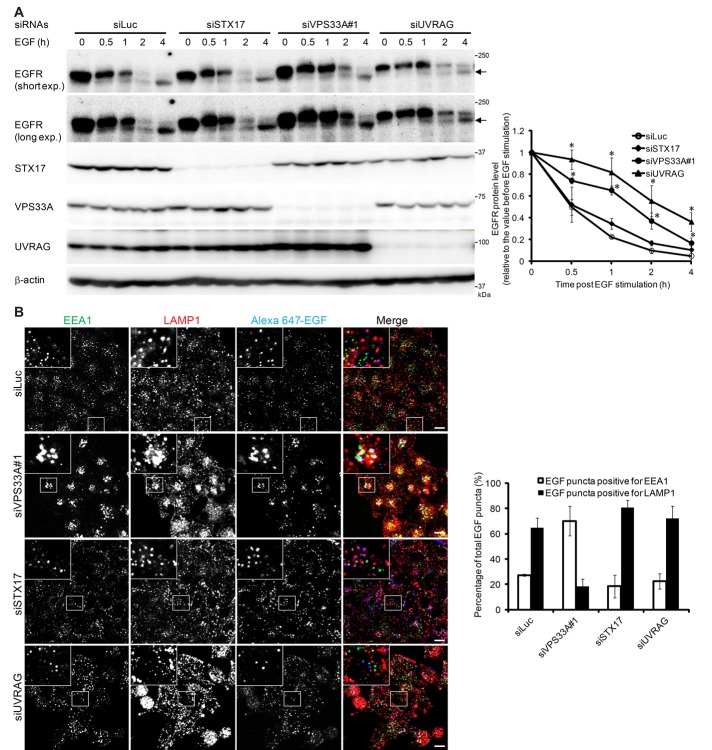
In addition to autophagosomes, we observed many electron-dense endolysosome-like structures in VPS33A-knockdown cells (Figure 7). These structures were rarely observed in STX17-knockdown cells (Figure 7). This finding is consistent with our LysoTracker staining data, which showed that LysoTracker signals increased in VPS33A-knockdown cells (Figure 4C). Also consistent with the well-established function of the HOPS complex in endolysosomal fusion (Balderhaar and Ungermann, 2013; Solinger and Spang, 2013), degradation of epidermal growth factor receptor (EGFR) after EGF stimulation was delayed in VPS33A-knockdown cells (Figure 8A). However, this delay was not observed in STX17-knockdown cells (Figure 8A). EGF was trapped in early endosome antigen 1 (EEA1)–positive endosomes in VPS33A-knockdown cells but was normally delivered to LAMP1-positive endolysosomes in STX17-knockdown cells (Figure 8B). Taken together, our data suggest that HOPS is required for both endocytosis and autophagy (autophagosome–lysosome fusion), whereas STX17 is specifically required only for the latter.
FIGURE 8:
Knockdown of VPS33A and UVRAG, but not STX17, impairs the endocytic pathway. (A) HeLa cells treated with siRNA against luciferase, VPS33A, STX17, or UVRAG were serum starved overnight and stimulated with 100 ng/ml EGF. The expression level of EGFR was determined at indicated time intervals. Arrows indicate a degradation product of EGFR. Short exp., short exposure; Long exp., long exposure. Protein levels of EGFR after 0, 0.5, 1, 2, and 4 h of EGF stimulation were quantified by densitometry. Quantification of three independent experiments is shown in the graph. *p < 0.05 compared with the luciferase siRNA–treated group, one-way analysis of variance followed by least significant difference post hoc tests. (B) siRNA-transfected HeLa cells were serum starved overnight and stimulated with 100 ng/ml Alexa 647–conjugated EGF for 1 h. Then cells were rinsed, fixed, and stained with antibodies against EEA1 and LAMP1. Scale bars, 10 μm. Quantification of the number of double EEA1- and EGF-positive or double LAMP1- and EGF-positive structures as a percentage of total EGF-positive puncta is shown in the graph. Data are presented as mean ± SEM values of 10 different cells.
UVRAG is not directly involved in autophagosome–lysosome fusion
A previous report suggested that UVRAG plays a role in promoting autophagosome maturation by recruiting the HOPS complex (Liang et al., 2008). The interaction between the HOPS component VPS33A and UVRAG was confirmed in our case, even though no UVRAG–STX17 interaction was detected (Figure 1, E and F). Autophagic flux (LC3 turnover) was also blocked in UVRAG-knockdown cells (Figure 3A). However, knockdown of UVRAG did not block the lysosomal quenching of GFP fluorescence in the mRFP-GFP-LC3 reporter assay (Figure 4). Moreover, knockdown of UVRAG did not significantly decrease the LC3–LAMP1 colocalization in the presence of lysosomal inhibitors (Figure 5). These results suggest that UVRAG is dispensable for autophagosome–lysosome fusion.
Furthermore, no STX17-positive structures accumulated in UVRAG-knockdown cells, whereas double LC3- and LAMP1-positive structures did accumulate (Figure 6). This result suggests that UVRAG is required for the recruitment of STX17 to completed autophagosomes or that STX17 is recycled from autolysosomes. To distinguish these possibilities, we performed double knockdown of both VPS33A and UVRAG and found that STX17- and LC3-positive structures accumulated in these cells (Figure 6). These data suggest that fusion of autophagosomes and lysosomes occurs in the absence of UVRAG and that STX17 dissociates from autolysosomes.
The role of UVRAG in the endocytic pathway has been extensively studied (Liang et al., 2008; Knaevelsrud et al., 2010; Sun et al., 2010; Lee et al., 2011). Consistent with previous findings, EGFR degradation was delayed in UVRAG-knockdown cells upon EGF stimulation (Liang et al., 2008; Knaevelsrud et al., 2010; Figure 8A). EGF was delivered to LAMP1-positive endolysosomes in UVRAG-knockdown cells (Figures 8B), indicating that UVRAG is not required in the early step of endocytosis. Taken together, these results suggest that UVRAG is not required for autophagosome–lysosome fusion but is required for the late step of endocytic degradation.
DISCUSSION
In the present study, we demonstrated that the HOPS complex is a bona fide autophagosomal fusion factor, which interacts with the autophagosomal SNARE, STX17. We noted that the interaction between transiently overexpressed HOPS subunits and STX17 seems to be relatively weak (Figure 1, A–D). STX17 may bind with high affinity to the whole HOPS complex (Sato et al., 2000; Lobingier and Merz, 2012). In fact, GFP-STX17 more efficiently binds to endogenous VPS33A and VPS16 (Figure 1E). Although STX17 is required for autophagosome–lysosome fusion but not for endocytic degradation, HOPS is required for both processes. Most of these findings are also observed in Drosophila (Takáts et al., 2014). Thus HOPS exerts multiple functions through interaction with different SNAREs. In the endocytic pathway, mammalian HOPS has been reported to interact with syntaxin 7 and other multiple SNAREs (Kim et al., 2001; Richardson et al., 2004). Yeast should have a similar mechanism for autophagosome-vacuole fusion. However, although the involvement of HOPS in the yeast autophagy pathway was proposed many years ago (Rieder and Emr, 1997), its binding partner, SNARE on the autophagosomal membrane, remains unknown.
Our current model for autophagosome–lysosome fusion is as follows. In normal growing cells, the HOPS complex is localized mainly to endolysosomes, whereas STX17 is present in the ER, mitochondria, and cytosol (Itakura et al., 2012b). On autophagy induction, the isolation membranes elongate to form completed autophagosomes. Then STX17 translocates to these autophagosomes, where HOPS is also recruited. HOPS bridges closely apposed autophagosomal and lysosomal membranes and facilitates the assembly of the trans-SNARE complex to mediate autophagosome–lysosome fusion. Recruitment of STX17 to autophagosomes is independent of HOPS because STX17-positive autophagosomes accumulate in HOPS-depleted cells (Figure 6).
It is intriguing that, although STX17 localizes not only to autophagosomes but also to the ER and mitochondria, HOPS does not associate with the ER or the mitochondria. This may explain why lysosomes can fuse with autophagosomes but not with the ER and mitochondria, even though they express STX17. Given that that HOPS can recognize phospholipids (Stroupe et al., 2006; Mima and Wickner, 2009), HOPS may recognize both STX17 (Figure 2B) and some specific lipids that are enriched in the autophagosomal membrane. HOPS is also important for prevention of disassembly of trans-SNARE complexes by Sec17 (yeast αSNAP) and Sec18 (yeast NSF) in yeast (Xu et al., 2010). More recently it was suggested that Munc18-1, a member of the Sec1-Munc18 (SM) protein family, prevents disassembly of syntaxin1–SNAP-25 heterodimers by NSF and SNAPs (Ma et al., 2013). Because VPS33 is also an SM protein, HOPS may be important for stabilizing STX17–SNAP29 heterodimers on autophagosomes.
One notable finding is that the STX17–HOPS complex does not interact with UVRAG (Figure 1E). UVRAG is considered to be involved in autophagosome formation and/or maturation steps (Liang et al., 2006, 2008). However, the present results demonstrate that, although UVRAG is indeed required for a late step in endocytic degradation, it is not directly involved in autophagosome formation or autophagosome–lysosome fusion. The apparent effect of UVRAG on autophagic degradation may be secondary to its role in endocytic degradation (Figure 3A). In fact, any defects in the endocytic pathway could also affect the autophagy pathway (Filimonenko et al., 2007; Platt et al., 2012). Furthermore, in a study by Liang et al. (2007), the involvement of UVRAG in autophagosome–lysosome fusion was determined mostly by observing the effect of overexpression rather than knockdown of UVRAG. Localization of UVRAG on the autophagic membrane is also controversial. Although Liang et al. (2007) showed colocalization between UVRAG and LC3, subsequent studies failed to confirm this finding (Itakura et al., 2008; Matsunaga et al., 2009).
On the other hand, the role of UVRAG–HOPS in the endocytic pathway has been demonstrated in a clearer manner. UVRAG has been reported to be present on both early endosomes (Liang et al., 2008; Sun et al., 2010) and late endosomes (Itakura et al., 2008; Matsunaga et al., 2009). Run domain–containing, Beclin 1–interacting, and cysteine-rich protein (Rubicon) negatively regulates endosome maturation by sequestering UVRAG from HOPS (Sun et al., 2010). Thus it is likely that the UVRAG–HOPS complex is required specifically for the fusion of late endosomes and lysosomes. In addition, UVRAG may be important for transport of lysosomal hydrolases from the Golgi, given that UVRAG shows partial homology to yeast Vps38 (Itakura et al., 2008). In yeast, Vps38 is required for endosome-to-Golgi retrograde transport (Burda et al., 2002). Consistently, Pichia pastoris UVRAG is required for transport of carboxypeptidase Y to the vacuole but not for any step of autophagy (Farre et al., 2010). The presence of UVRAG on Rab9-positive endosomes (Itakura et al., 2008), which are involved in retrograde transport in mammalian cells, supports that this is also the case in mammals. In this scenario, the interaction of UVRAG with HOPS may be important for trafficking between the Golgi and late endosomes.
MATERIALS AND METHODS
Plasmids
Full-length cDNAs encoding human VPS11, VPS16, VPS18, VPS33A, VPS39, and VPS41 were amplified by PCR from the total cDNA of HeLa cells and subcloned into pMRXIP (provided by Shoji Yamaoka, Tokyo Medical and Dental University, Tokyo, Japan; Saitoh et al., 2002) together with enhanced GFP (Clontech, Mountain View, CA). mRFP-GFP-LC3 was introduced into pMXs-puro (provided by Toshio Kitamura, University of Tokyo, Tokyo, Japan; Kitamura et al., 2003). CMV-Myc-VPS33A, CMV-Myc-VPS18, CMV-EGFP-VPS11, and CMV-EGFP-VPS16 were gifts from Chihiro Akazawa (Tokyo Medical and Dental University; Kim et al., 2001). pMRXIP-GFP-STX18, CMV-FLAG-STX17, pMRXIP-GFP-STX17, and pMRXIP-Myc-STX17 were previously described (Itakura et al., 2012b). An siRNA-resistant VPS33A silent mutant was created by substituting eight nucleotides in the VPS33A siRNA #1–targeting region (G524A, G527A, G530A, C533T, C536T, A539G, T542C, and C545G).
Antibodies
Mouse monoclonal anti-LC3 (clone 1703; Cosmo Bio Co., Tokyo, Japan), anti–β-actin (clone AC74; Sigma-Aldrich, Tokyo, Japan), anti-FLAG (clone M2; Sigma-Aldrich), anti-Myc (9E10; Covance Research Products, Denver, PA), anti-EGF receptor (610017; BD Biosciences, Franklin Lakes, NJ), anti-EEA1 (M176-3; MBL, Tokyo, Japan), rat monoclonal anti-GFP antibody (clone GF090R; Nacalai Tesque, Kyoto, Japan), rabbit polyclonal anti-GFP (598; MBL), anti-p62 (PM045; MBL), anti-STX17 (HPA001204; Sigma-Aldrich), anti-VPS33A (C1C3; GeneTex, Irvine, CA), anti-VPS39 (ab107570; Abcam, Cambridge, MA), and anti-UVRAG (A301-996A; Bethyl Laboratories, Montgomery, TX) antibodies were used. The rabbit polyclonal antibodies against LC3 (LC3 #1) for immunoblotting (Hosokawa et al., 2006) and Atg16L1 (Mizushima et al., 2003) have been described previously. Rabbit polyclonal anti-LAMP1 antibody was provided by Yoshitaka Tanaka (Kyushu University, Fukuoka, Japan). Rabbit polyclonal anti-VPS16 antibody was provided by Chihiro Akazawa (Tokyo Medical and Dental University; Kim et al., 2001). Alexa 488–conjugated anti-mouse, rabbit, and rat immunoglobulin G (IgG), Alexa 568–conjugated anti-mouse and rabbit IgG, and Alexa 660–conjugated anti-mouse and rabbit IgG secondary antibodies were purchased from Molecular Probes (Eugene, OR).
Reagents
E-64-d (4321-v), pepstatin A (4397), and leupeptin (4041) were purchased from the Peptide Institute (Osaka, Japan). LysoTracker Red DND-99 (L-7528) and Alexa 647–conjugated EGF (E-35351) were purchased from Molecular Probes. Bafilomycin A1 was purchased from Wako Chemicals (Osaka, Japan). Puromycin (P8333) was purchased from Sigma-Aldrich.
Cell culture and transfection
HeLa cells, HEK 293T cells, and mouse embryonic fibroblasts (MEFs) were cultured in DMEM supplemented with 10% fetal bovine serum, 50 μg/ml penicillin, and streptomycin (regular medium) in a 5% CO2 incubator. For starvation treatment, cells were washed with phosphate-buffered saline (PBS) and incubated in amino acid–free DMEM without serum (starvation medium).
MEFs stably expressing Myc-STX17 and GFP-VPS33A or GFP-VPS16, and HeLa cells stably expressing GFP-STX17, GFP-VPS33A, or mRFP-GFP-LC3, were generated as follows: HEK 293T cells were transiently transfected using Lipofectamine 2000 reagent (11668-019; Invitrogen, Carlsbad, CA) with the indicated plasmids together with pCG-VSV-G and pCG-gag-pol. After 72 h of culture, the growth medium containing retroviruses was collected by centrifugation, and cells were incubated with the collected virus-containing medium with 8 μg/ml hexadimethrine bromide (H9268; Sigma-Aldrich) for 24 h. Uninfected cells were removed by puromycin selection.
RNA interference
Stealth RNA interference oligonucleotides were used for siRNA experiments (Invitrogen). The sequences used were as follows: luciferase siRNA antisense, 5′-CGCGGUCGGUAAAGUUGUUCCAUUU-3′, sense, 5′-AAAUGGAACAACUUUACCGACCGCG-3′; human STX17 siRNA antisense, 5′-AAUUAAGUCCGCUUCUAAGGUUUCC-3′, sense, 5′-GGAAACCUUAGAAGCGGACUUAAUU-3′; human UVRAG-siRNA antisense, 5′-UAAAGUAGGUAUCAAGGAGCUGAUG-3′, sense, 5′-CAUCAGCUCCUUGAUACCUACUUUA-3′; human VPS16 siRNA #1 antisense, 5′-ACAAUACGAGGCUCAGCUCAGCCUC-3′, sense, 5′-GAGGCUGAGCUGAGCCUCGUAUUGU-3′; human VPS16 siRNA #2 antisense, 5′-ACUUCUUCUGGGCUUGUGCCCUGGC-3′, sense, 5′-GCCAGGGCACAAGCCCAGAAGAAGU-3′; human VPS33A siRNA #1 antisense, 5′-AUGAGAUCUAAGCUGUACUCCUCCC-3′, sense, 5′-GGGAGGAGUACAGCUUAGAUCUCAU-3′; human VPS33A siRNA #2 antisense, 5′-UGAAGUUCUUAUCUCGGAUCUCAGC-3′, sense, 5′-GCUGAGAUCCGAGAUAAGAACUUCA-3′; human VPS39 siRNA #1 antisense, 5′-AUCAGAGUCAUUCAGCUUCUUUACC-3′, sense, 5′-GGUAAAGAAGCUGAAUGACUCUGAU-3′; and human VPS39 siRNA #2 antisense, 5′-AACAUAUCGAGGCAACACUGCAAUG-3′, sense, 5′-CAUUGCAGUGUUGCCUCGAUAUGUU-3′.
Two different siRNAs were used for VPS16, VPS33A, and VPS39. The Stealth RNAi oligonucleotides were transfected into cells using Lipofectamine RNAi MAX (13778-150; Invitrogen) according to the manufacturer's instructions. After 2 d, the cells were again transfected with the same siRNA and cultured for an additional 3 d before analysis.
Immunoprecipitation and immunoblotting
For immunoprecipitation analysis, HEK 293T cells were transiently transfected with plasmids using FuGENE 6 reagent (1814443; Roche Applied Science, Penzberg, Germany). Cells were collected in ice-cold PBS by scraping and sedimented by centrifugation at 5000 × g for 3 min; precipitated cells were suspended in lysis buffer containing CHAPS (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.3% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 0.4 mM Na3VO4, 10 mM NaF, 10 mM sodium pyrophosphate, 1 mM phenylmethanesulfonyl fluoride, and a protease inhibitor cocktail [Complete EDTA-free protease inhibitor; Roche Applied Science]). Cell lysates were then clarified by centrifugation at 12,000 × g for 20 min and analyzed by immunoprecipitation using anti-FLAG M2 affinity gel (50% slurry; Sigma-Aldrich) or anti-GFP antibody in combination with protein G–Sepharose (GE Healthcare, Chalfont St Giles, United Kingdom). The immunoprecipitated complexes were washed five times in the lysis buffer and boiled in sample buffer (46.7 mM Tris HCl, pH 6.8, 5% glycerol, 1.67% SDS, 1.55% dithiothreitol, and 0.003% bromophenol blue). The supernatant was then pelleted to beads by centrifugation for use in subsequent experiments. For immunoblotting analysis, cells were lysed with lysis buffer containing 1% Triton X-100. Samples were subsequently separated by SDS–PAGE and transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore, Billerica, MA). Immunoblot analysis was performed and visualized with Super-Signal West Pico Chemiluminescent substrate (Pierce Chemical Co., Rockford, IL) or Immobilon Western Chemiluminescent HRP substrate (Millipore). Signal intensities were analyzed using a LAS-3000 mini imaging analyzer and Multi Gauge software, version 3.0 (Fujifilm, Tokyo, Japan). Contrast and brightness adjustment was applied to the images in Photoshop CS3 (Adobe, San Jose, CA).
Immunocytochemistry
Cells grown on coverslips were washed with PBS and fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. Fixed cells were permeabilized with 50 μg/ml digitonin in PBS for 5 min, blocked with 3% bovine serum albumin in PBS for 30 min at room temperature, and incubated with primary antibodies for 1 h. After PBS washing, cells were incubated with fluorescence-conjugated secondary antibodies for another 1 h. Images were acquired on a confocal laser microscope (FV1000D IX81; Olympus, Tokyo, Japan) using a 60× PlanApoN oil immersion lens (1.42 NA; Olympus) and captured with FluoView software (Olympus). For final output, images were processed using Photoshop CS3. Colocalization analysis was performed in MetaMorph software (Molecular Devices, Sunnyvale, CA) after binarizing images of each channel as previously described (Itakura et al., 2012a).
EGFR degradation assay
HeLa cells were plated at 80% confluence, starved in serum-free DMEM overnight, and then incubated with DMEM supplemented with 100 ng/ml EGF (059-07873; Wako Chemicals). Cells were rinsed with PBS twice and lysed in lysis buffer containing 1% Triton X-100.
EGF trafficking assay
HeLa cells grown on glass coverslips were starved in serum-free DMEM overnight. Alexa 647–conjugated EGF (100 ng/ml) was added and incubated for 1 h, then fixed and observed as before.
Electron microscopy
HeLa cells were cultured on collagen-coated plastic coverslips (Cell tight C-1 Cell disk; Sumitomo Bakelite, Tokyo, Japan) and fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 2 h. The cells were washed in the same buffer three times, postfixed in 1.5% osmium tetroxide in 0.1 M phosphate buffer for 2 h, dehydrated, and embedded in Epon 812. Ultrathin sections were stained with uranyl acetate and lead citrate and observed using a Hitachi H-7100 electron microscope.
Statistical analysis
Numerical data are presented as mean ± SEM. Statistical comparisons were made using the two-tailed Student's t test, unless otherwise indicated.
Supplementary Material
Acknowledgments
We thank Chihiro Akazawa for the cDNAs of the HOPS components and the VPS16 antibody, Shoji Yamaoka and Toshio Kitamura for the retroviral vectors, Roger Y. Tsien for the mRFP cDNA, and Teruhito Yasui for the pCG-VSV-G and pCG-gag-pol plasmids. This work was supported in part by the Funding Program for Next Generation World-Leading Researchers and Japan Society for the Promotion of Science KAKENHI Grants-in-Aid for Scientific Research on Innovative Areas (Grant 25111005; to N.M.) and the Naito Foundation (to T.N.). P.J. is supported by a scholarship from the China Scholarship Council in China and by the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Abbreviations used:
- EEA1
early endosome antigen 1
- GFP
green fluorescent protein
- HOPS
homotypic fusion and protein sorting
- LAMP1
lysosomal-associated membrane protein 1
- LC3
microtubule-associated protein light chain 3
- mRFP
monomeric red fluorescent protein
- SNARE
soluble N-ethylmaleimide–sensitive factor attachment protein receptor
- STX17
syntaxin 17
- UVRAG
ultraviolet irradiation resistance–associated gene
- VPS
vacuolar protein sorting
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0447) on February 19, 2014.
*Present address: MRC Laboratory of Molecular Biology, Cambridge CB2 0QH, United Kingdom.
REFERENCES
- Akbar MA, Ray S, Kramer H. The SM protein Car/Vps33A regulates SNARE-mediated trafficking to lysosomes and lysosome-related organelles. Mol Biol Cell. 2009;20:1705–1714. doi: 10.1091/mbc.E08-03-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes—coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- Burda P, Padilla SM, Sarkar S, Emr SD. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J Cell Sci. 2002;115:3889–3900. doi: 10.1242/jcs.00090. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yu L. Autophagic lysosome reformation. Exp Cell Res. 2013;319:142–146. doi: 10.1016/j.yexcr.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Mathewson RD, Manjithaya R, Subramani S. Roles of Pichia pastoris Uvrag in vacuolar protein sorting and the phosphatidylinositol 3-kinase complex in phagophore elongation in autophagy pathways. Autophagy. 2010;6:86–99. doi: 10.4161/auto.6.1.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Stevens TH. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol Biol Cell. 1999;10:1719–1732. doi: 10.1091/mbc.10.6.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell. 2011;42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- Hamasaki M, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Koyama-Honda I, Mizushima N. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J Cell Sci. 2012a;125:1488–1499. doi: 10.1242/jcs.094110. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012b;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen E-L. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Kramer H, Yamamoto A, Kominami E, Kohsaka S, Akazawa C. Molecular characterization of mammalian homologues of class C Vps proteins that interact with syntaxin-7. J Biol Chem. 2001;276:29393–29402. doi: 10.1074/jbc.M101778200. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- Knaevelsrud H, Ahlquist T, Merok MA, Nesbakken A, Stenmark H, Lothe RA, Simonsen A. UVRAG mutations associated with microsatellite unstable colon cancer do not affect autophagy. Autophagy. 2010;6:863–870. doi: 10.4161/auto.6.7.13033. [DOI] [PubMed] [Google Scholar]
- Lee G, Liang C, Park G, Jang C, Jung JU, Chung J. UVRAG is required for organ rotation by regulating Notch endocytosis in Drosophila. Dev Biol. 2011;356:588–597. doi: 10.1016/j.ydbio.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Oh BH, Jung JU. UVRAG: a new player in autophagy and tumor cell growth. Autophagy. 2007;3:69–71. doi: 10.4161/auto.3437. [DOI] [PubMed] [Google Scholar]
- Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A. Autophagy in lysosomal storage disorders. Autophagy. 2012;8:719–730. doi: 10.4161/auto.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobingier BT, Merz AJ. Sec1/Munc18 protein Vps33 binds to SNARE domains and the quaternary SNARE complex. Mol Biol Cell. 2012;23:4611–4622. doi: 10.1091/mbc.E12-05-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Mima J, Wickner W. Complex lipid requirements for SNARE- and SNARE chaperone-dependent membrane fusion. J Biol Chem. 2009;284:27114–27122. doi: 10.1074/jbc.M109.010223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Natsume T, Yamauchi Y, Nakayama H, Shinkawa T, Yanagida M, Takahashi N, Isobe T. A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal Chem. 2002;74:4725–4733. doi: 10.1021/ac020018n. [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21:3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, et al. Ablation of vacuole protein sorting 18 (Vps18) gene leads to neurodegeneration and impaired neuronal migration by disrupting multiple vesicle transport pathways to lysosomes. J Biol Chem. 2012;287:32861–32873. doi: 10.1074/jbc.M112.384305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinar M, Pantazopoulou A, Penalva MA. Live-cell imaging of Aspergillus nidulans autophagy: RAB1 dependence, Golgi independence and ER involvement. Autophagy. 2013;9:1024–1043. doi: 10.4161/auto.24483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt FM, Boland B, van der Spoel AC. The cell biology of disease: lysosomal storage disorders: the cellular impact of lysosomal dysfunction. J Cell Biol. 2012;199:723–734. doi: 10.1083/jcb.201208152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SC, Winistorfer SC, Poupon V, Luzio JP, Piper RC. Mammalian late vacuole protein sorting orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol Biol Cell. 2004;15:1197–1210. doi: 10.1091/mbc.E03-06-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Simonsen A. ESCRT functions in autophagy and associated disease. Cell Cycle. 2008;7:1166–1172. doi: 10.4161/cc.7.9.5784. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Nakano H, Yamamoto N, Yamaoka S. Lymphotoxin-beta receptor mediates NEMO-independent NF-kappaB activation. FEBS Lett. 2002;532:45–51. doi: 10.1016/s0014-5793(02)03622-0. [DOI] [PubMed] [Google Scholar]
- Sato TK, Darsow T, Emr SD. Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol Cell Biol. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Rehling P, Peterson MR, Emr SD. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol Cell. 2000;6:661–671. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Spang A. Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 2013;280:2743–2757. doi: 10.1111/febs.12151. [DOI] [PubMed] [Google Scholar]
- Stroupe C, Collins KM, Fratti RA, Wickner W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25:1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci USA. 2010;107:19338–19343. doi: 10.1073/pnas.1010554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S, Nagy P, Varga A, Pircs K, Kárpáti M, Varga K, Kovács AL, Hegedu˝s K, Juhász G. Autophagosomal syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013;201:531–539. doi: 10.1083/jcb.201211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S, Pircs K, Nagy P, Varga A, Kárpáti M, Hegedu˝s K, Kramer H, Kovács AL, Sass M, Juhász G. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell. 2014;25:1338–1354. doi: 10.1091/mbc.E13-08-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jun Y, Thompson J, Yates J, Wickner W. HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J. 2010;29:1948–1960. doi: 10.1038/emboj.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, et al. UV irradiation resistance-associated gene suppresses apoptosis by interfering with BAX activation. EMBO Rep. 2011;12:727–734. doi: 10.1038/embor.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, et al. A dual role for UVRAG in maintaining chromosomal stability independent of autophagy. Dev Cell. 2012;22:1001–1016. doi: 10.1016/j.devcel.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.