Numerous muscle lineages are formed during myogenesis within both slow- and fast-specific cell groups. New muscle domains are identified along the anteroposterior axis in zebrafish and are defined by individual nonoverlapping expression of myosin heavy chain isoforms differentially regulated by retinoic acid and wnt.
Abstract
Numerous muscle lineages are formed during myogenesis within both slow- and fast-specific cell groups. In this study, we show that six fast muscle–specific myosin heavy chain genes have unique expression patterns in the zebrafish embryo. The expression of tail-specific myosin heavy chain (fmyhc2.1) requires wnt signaling and is essential for fast muscle organization within the tail. Retinoic acid treatment results in reduced wnt signaling, which leads to loss of the fmyhc2.1 domain. Retinoic acid treatment also results in a shift of muscle identity within two trunk domains defined by expression of fmyhc1.2 and fmyhc1.3 in favor of the anteriormost myosin isoform, fmyhc1.2. In summary, we identify new muscle domains along the anteroposterior axis in the zebrafish that are defined by individual nonoverlapping, differentially regulated expression of myosin heavy chain isoforms.
INTRODUCTION
Patterning of the skeletal muscle tissue is a developmental process by which precursor cells are allocated to a certain muscle lineage. In vertebrates, muscle fibers are traditionally divided into two major classes— slow- and fast-twitch fibers—depending on their biochemistry and metabolism. The slow- and fast-twitch fibers also express genes respectively encoding specific forms of sarcomeric components, such as the troponins and myosin light and heavy chains, by which they can be categorized (Schiaffino and Reggiani, 2011). In particular, the different forms of myosin heavy chain genes are used to define the various fiber subtypes. In mammals, at least 13 different genes encoding myosin heavy chain (MyHC) isoforms are specifically expressed in muscle lineages ranging from heart to extraocular muscle or specifically during certain embryonic and juvenile life stages (Schiaffino and Reggiani, 2011). In mammals, the broadest heterogeneity can be found among the skeletal fast fibers, among which three isoforms, MyHC 2a, 2b, and 2x, have been identified using monoclonal antibodies, in addition to extraocular and embryonic forms (Edman et al., 1988; Schiaffino et al., 1989). In zebrafish, slow fibers are mononucleated, whereas fast fibers are multinucleated, which facilitates their initial classification. The slow and fast fibers also occupy distinct domains in the zebrafish myotome (Devoto et al., 1996; Elworthy et al., 2008; Burguiere et al., 2011). The progenitors that contribute to formation of slow and fast fibers are defined already at gastrula stages, even though they remain competent to differentiate into either type until they have terminally differentiated (Hirsinger et al., 2004). Myogenesis requires the expression of myogenic regulatory factors (MRFs) such as MyoD, Myf5, and myogenin, which will turn precursors into differentiation mode (Tapscott et al., 1988; Buckingham and Vincent, 2009), but additional factors are required to define the fiber type. Slow fibers are formed from the adaxial cells, which are in direct contact with the notochord (Currie and Ingham, 1996; Blagden et al., 1997; Du et al., 1997), whereas the remaining part of the myotome consists of fast-specific progenitors that will differentiate and eventually become multinucleated fibers. Myogenic precursors within the transient somite are separated into lineage-restricted domains, each of which will contribute to specific cell types in the mature myotome. A subset of these cells—the anterior border cells within the myotome that give rise to the Pax3- and Pax7-positive dermomyotome cells (Devoto et al., 2006; Feng et al., 2006; Hollway et al., 2007)—will subsequently migrate into position, at least in part controlled by the action of the Sdf family of secreted cytokines and the Cxcr4 receptors (Hollway et al., 2007). The high levels of Hedgehog signaling from the notochord will induce Engrailed expression in a subset of fast fibers—the medial fast fibers—situated in the most medial part of the fast-specific domain (Hatta et al., 1991; Devoto et al., 1996; Wolff et al., 2003). Interplay between Hedgehog and bone morphogenic protein (BMP) signaling creates morphogen gradients that orchestrate the specification of these fibers (Maurya et al., 2011). However, the majority of the fast-specific myoblasts initially also require fibroblast growth factor (FGF) signaling, which is essential for the expression of myod (Groves et al., 2005). Recent studies also indicate that zebrafish skeletal muscle can be further divided into subclasses based on the expression of different versions of myosins (Elworthy et al., 2008) and different requirements of MRFs in different domains of the embryo (Hinits et al., 2009, 2011). The slow-twitch fibers express specific forms of slow-myosin heavy chain genes in different muscle fibers, where primary slow fibers predominantly express smyhc1 and secondary slow fibers, which are formed later in development, express smyhc2 and smyhc3 (Elworthy et al., 2008). The zebrafish fast-specific myosin heavy chain genes are less characterized, however, and their individual expression and regulation have not been determined.
Similar to other vertebrates, zebrafish body segments are formed starting with the anteriormost somite, after which new somites are formed from paraxial mesoderm cells in a clockwise manner toward the tail of the developing embryo (Mara and Holley, 2007). The regionalization of the body is specified by hox patterns, which in many cases are orchestrated or at least influenced by signaling molecules such as retinoic acid, Wnt, and FGFs (Kessel and Gruss, 1991; Marshall et al., 1992; Lumsden and Krumlauf, 1996; Grandel et al., 2002; Hashiguchi and Mullins, 2013). Previous studies showed that regional specification within mesodermal progenitors, including fast muscle, subdivide the body musculature into defined domains along the anteroposterior axis (Szeto and Kimelman, 2006). By use of transplants and cell tracing experiments, Szeto and Kimelman (2006) showed that the anterior and posterior trunk domains are both established through Nodal signaling, whereas the tail domain requires BMP. Even though the mesodermal progenitors are committed to a certain somite identity, any difference between muscles formed within the different domains has not been described.
In this study we analyze the formation of fast-twitch muscle fibers in zebrafish trunk and tail domains and show that retinoic acid and wnt coordinate the differential expression of myosin heavy chains that defines distinct muscle identities along the anteroposterior axis.
RESULTS
Tandem triplets of fast myosin heavy chain genes
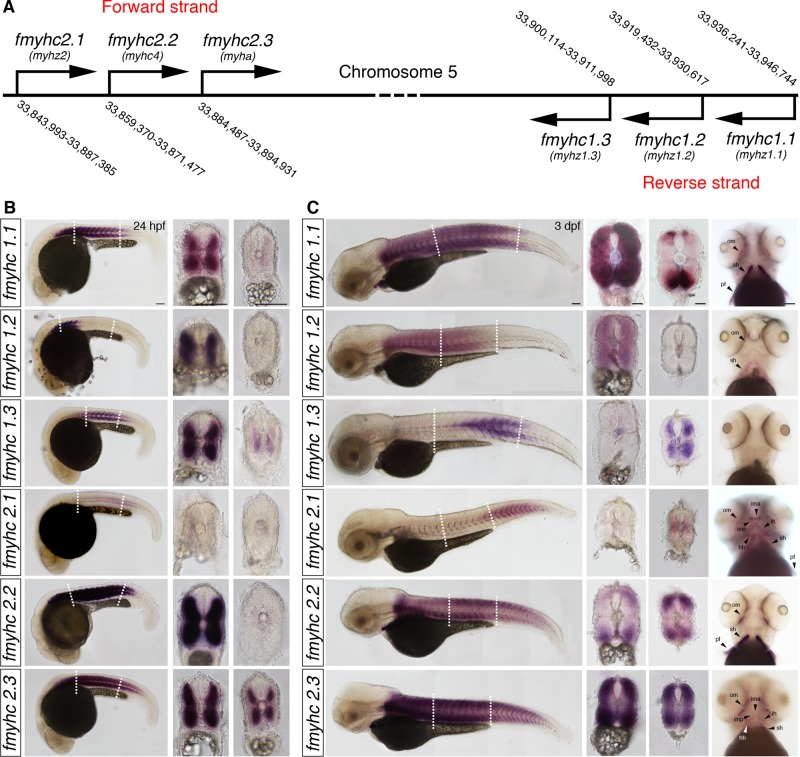
Six fmyhc genes, arranged as triple repeats, are located in a narrow region on opposite strands of chromosome 5 in the zebrafish genome (Figure 1A). The overall sequence similarity within the group of six is high (96–98%) on the mRNA level. To analyze the mRNA expression patterns and to avoid cross-hybridization, we designed cRNA probes corresponding to the 5′- and 3′-untranslated regions (UTRs), where the sequence similarities are significantly lower. Of interest, we found that the expression patterns of the fmyhc genes showed distinct differences—the genes in group 1 (fmyhc1.1, fmyhc1.2, and fmyhc1.3) all were excluded from the tail and the majority of the cranial muscle, whereas the genes in group 2 (fmyhc2.1, fmyhc2.2, and fmyhc2.3) generally were highly expressed in the cranial muscle. A number of individual differences were observed in particular at later developmental stages and early juvenile stages but were evident already at 24 h postfertilization (hpf; Figure 1B). At 3 d postfertilization (dpf), all six fmyhc genes showed unique individual expression patterns in restricted, and well-defined, regions of the skeletal muscle (Figure 1C). Expression of fmyhc1.1 was found in somites 1–22, fmyhc1.2 was expressed in somites 1–12, and fmyhc1.3 was expressed in a medial subsection of somites 11–20 (Figure 1C). When analyzing cross sections at somite 20, we found that even though fmyhc1.1 and fmyhc1.3 both were expressed at this level at the anteroposterior axis, they did not overlap. At this somite level, fmyhc1.1 was restricted to the dorsomedial and ventromedial domains, whereas fmyhc1.3 was expressed more medially (Figure 1C). Both fmyhc1.1 and fmyhc1.2 were expressed in the sternohyoideus and the ocular muscle, whereas fmyhc1.3 was excluded from all cranial muscle. At 3 dpf, fmyhc2.1 was expressed in the cranial muscle, the pectoral fins, and the somites of the posterior trunk and tail. In addition, fmyhc2.1 was expressed in a subset of cells along the midline (Figures 1B and 2B). fmyhc2.2 and fmyhc2.3 were both expressed throughout the whole trunk and tail, but fmyhc2.2 was excluded from the medial parts of the somite and most of the cranial muscle (Figure 1).
FIGURE 1:
Expression pattern of different fast myosin heavy chain genes. (A) Location of fmyhc1.1, fmyhc1.2, fmyhc1.3, fmyhc2.1, fmyhc2.2, and fmyhc2.3 genes on zebrafish chromosome 5. In situ hybridization showing mRNA expression of fmyhc1.1, fmyhc1.2, fmyhc1.3, fmyhc2.1, fmyhc2.2, and fmyhc2.3 at (B) 24 hpf and (C) 3 dpf. Dashed lines indicate levels of cross sections. hh, hyohyoideus; ih, interhyoideus; ima, intermandibular anterior; imp, intermandibular posterior; om, ocular muscles; pf, pectoral fin; sh, sternohyoideus. Scale bar, 100 μm.
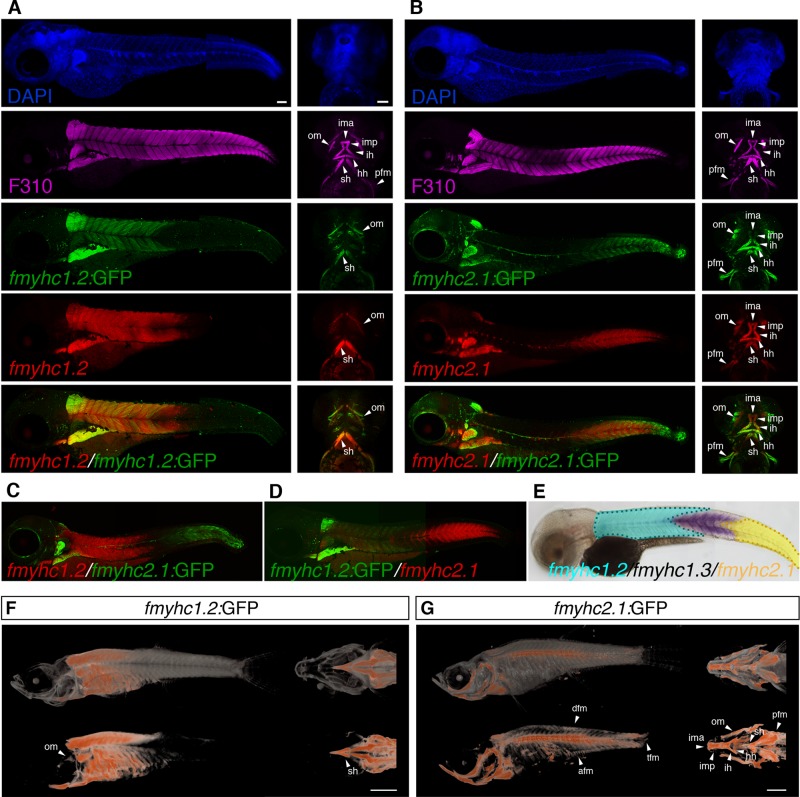
FIGURE 2:
fmyhc1.2:GFP and fmyhc2.1:GFP transgenic lines phenocopy fmyhc1.2 and fmyhc2.1 mRNA expression and have different expression domains in the embryo and juvenile zebrafish. (A) Lateral and ventral views of 3-dpf fmyhc1.2:GFP (green) embryo stained with F310 (magenta) and fmyhc1.2 (red) showing coexpression of fmyhc1.2:GFP and fmyhc1.2 mRNA. (B) Lateral and ventral views of 3-dpf fmyhc2.1:GFP (green) embryo stained with F310 (magenta) and fmyhc2.1 (red) showing coexpression of fmyhc2.1:GFP and fmyhc2.1 mRNA. (C) Expression of fmyhc1.2 (red) in fmyhc2.1:GFP (green) embryo 3-dpf showing separate expression domains. (D) Expression of fmyhc2.1 (red) in fmyhc1.2:GFP (green) embryo 3-dpf showing separate expression domains. (E) Schematic showing expression domains of fmyhc1.2 (turquoise), fmyhc1.3 (purple), and fmyhc2.1 (yellow) in a 3-dpf embryo. Lateral view of entire fish and ventral view of head showing expression of (F) fmyhc1.2:GFP and (G) fmyhc2.1:GFP in juvenile zebrafish visualized using optical projection tomography; top, volume rendering of anatomy (in gray) and GFP (in red); bottom, GFP (red) only. afm, anal fin muscle; dfm, dorsal fin muscle; hh, hyohyoideus; ih, interhyoideus; ima, intermandibular anterior; imp, intermandibular posterior; om, ocular muscles; pfm, pectoral fin muscle; sh, sternohyoideus; tfm, tail fin muscle. Scale bar, 100 μm (A–E), 1000 μm (F, G).
The two genes that differed the most in their expression patterns within the trunk and tail were fmyhc1.2 and fmyhc2.1, as they were expressed, respectively, in exclusive anterior and posterior domains at 3 dpf. To further examine these two opposing gene expression patterns, we generated stable transgenic lines expressing green fluorescent protein (GFP) under the control of a 7- to 8-kb genomic sequence upstream of the translation start site of both of these genes. To verify that GFP expression phenocopied the endogenous mRNA expression pattern, we analyzed the in situ expression of both fmyhc genes in conjunction with the fast myosin marker F310 and GFP in the transgenic strains (Figure 2). This analysis confirmed that the in situ mRNA expression pattern coincided with the transgenic expression of GFP and also that the genes indeed were fast fiber specific. The fmyhc2.1:GFPumu153 ectopically expressed GFP in a subset of motor neurons, which we also found in transient experiments and in all individual fmyhc2.1:GFP lines we generated. This did not affect, however, the analysis of the muscle expression. Of interest, when we analyzed the expression of fmyhc1.2 in the fmyhc2.1:GFPumu153 transgenic line or vice versa, we found that there was a gap between the anterior fmyhc1.2 and the posterior fmyhc2.1 expression domains (Figure 2, C and D). This gap could be perfectly filled, however, by the expression of fmyhc1.3, as shown in our model (Figure 2E), and indicates that these three domains constitute distinct muscle domains with unique myosin heavy chain expression profiles.
Muscle domains are consistent in juveniles and adults
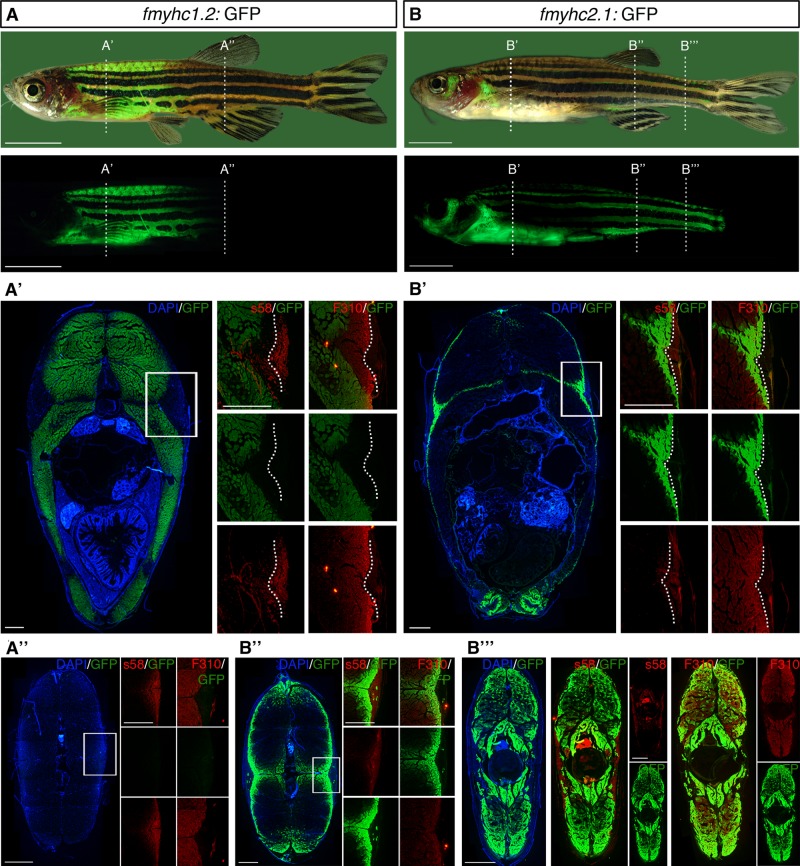
To study whether the expression domains of fmyhc1.2 and fmyhc2.1 represent distinct muscle domains in juvenile and older zebrafish, we analyzed transgenic GFP expression using optical projection tomography (OPT) on zebrafish with body length of up to 10 mm. In this analysis we were able to verify that the anterior trunk muscle domain and the tail domain still express the fmyhc1.2 and fmyhc2.1 genes, respectively, during juvenile stages (Figure 2, F and G, and Supplemental Movies S1 and S2). Whereas fmyhc1.2:GFPumu160 remained to be expressed in the same muscle domains as during the 3-dpf stage (Figure 2F), the expression of fmyhc2.1:GFPumu153 expanded in the subset of muscle fibers along the midline (Figure 2G and Supplemental Movie S2). The subsets of craniofacial and ocular muscles that were identified already at 3 dpf were also still respectively expressing the two different fmyhc isoforms (Figure 2, F and G, and Supplemental Movies S1 and S2). fmyhc2.1:GFPumu153 was also expressed in the fin musculature. Expression in the pectoral fins was observed already at embryonic stages, where the pectoral fins are the first fins to have formed. It is evident, however, that fmyhc2.1 is expressed in all fins at later life stages in the zebrafish (Figure 2G and Supplemental Movie S2). In adult zebrafish, we first analyzed expression of the fmyhc1.2:GFPumu160 and fmyhc2.1:GFPumu153 reporters by detecting GFP fluorescence in live fish up to 45 mm. These analyses confirmed that the anterior trunk muscle domain, as marked by expression of fmyhc1.2, and the midline and tail domains, as marked by fmyhc2.1, are still distinct in the adult zebrafish (Figure 3, A and B). To examine fiber-type specificity, we stained serial tissue sections using slow fiber–specific (S58) and fast fiber–specific (F310) antibodies. We found that both fmyhc1.2 and fmyhc2.1 remain fast fiber-specific, as both reporter genes are coexpressed in the fast muscle region and excluded from the slow fiber domain (Figure 3, A and B). We also found that fmyhc1.2 is excluded from the lateralmost region of the fast fiber domain (Figure 3A′), whereas fmyhc2.1 is expressed exclusively in the lateralmost region of the fast fiber domain (Figure 3B′), indicating that the fmyhc1.2 and fmyhc2.1 genes are expressed in separate subtypes of fast muscle fibers, even in adult zebrafish. In more posterior regions, the expression of fmyhc2.1 becomes increasingly widespread (Figure 3, B′′ and B′′′), and fmyhc1.2 is excluded (Figure 3A′′).
FIGURE 3:
fmyhc1.2:GFP and fmyhc2.1:GFP are expressed in restricted domains in adult zebrafish. Bright-field and fluorescence lateral view of adult (A) fmyhc1.2:GFP and (B) fmyhc2.1:GFP zebrafish; dashed lines indicate levels of cross sections in A′, A′′, B′, B′′, and B′′′. (A′, A′′) Cross section of fmyhc1.2:GFP; square indicate area of magnifications showing sections stained with S58 or F310. Dashed line indicates border between S58 and F310 staining. (B′, B′′, B′′′) Cross section of fmyhc2.1:GFP; square indicates area of magnifications showing sections stained with S58 or F310. Dashed line indicates border between S58 and F310 staining. Scale bar, 5 mm (whole fish), 500 μm (sections).
Inhibition of fmyhc2.1 results in striation defects
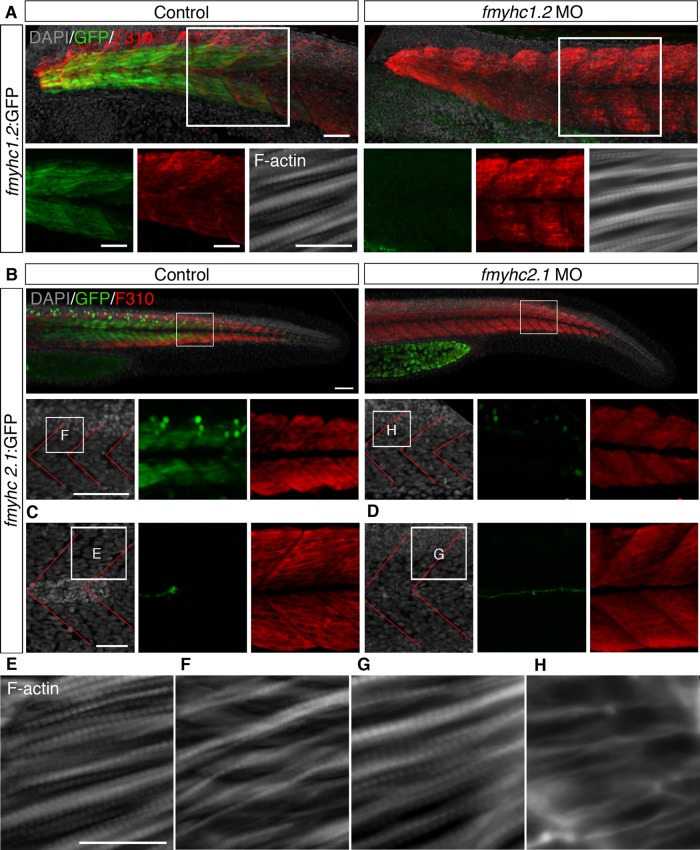
To examine functional aspects of the anterior and posterior myosin heavy chain expression domains, we generated morphant embryos in which the antisense morpholinos were designed to inhibit translation of the fmyhc1.2 and fmyhc2.1 transcripts, respectively. The morpholinos were designed to bind upstream of the translational start site, which enabled us to use the GFP-expressing transgenes as knockdown efficiency control.
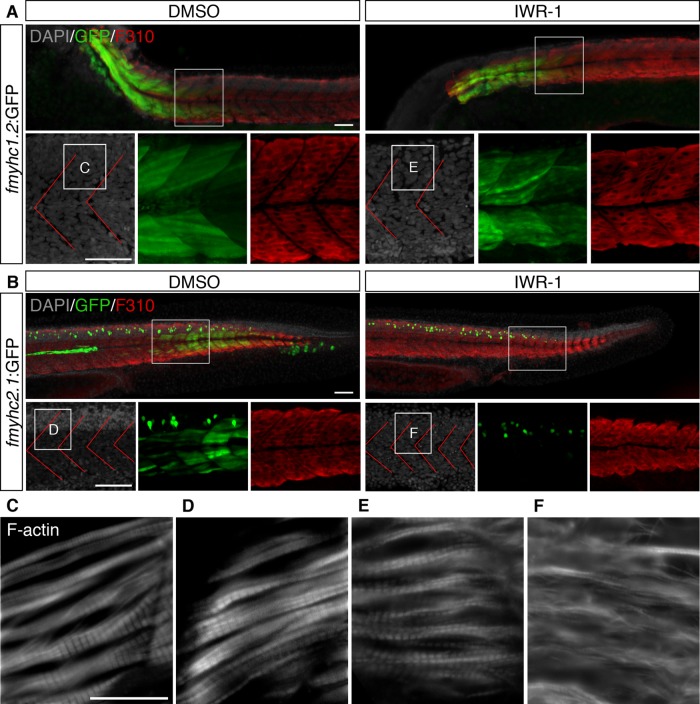
The fmyhc1.2 morphants developed normally and did not show any phenotype, even when fmyhc1.2:GFP expression was totally blocked (Figure 4A). On the other hand, the fmyhc2.1 morphants generally developed normally, with the exception of the muscle cells in the most-posterior somites, which were misshaped or failed to form properly (Figure 4). The most striking morphant phenotype was the failure of proper muscle organization when analyzed at 30 hpf (Figure 4, B–H). This phenotype was observed in 77 of 77 embryos, in which GFP expression also was significantly reduced. Of interest, the malformation was confined to the muscle cells, as the 4′,6-diamidino-2-phenylindole (DAPI) pattern indicated that somite formation was unchanged in the tails of these embryos (Figure 4). Even though the fast muscle fibers appeared to form normally, as detected by the fast fiber–specific F310 antibody (Figure 4B), subcellular sarcomeric organization was severely affected (Figure 4B–H). The sarcomeric striation, as detected by phalloidin, α-actin, and myomesin, which label F-actin, the sarcomeric Z-disk, and the M-band, respectively, was affected only in the tail domain, however, in congruence with the expression domain of the fmyhc2.1 transcript (Figure 4, E–H, and Supplemental Figure S1).
FIGURE 4:
Expression of fmyhc1.2:GFP and fmyhc2.1:GFP is down-regulated in fmyhc1.2 and fmyhc2.1 morphants. (A) Expression of GFP (green), F310 (red), and F-actin (white) in uninjected control fmyhc1.2:GFP embryos and fmyhc1.2:GFP embryos injected with fmyhc1.2 morpholino at 30 hpf; white squares indicate area of separate GFP and F310 channels below and enlargement of F-actin staining. (B) Expression of GFP (green) and F310 (red) in uninjected control fmyhc2.1:GFP embryos and fmyhc2.1:GFP embryos injected with fmyhc2.1 morpholino at 30 hpf; white squares indicate area of enlargements below. Expression of GFP (green) and F310 (red) in somites 8–9 in (C) uninjected control fmyhc2.1:GFP embryos and (D) fmyhc2.1:GFP embryos injected with fmyhc2.1 morpholino at 30 hpf. Expression of F-actin in (E, F) uninjected control embryo and (G, H) embryos injected with fmyhc2.1 morpholino at 30 hpf; area of view is indicated in B–D. Red lines, myosepta. Scale bar, 50 μm, 25 μm (F-actin).
Wnt signaling defines the tail domain
To assay which signaling pathways regulate the expression of fmyhc1.2 in the anterior trunk and fmyhc2.1 in the tail domain, we treated embryos with inhibitory chemicals from the 21-somite stage and analyzed them for GFP expression at the 48-hpf stage. We found that inhibition of the canonical wnt pathway by 10–100 μM IWR-1 treatment reduced fmyhc2.1:GFP expression with increasing concentration (Figure 5). Inhibition of FGF signaling through SU5401, BMP (dorsomorphin), retinoic acid (diethylaminobenzaldehyde [DEAB]), Notch (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester [DAPT]), and Hedgehog (cyclopamine) did not lead to any significant alterations of fmyhc2.1:GFP expression (Supplemental Figure S2). By using IWR-1–mediated wnt inhibition we could also determine that sarcomeric striation was disturbed, in particular in the tail fibers, where fmyhc2.1 normally is expressed (Figure 5, B–F, and Supplemental Figure S1), leading to the conclusion that the lack of tail fiber striation results from inhibited fmyhc2.1 expression. In contrast, even though the organization of the fibers in the anterior part of the trunk was mildly affected, IWR-1 treatment did not influence the expression of fmyhc1.2:GFP or the striation in the anterior part of the trunk (Figure 5, A and E, and Supplemental Figure S1).
FIGURE 5:
Expression of fmyhc2.1:GFP is down-regulated upon inhibition of wnt signaling. (A, B) Expression of GFP (green) and F310 (red) in (A) fmyhc1.2:GFP embryos and (B) fmyhc2.1:GFP embryos treated with DMSO or 100 μM IWR-1 from 20 ss to 30 hpf. White square indicates area of enlargements below. (C–F) Expression of F-actin in embryos treated with DMSO or 100 μM IWR-1 from 20 ss to 30 hpf; areas of enlargements are indicated in A and B. Red lines, myosepta. Scale bar, 50 μm (A, B), 25 μm (C–F).
Retinoic acid signaling acts upstream of wnt and is required for the anterior trunk domain
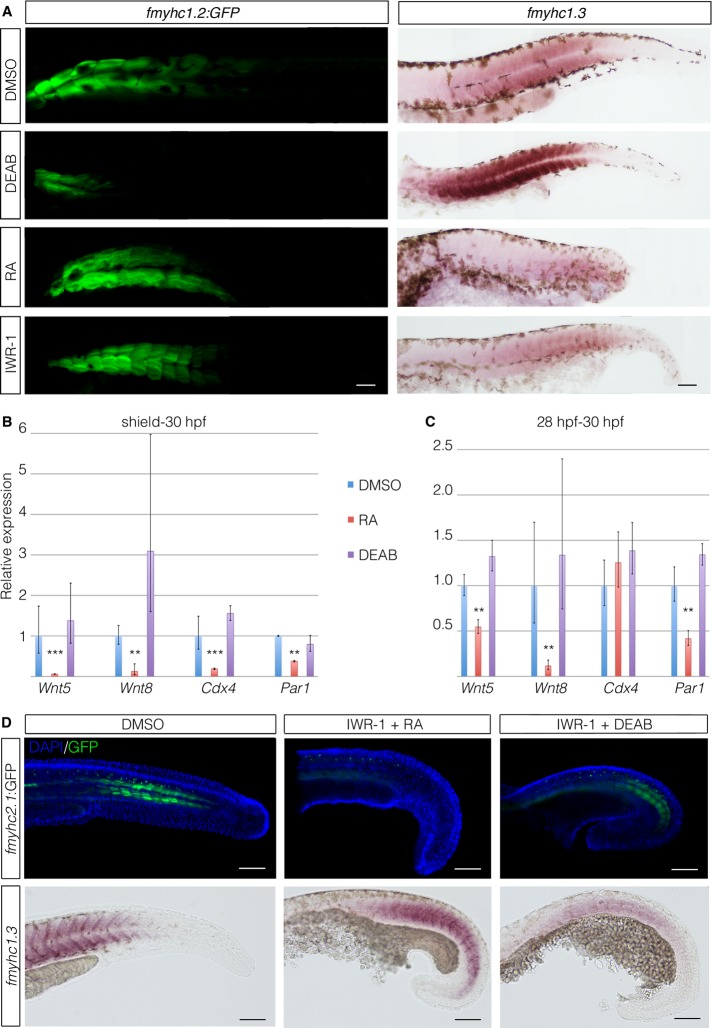
To analyze whether retinoic acid (RA) signaling played a part in the establishment of the muscle domains in the trunk, we treated fmyhc1.2:GFP embryos with DEAB or RA and examined the expression domains of fmyhc1.2 and fmyhc1.3. DEAB treatment efficiently inhibited expression of the fmyhc1.2:GFP in favor of the more-posterior fmyhc1.3 domain, which was expanded anteriorly (Figure 6A). The expression level of fmyhc1.3 also increased in DEAB-treated embryos. RA treatment did not significantly affect the expression level of fmyhc1.2:GFP, which in RA-treated embryos was detected throughout most of the muscle tissue in the trunk (Figure 6A). These embryos completely lacked the tail domain and expressed low levels of fmyhc1.3 in only the most-posterior part of the remaining trunk muscle. Treatment with IWR-1 to block Wnt signaling did not lead, however, to any significant change in the fmyhc1.2:GFP or the fmyhc1.3 expression domains (Figures 5A and 6A). We also examined whether RA, DEAB, cyclopamine, dorsomorphin, DAPT, or SU5401 treatment influenced expression of fmyhc1.2:GFP when initiated at 20 somites (ss), after the formation of the anterior somites, but found no significant changes in fmyhc1.2:GFP expression levels (Supplemental Figure S3), leading to the conclusion that the fmyhc1.2 domain is defined before 20 ss.
FIGURE 6:
Retinoic acid and wnt signaling determines fmyhc expression domains. (A) fmyhc1.3 in situ hybridization in fmyhc1.2:GFP embryos treated with DMSO, 0.2 μM RA, 200 μM DEAB, or 100 μM IWR-1 from shield stage to 30 hpf. (B) qPCR analysis of genes in the wnt signaling pathway on 30-hpf embryos treated with RA from shield stage. (C) qPCR analysis of genes in the wnt signaling pathway on 30-hpf embryos treated with RA from 28 hpf. (D) Top, fmyhc1.2:GFP embryos treated with DMSO, 100 μM IWR-1 in combination with 0.2 μM RA, or 200 μM DEAB from 18 ss to 36 hpf. Bottom, in situ hybridization showing mRNA expression of fmyhc1.3. Scale bar, 50 μm.
Because of known interactions between RA and Wnt signaling, we examined their relationship under our experimental conditions by analyzing the expression levels of a number of genes in the wnt signaling pathway in embryos treated with RA or DEAB using quantitative reverse transcriptase-PCR. The relative expression levels of wnt5, wnt8, cdx4, and paraxis1 (par1) were all significantly down-regulated in 30-hpf embryos treated with RA from the shield stage (Figure 6B). Treatment with DEAB did not lead to any significant change in expression levels, even though a general up-regulation of these genes was observed. We also conducted the same experiment using only the tails of embryos treated with RA or DEAB for 2 h between 28 and 30 hpf, which again resulted in significant down-regulation of wnt5, wnt8, and par1 (Figure 6C). Further, we observed nonsignificant up-regulation of these genes in DEAB- treated embryos. The expression of cdx4 did not show any significant change in either RA or DEAB treatment in these experiments. Taken together, the results indicate that RA signaling acts upstream of wnt in the establishment of the anteroposterior trunk muscle domains.
To further explore the putative epistatic relationship between the wnt and RA pathways, we treated fmyhc2.1:GFP embryos with IWR-1 in combination with either RA or DEAB from 18 ss until ∼36 hpf. Adding RA did not alter the inhibitory effect that the IWR-1 treatment had on fmyhc2.1 expression in these embryos (Figure 6D), but the expression of fmyhc1.3 was expanded into tail myotomes. In contrast, inhibition of both wnt and RA by combined IWR-1 and DEAB treatment resulted in rescue of fmyhc2.1 expression, and fmyhc1.3 expression domain returned to a more intermediate position along the anteroposterior axis (Figure 6D), leading to the conclusion that these muscle domains are determined by levels of RA and wnt during myogenesis.
DISCUSSION
Vertebrate skeletal muscle is composed of a heterogeneous group of cells with similar structural and functional properties, with individual subtypes specialized to respond to unique functional demands. Genes encoding numerous myosin heavy chain subtypes are individually expressed in specific cell lineages defining fiber identity. Fast myosin heavy chain expression in zebrafish embryos is very complex, as shown in Figure 1. Even though the transcript sequence homology is very high between the six isoforms that we analyzed, they are all differentially expressed, which indicates that the individual muscle domains have different functional requirements. Previous studies in zebrafish identified fast-fiber subtypes in the medial versus lateral parts of the myotome (Wolff et al., 2003; Groves et al., 2005; Hinits et al., 2009). The existence of distinct muscle domains along the anteroposterior axis was not described previously, even though tail-, fin-, and cranial muscle–specific expression of myhz2 (fmyhc2.1) was originally identified in Peng et al. (2002). In spite of this, RNA probes targeting fast myosin heavy chains have been used in many cases to identify fast-specific muscle cells in zebrafish, indicating that fast myosin heavy chains are uniformly expressed throughout the fast muscle domain. This is likely due to cross-hybridization between the highly similar myosin heavy chain isoforms. However, using more-stringent analyses targeting the untranslated regions of these genes, we find that the fast myosin heavy chains are indeed expressed in very distinct subdomains. We also confirmed this by generating reporter constructs using regulatory regions of the two extremes— anteriorly expressed fmyhc1.2 and posteriorly expressed fmyhc2.1—which both phenocopied endogenous mRNA expression as detected using in situ hybridization. Of interest, fmyhc1.2 and fmyhc2.1 expression domains do not overlap in the zebrafish trunk, even in adults, where fmyhc2.1-positive fibers that initially resided along the midline spread to more-lateral domains (Figure 3), supporting the idea that fmyhc1.2+ and fmyhc2.1+ fibers are indeed distinct fiber subtypes.
We found redundancy between the myosin heavy chains in the most-anterior muscle cells, since fmyhc1.2-morphant embryos lacked phenotype. The fmyhc1.2 gene is coexpressed with fmyhc1.1 and to some extent also with fmyhc 2.2 and fmyhc2.3 (Figure 1), and hence knockdown of fmyhc1.2 alone does not result in sarcomeric disorganization (Figure 4). However, expression of fmyhc2.1 is confined to the medial part of the myotome in the tail at 3 dpf. This is in contrast to the more-lateral expression of fmyhc2.2 and fmyhc2.3. Thus fmyhc2.1 is the only myosin heavy chain expressed in this domain. In congruence with this observation, knockdown of fmyhc2.1 did lead to altered cell morphology and disturbed sarcomeric striation, specifically in the tail region, as detected by phalloidin staining (Figure 4). This indicates that fmyhc2.1 is essential for formation of a subset of fast fibers in the tail and further supports our previous observations.
To study the transcriptional regulation of the fmyhc2.1 gene, we blocked numerous signaling pathways, using established chemical inhibitors. We chose this approach rather than using mutants since we wanted to avoid early effects on the initial formation of the tail somites, as mutations in the BMP (piggytail, pgy) and wnt (pipetail, ppt) signaling pathways affects somitogenesis in the tail (Hammerschmidt et al., 1996; van Eeden et al., 1996) or causes blocking of myod expression in the lateral fast domain by early FGF inhibition (Groves et al., 2005). Therefore we initiated the chemical treatments after tail formation but before fmyhc2.1 transcriptional onset, which began at around 24 hpf (Figure 1), and found that inhibition of wnt signaling efficiently blocked fmyhc2.1 expression in tail-specific fast fibers (Figure 5). None of the other pathways leads to significant changes in fmyhc2.1 expression in the tail fibers (Supplemental Figure S2), leading to the conclusion that wnt signaling is required for fmyhc2.1 expression. Consequently, wnt inhibition also resulted in disturbed sarcomeric striation specifically in tail fibers, which supports our conclusion that the fmyhc2.1 gene is essential for formation of the sarcomeric structure in tail fibers.
Both the Wnt and the RA signaling pathways establish cellular fates in the anteroposterior axis and account for graded activity within the paraxial mesoderm in many different species (Onai et al., 2009; Petersen and Reddien, 2009; Aulehla and Pourquié, 2010). It has also been shown that RA activity has its peak at the level of the anteriormost somites in the zebrafish embryo (Shimozono et al., 2013), whereas the level of wnt signaling is highest in the zebrafish posterior tail and tail bud regions (Shimizu et al., 2012). Wnt signaling is important for proper tail formation in zebrafish since mutations in the wnt pathway (ppt) result in failure of tail somite formation (van Eeden et al., 1996), but we found that even after formation of these somites, wnt signaling is required for expression of the fmyhc2.1 gene and hence formation and differentiation of proper tail-specific muscle cell type. Of interest, RA treatment also has an effect on fmyhc2.1 expression, suggesting that RA may inhibit wnt signaling in this region. However, if RA treatment is initiated at an early stage, before somitogenesis, the fmyhc2.1-expressing region does not form at all, and if RA treatment is initiated at the 20-somite stage or later, expression of fmyhc2.1 will still be present, albeit reduced (Supplemental Figure S2). We used a quantitative PCR (qPCR) strategy to confirm that RA indeed affects the wnt pathway, where the expression of wnt5 and wnt8, as well as of cdx4 and par1, were all significantly inhibited in RA-treated embryos. This observation is supported by a study by Martin and Kimelman (2010), who showed that RA inhibits wnt3a in the tail bud.
We propose that a balance between levels of RA and wnt strongly influence the determination of fast muscle domains along the anteroposterior axis. Our data show that the expression domains of the fast myosin genes can be shifted by pharmacological manipulation of RA and wnt levels. In particular, expression of fmyhc1.3 is shifted into more-anterior somites when RA is inhibited (Figure 6A) and becomes ectopically expressed in tail somites when wnt is inhibited in combination with RA treatment (Figure 6D). Of interest, the fast-muscle domains appear to be restored to wild type when both RA and wnt are inhibited (Figure 6D), which suggests that the balance between the two pathways is more important than the absolute levels. It is also not unlikely that presence of RA potentiates the effect of IWR-1 treatment. Early combinatory treatments were lethal (unpublished data), which excluded the possibility of examining how the anteriormost domain, defined by fmyhc1.2, was affected. Furthermore, we do not exclude the possibility that the outcome of both RA and wnt treatments are stage dependent.
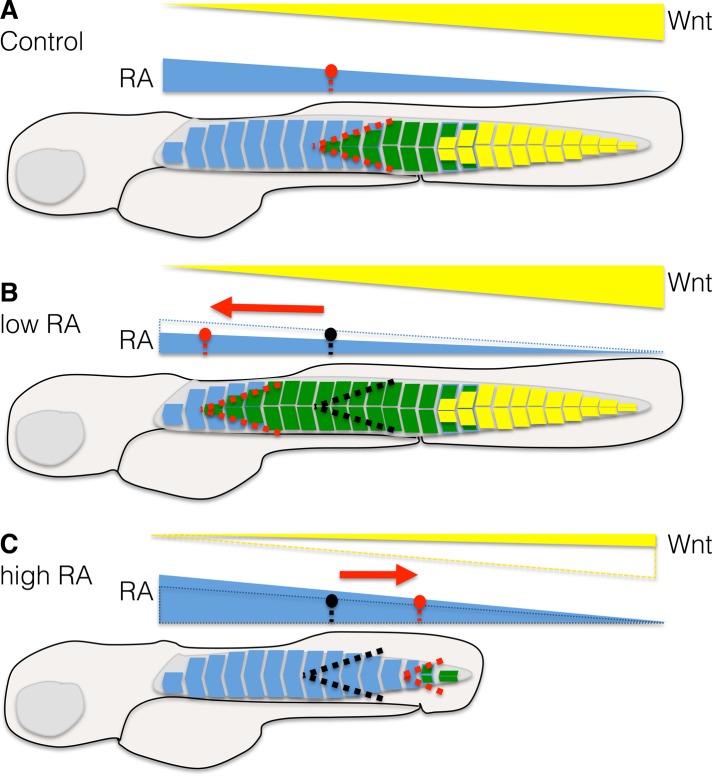
On the basis of this, we propose a model in which the genes fmyhc1.2, fmyhc1.3, and fmyhc2.1 define three distinct fast-muscle domains in the anteroposterior axis, where the levels of RA and wnt are instructive (Figure 7). The anterior border of the intermediate domain, defined by fmyhc1.3, moves anteriorly with decreased RA levels and posteriorly when RA levels are increased. The posterior border of the fmyhc1.3, however, does not move into the tail region, defined by fmyhc2.1, unless wnt inhibition is combined with RA treatment from the 18 ss. The tail region does require wnt signaling, which we found to be affected negatively by RA treatment. Of interest, inhibition of wnt and subsequent loss of fmyhc2.1 expression result in disturbed sarcomeric striation of muscle in this domain (Figure 5), which further strengthens our observation that, without additional RA treatment, none of the other myosins is interchanged into the tail domain.
FIGURE 7:
Model of the roles of RA and wnt in the establishment of muscle domains. (A) In the wild-type situation, there is a gradient of RA from anterior to posterior (blue) and wnt from posterior to anterior (yellow), by which the levels of the respective signaling pathways determine the anterior domain (defined by fmyhc1.2, as illustrated with blue somites), the intermediate domain (defined by fmyhc1.3 as green somites), and the posterior/tail domain (defined by fmyhc2.1 as yellow somites). The anterior border of the intermediate (green) domain is illustrated with a red dashed line. (B) In an embryo with reduced RA, the anterior border (red dashed line) of the intermediate domain (green somites) is shifted anteriorly along the RA threshold. (C) In an embryo treated with RA the anterior border (red dashed line) of the intermediated domain (green somites) is shifted posteriorly along the RA threshold. In addition, these embryos lose the tail domain (yellow somites) and subsequently the expression of fmyhc2.1 if the treatment is initiated at an early embryonic stage.
A study by Szeto and Kimelman (2006) elegantly showed how nodal and BMP signaling organize different populations of mesodermal progenitor cells, which will divide the somites into tail and anterior and posterior trunk domains before somitogenesis. These early developmental events likely play a role in the specification of somite identity, and the domains described correspond well to our observations regarding fast-muscle domains. Of interest, these authors found that wnt inhibitory factor-1 (WIF1) is strongly expressed in the trunk domain but not in the tail bud, which supports our finding that wnt is required for myosin expression in the tail region.
The role of hox genes in patterning along the anteroposterior axis is well established, in particular for patterning of the CNS. Even if we exclude experiments examining the role of hox genes in this study, we find it likely that hox genes contribute to the patterning of trunk musculature. Our data do not exclude the possibility that the RA and wnt gradients act indirectly, via activation and repression of genes, including hox genes, which would account for the expression of the individual fmyhc genes. The tail-specific expression of fmyhc2.1 may very well be regulated by cdx or posterior hox genes. Of interest, cdx2/4 and hox genes belonging to posterior group 13 are important for maintenance of wnt in the tail region during mouse development (Young et al., 2009). In summary, fast-specific myosin heavy chains are differentially expressed and regulated in the zebrafish trunk, and spatially restricted muscle domains can be defined according to the subtype they express. The tail fibers require wnt-regulated expression of fmyhc2.1, which, when inhibited, causes sarcomeric striation defects, whereas the identity of anterior somites requires RA for expression of fmyhc1.2. Collectively our data illustrate how the myosin heavy chain genes define novel fast-fiber subtypes in the zebrafish anteroposterior axis and that RA and wnt determine their expression domains.
MATERIALS AND METHODS
Zebrafish strains and maintenance
Embryos were obtained from wild-type zebrafish (Danio rerio, London wild type [LWT] and tupfel long fin [TL]). Zebrafish were maintained by standard procedures in the Umeå University Zebrafish Facility. Embryos were staged up to the 15-somite stage by counting the number of somites, as described previously (Kimmel et al., 1995).
To generate fmyhc:GFP lines, genomic DNA corresponding to 8074 base pairs upstream of the fmyhc2.1 translational start site and 6969 base pairs upstream of the fmyhc1.2 translational start site was cloned from genomic zebrafish DNA and ligated into the translational start site of enhanced GFP in a vector flanked with I-Sce-I sites. Zebrafish embryos at the one-cell stage were injected with the DNA constructs together with the I-Sce-I enzyme (New England Biolabs, Ipswich, MA). Injected embryos were screened for transient GFP expression before being allowed to grow into adulthood and crossed to establish transgenic founders. Four founders with identical expression patterns to fmyhc2.1:GFPumu153 and two founders identical to fmyhc1.2:GFPumu160 were identified.
Genes and probe design
Based on Ensembl sequence data, 5′-UTR fragments of fast myosin heavy chain (fmyhc) 1.1 (myhz1; ENSDARG00000067990), fmyhc1.2 (ENSDARG00000067995), fmyhc1.3 (fmyhcx; ENSDARG00000067997), fmyhc2.1 (myhz2; ENSDARG00000012944), and fmyhc2.2 (myhc4; ENSDARG00000035438) and 3′-UTR fragments of fmyhc2.3 (myha; ENSDARG00000095930) cDNA were amplified using PCR. The primers used are listed in Table 1. The resulting PCR fragments were cloned into pCRII-TOPO TA vectors from Invitrogen (Carlsbad, CA) and inserts confirmed by sequencing before digoxigenin (DIG)-labeled probes were synthesized using standard methods.
TABLE 1:
Primers for amplification of cDNA fragments used for probe synthesis.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| fmyhc1.1 | 5′-aagagcttgagctggactgg-3′ | 5′-aggtaaatggcagccttgc-3′ |
| fmyhc1.2 | 5′-gatcagccagggttgactgt-3′ | 5′-tggcggcttacttcttacca-3′ |
| fmyhc1.3 | 5′-gcgcttggaagaccagcc-3′ | 5′-ggcggcttaccgtacttctt-3′ |
| fmyhc2.1 | 5′-aatcattcatctggtctcaagga-3′ | 5′-gccatctccgcgtcagta-3′ |
| fmyhc2.2 | 5′-ctcaaggatttccaccaagc-3′ | 5′-ggtggacgtcatcctcctta-3′ |
| fmyhc2.3 | 5′-gctgcaggatctggttgata-3′ | 5′-aagcacagatttatttgaaaagca-3′ |
Whole-mount in situ hybridization
Zebrafish embryos were fixed in 4% paraformaldehyde (PFA) overnight. Whole-mount in situ hybridization was performed as described previously (Thisse et al., 1993) with minor changes; 1% blocking reagent (Roche, Indianapolis, IN) was used instead of 2% sheep serum and 2 mg/ml bovine serum albumin. DIG-labeled and fluorescein-labeled RNA probes were detected using Fast Red (Roche) and NBT/BCIP (Roche). RNA probes used were fmyhc1.1, fmyhc1.2, fmyhc1.3, fmyhc2.1, fmyhc2.2, and fmyhc2.3.
Immunohistochemistry
Zebrafish embryos were fixed in 4% PFA overnight. For sections, adult zebrafish were fixed in 4% PFA overnight, then stepwise treated with 10, 20, and 30% sucrose, frozen in Tissue-Tek (O.C.T. compound; Sakura, Alphen aan den Rijn, Netherlands), and serially sectioned at 10 μm. Immunohistochemistry was performed using standard procedures. Primary antibodies used were S58 (1:10; slow muscle fibers; Developmental Studies Hybridoma Bank [DSHB], Iowa City, IA) and F310 (1:50; myosin light chain 1 and 3F; DSHB), mMaC myomesin B4 (1:5; DSHB), and anti α-actinin (1:300; Sigma-Aldrich, St. Louis, MO). F-actin was visualized using rhodamine– phalloidin (1:100; Molecular Probes, Eugene, OR). Nuclei were stained using DAPI (Sigma-Aldrich).
Optical projection tomography
Juvenile zebrafish were immunohistochemically processed and prepared for OPT scanning essentially as described previously (Alanentalo et al., 2007). Antibodies used were chicken anti-GFP (1:200; Aves, Tigard, OR) and Alexa Fluor 594 goat anti-chicken (1:500; Molecular Probes). The specimens were scanned using the Bioptonics 3001 OPT scanner (Bioptonics, Edinburgh, United Kingdom), and data were processed as described previously (Cheddad et al., 2012). Drishti software (version 2.2; ANUSF VizLab, Australian National University, Canberra, Australia) was used to render and visualize three-dimensional volumes of reconstructed data sets.
Embryo treatments
Dechorionated embryos were incubated at desired stages in 100 μM IWR-1, 50 μM cyclopamine, 100 μM DAPT, 0.2 μM RA, 40 μM SU5402, 200 μM dorsomorphin, 200–500 μM DEAB, or dimethyl sulfoxide (DMSO) dissolved in embryo medium and subsequently fixed with 4% PFA.
Morpholino injections
For morpholino treatment, zebrafish embryos were injected at the one- to four-cell stage with 4 nl of morpholinos targeting fmyhc1.2 (tggcggcttacttcttaccacgctc) and fmyhc2.1 (ctctacccaaaatcttaccttgcgt; GeneTools, Philomath, OR) at a concentration of 1 nmol/μl.
Quantitative PCR
Embryos were treated with DEAB or RA starting from shield stage or 28 hpf. At 30 hpf, RNA was extracted from whole embryos (treatment at shield stage) or the tail region (treatment at 28 hpf), respectively. RNA extraction and cDNA generation were performed as described previously (Peterson and Freeman, 2009). qPCR was performed using FastStartUniversal qPCR Mastermix (Roche). Primer sequences were taken from Lu et al. (2011).
Supplementary Material
Acknowledgments
This study was initiated at the MRC Center for Developmental and Biomedical Genetics at the University of Sheffield (Sheffield, United Kingdom) in the lab of Philip W. Ingham, whom we acknowledge for support during the early phase of this project. We also acknowledge Edward Samuel Inbaraj and Alexandra Haas for technical assistance. Financial support was provided by the Kempe Foundation, the Jeansson Foundation, the Åke Wiberg Foundation, the Magnus Bergvall Foundation, and Umeå University.
Abbreviations used:
- afm
anal fin muscle
- BMP
bone morphogenic protein
- DAPI
4′,6-diamidino-2-phenylindole
- DAPT
N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester
- DEAB
diethylaminobenzaldehyde
- dfm
dorsal fin muscle
- DMSO
dimethyl sulfoxide
- dpf
days postfertilization
- FGF
fibroblast growth factor
- GFP
green fluorescent protein
- hh
hyohyoideus
- hpf
hours postfertilization
- ih
interhyoideus
- ima
intermandibular anterior
- imp
intermandibular posterior
- LWT
London wild type
- MRFs
myogenic regulatory factors
- MyHC
myosin heavy chain
- om
ocular muscles
- OPT
optical projection tomography
- pf
pectoral fin
- PFA
paraformaldehyde
- pfm
pectoral fin muscle
- pgy
piggytail
- ppt
pipetail
- qPCR
quantitative PCR
- RA
retinoic acid
- sh
sternohyoideus
- ss
somite stage
- TL
tupfel long fin
- tfm
tail fin muscle
- WIF1
wnt inhibitory factor 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0486) on February 12, 2014.
REFERENCES
- Alanentalo T, Asayesh A, Morrison H, Lorén CE, Holmberg D, Sharpe J, Ahlgren U. Tomographic molecular imaging and 3D quantification within adult mouse organs. Nat Methods. 2007;4:31–33. doi: 10.1038/nmeth985. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Pourquié O. Signaling gradients during paraxial mesoderm development. Cold Spring Harb Perspect Biol. 2010;2:a000869. doi: 10.1101/cshperspect.a000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden CS, Currie PD, Ingham PW, Hughes SM. Notochord induction of zebrafish slow muscle mediated by Sonic Hedgehog. Genes Dev. 1997;11:2163–2175. doi: 10.1101/gad.11.17.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Vincent S. Distinct and dynamic myogenic populations in the vertebrate embryo. Curr Opin Genet Dev. 2009;19:444–453. doi: 10.1016/j.gde.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Burguiere AC, Nord H, von Hofsten J. Alkali-like myosin light chain-1 (myl1) is an early marker for differentiating fast muscle cells in zebrafish. Dev Dyn. 2011;240:1856–1863. doi: 10.1002/dvdy.22677. [DOI] [PubMed] [Google Scholar]
- Cheddad A, Svensson C, Sharpe J, Georgsson F, Ahlgren U. Image processing assisted algorithms for optical projection tomography. IEEE Trans Med Imaging. 2012;31:1–15. doi: 10.1109/TMI.2011.2161590. [DOI] [PubMed] [Google Scholar]
- Currie P, Ingham PW. Induction of a specific muscle cell type by a Hedgehog-like protein in zebrafish. Nature. 1996;382:452–455. doi: 10.1038/382452a0. [DOI] [PubMed] [Google Scholar]
- Devoto SH, Melancon E, Eisen JS, Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–3380. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- Devoto SH, Stoiber W, Hammond CL, Steinbacher P, Haslett JR, Barresi MJ, Patterson SE, Adiarte EG, Hughes SM. Generality of vertebrate developmental patterns: evidence for a dermomyotome in fish. Evol Dev. 2006;8:101–110. doi: 10.1111/j.1525-142X.2006.05079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SJ, Devoto SH, Westerfield M, Moon RT. Positive and negative regulation of muscle cell identity by members of the Hedgehog and TGF-beta gene families. J Cell Biol. 1997;139:145–156. doi: 10.1083/jcb.139.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman KA, Reggiani C, Schiaffino S, te Kronnie G. Maximum velocity of shortening related to myosin isoform composition in frog skeletal muscle fibres. J Physiol. 1988;395:679–694. doi: 10.1113/jphysiol.1988.sp016941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elworthy S, Hargrave M, Knight R, Mebus K, Ingham PW. Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development. 2008;135:2115–2126. doi: 10.1242/dev.015719. [DOI] [PubMed] [Google Scholar]
- Feng X, Adiarte EG, Devoto SH. Hedgehog acts directly on the zebrafish dermomyotome to promote myogenic differentiation. Dev Biol. 2006;300:736–746. doi: 10.1016/j.ydbio.2006.08.056. [DOI] [PubMed] [Google Scholar]
- Grandel H, et al. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development. 2002;129:2851–2865. doi: 10.1242/dev.129.12.2851. [DOI] [PubMed] [Google Scholar]
- Groves JA, Hammond CL, Hughes SM. Fgf8 drives myogenic progression of a novel lateral fast muscle fibre population in zebrafish. Development. 2005;132:4211–4222. doi: 10.1242/dev.01958. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, et al. Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development. 1996;123:143–151. doi: 10.1242/dev.123.1.143. [DOI] [PubMed] [Google Scholar]
- Hashiguchi M, Mullins MC. Anteroposterior and dorsoventral patterning are coordinated by an identical patterning clock. Development. 2013;140:1970–1980. doi: 10.1242/dev.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta K, Bremiller R, Westerfield M, Kimmel CB. Diversity of expression of engrailed-like antigens in zebrafish. Development. 1991;112:821–832. doi: 10.1242/dev.112.3.821. [DOI] [PubMed] [Google Scholar]
- Hinits Y, Osborn DP, Hughes SM. Differential requirements for myogenic regulatory factors distinguish medial and lateral somitic, cranial and fin muscle fibre populations. Development. 2009;136:403–414. doi: 10.1242/dev.028019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Williams VC, Sweetman D, Donn TM, Ma TP, Moens CB, Hughes SM. Defective cranial skeletal development, larval lethality and haploinsufficiency in Myod mutant zebrafish. Dev Biol. 2011;358:102–112. doi: 10.1016/j.ydbio.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsinger E, Stellabotte F, Devoto SH, Westerfield M. Hedgehog signaling is required for commitment but not initial induction of slow muscle precursors. Dev Biol. 2004;275:143–157. doi: 10.1016/j.ydbio.2004.07.030. [DOI] [PubMed] [Google Scholar]
- Hollway GE, Bryson-Richardson RJ, Berger S, Cole NJ, Hall TE, Currie PD. Whole-somite rotation generates muscle progenitor cell compartments in the developing zebrafish embryo. Dev Cell. 2007;12:207–219. doi: 10.1016/j.devcel.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Kessel M, Gruss P. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell. 1991;67:89–104. doi: 10.1016/0092-8674(91)90574-i. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Lu FI, Thisse C, Thisse B. Identification and mechanism of regulation of the zebrafish dorsal determinant. Proc Natl Acad Sci USA. 2011;108:15876–15880. doi: 10.1073/pnas.1106801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mara A, Holley SA. Oscillators and the emergence of tissue organization during zebrafish somitogenesis. Trends Cell Biol. 2007;17:593–599. doi: 10.1016/j.tcb.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Marshall H, Nonchev S, Sham MH, Muchamore I, Lumsden A, Krumlauf R. Retinoic acid alters hindbrain Hox code and induces transformation of rhombomeres 2/3 into a 4/5 identity. Nature. 1992;360:737–741. doi: 10.1038/360737a0. [DOI] [PubMed] [Google Scholar]
- Martin BL, Kimelman D. Brachyury establishes the embryonic mesodermal progenitor niche. Genes Dev. 2010;24:2778–2783. doi: 10.1101/gad.1962910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya AK, Tan H, Souren M, Wang X, Wittbrodt J, Ingham PW. Integration of Hedgehog and BMP signalling by the engrailed2a gene in the zebrafish myotome. Development. 2011;138:755–765. doi: 10.1242/dev.062521. [DOI] [PubMed] [Google Scholar]
- Onai T, Lin HC, Schubert M, Koop D, Osborne PW, Alvarez S, Alvarez R, Holland ND, Holland LZ. Retinoic acid and Wnt/beta-catenin have complementary roles in anterior/posterior patterning embryos of the basal chordate amphioxus. Dev Biol. 2009;332:223–233. doi: 10.1016/j.ydbio.2009.05.571. [DOI] [PubMed] [Google Scholar]
- Peng MY, Wen HJ, Shih LJ, Kuo CM, Hwang SP. Myosin heavy chain expression in cranial, pectoral fin, and tail muscle regions of zebrafish embryos. Mol Reprod Dev. 2002;63:422–429. doi: 10.1002/mrd.10201. [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Wnt signaling and the polarity of the primary body axis. Cell. 2009;139:1056–1068. doi: 10.1016/j.cell.2009.11.035. [DOI] [PubMed] [Google Scholar]
- Peterson SM, Freeman JL. RNA isolation from embryonic zebrafish and cDNA synthesis for gene expression analysis. J Vis Exp. 2009. p. , e1470. [DOI] [PMC free article] [PubMed]
- Schiaffino S, Gorza L, Sartore S, Saggin L, Ausoni S, Vianello M, Gundersen K, Lømo T. Three myosin heavy chain isoforms in type 2 skeletal muscle fibres. J Muscle Res Cell Motil. 1989;10:197–205. doi: 10.1007/BF01739810. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscle. Physiol Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- Shimizu N, Kawakami K, Ishitani T. Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/β-catenin signaling-reporter transgenic zebrafish. Dev Biol. 2012;370:71–85. doi: 10.1016/j.ydbio.2012.07.016. [DOI] [PubMed] [Google Scholar]
- Shimozono S, Iimura T, Kitaguchi T, Higashijima S, Miyawaki A. Visualization of an endogenous retinoic acid gradient across embryonic development. Nature. 2013;496:363–366. doi: 10.1038/nature12037. [DOI] [PubMed] [Google Scholar]
- Szeto DP, Kimelman D. The regulation of mesodermal progenitor cell commitment to somitogenesis subdivides the zebrafish body musculature into distinct domains. Genes Dev. 2006;20:1923–1932. doi: 10.1101/gad.1435306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- van Eeden FJ, et al. Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development. 1996;123:153–164. doi: 10.1242/dev.123.1.153. [DOI] [PubMed] [Google Scholar]
- Wolff C, Roy S, Ingham PW. Multiple muscle cell identities induced by distinct levels and timing of Hedgehog activity in the zebrafish embryo. Curr Biol. 2003;13:1169–1181. doi: 10.1016/s0960-9822(03)00461-5. [DOI] [PubMed] [Google Scholar]
- Young T, et al. Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev Cell. 2009;17:516–526. doi: 10.1016/j.devcel.2009.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.