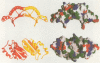Abstract
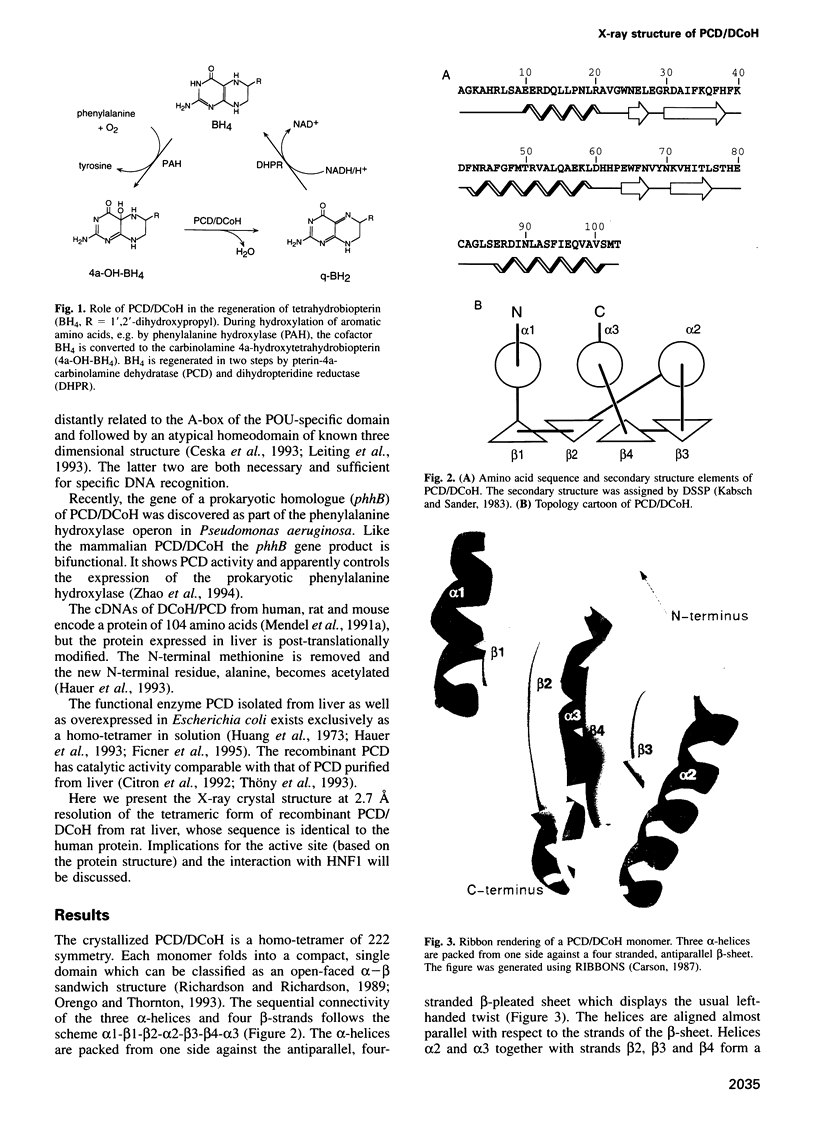
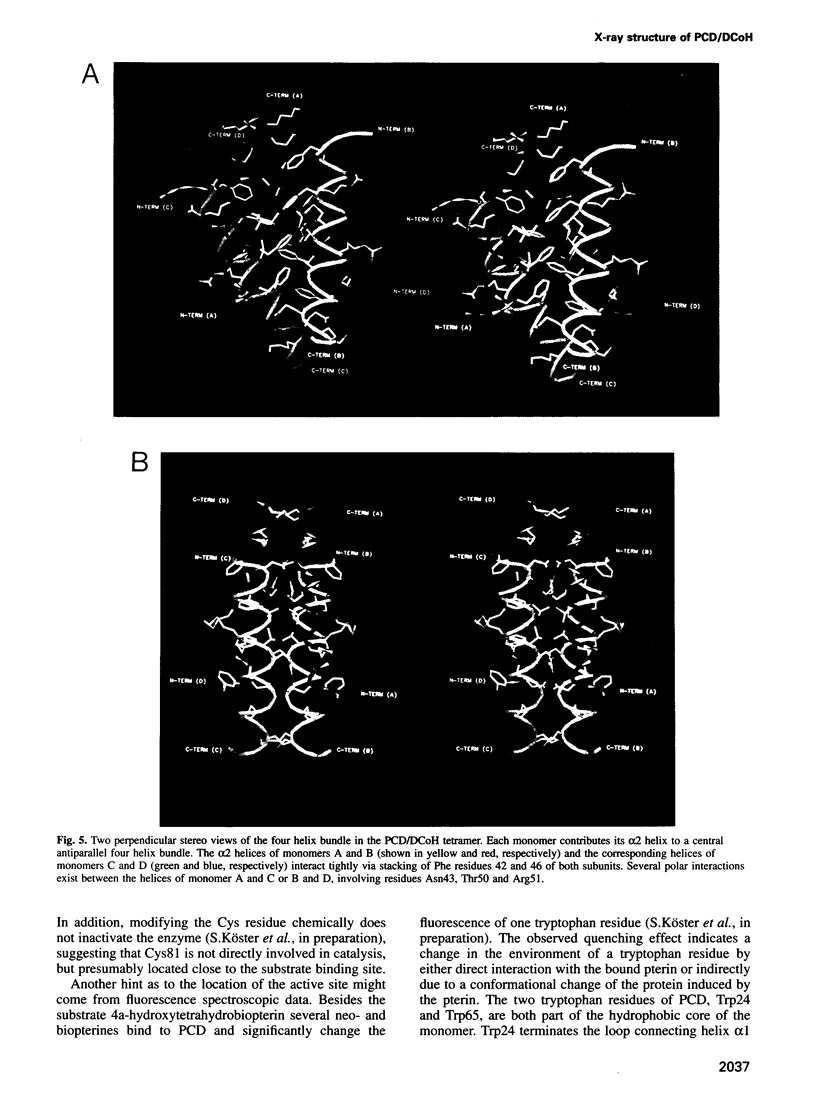
The bifunctional protein pterin-4a-carbinolamine dehydratase (PCD)/dimerization cofactor of HNF1 (DCoH) is a cytoplasmic enzyme involved in the tetrahydrobiopterin regeneration and is found in complex with the transcription factor HNF1 in liver cell nuclei. An atypical hyperphenylalaninemia and the depigmentation disorder vitiligo are related to a deficiency of PCD/DCoH activity. The crystal structure of PCD/DCoH was solved by multiple isomorphous replacement and refined to a crystallographic R-factor of 20.5% at 2.7 A resolution. The single domain monomer comprises three alpha-helices packed against one side of a four-stranded, antiparallel beta-sheet. The functional enzyme is a homo-tetramer of 222 symmetry where each of the monomers contributes one helix to a central four helix bundle. In the tetramer two monomers form an eight-stranded, antiparallel beta-sheet with six helices packing against it from one side. The concave, hydrophobic surface of the eight-stranded beta-sheet with its two protruding loops at either end is reminiscent of the saddle-like shape seen in the TATA-box binding protein. PCD/DCoH binds as a dimer to the helical dimerization domain of dimeric HNF1 forming a hetero-tetramer possibly through a mixed four helix bundle.
Full text
PDF

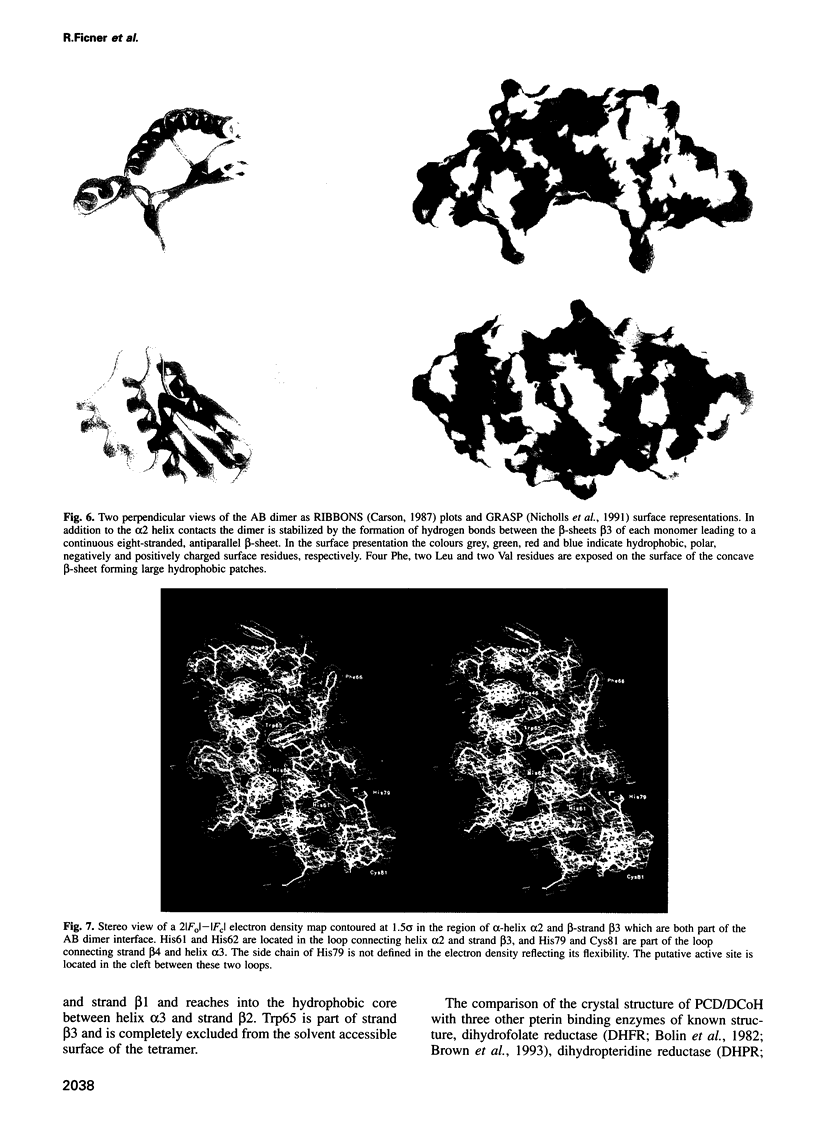
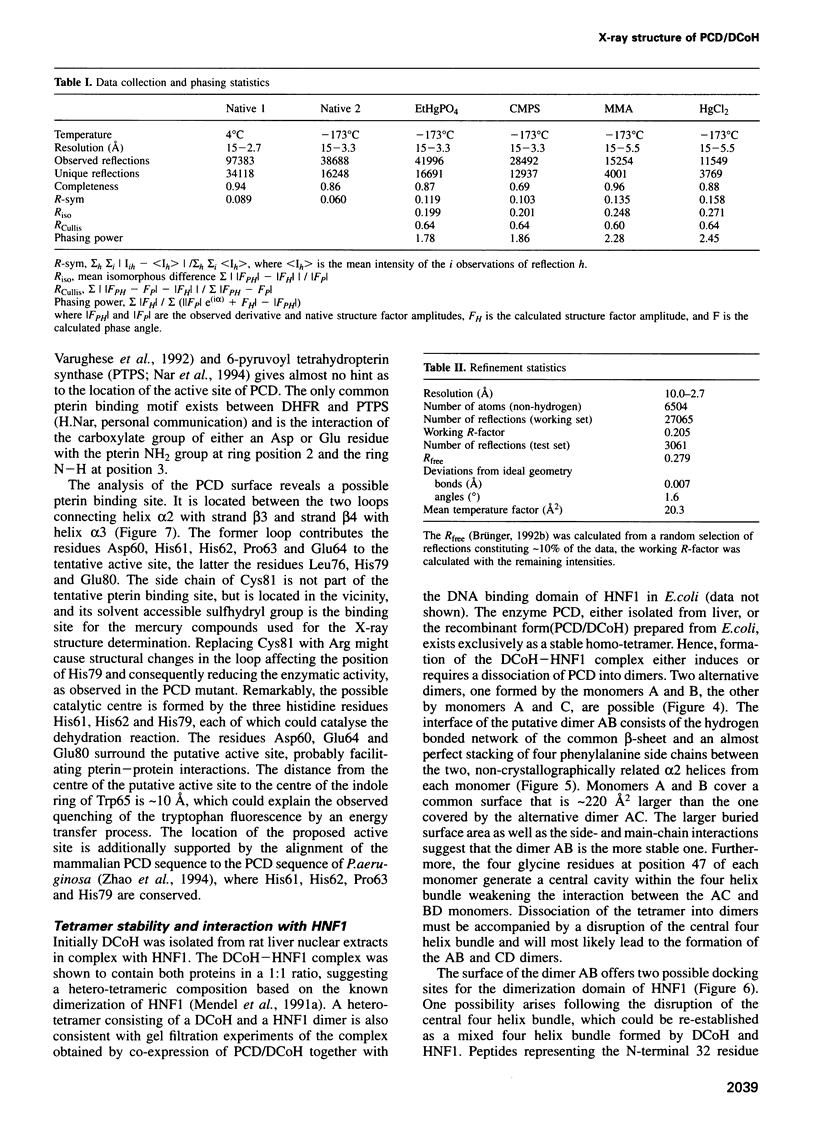



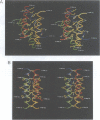
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler C., Ghisla S., Rebrin I., Haavik J., Heizmann C. W., Blau N., Kuster T., Curtius H. C. 7-substituted pterins in humans with suspected pterin-4a-carbinolamine dehydratase deficiency. Mechanism of formation via non-enzymatic transformation from 6-substituted pterins. Eur J Biochem. 1992 Aug 15;208(1):139–144. doi: 10.1111/j.1432-1033.1992.tb17167.x. [DOI] [PubMed] [Google Scholar]
- Bailey S. W., Boerth S. R., Dillard S. B., Ayling J. E. The mechanism of cofactor regeneration during phenylalanine hydroxylation. Adv Exp Med Biol. 1993;338:47–54. doi: 10.1007/978-1-4615-2960-6_9. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bolin J. T., Filman D. J., Matthews D. A., Hamlin R. C., Kraut J. Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. I. General features and binding of methotrexate. J Biol Chem. 1982 Nov 25;257(22):13650–13662. [PubMed] [Google Scholar]
- Bredt D. S., Snyder S. H. Nitric oxide: a physiologic messenger molecule. Annu Rev Biochem. 1994;63:175–195. doi: 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- Brown K. A., Howell E. E., Kraut J. Long-range structural effects in a second-site revertant of a mutant dihydrofolate reductase. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11753–11756. doi: 10.1073/pnas.90.24.11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceska T. A., Lamers M., Monaci P., Nicosia A., Cortese R., Suck D. The X-ray structure of an atypical homeodomain present in the rat liver transcription factor LFB1/HNF1 and implications for DNA binding. EMBO J. 1993 May;12(5):1805–1810. doi: 10.2210/pdb1lfb/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouard T., Blumenfeld M., Bach I., Vandekerckhove J., Cereghini S., Yaniv M. A distal dimerization domain is essential for DNA-binding by the atypical HNF1 homeodomain. Nucleic Acids Res. 1990 Oct 11;18(19):5853–5863. doi: 10.1093/nar/18.19.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron B. A., Davis M. D., Milstien S., Gutierrez J., Mendel D. B., Crabtree G. R., Kaufman S. Identity of 4a-carbinolamine dehydratase, a component of the phenylalanine hydroxylation system, and DCoH, a transregulator of homeodomain proteins. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11891–11894. doi: 10.1073/pnas.89.24.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron B. A., Kaufman S., Milstien S., Naylor E. W., Greene C. L., Davis M. D. Mutation in the 4a-carbinolamine dehydratase gene leads to mild hyperphenylalaninemia with defective cofactor metabolism. Am J Hum Genet. 1993 Sep;53(3):768–774. [PMC free article] [PubMed] [Google Scholar]
- Curtius H. C., Adler C., Rebrin I., Heizmann C., Ghisla S. 7-Substituted pterins: formation during phenylalanine hydroxylation in the absence of dehydratase. Biochem Biophys Res Commun. 1990 Nov 15;172(3):1060–1066. doi: 10.1016/0006-291x(90)91554-6. [DOI] [PubMed] [Google Scholar]
- Davis M. D., Kaufman S., Milstien S. Conversion of 6-substituted tetrahydropterins to 7-isomers via phenylalanine hydroxylase-generated intermediates. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):385–389. doi: 10.1073/pnas.88.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. D., Kaufman S., Milstien S. Distribution of 4a-hydroxytetrahydropterin dehydratase in rat tissues. Comparison with the aromatic amino acid hydroxylases. FEBS Lett. 1992 May 4;302(1):73–76. doi: 10.1016/0014-5793(92)80288-r. [DOI] [PubMed] [Google Scholar]
- Davis M. D., Ribeiro P., Tipper J., Kaufman S. "7-tetrahydrobiopterin," a naturally occurring analogue of tetrahydrobiopterin, is a cofactor for and a potential inhibitor of the aromatic amino acid hydroxylases. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10109–10113. doi: 10.1073/pnas.89.21.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone V., Cortese R. Transcriptional regulation of liver-specific gene expression. Curr Opin Cell Biol. 1991 Dec;3(6):960–965. doi: 10.1016/0955-0674(91)90114-e. [DOI] [PubMed] [Google Scholar]
- Ficner R., Sauer U. H., Ceska T. A., Stier G., Suck D. Crystallization and preliminary crystallographic studies of recombinant dimerization cofactor of transcription factor HNF1/pterin-4 alpha-carbinolamine dehydratase from liver. FEBS Lett. 1995 Jan 2;357(1):62–64. doi: 10.1016/0014-5793(94)01325-u. [DOI] [PubMed] [Google Scholar]
- Hansen L. P., Crabtree G. R. Regulation of the HNF-1 homeodomain proteins by DCoH. Curr Opin Genet Dev. 1993 Apr;3(2):246–253. doi: 10.1016/0959-437x(93)90030-s. [DOI] [PubMed] [Google Scholar]
- Harris N. L., Presnell S. R., Cohen F. E. Four helix bundle diversity in globular proteins. J Mol Biol. 1994 Mar 11;236(5):1356–1368. doi: 10.1016/0022-2836(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Hauer C. R., Rebrin I., Thöny B., Neuheiser F., Curtius H. C., Hunziker P., Blau N., Ghisla S., Heizmann C. W. Phenylalanine hydroxylase-stimulating protein/pterin-4 alpha-carbinolamine dehydratase from rat and human liver. Purification, characterization, and complete amino acid sequence. J Biol Chem. 1993 Mar 5;268(7):4828–4831. [PubMed] [Google Scholar]
- Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993 Sep 5;233(1):123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Huang C. Y., Max E. E., Kaufman S. Purification and characterization of phenylalanine hydroxylase-stimulating protein from rat liver. J Biol Chem. 1973 Jun 25;248(12):4235–4241. [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kaufman S., Pollock R. J., Summer G. K., Das A. K., Hajra A. K. Dependence of an alkyl glycol-ether monooxygenase activity upon tetrahydropterins. Biochim Biophys Acta. 1990 Aug 1;1040(1):19–27. doi: 10.1016/0167-4838(90)90141-2. [DOI] [PubMed] [Google Scholar]
- Kim J. L., Burley S. K. 1.9 A resolution refined structure of TBP recognizing the minor groove of TATAAAAG. Nat Struct Biol. 1994 Sep;1(9):638–653. doi: 10.1038/nsb0994-638. [DOI] [PubMed] [Google Scholar]
- Lee W., Harvey T. S., Yin Y., Yau P., Litchfield D., Arrowsmith C. H. Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol. 1994 Dec;1(12):877–890. doi: 10.1038/nsb1294-877. [DOI] [PubMed] [Google Scholar]
- Leiting B., De Francesco R., Tomei L., Cortese R., Otting G., Wüthrich K. The three-dimensional NMR-solution structure of the polypeptide fragment 195-286 of the LFB1/HNF1 transcription factor from rat liver comprises a nonclassical homeodomain. EMBO J. 1993 May;12(5):1797–1803. doi: 10.1002/j.1460-2075.1993.tb05827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. W. Solvent content of protein crystals. J Mol Biol. 1968 Apr 28;33(2):491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Mendel D. B., Hansen L. P., Graves M. K., Conley P. B., Crabtree G. R. HNF-1 alpha and HNF-1 beta (vHNF-1) share dimerization and homeo domains, but not activation domains, and form heterodimers in vitro. Genes Dev. 1991 Jun;5(6):1042–1056. doi: 10.1101/gad.5.6.1042. [DOI] [PubMed] [Google Scholar]
- Mendel D. B., Khavari P. A., Conley P. B., Graves M. K., Hansen L. P., Admon A., Crabtree G. R. Characterization of a cofactor that regulates dimerization of a mammalian homeodomain protein. Science. 1991 Dec 20;254(5039):1762–1767. doi: 10.1126/science.1763325. [DOI] [PubMed] [Google Scholar]
- Miller S., Lesk A. M., Janin J., Chothia C. The accessible surface area and stability of oligomeric proteins. 1987 Aug 27-Sep 2Nature. 328(6133):834–836. doi: 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]
- Nar H., Huber R., Heizmann C. W., Thöny B., Bürgisser D. Three-dimensional structure of 6-pyruvoyl tetrahydropterin synthase, an enzyme involved in tetrahydrobiopterin biosynthesis. EMBO J. 1994 Mar 15;13(6):1255–1262. doi: 10.1002/j.1460-2075.1994.tb06377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol C. A., Smith G. K., Duch D. S. Biosynthesis and metabolism of tetrahydrobiopterin and molybdopterin. Annu Rev Biochem. 1985;54:729–764. doi: 10.1146/annurev.bi.54.070185.003501. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp K. A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Nicosia A., Monaci P., Tomei L., De Francesco R., Nuzzo M., Stunnenberg H., Cortese R. A myosin-like dimerization helix and an extra-large homeodomain are essential elements of the tripartite DNA binding structure of LFB1. Cell. 1990 Jun 29;61(7):1225–1236. doi: 10.1016/0092-8674(90)90687-a. [DOI] [PubMed] [Google Scholar]
- Nikolov D. B., Hu S. H., Lin J., Gasch A., Hoffmann A., Horikoshi M., Chua N. H., Roeder R. G., Burley S. K. Crystal structure of TFIID TATA-box binding protein. Nature. 1992 Nov 5;360(6399):40–46. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- Orengo C. A., Thornton J. M. Alpha plus beta folds revisited: some favoured motifs. Structure. 1993 Oct 15;1(2):105–120. doi: 10.1016/0969-2126(93)90026-d. [DOI] [PubMed] [Google Scholar]
- Pastore A., De Francesco R., Barbato G., Castiglione Morelli M. A., Motta A., Cortese R. 1H resonance assignment and secondary structure determination of the dimerization domain of transcription factor LFB1. Biochemistry. 1991 Jan 8;30(1):148–153. doi: 10.1021/bi00215a022. [DOI] [PubMed] [Google Scholar]
- Pastore A., De Francesco R., Castiglione Morelli M. A., Nalis D., Cortese R. The dimerization domain of LFB1/HNF1 related transcription factors: a hidden four helix bundle? Protein Eng. 1992 Dec;5(8):749–757. doi: 10.1093/protein/5.8.749. [DOI] [PubMed] [Google Scholar]
- Schallreuter K. U., Wood J. M., Pittelkow M. R., Gütlich M., Lemke K. R., Rödl W., Swanson N. N., Hitzemann K., Ziegler I. Regulation of melanin biosynthesis in the human epidermis by tetrahydrobiopterin. Science. 1994 Mar 11;263(5152):1444–1446. doi: 10.1126/science.8128228. [DOI] [PubMed] [Google Scholar]
- Tomei L., Cortese R., De Francesco R. A POU-A related region dictates DNA binding specificity of LFB1/HNF1 by orienting the two XL-homeodomains in the dimer. EMBO J. 1992 Nov;11(11):4119–4129. doi: 10.1002/j.1460-2075.1992.tb05505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F., Yaniv M. HNF1, a homeoprotein member of the hepatic transcription regulatory network. Bioessays. 1992 Sep;14(9):579–587. doi: 10.1002/bies.950140902. [DOI] [PubMed] [Google Scholar]
- Varughese K. I., Skinner M. M., Whiteley J. M., Matthews D. A., Xuong N. H. Crystal structure of rat liver dihydropteridine reductase. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):6080–6084. doi: 10.1073/pnas.89.13.6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. P., Shewchuk L. M., Brennan R. G., Otsuka A. J., Matthews B. W. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Xia T., Song J., Jensen R. A. Pseudomonas aeruginosa possesses homologues of mammalian phenylalanine hydroxylase and 4 alpha-carbinolamine dehydratase/DCoH as part of a three-component gene cluster. Proc Natl Acad Sci U S A. 1994 Feb 15;91(4):1366–1370. doi: 10.1073/pnas.91.4.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]