Abstract
Cyclic diadenosine monophosphate (c-di-AMP) has emerged as an important dinucleotide that is involved in several processes in bacteria, including cell wall remodeling (and therefore resistance to antibiotics that target bacterial cell wall). Small molecules that target c-di-AMP metabolism enzymes have the potential to be used as antibiotics. Coralyne is known to form strong complexes with polyadenine containing eight or more adenine stretches but not with short polyadenine oligonucleotides. Using a panel of techniques (UV, both steady state fluorescence and fluorescence lifetime measurements, circular dichroism (CD), NMR, and Job plots), we demonstrate that c-di-AMP, which contains only two adenine bases is an exception to this rule and that it can form complexes with coralyne, even at low micromolar concentrations. Interestingly, pApA (the linear analog of c-di-AMP that also contains two adenines) or cyclic diguanylate (c-di-GMP, another nucleotide second messenger in bacteria) did not form any complex with coralyne. Unlike polyadenine, which forms a 2:1 complex with coralyne, c-di-AMP forms a higher order complex with coralyne (≥6:1). Additionally, whereas polyadenine reduces the fluorescence of coralyne when bound, c-di-AMP enhances the fluorescence of coralyne. We use the quenching property of halides to selectively quench the fluorescence of unbound coralyne but not that of coralyne bound to c-di-AMP. Using this simple selective quenching strategy, the assay could be used to monitor the synthesis of c-di-AMP by DisA or the degradation of c-di-AMP by YybT. Apart from the practical utility of this assay for c-di-AMP research, this work also demonstrates that, when administered to cells, intercalators might not only associate with polynucleotides, such as DNA or RNA, but also could associate with cyclic dinucleotides to disrupt or modulate signal transduction processes mediated by these nucleotides.
Cyclic dinucleotides have come to the forefront of microbial research due to the impressive arrays of processes in bacteria that they regulate.1,2 For example, cyclic diguanylate (c-di-GMP, the first cyclic dinucleotide to be discovered by Benziman almost three decades ago) has been shown to regulate motility, sessility and biofilm formation, virulence, cell cycle progression, heavy metal resistance, phage resistance, antibiotic resistance, and quorum sensing among others in myriads of bacteria, including bacteria of clinical and military relevance.3−5 Due to the varied processes in bacteria that c-di-GMP regulates, there has been an explosion of investigations aimed at unraveling processes, which are regulated by this nucleotide.6 Recently, another cyclic dinucleotide, cyclic diadenosine monophosphate (c-di-AMP),7,8 has also stoked the interests of microbiologists as it has emerged that c-di-AMP is as important a second messenger in bacteria as c-di-GMP. c-di-AMP, originally discovered as a signaling molecule that controls DNA integrity in B. subtilis, has now been shown to regulate bacterial cell size and/or cell wall formation,7,9 and hence, receptors involved in c-di-AMP signaling could become important antibacterial targets.10 In S. aureus, L. monocytogens, or B. subtilis, it has been shown that increased intracellular c-di-AMP endowed these pathogens with the ability to resist β-lactam antibiotics whereas decreased intracellular c-di-AMP concentrations made the bacteria susceptible to antibiotics that target bacterial cell wall synthesis.11−15 In some bacteria, c-di-AMP also modulates ion transport across bacterial membranes and is hypothesized to modulate bacterial physiology as the concentration of metals in the environment changes.16 Apart from the aforementioned processes, c-di-AMP is also involved in resistance to acid17 and heat stress18 in some bacteria. In addition to controlling bacterial physiology, c-di-AMP also affects eukaryotic host cells and elicits type I interferon response.19−21 Unlike c-di-GMP, for which many of the protein and RNA receptors have been biochemically and biophysically characterized, the metabolism proteins and “adaptor/effector” proteins for c-di-AMP remain largely uncharacterized and the coming years will undoubtedly witness an explosion of biochemical and structural characterizations of c-di-AMP-related proteins and RNA. In this regard, tools that would aid the characterization of c-di-AMP receptors would help delineate the details of c-di-AMP signaling in bacteria. Herein, we provide a surprisingly simple fluorescent detection of c-di-AMP, using readily available coralyne. We then demonstrate that this new assay can be used to monitor the synthesis c-di-AMP by diadenyl cyclase (DAC) DisA7 as well as the degradation of c-di-AMP by the phosphodiestearses YybT17 and SVPD (snake venom phosphodiesterase). In addition to providing a practical detection of c-di-AMP, this work also suggests that coralyne associates with cyclic adenine nucleotides in a mode that is markedly different from the well-characterized complex formation between coralyne and linear polyadenine oligonucleotides.22 The interactions between aromatic heterocycles and DNA/RNA have been intensively studied for several decades due to the link between cancer and these planar intercalators.23 Many of these heterocyclic molecules are known to intercalate into Watson–Crick duplexes, whereas a few have also been reported to intercalate into non-Watson–Crick duplexes24−26 and higher order structures, such as triplexes27−32 and G-quadruplexes.33−38 In the past few years, efforts to design small molecules that could stabilize adenine-rich oligonucleotides have intensified due to potential biotechnological and medical applications of these molecules.39−47 For example, it has been demonstrated that adenine-rich oligonucleotides can form hydrogels in the presence of metals and that these hydrogels are responsive to pH changes.48 Polyadenylation is also known to play a role in the progression of cancer, and it has been known for more than a decade that tumor cells overexpress poly rA polymerases,42 suggesting that perhaps targeting poly rA could be a viable anticancer strategy.49−52 Poly A duplex formation in the presence of a π-system is well-known; however, formation of higher order structure with poly A in the presence of heterocyclic intercalators has not been well explored.53,54
Experimental Section
Sample Preparation for Spectrometric Measurements
c-di-AMP or pApA, water, buffer solution, and metal solution were mixed, heated, kept at 95 °C for 5 min, and cooled back to room temperature, and coralyne was added. The samples were then incubated at 4 °C for 12 h.
Optical Measurements
NMRs were measured on a Bruker AVANCE II 600 MHz spectrometer equipped with BBI probe at 20 °C.
UV absorbance spectra were obtained on a JASCO V-630 spectrophotometer with a 1 cm path length cuvette. The concentration of each stock solution of c-di-AMP and pApA was determined by measuring the absorbance at 259 nm for c-di-AMP and pApA, using 27,000 M–1cm–1 as a molar extinction coefficient for both compounds.
Fluorescence measurements were carried out on a Cary Eclipse fluorescence spectrophotometer at 10 °C, with λex = 420 nm (slit 5 nm) and λem = 430–700 nm (slit 5 nm). The concentration of c-di-AMP or pApA was 40 μM; coralyne was 10 μM, and buffer was 50 mM Tris-H3PO4 (pH 4.5, 7.5, or 9.2) containing 250 mM KBr.
Fluorescence lifetime was measured using a time-domain system integrated with a fluorescence lifetime imaging microscope (FLIM) system Alba V (ISS, Urbana, IL). The system is equipped with a SPC-830 TCSPC module and pulsed laser system (Becker and Hickl GmbH). Laser BHL-445 nm and observation through band-pass filter 485/30 nm was used. Data analysis was performed using Vista Vision software v. 218 from ISS.
Circular dichroism (CD) experiments were performed on a JASCO J-81 spectropolarimeter with 1 cm path length cuvette. The concentration of c-di-AMP or pApA was 40 μM; coralyne was 10 μM, and buffer was 50 mM Tris-H3PO4 (pH 7.5) containing 250 mM KBr.
Enzymatic Assay
DisA and YybT were expressed in BL21(DE3) and purified by Nickel-affinity chromatography column (GE Healthcare). DisA was dialyzed into a 10 mM Tris-HCl, pH 8.0, and 100 mM NaCl solution, and YybT was dialyzed into a 50 mM Tris-HCl, pH 8.0, and 150 mM NaCl solution. Phosphodiesterase I from Crotalus adamanteus venom (snake venom phosphodiesterase, SVPD) was purchased from Sigma-Aldrich. For the c-di-AMP synthesis assay, DisA (10 μM) was added to 100 μM ATP in 40 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 10 mM MgCl2 at 30 °C.7 For the c-di-AMP cleavage assay, YybT (10 μM) in 100 mM Tris-HCl, pH 8.3, 20 mM KCl, 500 μM MnCl2, and 1 mM DTT17 or SVPD (1 mg/mL) in 50 mM Tris-HCl, pH 8.8, and 15 mM MgCl2 was used to cleave c-di-AMP (100 μM) at 37 °C. Reactions were stopped by heating up to 95 °C for 5 min, and the precipitated proteins were removed by centrifugation. KBr and coralyne were added to the sample to give final concentrations of 250 mM and 10 μM for KBr and coralyne, respectively. The sample was incubated at 4 °C for 12 h.
Results and Discussion
Isoquinoline alkaloids, such as coralyne (Figure 1), are known to bind to adenine-rich oligonucleotides and stabilize adenine–adenine duplexes.24,28,29,55−58 In an important paper by Hud and co-workers, it was demonstrated that oligonucleotides containing long tracts of adenine, such as (dA)16 but not (dA)4, could bind to coralyne and form fibers.55 Subsequent works by others have revealed that the fluorescence of coralyne is quenched when bound to poly dA59 or rA.24 In line with Hud’s observation that short polyadenines do not bind coralyne,55 when coralyne was incubated with pApA (the degradation product of c-di-AMP by phosphodiesterases) or cAMP in a buffer containing KCl, there was no significant change in both the UV and fluorescence profiles of coralyne (see Figure 2a,b). On the other hand, when c-di-AMP (which contains the same number of adenine as pApA) was incubated with coralyne in the presence of KCl, both the UV and fluorescence intensities of coralyne increased (see Figure 2a,b). It is known that, when coralyne binds to polyadenine, its fluorescence and UV absorbance is decreased,40 but here c-di-AMP enhanced the fluorescence and absorbance of coralyne; this suggests that the binding mode between c-di-AMP and coralyne might be different from the proposed duplex intercalation model.40
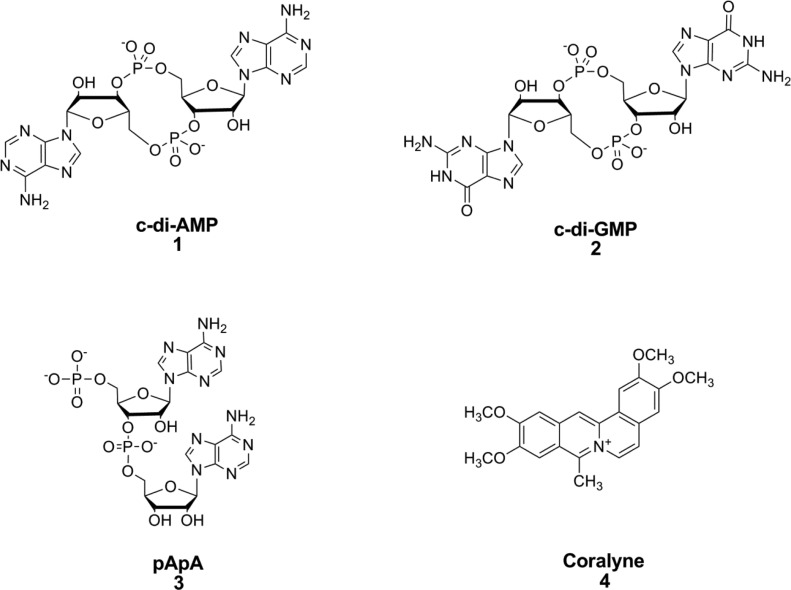
Figure 1.
Structures of c-di-AMP, c-di-GMP, pApA, and coralyne.
Figure 2.
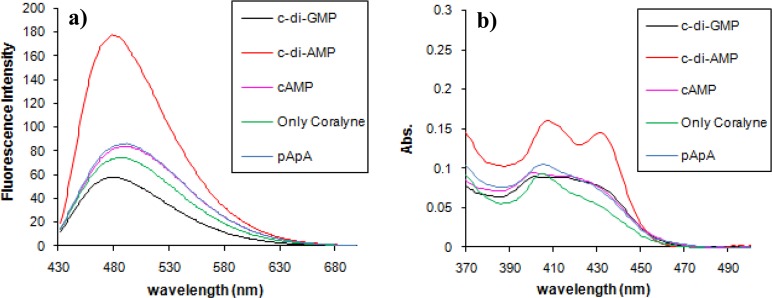
Fluorescence (a) and UV (b) profiles of coralyne in the presence of various nucleotides. Condition: [coralyne] = 10 μM, [nucleotides] = 40 μM; buffer: 50 mM Tris-H3PO4 (pH 7.5) containing 250 mM KCl. Temperature = 10 °C. ex. 420 nm, em. 475 nm.
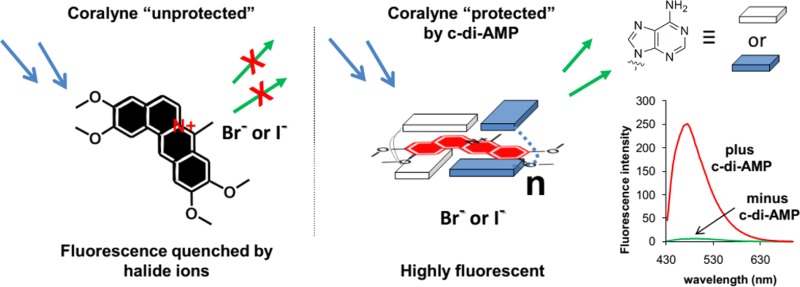
Although the fluorescence of coralyne was enhanced by c-di-AMP (∼3-fold fluorescence increase), in the presence of 250 mM KCl, the high fluorescence of free coralyne impeded our initial efforts to use coralyne to detect c-di-AMP concentrations lower than 20 μM (see Supporting Information, Figure S1). We therefore sought ways to reduce the fluorescence of the unbound coralyne. We hypothesized that the fluorescence enhancement of coralyne, in the presence of c-di-AMP, was the result of coralyne intercalating between the two adenine bases of c-di-AMP. If this was the case, then it was expected that the bound coralyne would be protected from fluorescence quenchers whereas the unbound coralyne could be readily quenched by anions. Therefore, it would be possible to increase the signal-to-noise ratio of c-di-AMP detection using an anion-quenching phenomenon.
The quenching of coralyne by KBr in the absence of c-di-AMP is dominantly static with Ks of 101.2 M–1 (from the intercept of the modified SV plot, 110.19–9.03 M–1 ≈ 101.2 M–1) compared with the dynamic constant of 9.03 M–1 (lifetime data, 0.00903 mM–1 = 9.03 M–1); see Table 1 and Figure 3. For the Stern–Volmer plot, we used lifetime data determined from a single exponential fit. This is because the long lifetime component is not well-defined and fitting the data required fixing a value of 30 ns. After complexing with c-di-AMP, the quenching is significantly reduced, from intensity measurements that include static and dynamic quenching Ksv = 0.918 M–1 (0.000918 mM–1 = 0.918 M–1) and from lifetime data by 0.521 M –1 (0.000512 mM–1 = 0.512 M–1); see Table 1 and Figure 4. For construction of the Stern–Volmer plot, we used average lifetime from two-component fit. There is a slightly better fit with three-exponential, which reveals a long lifetime component of about 30 ns which is in agreement with previous reports on coralyne dimers.60
Table 1. Fluorescence Quenching of Coralyne with KBra.
| in absence
of c-di-AMP |
in presence of 40 μM of c-di-AMP |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| quencher KBr [mM] | τI (νσ) | fi | ταω̅γ (νσ) | χ2 | το/τb (I0/I) | I | τI (νσ) | fi | ταω̅γ (νσ) | χ2 | το/τb (I0/I) | I |
| 0 | 14.22 | 1 | 14.2 | 1.85 | 1 | 242 | n/a | |||||
| 9.82 | 0.317 | (1) | n/a | n/a | n/a | n/a | n/a | |||||
| <30> | 0.683 | 24.97 | 1.16 | |||||||||
| 10 | 11.61 | 1 | 11.61 | 1.42 | 1.22 | 96 | 12.13 | 1 | 12.13 | 30 | 428 | |
| 9.83 | 0.592 | (2.38) | 2.33 | 0.137 | ||||||||
| <30> | 0.408 | 18.06 | 1.21 | 19 | 0.863 | 16.73 | 1.86 | 1 | ||||
| 1.29 | 0.072 | |||||||||||
| 5.92 | 0.131 | |||||||||||
| 28.73 | 0.797 | 23.76 | 1.04 | |||||||||
| 25 | 9.86 | 1 | 9.86 | 1.27 | 1.44 | 48 | 12.39 | 1 | 12.04 | 31 | 411 | |
| 8.86 | 0.731 | (4.75) | 2.5 | 0.136 | ||||||||
| <30> | 0.269 | 14.55 | 1.10 | 19.43 | 0.864 | 17.11 | 1.4 | 0.98 | ||||
| 1.46 | 0.075 | (1.04) | ||||||||||
| 6.79 | 0.144 | |||||||||||
| 30.52 | 0.781 | 24.52 | 1.21 | |||||||||
| 50 | 8.52 | 1 | 8.52 | 1.10 | 1.67 | 14.3 | 12.04 | 1 | 12.04 | 25 | 420 | |
| 3.50 | 0.063 | (15.94) | 2.31 | 0.139 | ||||||||
| 8.99 | 0.937 | 8.64 | 1.04 | 18.58 | 0.861 | 16.32 | 1.54 | 1.02 | ||||
| 1.48 | 0.073 | (1.02) | ||||||||||
| 7.04 | 0.148 | |||||||||||
| 31,3 | 0.779 | 25.52 | 1.21 | |||||||||
| 75 | 7.11 | 1 | 7.11 | 1.37 | 2.0 | 12.3 | 11.9 | 1 | 11.9 | 26 | 386 | |
| 4.11 | 0.228 | (18.54) | 2.49 | 0.141 | ||||||||
| 8.22 | 0.772 | 7.29 | 1.12 | 18.63 | 0.859 | 16.35 | 1.6 | 1.02 | ||||
| 1.4 | 0.064 | (1.11) | ||||||||||
| 6.82 | 0.154 | |||||||||||
| 33.67 | 0.782 | 27.46 | 1.14 | |||||||||
| 100 | 6.67 | 1 | 6.67 | 1.35 | 2.13 | 6.6 | 11.89 | 1 | 11.89 | 27 | 387 | |
| 2.18 | 0.131 | (34.55) | 2.5 | 0.142 | ||||||||
| 7.19 | 0.869 | 6.53 | 1.03 | 18.7 | 0.858 | 16.4 | 1.51 | 1.02 | ||||
| 1.37 | 0.067 | (1.11) | ||||||||||
| 6.33 | 0.149 | |||||||||||
| 30.39 | 0.784 | 24.86 | 1.06 | |||||||||
| 250 | 4.38 | 1 | 4.38 | 3.77 | 3.25 | 3.9 | 11.36 | 1 | 1 | 28 | 317 | |
| 6.19 | 0.506 | (58.46) | 2.37 | 0.152 | ||||||||
| 2.02 | 0.494 | 4.13 | 1.03 | 16.96 | 0.848 | 14.75 | 1.69 | 1.13 | ||||
| 1.07 | 0.088 | (1.35) | ||||||||||
| 5.14 | 0.153 | |||||||||||
| 23.05 | 0.759 | 18.37 | 1.09 | |||||||||
Ex. 445 nm, Em 480/30 nm. Condition: [coralyne] = 10 μM, [c-di-AMP] = 40 μM; buffer: 50 mM Tris-H3PO4 (pH 7.5) at 25 °C; average lifetime τavg = Σ fiτi.
For relative lifetime, single lifetime fit was used for KBr and average lifetimes from double-exponential fit in the presence of c-di-AMP.
Figure 3.
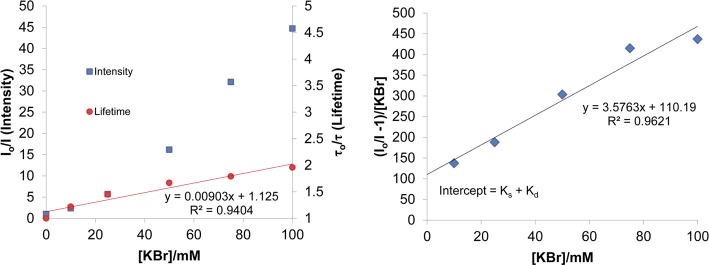
(Left) KBr quenching of Coralyne in 50 mM Tris-H3PO4 buffer (pH 7.5) at 25 °C. The squares show the values of relative intensity and circles of lifetimes. (Right) Modified Stern–Volmer plot for quenching of Coralyne with KBr.
Figure 4.
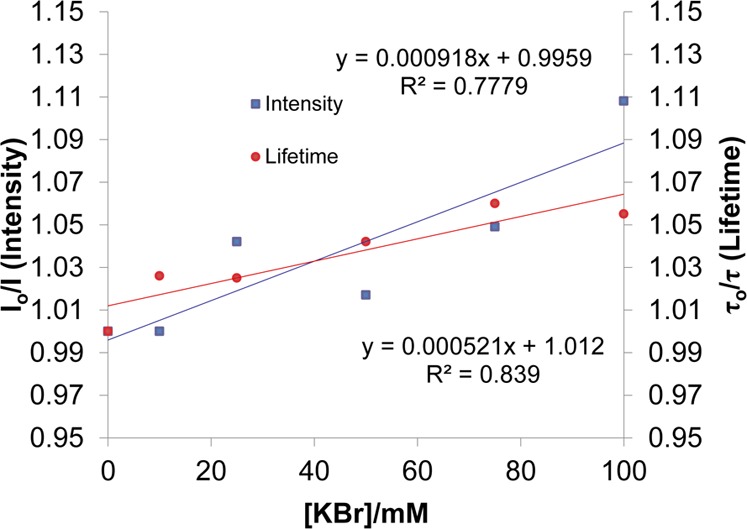
KBr quenching of Coralyne in the presence of 40 μM of c-di-AMP at 25 °C. Buffer: 50 mM Tris-H3PO4 (pH 7.5). Squares are intensity, and circles are lifetime data.
The quenching of coralyne by KI in the absence of c-di-AMP is also dominantly static with Ks of 35,860 M–1 (intensity data, 35.86 mM–1 = 35,860 M–1) compared with dynamic constant of 912.0 M–1 (lifetime data, 0.912 mM–1 = 912.0 M–1); see Supporting Information, Figure S3. After complexing with c-di-AMP, the quenching by KI is significantly reduced (see Supporting Information, Figure S3); from intensity measurements that include static and dynamic quenching, Ksvc = 25.6 M–1 (0.0256 mM–1 = 25.6 M–1) and from lifetime data, Ksv =6.4 M–1 (0.0064 mM–1 = 6.4 M–1). Overall, KI is a significantly stronger quencher than KBr for coralyne, and similar quenching effect can be obtained with ∼100 times lower concentration compared to KBr. However, KI also partially quenches coralyne even when c-di-AMP is present, so for our c-di-AMP detection, we decided to use KBr as the quencher. Lifetime measurements (Supporting Information, Tables S1 and S2) show very similar components as those observed with KBr quenching.
At higher temperature (60 °C), c-di-AMP did not “protect” coralyne from fuorescence quenching by bromide and the value of I0/I was ∼1. (see Supporting Information, Figures S4 and S5) This observation suggests that coralyne forms a supramolecular inclusion complex with c-di-AMP, but at higher temperatures, the complex is not stable and hence coralyne would no longer be protected from halide quenching at high temperatures.
c-di-AMP could be detected with coralyne at both acidic and basic pH (see Supporting Information, Figure S6), although the fold fluorescence increase at basic pH 9.2 (22.4) was slightly better than at pH 7.5 or 4.5 (13 and 14.5, respectively). Having established that the inclusion of a bromide quencher and conducting the c-di-AMP detection assay at pH 9.2 was optimal for sensitive detection, we proceeded to investigate if c-di-AMP concentrations lower than 40 μM could be detected with our new system. Pleasing lower concentrations of c-di-AMP (down to 5 μM) could be detected using our system (see Figure 5a). This simple fluorescent detection system could therefore be suitable for determining the enzymatic proficiencies of c-di-AMP synthases or phosphodiesrases, vide infra.
Figure 5.
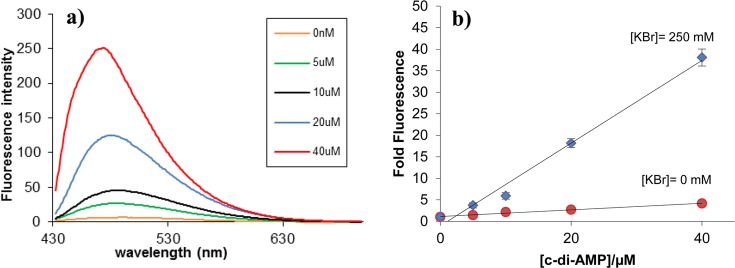
The fluorescence of coralyne is proportional to the concentration of c-di-AMP. Condition: [Coralyne] = 10 μM, [c-di-AMP] = 0, 5, 10, 20, and 40 μM; buffer: 50 mM Tris-H3PO4 (pH 9.2) containing 250 mM KBr, showing c-di-AMP concentration dependence on fluorescence enhancement. Temperature = 10 °C. ex. 420 nm, em. 475 nm.
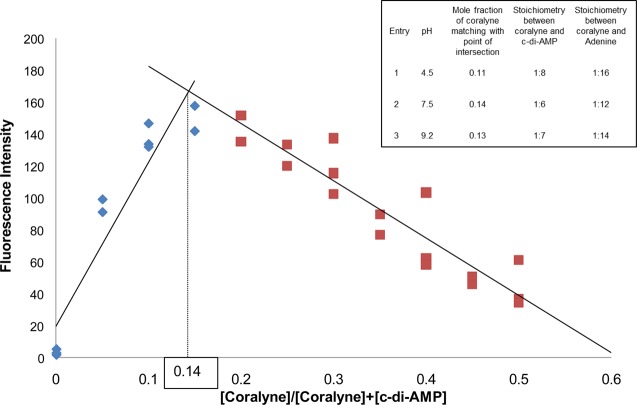
Circular dichroism (CD) is an excellent tool to study structural perturbations that result from the association of a chiral molecule with another molecule. The CD spectrum of c-di-AMP in the absence of coralyne is different from that in the presence of coralyne (Figure 6). Upon the addition of coralyne to c-di-AMP, both the negative and positive CD bands of c-di-AMP increase, indicating that both the helicity and π–π stacking interactions in c-di-AMP changes upon the addition of coralyne. An increase in the CD band is indicative of increased π–π stacking (presumably due to coralyne-adenine π–π stacking). For pApA however, there is no difference between the presence and absence of coralyne (see Figure 6 and Supporting Information, Figure S7). This CD data augments the UV and fluorescence data, which suggested that coralyne associates with c-di-AMP but not with pApA (refer to Figure 2a,b). Whereas the inability of pApA to form a complex with coralyne, at the tested micromolar concentrations, was in line with earlier studies by Hud et al.,55 the complex formation between the cyclic dinucleotide, c-di-AMP, and coralyne was unexpected. To gain some insights into the stiochiometry of the c-di-AMP/coralyne complex, we performed a Job plot analysis (Figure 7). This analysis revealed that c-di-AMP forms a higher order complex with coralyne (not the expected 2:1 complex that would have been predicted from the polyadenine-coralyne model).
Figure 6.
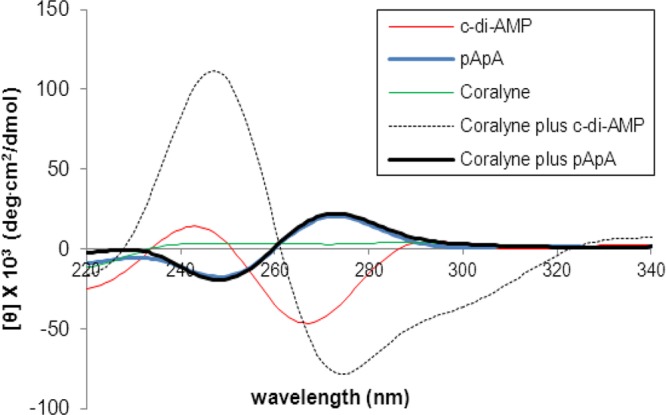
CD of coralyne-c-di-AMP or pApA complex. Condition: [coralyne] = 10 μM, [c-di-AMP] = 40 μM; buffer: 50 mM Tris-H3PO4 (pH 7.5) containing 250 mM KBr. Coralyne plus c-di-AMP or pApA indicates coralyne incubated with c-di-AMP or pApA, with incubation conditions listed in the Experimental Section.
Figure 7.
Job plot of coralyne and c-di-AMP interaction. [Coralyne] + [c-di-AMP] was fixed at 50 μM. The experiment was done in triplicate and plotted together on the graphs. Buffer: 50 mM Tris-H3PO4 (pH 7.5) containing 250 mM KBr. Job plots of pH 4.2 and 9.2 are in Supporting Information (see Figure S6). Temperature = 10 °C. ex. 420 nm, em. 475 nm.
To corroborate the Job plot data, which indicated that c-di-AMP forms higher order supramolecular aggregate in the presence of coralyne, we proceeded to conduct the NMR titration experiment. The addition of only 0.1 equivalence of coralyne to c-di-AMP, in the presence of potassium cations (100 mM), resulted in the complete disappearance of the c-di-AMP 1H NMR peaks around 8.24, 7.98, and 6.01 ppm (see Figure 8). The complete disappearance of the proton NMR peaks is either indicative of polymer formation or intermediates in fast exchange. The complete disappearance of the c-di-AMP peaks in the proton NMR (Figure 8) when only 0.1 equivalence of coralyne was added cannot be explained by a 2:1 complex between c-di-AMP and coralyne; a 2:1 complex between c-di-AMP and coralyne would have expected molecular weight of 1676.34, which should be visible by NMR. Second, the Job plot revealed a c-di-AMP/coralyne stiochiometry that is or greater than 6:1. Adenine is known to form hydrogen bonds with itself or other nucleobases to form various higher order structures, including quartets,61 pentads,62 hexads,63,64 and heptad.65 Adenine tetrads, which usually contain four hydrogen bonds per tetrad, are not as stable as G-tetrads (which contain eight hydrogen bonds per tetrad). Could coralyne promote A-tetrad formation by c-di-AMP and are these A-tetrads stacking on top of each other to form polymers? It has been shown that the formation adenine tetrads could be facilitated if the A-tetrad could π-stack with a proximal G-quadruplex.66 Analogously, it is plausible that aromatic ligands could be used to promote the formation of adenine higher order structures.
Figure 8.
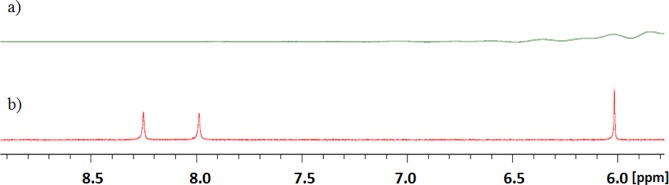
1H NMR spectra of c-di-AMP with (a) or without (b) coralyne. Addition of coralyne (0.1 equiv.) to c-di-AMP caused complete disappearance of the c-di-AMP peaks in the 1H NMR spectrum (spectrum a). Condition: [coralyne] = 0 or 40 μM, [c-di-AMP] = 400 μM in D2O containing 100 mM KBr. Temperature = 20 °C.
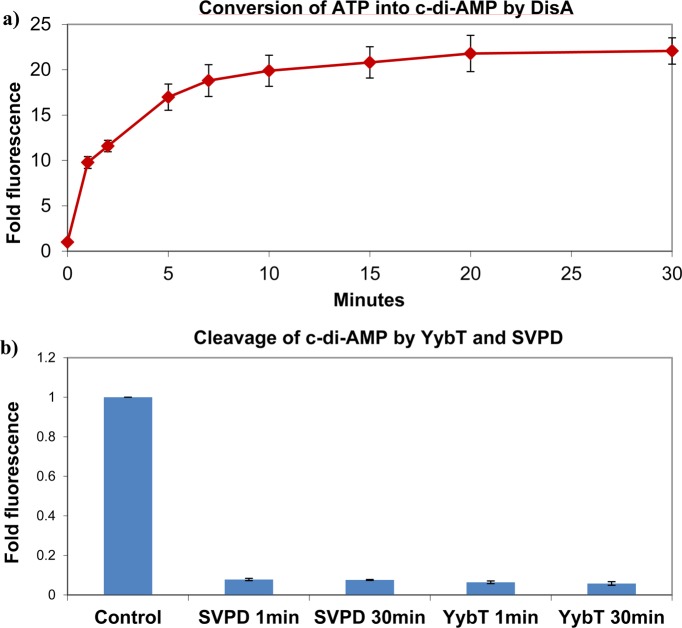
Because c-di-AMP plays an important role in bacterial cell wall formation and susceptibility to antibiotics, which target cell wall formation, it is of interest to identify c-di-AMP synthases and phosphodiesterases in pathogenic bacteria, with the ultimate goal that the inhibition of the synthases or phosphodiesterases could potentiate the activities of antibiotics, such as the β-lactams. We therefore investigated if coralyne could be used to assay the activities of c-di-AMP metabolism enzymes. It has been demonstrated that the PDE enzyme, YybT, is a c-di-AMP phosphodiesterase17 whereas DisA is a c-di-AMP synthase.7 We incubated YybT with c-di-AMP (40 μM) and stopped the reaction at 1 and 30 min. We then used our newly developed c-di-AMP detection assay to investigate the c-di-AMP cleavage reaction, Figure 8a. As a control, c-di-AMP (40 μM) was also treated with snake venom phosphodiesterase and the cleavage reaction analyzed with our coralyne assay. Pleasingly, our assay revealed that, under the reaction conditions, YybT (10 μM) cleaved the majority of the c-di-AMP (40 μM) within 1 min; see Figure 9. Coralyne can also be used to monitor the synthesis of c-di-AMP from ATP by synthases, such as DisA; see Figure 9b.
Figure 9.
(a) Conversion of ATP into c-di-AMP by DisA. DisA (10 μM) was added to ATP (100 μM) in 200 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 10 mM MgCl2 at 30 °C. Reactions were stopped at 1, 2, 5, 7, 10, 15, 20, and 30 min and incubated with conditions stated in the experimental part. The fluorescence was subsequently measured. (b) Cleavage of c-di-AMP by YybT and SVPD. YybT (10 μM) in 100 mM Tris-HCl, pH 8.3, 20 mM KCl, 500 μM MnCl2, and 1 mM DTT or SVPD (1 mg/mL) in 50 mM Tris-HCl, pH 8.8, and 15 mM MgCl2 were used to cleave c-di-AMP (100 μM) at 37 °C. Reactions were stopped at 1 and 30 min and incubated with conditions stated in the Experimental Section. Fluorescence measurements were taken after 1 and 30 min.
Conclusion
In our continuing efforts to investigate the interactions of heterocycles with bacterial dinucleotide second messengers, we uncovered an unexpected interaction of coralyne with c-di-AMP but not the linear analog pApA, which also contains two rA. Although we have been unable to define the exact nature of the complex between coralyne and c-di-AMP, due to possible polymer formation, the optical properties of this supramolecular complex has facilitated the detection of c-di-AMP, which could be useful for c-di-AMP research. Nucleotide signaling has emerged as important in bacteria and regulates diverse bacterial phenotypes. Currently, there are efforts to identify and characterize both synthase and phosphodiesterases of c-di-AMP, and it is believed that c-di-AMP metabolism enzymes could become new drugable targets that could potentiate the effects of cell wall modifying antibiotics. Herein, we demonstrate an alternative nonradioactive assay to study the enzymatic proficiencies of c-di-AMP metabolism proteins (DAC and PDE). This simple fluorescent assay could also be adapted for a high-throughput screen for c-di-AMP phosphodiesterase inhibitors, which are expected to potentiate the killing effects of peptidoglycan inhibition drugs. This work also demonstrates that, although at low micromolar concentrations, small nucleotides are not known to readily associate with heterocyclic intercalators (unlike polynucleotides), the circularization of dinucleotides appear to enhance aggregate formation. Plausibly, the high entropic cost, associated with bringing many small nucleotides together for complex formation, is reduced upon circularization because of reduction in degree of freedom. Circularization of oligonucleotides could be an under-utilized strategy, which might improve oligonucleotide association kinetics or stability.
Acknowledgments
NSF (CHE 1307218), NIH S10, Camille Dreyfus foundation, American Heart Association (11PRE7890040 fellowship to J.Z.), and University of Maryland Graduate Dean’s Dissertation Fellowship (J.Z.) supported this work. We thank Prof. Liang Zhao-Xun for a plasmid encoding YybT and Prof. Karl-Peter Hopfner and Dr. Angelika Grundling for plasmids encoding DisA.
Supporting Information Available
Stern–Volmer titration with different salts, Job plot at different pH values, and CD.This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Kalia D.; Merey G.; Nakayama S.; Zheng Y.; Zhou J.; Luo Y.; Guo M.; Roembke B. T.; Sintim H. O. Chem. Soc. Rev. 2013, 42, 305. [DOI] [PubMed] [Google Scholar]
- Gomelsky M. Mol. Microbiol. 2011, 79, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U.; Gomelsky M.; Galperin M. Y. Mol. Microbiol. 2005, 57, 629. [DOI] [PubMed] [Google Scholar]
- Sondermann H.; Shikuma N. J.; Yildiz F. H. Curr. Opin. Microbiol. 2012, 15, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R. P.; Fouhy Y.; Lucey J. F.; Dow J. M. J. Bacteriol. 2006, 188, 8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer T.; Jenal U. Nat. Rev. Microbiol. 2009, 7, 724. [DOI] [PubMed] [Google Scholar]
- Witte G.; Hartung S.; Büttner K.; Hopfner K. P. Mol. Cell 2008, 30, 167. [DOI] [PubMed] [Google Scholar]
- Romling U. Sci. Signaling 2008, 1, pe39. [DOI] [PubMed] [Google Scholar]
- Bejerano-Sagie M.; Oppenheimer-Shaanan Y.; Berlatzky I.; Rouvinski A.; Meyerovich M.; Ben-Yehuda S. Cell 2006, 125, 679. [DOI] [PubMed] [Google Scholar]
- Corrigan R. M.; Gründling A. Nat. Rev. Microbiol. 2013, 11, 513. [DOI] [PubMed] [Google Scholar]
- Luo Y.; Helmann J. D. Mol. Microbiol. 2012, 83, 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi C.; Waters E. M.; Rudkin J. K.; Schaeffer C. R.; Lohan A. J.; Tong P.; Loftus B. J.; Pier G. B.; Fey P. D.; Massey R. C.; O’Gara J. P. PLoS Pathog. 2012, 8, e1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J. M.; O’Neill A. J. Antimicrob. Agents Chemother. 2012, 56, 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R.; Gretes M.; Harlem C.; Basuino L.; Chambers H. F. Antimicrob. Agents Chemother. 2010, 54, 4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte C. E.; Whiteley A. T.; Burke T. P.; Sauer J. D.; Portnoy D. A.; Woodward J. J. MBio 2013, 4, e00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan R. M.; Campeotto I.; Jeganathan T.; Roelofs K. G.; Lee V. T.; Gründling A. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 9084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F.; See R. Y.; Zhang D.; Toh D. C.; Ji Q.; Liang Z. X. J. Biol. Chem. 2010, 285, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W. M.; Pham T. H.; Lei L.; Dou J.; Soomro A. H.; Beatson S. A.; Dykes G. A.; Turner M. S. Appl. Environ. Microbiol. 2012, 78, 7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward J. J.; Iavarone A. T.; Portnoy D. A. Science 2010, 328, 1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J. R.; Koestler B. J.; Carpenter V. K.; Burdette D. L.; Waters C. M.; Vance R. E.; Valdivia R. H. MBio 2013, 4, e00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L.; Hill K. K.; Filak H.; Mogan J.; Knowles H.; Zhang B.; Perraud A. L.; Cambier J. C.; Lenz L. L. J. Immunol. 2011, 187, 2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung I. S.; Çetinkol Ö. P.; Hud N. V.; Cheatham T. E. III Nucleic Acids Res. 2009, 37, 7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G.; Ren J. Chem. Commun. (Cambridge, U. K.) 2010, 46, 7283. [DOI] [PubMed] [Google Scholar]
- Islam M. M.; Chowdhury S. R.; Kumar G. S. J. Phys. Chem. B 2009, 113, 1210. [DOI] [PubMed] [Google Scholar]
- Wang A. H.; Ughetto G.; Quigley G. J.; Rich A. Biochemistry 1987, 26, 1152. [DOI] [PubMed] [Google Scholar]
- Kopka M. L.; Yoon C.; Goodsell D.; Pjura P.; Dickerson R. E. Proc. Natl. Acad. Sci. U. S. A. 1985, 82, 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraru-Allen A. A.; Cassidy S.; Alvarez J. L. A.; Fox K. R.; Brown T.; Lane A. N. Nucleic Acids Res. 1997, 25, 1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak M.; Hud N. V. Nucleic Acids Res. 2002, 30, 983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S. S.; Polak M.; Hud N. V. Nucleic Acids Res. 2003, 31, 4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biver T.; Boggioni A.; García B.; Leal J. M.; Ruiz R.; Secco F.; Venturini M. Nucleic Acids Res. 2010, 38, 1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström K.; Wärmländer S.; Bergman J.; Engqvist R.; Leijon M.; Gräslund A. J. Mol. Recognit. 2004, 17, 277. [DOI] [PubMed] [Google Scholar]
- Lee J. S.; Latimer L. J.; Hampel K. J. Biochemistry 1993, 32, 5591. [DOI] [PubMed] [Google Scholar]
- Bhadra K.; Kumar G. S. Biochim. Biophys. Acta 2011, 1810, 485. [DOI] [PubMed] [Google Scholar]
- Bertrand H.; Granzhan A.; Monchaud D.; Saettel N.; Guillot R.; Clifford S.; Guédin A.; Mergny J. L.; Teulade-Fichou M. P. Chemistry 2011, 17, 4529. [DOI] [PubMed] [Google Scholar]
- Monchaud D.; Granzhan A.; Saettel N.; Guédin A.; Mergny J. L.; Teulade-Fichou M. P. J. Nucleic Acids 2010, 2010, 525862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monchaud D.; Teulade-Fichou M. P. Org. Biomol. Chem. 2008, 6, 627. [DOI] [PubMed] [Google Scholar]
- Alzeer J.; Vummidi B. R.; Roth P. J.; Luedtke N. W. Angew. Chem., Int. Ed. Engl. 2009, 48, 9362. [DOI] [PubMed] [Google Scholar]
- Luedtke N. Chimia 2009, 63, 134. [Google Scholar]
- Xing F.; Song G.; Ren J.; Chaires J. B.; Qu X. FEBS Lett. 2005, 579, 5035. [DOI] [PubMed] [Google Scholar]
- Cetinkol O. P.; Hud N. V. Nucleic Acids Res. 2009, 37, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G. J. Biosci. 2012, 37, 539. [DOI] [PubMed] [Google Scholar]
- Topalian S. L.; Kaneko S.; Gonzales M. I.; Bond G. L.; Ward Y.; Manley J. L. Mol. Cell. Biol. 2001, 21, 5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian S. L.; Gonzales M. I.; Ward Y.; Wang X.; Wang R. F. Cancer Res. 2002, 62, 5505. [PubMed] [Google Scholar]
- Chaires J.; Waring M. Top. Curr. Chem. DNA Binders 2005, 253, 33. [Google Scholar]
- Wilson W. D.; Gough A. N.; Doyle J. J.; Davidson M. W. J. Med. Chem. 1976, 19, 1261. [DOI] [PubMed] [Google Scholar]
- Dower K.; Kuperwasser N.; Merrikh H.; Rosbash M. RNA 2004, 10, 1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M.; Anderson P.; Jackson R. J. Curr. Opin. Genet. Dev. 1997, 7, 220. [DOI] [PubMed] [Google Scholar]
- Sukul P.; Malik S. Soft Matter 2011, 7, 4234. [Google Scholar]
- Martins S. B.; Rino J.; Carvalho T.; Carvalho C.; Yoshida M.; Klose J. M.; de Almeida S. F.; Carmo-Fonseca M. Nat. Struct. Mol. Biol. 2011, 18, 1115. [DOI] [PubMed] [Google Scholar]
- Rask-Andersen M.; Almén M. S.; Schiöth H. B. Nat. Rev. Drug Discovery 2011, 10, 579. [DOI] [PubMed] [Google Scholar]
- Das A.; Suresh Kumar G. Mol. Biosyst. 2012, 8, 1958. [DOI] [PubMed] [Google Scholar]
- Balatsos N. A.; Havredaki M.; Tsiapalis C. M. Int. J. Biol. Markers 2000, 15, 171. [DOI] [PubMed] [Google Scholar]
- Xu X.; Wang J.; Yang F.; Jiao K.; Yang X. Small 2009, 5, 2669. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Wang J.; Yang F.; Yang X. Anal. Chem. 2012, 84, 924. [DOI] [PubMed] [Google Scholar]
- Persil O.; Santai C. T.; Jain S. S.; Hud N. V. J. Am. Chem. Soc. 2004, 126, 8644. [DOI] [PubMed] [Google Scholar]
- Buckley R.; Enekwa C. D.; Williams L. D.; Hud N. V. ChemBioChem 2011, 12, 2155. [DOI] [PubMed] [Google Scholar]
- Giri P.; Kumar G. S. Mol. Biosyst. 2008, 4, 341. [DOI] [PubMed] [Google Scholar]
- Islam M.; Kumar G. S. Biochim. Biophys. Acta 2009, 1790, 829. [DOI] [PubMed] [Google Scholar]
- Sinha R.; Saha I.; Kumar G. Chem. Biodiversity 2011, 8, 1512. [Google Scholar]
- Megyesi M.; Biczók L.; Görner H. Photochem. Photobiol. Sci. 2009, 8, 556. [DOI] [PubMed] [Google Scholar]
- Patel P. K.; Koti A. S.; Hosur R. V. Nucleic Acids Res. 1999, 27, 3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N.; Gorin A.; Majumdar A.; Kettani A.; Chernichenko N.; Skripkin E.; Patel D. J. J. Mol. Biol. 2001, 311, 1063. [DOI] [PubMed] [Google Scholar]
- Liu H.; Matsugami A.; Katahira M.; Uesugi S. J. Mol. Biol. 2002, 322, 955. [DOI] [PubMed] [Google Scholar]
- Kettani A.; Gorin A.; Majumdar A.; Hermann T.; Skripkin E.; Zhao H.; Jones R.; Patel D. J. J. Mol. Biol. 2000, 297, 627. [DOI] [PubMed] [Google Scholar]
- Matsugami A.; Ouhashi K.; Kanagawa M.; Liu H.; Kanagawa S.; Uesugi S.; Katahira M. J. Mol. Biol. 2001, 313, 255. [DOI] [PubMed] [Google Scholar]
- Pan B.; Xiong Y.; Shi K.; Deng J.; Sundaralingam M. Structure 2003, 11, 815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.