Abstract
Bdellovibrio bacteriovorus invade Gram-negative bacteria in a predatory process requiring Type IV pili (T4P) at a single invasive pole, and also glide on surfaces to locate prey. Ras-like G-protein MglA, working with MglB and RomR in the deltaproteobacterium Myxococcus xanthus, regulates adventurous gliding and T4P-mediated social motility at both M. xanthus cell poles. Our bioinformatic analyses suggested that the GTPase activating protein (GAP)-encoding gene mglB was lost in Bdellovibrio, but critical residues for MglABd GTP-binding are conserved. Deletion of mglABd abolished prey-invasion, but not gliding, and reduced T4P formation. MglABd interacted with a previously uncharacterised tetratricopeptide repeat (TPR) domain protein Bd2492, which we show localises at the single invasive pole and is required for predation. Bd2492 and RomR also interacted with cyclic-di-GMP-binding receptor CdgA, required for rapid prey-invasion. Bd2492, RomRBd and CdgA localize to the invasive pole and may facilitate MglA-docking. Bd2492 was encoded from an operon encoding a TamAB-like secretion system. The TamA protein and RomR were found, by gene deletion tests, to be essential for viability in both predatory and non-predatory modes. Control proteins, which regulate bipolar T4P-mediated social motility in swarming groups of deltaproteobacteria, have adapted in evolution to regulate the anti-social process of unipolar prey-invasion in the “lone-hunter” Bdellovibrio. Thus GTP-binding proteins and cyclic-di-GMP inputs combine at a regulatory hub, turning on prey-invasion and allowing invasion and killing of bacterial pathogens and consequent predatory growth of Bdellovibrio.
Author Summary
Bacterial cell polarity control is important for maintaining asymmetry of polar components such as flagella and pili. Bdellovibrio bacteriovorus is a predatory deltaproteobacterium which attaches to, and invades, other bacteria using Type IV pili (T4P) extruded from the specialised, invasive, non-flagellar pole of the cell. It was not known how that invasive pole is specified and regulated. Here we discover that a regulatory protein-hub, including Ras-GTPase-like protein MglA and cyclic-di-GMP receptor-protein CdgA, control prey-invasion. In the deltaproteobacterium, Myxococcus xanthus, MglA, with MglB and RomR, was found by others to regulate switching of T4P in social ‘swarming’ surface motility by swapping the pole at which T4P are found. In contrast, in B. bacteriovorus MglA regulates the process of prey-invasion and RomR, which is required for surface motility regulation in Myxococcus, is essential for growth and viability in Bdellovibrio. During evolution, B. bacteriovorus has lost mglB, possibly as T4P-pole-switching is not required; pili are only required at the invasive pole. A previously unidentified tetratricopeptide repeat (TPR) protein interacts with MglA and is essential for prey-invasion. This regulatory protein hub allows prey-invasion, likely integrating cyclic-di-GMP signals, pilus assembly and TamAB secretion in B. bacteriovorus.
Introduction
Bdellovibrio bacteriovorus is a small, predatory deltaproteobacterium which invades other Gram-negative bacteria wherein it replicates. Bdellovibrio can encounter their prey by fast motility, driven by rotation of a single flagellum in liquid environments [1], [2], or by slow gliding motility on solid surfaces [3], but do not show social- or S-motility a process that is shown by other deltaproteobacteria (discussed below).
In Bdellovibrio invasion into the prey cell periplasm requires T4P, thus pilus-minus cells are incapable of host/prey-dependent (HD) growth and must be cultivated on artificial media as HI - host/prey-independent - cells [4], [5]. In flagellate HD Bdellovibrio the T4P are at the non-flagellar pole and prey-invasion occurs only from that anterior pole. On surfaces a flagellum is not present and the Bdellovibrio glide bidirectionally. Both HD and HI Bdellovibrio can glide and invade prey on surfaces. Our study began by examining the genetics of surface motility control in Bdellovibrio. This work led us to find that proteins known for surface motility control in a second deltaproteobacterium, Myxococcus xanthus, have evolved to control predatory invasion of bacteria by Bdellovibrio.
Regulation of surface motility in the deltaproteobacterium M. xanthus (which is always non-flagellate), has been well characterised by pioneering work of the Søgaard-Andersen [6], Mignot [7], Zusman [8], Hartzell [9] and Kaiser [10] groups for its two types of bidirectional surface motility. These are social (S)-motility, swarming movement of streams of cells using retraction of T4P at alternate poles of the cells; and adventurous (A)-motility, characterised by the movement of individual cells on a surface. A-motility (or gliding), is thought to be powered by cell envelope-spanning motor-protein complexes, [11], [12], though the precise mechanism of movement is still being revealed [13]–[15]. In M. xanthus, T4P localize to one pole at a time. Occasionally, M. xanthus cells reverse direction; this involves a switch in the polarity of the two motility systems, including a switch in the pole at which T4P assembly occurs. Thus, M. xanthus cells can assemble T4P at both poles but at any one time, T4P are found only at one pole [16].
Recent data suggest that the four putative gliding motor-gene operons in the B. bacteriovorus HD100 genome are evolutionarily linked to those A-motility gene clusters in Myxococcus [17], with subtle distinctive absences and additions likely reflecting Bdellovibrio morphology and gliding differences.
Bdellovibrio exhibits A-motility on surfaces in a gliding process that does not use T4P [3]. In this gliding, A-motility, individual Bdellovibrio cells move bidirectionally, cells can follow each other along previous paths and reversals of individual cells and re-orientations are seen. Gliding may be a particularly important mechanism by which Bdellovibrio explores biofilms and locates bacteria to prey upon [3], [18]. It is critical for HD Bdellovibrio to be able to explore or leave solid surfaces by gliding (when its flagellum cannot operate). Unlike other non-predatory bacteria, Bdellovibrio HD cells cannot replicate outside prey without acquiring “HI mutations” to do so [19], [20], thus without surface motility they could be trapped and starve. B. bacteriovorus gliding motility is slow, with cells moving, on average, 16 µm hr−1 [3] compared to the 24–36 µm hr−1 of Myxococcus [21]. Both B. bacteriovorus and M. xanthus show reversals in gliding direction. In Myxococcus, reversals during surface motility are known, from the work of the Søgaard-Andersen and Mignot labs, to be regulated by a Ras-like GTPase, MglA, which polarises the cell during gliding [6], [7], and GTPase-activating protein (GAP) protein MglB, which activates the GTPase activity of MglA to inhibit cellular reversals [6], [7]. MglA is important for activation of both the A- and S- motility “engines” (S motility engines are T4P), at the alternating leading pole, during bidirectional movements [6], [7]. In the absence of MglA, Myxococcus is both A- and S- non-motile. This means that MglA in M. xanthus, in conjunction with interacting partner RomR, regulates the localization/pole-switching activity of both T4P and gliding engine component proteins, in this bipolar bacterium. Chemotactic signals via the Frz system control cellular reversals in M. xanthus [22] via the RomR response regulator; RomR receives signals from the chemosensory Frz system and this modulates MglA activity [23], [24]. Although romR is conserved in Bdellovibrio, the genes encoding the Frz apparatus are not.
Bdellovibrio gliding is controlled by the bacterial secondary messenger cyclic-di-GMP. A diguanylyl cyclase (dgcA) mutation abolishes gliding, rendering Bdellovibrio cells unable to glide out of a consumed prey cell bdelloplast on a surface, even 2 hours after making lytic pores in it [25].The c-di-GMP receptor CdgA (GVNEF – a degenerate GGDEF protein) was found to be present at the predatory pole of B. bacteriovorus and deletion of cdgA slowed prey-invasion significantly, showing a link between c-di-GMP signalling and predation [25].
Whilst the B. bacteriovorus HD100 genome encodes MglA (Bd3734; accession: NP_970444.1), it does not encode an MglB homologue [23]. This report caused us to ask how bipolar switching might be achieved during Bdellovibrio gliding on surfaces; and whether the non-equivalent poles of the monoflagellate Bdellovibrio in liquids might correlate with an alternative role for MglABd. Here we show that MglABd is required for predatory invasion, as well as being associated with changes in gliding reversal behaviour in B. bacteriovorus, but is not required for gliding motility per se. This activity of MglABd occurs without an MglB partner, but in a cell with a RomRBd homologue. Both of these latter proteins are important to the control of bipolar motility in Myxobacteria. However we show that RomRBd has an essential role for growth in Bdellovibrio. We also report a previously undescribed interacting protein partner of MglA, and show that MglABd and RomRBd interact with this tetratricopeptide repeat protein (TPR) which is also required for predation. TPR is expressed from an operon that encodes a TamAB transport system and again TamA was essential for growth. Implications of this for predation and the onset of predatory growth upon prey-invasion are discussed.
Whilst MglAMx is involved in regulation of T4P-mediated social motility in M. xanthus, we show that MglABd is involved in Bdellovibrio in the control of pilus extrusion for the process of T4P-mediated invasion of prey cells at the single predatory pole. We show that a complex of proteins, additional to the T4P, is required at the ‘biting’ pole to organise the prey-entry machinery.
Results
MglA is required for predatory invasion by B. bacteriovorus HD100
To investigate the role of MglABd, a deletion strategy was adopted screening for possible Bdellovibrio mutants in both prey/host-dependent (HD) and host-independent (HI) growth modes. All attempts to inactivate mglA in host/prey-dependent B. bacteriovorus HD100 were unsuccessful, despite screening many more cells than required to generate other Bdellovibrio deletion strains [26] (364 revertants obtained from second crossover events, but no deletion mutants, from three separate conjugations); suggesting that MglABd is essential for an aspect of the predatory life cycle.
Three host-independent (HI) ΔmglABd strains were obtained through sucrose-suicide counter-selection from a total of 76 screened. When challenged with prey, ΔmglABd HI B. bacteriovorus strains were unable to lyse E. coli in either a soft agar prey-lawn on the surface of YPSC plates, or in liquid culture (Figure 1A). Introduction of wild type mglABd by in cis complementation method (as described previously [25]) restored predation (Figure 1B) confirming that MglABd is essential for predatory growth.
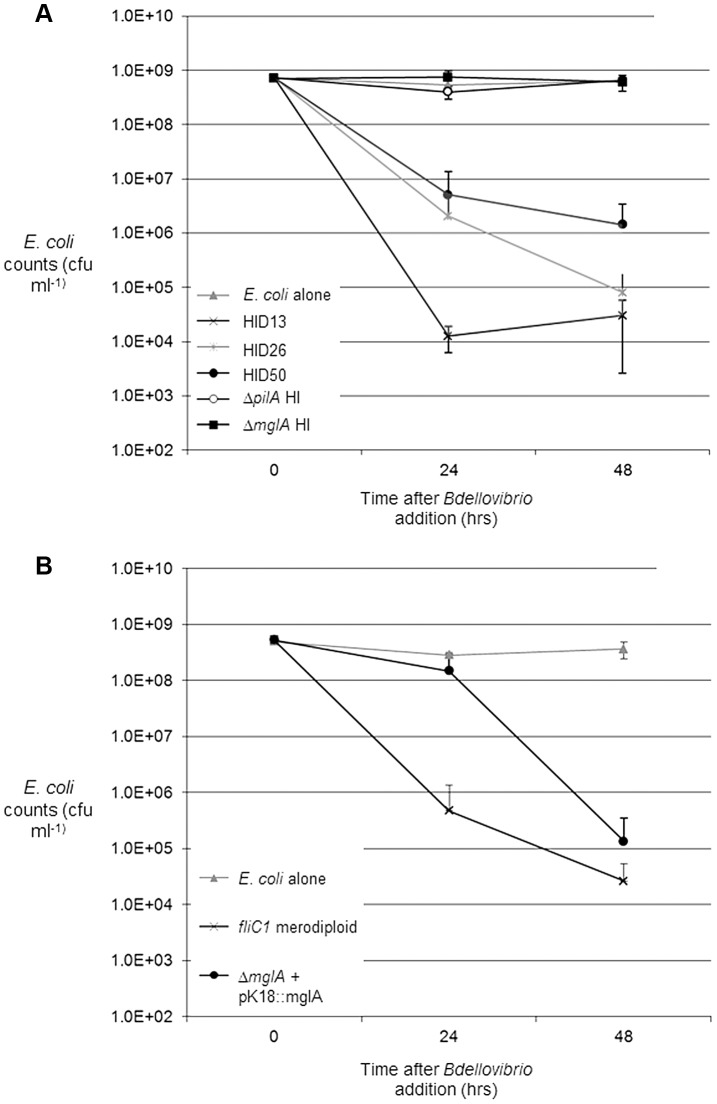
Figure 1. Predation and in cis complementation of B. bacteriovorus ΔmglA HI strains, on E. coli prey.
(A) Predation efficiency of the ΔmglA HI strain was assayed against predatory and non-predatory controls by the reduction of E. coli numbers over 48 hours. Three wild-type HI strains (HID13, HID26 and HID50) reduced E. coli numbers in liquid cultures by up to four logs (grey region shows known natural variation in predation rate between different wild-type HI isolates). The ΔmglA HI strain showed no reduction in E. coli numbers, comparable to a previously-studied, non-predatory ΔpilA HI strain, and to E. coli with no added B. bacteriovorus. (B) Reintroduction of the mglA ORF in cis to the ΔmglA HI strain in plasmid pK18::mglA restored predatory growth. Error bars represent 1 SD from the mean (for predation-testing of Δ bd2492 strain see Figure S5).
The ΔmglA HI B. bacteriovorus strain could not reduce E. coli numbers in liquid culture, though this strain could still attach to the exterior of potential prey cells (Figure 2). A parallel assay showed that 43.5% of wild-type B. bacteriovorus HI cells attached to, or had entered, E. coli prey cells after 1 hour (Figure 2A) but no ΔmglA HI strain formed prey-bdelloplasts (Bdellovibrio cause the prey to round-up into ‘bdelloplast’ structures after invasion), even after 22 hours. Figure 2 also shows that both the ΔmglA HI and ΔpilA HI (Δbd1290, which is known to lack pili and is obligately host-independent [4]) could still attach to E. coli prey cells, albeit at a lower frequency. This suggests that pili are not a prerequisite for attachment, (although they are required for prey-invasion [4], [5]), and suggests that the ΔmglA HI predatory defect is not due to the inability of the Bdellovibrio cell to attach to prey cells.
Figure 2. Host-independent invasion and attachment assays of ΔmglA strain and wild-type controls.
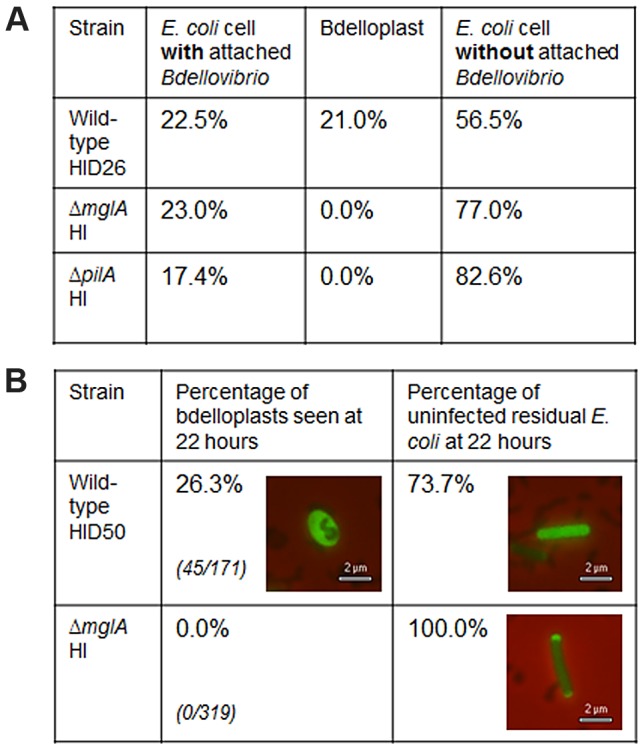
(A) Attachment assay: After 1 hour, 21.0% of E. coli cells were attached to and invaded by wild-type Bdellovibrio HI strain HID26 cells. A further 22.5% of E. coli cells were attached to, but not invaded by HID26 cells. 23.0% and 17.4% of E. coli cells were attached to by ΔmglA HI and ΔpilA HI, respectively. The ΔmglA HI and ΔpilA HI strains never invaded to form bdelloplasts. The attachment assay has the following variability: Percentage points (pp): WT 43.5%±15.5 pp; ΔmglA 23.0%±4.2 pp; ΔpilA 17.4%±7.3 pp. (B) Invasion assay: HI wild-type Bdellovibrio control HID50 was able to infect E. coli prey cells (26.3% of E. coli cells invaded to form bdelloplasts after 22 hours, 45/171 E. coli cells). Bdellovibrio ΔmglA HI strain could not invade E. coli prey (0.0% of E. coli cells invaded to form bdelloplasts after 22 hours, 0/319 E. coli cells). Fluorescent images show representative fluorescent E.coli S17-1::pMAL_p2-mCherry cells, either uninfected or rounded to form bdelloplasts.
The nature of the predatory defect of the ΔmglA HI strain was analysed further by microscopy, using a fluorescent E.coli S17-1::pMAL_p2-mCherry prey strain [27]. Addition of the ΔmglA HI strain to E.coli S17-1::pMAL_p2-mCherry and incubation for 22 hours demonstrated that although ΔmglA HI cells could attach to the outside of a prey cell, they could not invade to form bdelloplasts (Figure 2B). A wild-type HI B. bacteriovorus strain (HID50) successfully invaded E. coli cells and killed them (as shown in Figure 1) and at the 22 hour stage was shown to have formed bdelloplasts from 26.3% of the remaining E. coli, compared to zero bdelloplasts for the ΔmglA HI strain. Thus the deletion of mglABd abolished a process required for prey-invasion.
The B. bacteriovorus mglA mutant is hypo-piliated
The ΔmglA HI strain showed a similar phenotype to that observed in a pilus-minus (ΔpilA) strain, which was known to be unable to invade prey cells [4]. We hypothesised that B. bacteriovorus ΔmglA might be defective in the synthesis or extrusion of pili, preventing prey cell invasion. This seemed plausible given that MglA regulates both the pole-switching of the A-motility and Type IV pilus-mediated S-motility systems in M. xanthus. Transmission electron microscopy of HI Bdellovibrio cultures grown to an OD600 of 0.2–0.3 showed that a wild type HI control had pili in 14.3% of cells, whilst ΔmglA HI had pili in only 2.3% of cells analysed (p = 0.02). These data suggested that MglABd regulates formation of pili; loss of mglA reduces the number of piliated cells. But, in contrast to the ΔpilA strain which completely lacks pilus fibres, the total inability of ΔmglA cells to invade, despite the presence of a low (but significant) frequency of piliated cells, suggests that these few pili present in the ΔmglA cells are not competent to facilitate invasion. This could be due to a defect in pilus retraction upon attachment to prey surfaces, or a requirement for another MglA-controlled factor to mediate invasion. Candidate MglABd-interacting proteins for invasive processes are discussed later.
MglABd controls gliding reversal frequency but not gliding per se
Knowing that MglABd controls pilus-mediated bacterial invasion in B. bacteriovorus, but that in M. xanthus both pilus-mediated S-motility and gliding A-motility are MglA controlled, we used time-lapse microscopy to observe ΔmglA and wild-type B. bacteriovorus strains for gliding motility on 1% agarose/CaHEPES. Surface motility in B. bacteriovorus begins after a period of incubation on an agarose surface and allows exploration of environments for potential prey.
In contrast to recent studies in Myxococcus xanthus which showed that a ΔmglAMx strain is non-motile on surfaces [7], and a mglA G21V strain displays hyper-reversals during A-motility [6], we found that Bdellovibrio ΔmglA cells showed sustained gliding runs on surfaces (Figure 3), indicating that MglABd is not absolutely required for Bdellovibrio cells to glide.
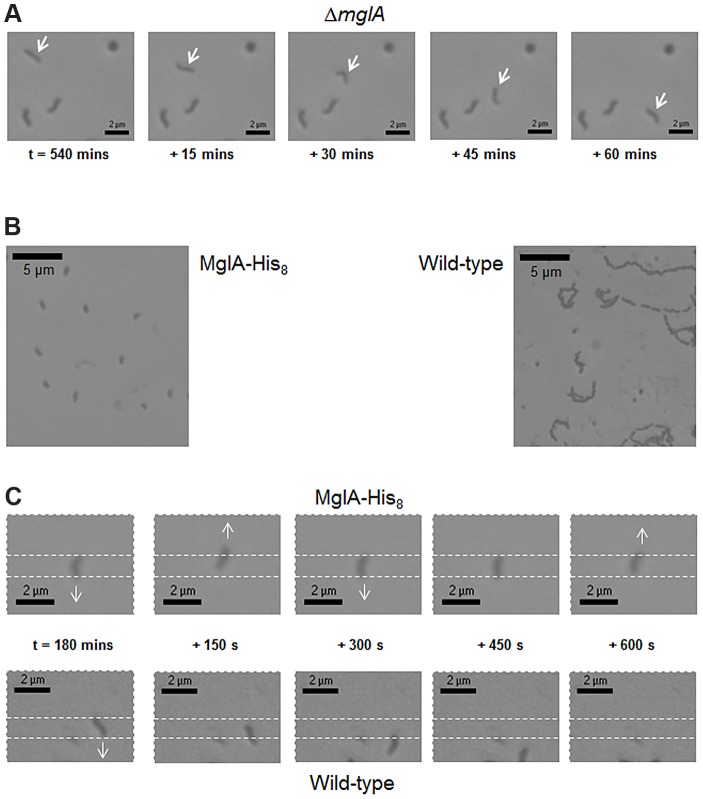
Figure 3. B. bacteriovorus ΔmglA HI cells show sustained gliding motility; MglA-His8 HD cells show hyper-reversals.
(A) B. bacteriovorus ΔmglA HI cells showed gliding motility on 1% agarose/CaHEPES. Gliding was sustained and progressive (cells were not hyper-reversing), as in the arrowed cell which moved at 9.49 µm hr−1. Each panel is a 15 minute timepoint, starting from 540 minutes after the cells were added to the agarose surface (three “bystander” cells that have not yet commenced gliding represent a stationary marker). (B) B. bacteriovorus MglA-His8 HD cells showed a high incidence of reversals during gliding motility on 1% agarose/CaHEPES compared to wild-type cells. Each larger panel shows a “trail-montage” of 60 minutes of gliding motility (150 second per frame): MglA-His8 cells show no progressive gliding motility (reversing rapidly), whilst wild-type cells show sustained runs of gliding (seen as curving trails with direction changes). (C) Smaller panels show individual wild-type and MglA-His8 HD cells gliding from an original start point (indicated by white dashed region), starting at 180 minutes after addition to the agarose surface and at 150 second intervals; arrow indicates direction of movement.
A Bdellovibrio strain with C-terminally His8-tagged MglABd, expressed from the endogenous bd3734 promoter in cis, with a plasmid promoter-driven wild-type copy of mglABd, could be grown predatorily, in contrast to the ΔmglA strain which was non-predatory. In a previous study in M. xanthus, the presence of tagged MglAMx protein in conjunction with wild-type MglAMx allowed gliding to remain fully functional [7]. In contrast to the sustained gliding motility of the ΔmglABd strain (Figure 3A), the predatory B. bacteriovorus HD100 MglA-His8 showed increased reversals during gliding: on average 9.0 reversals hr−1 (n = 28), significantly more than wild-type HD100 cells with an average of 3.2 reversals hr−1 (n = 21) (p<0.001) (Figure 3B,C). The same hyper-reversal phenotype was also observed in B. bacteriovorus HD100 MglA-mCherry cells (data not shown).
Differences between MglABd and MglAMX sequences may reflect diverse functions in monopolar predation versus bipolar surface motility
MglABd (Bd3734) shares significant sequence similarity (Figure 4) with MglAMx (MXAN1925 accession: YP_630169.1), with 64% protein identity and 82% similarity (NEEDLE global alignment). The majority of residues shown to be important for MglAMx function [28], [29] are conserved in MglABd (Figure 4A–D).
Figure 4. Protein alignment of MglAMx and MglABd.
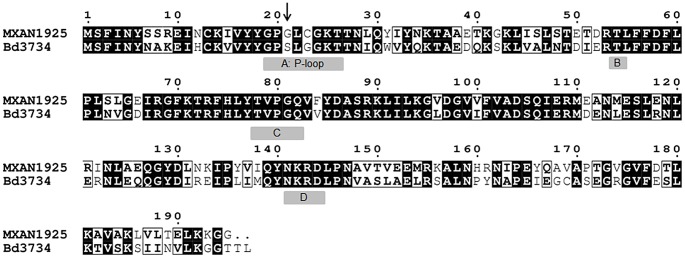
MglAMx (MXAN1925) alignment with MglABd (Bd3734) shows significant sequence similarity between the two proteins. (A) The P-loop is conserved in B. bacteriovorus, although a serine is present in place of a glycine residue at position 21 (signified by arrow). The PM1/G1 threonine residue (B) and PM3 (C); and G2 (D) motifs are all conserved between the two proteins (for mglA genes, encoding MglA G21, co-occurring with mglB genes see Figure S1).
The P-loop region (19GXXXXGKT26) of MglAMx was shown by Søgaard Andersen and co-workers to be important for GTP hydrolysis, and for MglA function [28], and substitutions in this region, such as G21V, were reported to decrease hydrolysis [6]. The P-loop region of MglABd contains a natural serine at residue 21; the corresponding G12S substitution in eukaryotic G protein Ras activates Ras protein [30], essentially locking the protein in a GTP-bound state, in the same way as a Ras G12V substitution. This suggests that MglABd exists in a permanently GTP-bound state. The G21-equivalent residue is a conserved glycine across 7 deltaproteobacterial genera (Figure S1A) which all also have a conserved mglB gene, though in Bdellovibrio the equivalent residue is a serine.
The difference at residue 21 in the MglABd sequence suggested to us a reason why we did not observe conservation of the gene encoding MglB in Bdellovibrio, as the GAP activity of an MglB would likely be ineffective on a permanently GTP-bound MglA protein such as that suggested by the MglABd sequence with S at position 21. We thought that it might also explain the lack of a Bdellovibrio Frz system [23], which stimulates motility reversals in M. xanthus, as a mutation, causing MglAMx G21V, bypasses the requirement for Frz for reversals in that deltaproteobacterium [6]. Thus we turned to examine the presence of mglB in the deltaproteobacterial relatives of Bdellovibrio. We also tested the conserved RomRBd protein, while also looking for other proteins, specific to Bdellovibrio, with which MglABd might interact. In M. xanthus, RomR is found at both poles of the cell and interacts with both MglA and MglB to link the Frz system to regulate polarity control [23], [24].
B. bacteriovorus has lost the mglB gene
The majority of sequenced deltaproteobacteria genomes contain both mglA and mglB, and these are often co-transcribed at the same locus, including in M. xanthus where the MglBMX protein has an important role in motility [6], [7], [31]. Although the mglA gene product in B. bacteriovorus HD100 shares extensive sequence similarity with other MglA proteins, there is no mglB homologue in the HD100 genome, despite neighbouring genes (dnaX, recR, mglA and a DUF149-encoding gene) showing conserved synteny to other deltaproteobacteria that do have an mglB. The closely related B. bacteriovorus Tiberius [32] also lacks an mglB homologue. The predatory, invasive, marine bacterium Bacteriovorax marinus is also closely related to B. bacteriovorus, although the Bdellovibrio and Bacteriovorax genera have diverged separately from Myxobacteria. A 16S rRNA phylogenetic tree of the deltaproteobacteria shows the ancestral lineage leading to Bdellovibrio and Bacteriovorax diverged from the ancestral lineage leading to the clade including Myxococcus xanthus [33] and in that divergent Bdellovibrio branch we detect mglB loss (Figure S1A). We found that in B. marinus, which also has an mglA gene (BMS_0054), there is an adjacent putative mglB homologue (BMS_0053), both genes lying downstream of recR (Figure S1B).
BMS_0053 shares only limited sequence similarity with other MglB Roadblock domain proteins (BMS_0053, 168 residues, shares 22% identity and 43% similarity (NEEDLE global alignment) with M. xanthus MglB protein, 159 residues, Figure S1C). This highly divergent MglB homologue in Bacteriovorax is likely still functional, since no frameshift or nonsense mutations have arisen in the B. marinus lineage, and protein sequence length is conserved; however, its function is unclear. We are unable to test whether mglB is under positive selection (dN/dS>1) in Bacteriovorax because synonymous substitution rates are saturated for available sequence comparisons (dS>2). The Bacteriovorax MglA homologue is much more conserved (66% identity and 83% similarity to MglABd) and may function in an analogous predatory role to that of B. bacteriovorus.
MglABd interacts with TPR- (tetratricopeptide repeat) domain protein Bd2492 in vivo
As MglABd had both similarities and differences to MglAMx, we sought to identify proteins that interact with MglA homologue Bd3734 in B. bacteriovorus as we reasoned that these proteins might have a predatory role. We used a pull-down co-purification assay with proteins from the predatory B. bacteriovorus strain producing MglABd with a C-terminal His8 tag from the endogenous mglABd promoter, mentioned above. For the co-purification assay, a host-independent isolate of the MglABd-His8 strain was used, as previous array data showed that mglABd transcription is up-regulated in wild type HI cells, (which remain predatory but are longer than attack phase Bdellovibrio). Whole cell lysates of this HI strain were used in the assay, in which the bait His-tagged protein MglABd binding to TALON-NX cobalt-charged resin allowed interacting proteins to be identified (Figure S2) that were not present in the control without the His-tag.
MglABd co-purified with Bd2492 (accession: NP_969302.1) (Figure S2) - a B. bacteriovorus protein with a hypothetical annotation, with predicted tetratricopeptide repeat (TPR) domains typically involved in protein-protein interactions. Bands were excised from the gel and analysed by LC-MS/MS. Corresponding regions of the wild-type HID13 control lane were also analysed, and neither MglABd nor Bd2492 were found in these regions, suggesting that MglABd and Bd2492 (TPRBd) interact in vivo.
Confirmation that MglABd and TPRBd proteins interact by bacterial two-hybrid and heterologous co-expression in E. coli
The mglA ORF and bd2492 ORF were cloned into pUT18C and pKT25 vectors containing T18 and T25 fragments of adenylate cyclase, respectively [34]. The bacterial two-hybrid assay for MglA and Bd2492 showed a strong signal (Figure S3A–B) suggesting that the two B. bacteriovorus proteins interact. This interaction was supported by the observation that MglA co-purifies with His6-tagged Bd2492 in nickel-affinity chromatography of E. coli lysates heterologously expressing these two proteins from plasmid pD2492N/3734 (Figure S4). Gel filtration and SDS-PAGE of purified MglA and Bd2492-His6 indicated that the MglA-Bd2492 complex has an Mw of approximately 63 kDa and exists predominantly as a heterodimeric complex of 1∶1 stoichiometry (data not shown).
TPRBd is required for predatory invasion by B. bacteriovorus HD100
As the B. bacteriovorus mglA mutant was non-predatory, we tested whether bd2492 (encoding TPRBd) was essential for predatory growth. All attempts to inactivate bd2492TPR in host-dependent B. bacteriovorus HD100 were unsuccessful (68 revertants obtained from second crossover events, but no deletion mutants). Two host-independent (HI) Δbd2492 strains were obtained through sucrose-suicide counter-selection from a total of 10 screened. When challenged with prey, Δbd2492TPR HI strains were unable to lyse E. coli in liquid culture (Figure S5). As with the ΔmglA HI strains, the Δbd2492TPR HI strains could still attach to E. coli prey cells (attachment assay; 26.6% of E. coli cells had attached Bdellovibrio), but could not invade to form bdelloplasts (invasion assay; 0/389 E. coli cells). The B. bacteriovorus Δbd2492TPR HI strain was still able to glide on 1% agarose CaHEPES (data not shown).
B. bacteriovorus genes bd2492, bd2494 and bd2495 are co-transcribed and syntenic in other deltaproteobacteria
TPR gene bd2492 is co-transcribed with bd2494 and bd2495 (Figure S6). The same gene synteny is also found in M. xanthus (MXAN_5763-5766) and B. marinus SJ (BMS_0137-140) (Figure 5) where the gene encoding a TPR domain protein is followed by genes encoding homologues of Bd2494 and Bd2495. In M. xanthus, the genes encoding homologues of Bd2492 and Bd2494 (MXAN_5766 and MXAN_5764) are interrupted by a gene encoding a putative Sec system ATPase, MXAN_5765.
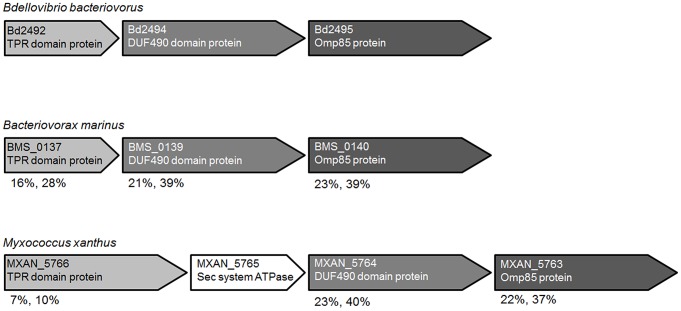
Figure 5. Gene synteny of bd2492–bd2495 homologues is conserved in B. bacteriovorus, M. xanthus and B. marinus.
Genes encoding a TPR domain protein are followed by genes encoding a DUF490 domain protein and an Omp85 superfamily protein in all three bacterial species. In M. xanthus, the three genes are interrupted by a gene encoding a putative Sec system ATPase, MXAN_5765. Percentage protein sequence identities and similarities with the B. bacteriovorus protein (NEEDLE global alignment) are shown underneath (for operon-confirmation of bd2492-2495 see Figure S6).
B. bacteriovorus gene bd2492 encodes a hypothetical 353 amino acid tetratricopeptide repeat (TPR) protein; TPRpred (http://tprpred.tuebingen.mpg.de/tprpred) was used to predict TPR domains [35]. TPRpred confirmed that both BMS_0137 (524 residues; accession: YP_005034048.1) and MXAN_5766 (1031 residues; accession: YP_633903.1) are also predicted to contain TPR domains. All three TPR domain proteins do not have predicted signal sequences, as predicted by SignalP [36].
Bd2494 is a predicted transmembrane protein with a DUF490 domain. Both BMS_0139 (accession: YP_005034049.1) and MXAN_5764 (accession: YP_633901.1) also contain predicted DUF490 domains. Bd2495 is a surface antigen variable number repeat domain protein of the (outer membrane protein) Omp85 (TamA/BamA/YaeT) superfamily, hereafter termed TamABd; homologues of which are conserved in both B. marinus (BMS_0140; accession: YP_005034050.1) and M. xanthus (MXAN_5763; accession: YP_633900.1).
RomRBd in B. bacteriovorus interacts with TPRBd by BTH and both proteins are located at the prey-invasion pole
As mentioned in the introduction, RomRMx interacts with the MglAMX signalling system to regulate surface motility in response to Frz system signals [23], [24], but the Frz system is not conserved in Bdellovibrio. We assessed the interaction of the RomRBd (Bd2761; accession: NP_969553.1) with the MglA-interacting protein TPRBd (Bd2492) by bacterial two-hybrid (Figure S3A). RomRBd shares homology with the REC domain and C-terminal region of RomRMx, whilst the remainder of the protein is less well conserved (Figure S7). RomRBd and TPRBd interact in the BTH assay (Figure S3A, C). We found that RomRBd and MglABd interacted weakly, but not significantly (p = 0.18) (Figure S3B–C).
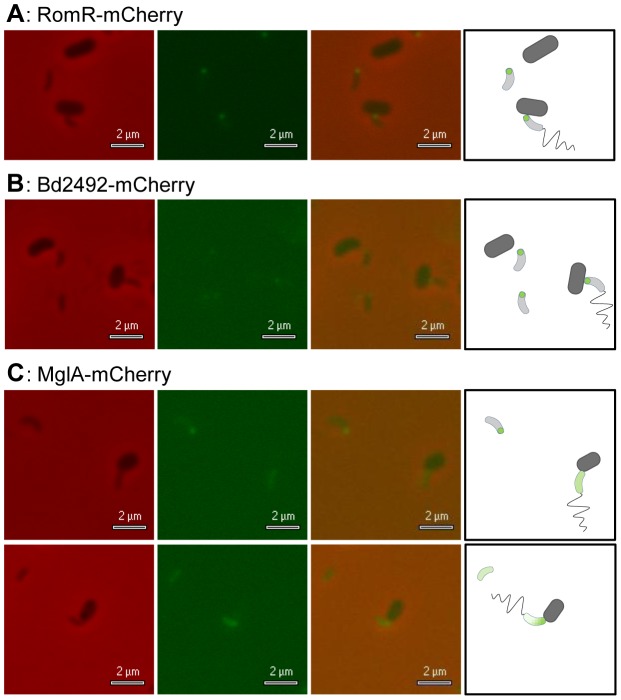
Fluorescent tagging of RomRBd and TPRBd with C-terminal mCherry revealed that both proteins are localised at only one pole of the cell. Co-incubation with E. coli prey cells confirmed that both RomRBd-mCherry and TPRBd-mCherry are found at the anterior, prey-interaction pole of B. bacteriovorus cells (Figure 6). Fluorescent tagging of MglABd with mCherry typically showed cells with diffuse fluorescence localization in cells directly after applying to 1% agarose/CaHEPES (i.e. not gliding) (Figure 6); 63% of HD100 MglA-mCherry Bdellovibrio had diffuse fluorescence, the remainder showing a unipolar focus (28.4%) or bipolar foci (8.6%).
Figure 6. B. bacteriovorus RomR-mCherry and Bd2492-mCherry localised at the prey-interaction pole; MglA-mCherry showed variable diffuse foci.
B. bacteriovorus cells were incubated with E. coli S17-1 prey cells for 5 minutes, allowing sufficient time for some of the Bdellovibrio cells to attach to prey. Panels- A: The lower prey-cell shows a typical attached Bdellovibrio cell, with a RomR-mCherry focus at the anterior (attached) pole of the Bdellovibrio. B: The rightmost prey-cell shows a typical attached Bdellovibrio cell, with a Bd2492-mCherry focus at the anterior (attached) pole of the Bdellovibrio. C: MglA-mCherry Bdellovibrio cells had variable foci, including diffuse and unipolar localisations. From left to right, all panels show brightfield, fluorescent, and merged images and a graphical representation. Fluorescent exposure = 2 seconds.
RomRBd and TPRBd interact with invasion-pole protein CdgA, a degenerate GVNEF c-di-GMP binding protein which is required for rapid prey-invasion by B. bacteriovorus
We found earlier that the Bdellovibrio ΔmglA strain does not show a hyper-reversal or non-motility phenotype (Figure 3). Thus, the regulation of MglABd localization in the control of gliding reversals (in the absence of MglB and Frz) is likely to employ an alternative signalling system to that of M. xanthus. Previous work suggested that this could be c-di-GMP as we have shown [25] that lack of GGDEF protein Bd0367 DgcA abolished gliding exit from bdelloplasts.
We had also had previously noted a link between a c-di-GMP binding protein and prey-invasion in Bdellovibrio [25]. Degenerate GGDEF (GVNEF) protein CdgA, Bd3125 (accession: NP_969891.1), is located at the prey invading pole of B. bacteriovorus and lack of this polar protein causes a very significant slowing of prey-invasion with bdelloplast formation taking 40–90 minutes compared to 30–40 minutes for wild type [25]. We concluded in that paper that “CdgA organises processes at the Bdellovibrio “nose” that are crucial to rapid prey-invasion”. In our current study, we found that both RomRBd and TPRBd (though not MglA) interacted with CdgA in the bacterial two-hybrid assay (Figure S3), supporting this idea. Whether RomRBd has a role in the regulation of gliding motility will be the subject of a subsequent study, but our interaction data suggested a link between RomRBd and predatory growth (as ΔcdgA was affected in predation [25]), so we tested for a romRBd deletion strain.
RomRBd is essential in B. bacteriovorus
Given the CdgA and TPRBd interactions found at the B. bacteriovorus invasive pole, we speculated that RomRBd would be required for prey-invasion. Attempts to delete romRBd, both predatorily (HD) and host-independently (HI), were unsuccessful (HD 104; HI 120 revertants screened), suggesting that RomRBd is required for both predatory and host-independent Bdellovibrio growth.
TamABd, encoded from the operon encoding TPRBd is also essential in B. bacteriovorus
As RomRBd interacted, by BTH, with TPRBd, encoded in an operon with the tamAB genes, we speculated that the TamAB complex would also be required for predatory growth. Attempts to delete tamABd also proved unsuccessful (HD 140; HI 97 revertants screened), suggesting that TamABd is also essential for both phases of Bdellovibrio growth.
Discussion
Here we report that B. bacteriovorus use homologues of adventurous/social motility-control proteins for the process of predatory invasion of other bacteria. Whilst non-invasive M. xanthus utilise the proteins MglA and MglB to control bipolar, bidirectional surface motility [6], [7] in Bdellovibrio MglABd has evolved to function without an MglB homologue (the mglB gene is absent) to regulate prey entry at a single pole.
A unipolar role for the B. bacteriovorus homologues of the M. xanthus motility proteins
There are three lines of evidence to suggest this: (1) The deletion of mglABd caused a non-prey-invasive phenotype (Figure 1) and severely reduced pilus formation on the cell surface; (2) the natural substitution in MglABd of serine for glycine (Figure 4) at the position equivalent to residue 21 in MglAMx suggests that MglABd exists in a permanently GTP-bound state, and is not involved in the GTPase cycle which is key to the alternate bi-polar switching of motility proteins in M. xanthus [6], [7]; (3) RomR-mCherry is unipolar in B. bacteriovorus (Figure 6), in contrast to its asymmetric bipolar localization in Myxococcus, controlling MglAMx positioning.
RomRBd is localised only at the prey-invasion pole and has a different phenotype to RomRMx
We had hypothesised that RomRBd might be involved in regulating pole activity to control gliding motility. As RomRBd was found at the predatory pole only, this suggested an alternative role. We could not detect a significant interaction between RomRBd and MglABd by BTH, but we did detect a significant interaction with Bd2492 TPR protein (Figure S3), which is also at the anterior pole (discussed later).
The RomRBd location at the anterior pole of B. bacteriovorus puts it where prey-invading T4P are located. Lotte Søgaard-Andersen's group showed that an mglA Mx deletion mutant resulted in unipolar RomRMx, with RomRMx and T4P, (used in that bacterium for bipolar social motility), found at the same pole [37]. Sequence- and localization- differences between unipolar RomRBd and MglABd (in the absence of an MglB) in B. bacteriovorus, versus those in M. xanthus (which has MglB), might explain why T4P are only found at the anterior Bdellovibrio pole where they control prey-invasion.
Deletion of romRBd abolished Bdellovibrio growth in both HI and predatory conditions, but in M. xanthus romR is viable with abolition of gliding motility and reduction of T4P-dependent social motility [23], [24]. Thus RomRBd, which does show some sequence divergence from RomRMx (Figure S7), could be reporting T4P activity and prey-invasion, at the anterior pole, back to initiate Bdellovibrio growth. It should be recalled that predatory “attack phase” Bdellovibrio do not replicate outside prey, but initiate replication when prey are entered [20].
Absence of MglB in Bdellovibrio is consistent with unipolar RomRBd
Our BTH interaction data were too weak to prove a significant interaction between RomRBd and MglABd. This could be interpreted to mean that RomRBd transiently docks with MglABd when RomRBd is complexed at the pole, that other partner proteins are required to contribute to this interaction, or that they do not interact, in contrast to published data for MglAMx [23], [24]. Our finding that RomRBd is unipolar fits with evidence in M. xanthus that MglBMX is required for bipolar localization of RomRMx [23] and the apparent loss of MglB from the prey-invasive Bdellovibrio lineage in evolution. The Bdellovibrio-like invasive B. marinus has a putative mglB gene, the product of which shows only limited sequence similarity to other MglB Roadblock domain proteins (Figure S1). This mglBBm gene is highly divergent from mglbMx but likely still functional. It may be undergoing selection to evolve an alternative function, while the B. marinus mglA gene is maintained for a predatory role analogous to that in B. bacteriovorus.
MglA and MglB were shown to be conserved by Keilberg and co-workers in many deltaproteobacteria but also occur in some evolutionarily distant bacteria such as the green non sulphur bacteria, Acidobacteria and Deinococcus-Thermus group, Figure S3 in Ref [23]. The authors calculated the following: out of a total of 70 species with (at least one) predicted MglA homologue 87% = 61/70 species have MglB and an MglA. Of the 9 without MglB, 4 bacteria had MglA G21 with no MglB; 5 had MglA A/S21 with no MglB. Of these 9 with no MglB, only B. bacteriovorus and one other species, (a soil Acidobacterium named Candidatus koribacteria versatilis), have predicted RomR homologues. Thus bacteria with RomR and MglA and B may have interacting protein complexes that move between poles; but our study on B. bacteriovorus is the first to examine the situation in a bacterium where MglA and RomR are present but MglB is not.
As mentioned above, we detected an interaction with an additional protein that could contribute to the localization of MglABd and RomRBd at the single prey-invasion pole of Bdellovibrio. This was with the unipolar tetratricopeptide repeat TPR protein, Bd2492 (TPRBd) shown using both His-tag pull-downs and BTH for MglA and BTH for RomR. TPRBd could sequester either MglABd or RomRBd at the prey-invasive pole, regulating their freedom to interact with each other, or promoting an interaction on the TPRBd surface. Deletion of bd2492TPRBd abolished prey-invasion in the same manner as ΔmglABd (Figure S5, Figure 1).
It was not possible to monitor localization of fluorescently tagged proteins informatively in the HI derivative strains of the non-predatory ΔmglABd and Δbd2492 mutants. This is because HI derivatives have pleomorphic cell morphotypes (HI cells naturally differ greatly in length and shape) [38], and indeed some long HI cells are predatory at both poles [25].
TPR gene bd2492 is in an operon with tamAB genes
The bd2492 gene is located upstream of, and is co-expressed (Figure 5, Figure S6) in an operon with, gene bd2494, which encodes a transmembrane protein with a C-terminal DUF490 domain, homologous to the TamB component of the TamAB autotransporter-secretion system [39]. Bd2494 might dock with TPRBd at the prey-invasive nose. The last gene in the operon (bd2495) encodes a 7-POTRA (polypeptide-transport-associated)-domain, outer membrane protein (OMP) member of the Omp85 superfamily. The Omp85 protein family includes the BamA component of the BAM complex, known to receive and assemble beta barrel proteins during outer membrane growth [40]. The family also includes the TamA component of the TamAB complex, which aids autotransporter secretion [39]; and two-protein secretion (TPS) proteins [41].
The TamA and TamB genes are typically adjacent in proteobacteria [39], suggesting that the adjacent B. bacteriovorus bd2494-2495 genes encode a TamAB-like transporter.
Thus our finding that MglABd and RomRBd interact with a TPR protein (Figure S3), encoded from the 5′ gene of a tamAB-like operon, suggests that the Bd2494-2495 TamAB-like transport activity might be required for OMP/autotransporter proteins involved in predation. This may account for our observation that some pili are present on the ΔmglA mutant but that despite this, it does not invade due to an effect on TamAB-dependent predatory protein transport. Similarly, the Δbd2492 mutant was also non-predatory (Figure S5), but attached to prey. This suggests that either: TPRBd and MglABd are important in the positioning of proteins (probably Bd2494-5 TamABBd) at the predatory pole of the B. bacteriovorus cell to facilitate prey entry; or that binding of RomRBd and MglABd to TPRBd affects its activity, and that of the TamABBd complex, regulating predatory protein secretion.
Reinforcing our observation (mentioned above) that RomRBd is essential, we found, by attempting to delete bd2495, that TamABd was also essential for both HD and HI growth of Bdellovibrio. This suggests that the activity of the TamAB complex (possibly involving a TPR-mediated interaction with RomRBd) is required for secretion of proteins required for prey-invasion and both predatory and HI growth. Potential candidates for TamAB export are proteins involved in the synthesis/secretion/maturation of extracellular polysaccharide (EPS) or polyelectrolytes; an earlier study proposed that RomR was responsible for stimulating polyelectrolyte secretion in M. xanthus [37]. We cannot yet define whether RomRBd activates a TamAB dependent process that is essential for predatory and HI growth, or whether it reports on the activity of a TamAB complex, via its interaction with TPRBd, to regulate Bdellovibrio growth. This will be the subject of a further extensive genetic study.
Conserved synteny of the TPR tamAB genes in Myxococcus
Although a TPR protein interaction with MglAMx or RomRMx has not been previously reported, the MXAN_5766 gene encoding a TPR domain protein, from a gene cluster with similar synteny to the B. bacteriovorus bd2492-2495 genes (Figure 5), has previously been implicated in M. xanthus S-motility by transposon studies carried out by the Hartzell group [42]. The low percentages of TPR ORF similarity/identity between MXAN_5766 and Bd2492 could reflect the greatly different protein sizes and may indicate interactions with additional protein partners in M. xanthus. However, in M. xanthus, similar TPR interactions with RomR, MglA and TamAB-like proteins could play a role in bipolar motility control. Whether or not this is the case in M. xanthus, it is clear that TPR, and likely TamABBd, proteins play an important role in defining the single, active, predatory pole of Bdellovibrio.
A protein hub controlling predatory invasion
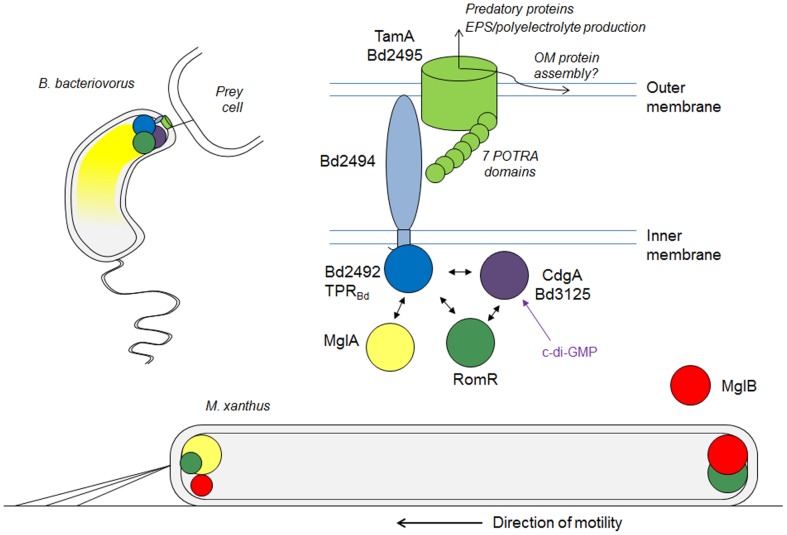
We propose a predatory regulatory ‘hub’ of proteins at the B. bacteriovorus prey-invasive pole (Figure 7), with the TamAB-associated Bd2492-5 TPRBd protein complex involved in the organisation/assembly of OMPs or autotransporters at the predatory pole. This is reflective of TamAB protein functions in other bacteria (discussed in [39]). Such protein secretion could facilitate predation directly or produce other extra-cellular compounds such as EPS or polyelectrolytes, as mentioned above, which contribute to predatory invasion. Predatory proteins could be secreted in outer membrane vesicles (OMVs); Sar and Arf GTPases (homologous to MglA) have functions in vesicle transport [43] and M. xanthus vesicles likely have an extra-cellular predatory role in the “wolf-pack” [44] hunting of M. xanthus [45]. Our studies show that the directed prey-invasion of Bdellovibrio requires a protein encoded by a tamAB operon, suggesting synergies in TamAB-mediated predation and cell interaction processes of B. bacteriovorus and M. xanthus which is worthy of further investigation. Regulatory protein hubs are reported to control pili and flagella in other bacteria [46].
Figure 7. Model for B. bacteriovorus predatory-pole regulation during prey-invasion and its relationship to M. xanthus bipolar motility-control proteins.
During prey-invasion; TamABd, RomRBd and CdgA protein interactions occur (see Figure S2, S3 S4) at the single B. bacteriovorus pole. This could control localization of the TamABd-like OMP at the prey-interaction pole or activate it to receive, (via its POTRA domains), and secrete predatory outer membrane or autotransporter proteins. The action of this secretion via Bd2495 TamA is essential to both predatory and HI lifestyles, and RomRBd, (which is also essential), may regulate or report the activity of the TamAB transport system, at the single predatory pole. Additional regulation of this activity could be influenced by c-di-GMP for which CdgA, (another hub protein that binds RomR and TPR Bd2492), is a receptor in Bdellovibrio. MglABd interacts with Bd2492 at the predatory pole but also is found more diffusely in the cell. MglA interactions may regulate prey entry via TPR Bd2492, as deletion of MglA or TPR Bd2492 abolishes prey-invasion but not prey attachment. MglA deletion in Bdellovibrio greatly reduces the level of Type IV pilus formation at the single anterior pole. In contrast Keilberg and co-workers showed that M. xanthus RomR and MglB localise bipolarly asymmetrically, while MglA typically localises at the leading cell pole, during surface movement, to regulate both A- and S- motility (M. xanthus after [23]).
Evolutionary comparisons in Myxococcus and Bdellovibrio
Considering evolutionary differences that led Bdellovibrio to prey-invasion via a single pole, we also suggest that the absence of mglB in B. bacteriovorus (and the high degree of divergence of this gene in B. marinus), is because MglB is no longer required for pole switching of pili: B. bacteriovorus pili are found at only one - the non-flagellar, prey-invasive pole [4]. This is also concordant with B. bacteriovorus cells being incapable of S-motility (which would require pole-switching of T4P) and instead using T4P at a single pole for prey-invasion. However, the absence of an MglB homologue does suggest that an alternative mechanism for regulating reversals during gliding motility is likely to exist.
The mechanism by which reduced incidence of pili or a change in their retraction state is caused, in the B. bacteriovorus ΔmglABd strain, remains to be determined. Capeness and coworkers have recently shown that regulation of Bdellovibrio pilus retraction status does correlate with prey-invasion [26]. Pilus retraction occurs through secretin PilQ [47], which is required for predation in B. bacteriovorus [18]. The OM-assembly of a pilus-biogenesis protein such as PilQ could be affected by the Bd2492-5 TamAB complex activity. Alternatively, OMPs required for secretion of EPS might be perturbed at the Bdellovibrio pole, preventing pilus retraction; EPS is required for pilus retraction in M. xanthus [48]. These considerations will be the subject of a subsequent study.
The MglA/RomR-TPR interactions reported in this paper may have evolved from ancient interactions common to ancestors of M. xanthus and Bdellovibrio, and are now used in B. bacteriovorus for prey-invasion control. They may also underlie the motility and “wolf-pack” predation of Myxobacteria, but the function of the M. xanthus TPR protein homologue remains to be explored. Pioneering work by Mignot/Theodoly has shown that adhesion during gliding motility is mediated by slime deposition [14], [15] on a solid surface and that gliding directionality is controlled by MglAMx [6], [7] and other interacting proteins. In nature gliding of M. xanthus may occur on top of prey bacterial biofilms and we hypothesise that the Bd2492-5 TamAB associated system may have a role in producing vesicles, not only for gliding, but to damage prey cells as part of the M. xanthus wolf-pack lytic process.
Cyclic-di-GMP signalling at the predation control hub
In M. xanthus, chemotactic phospho-transfer signalling, involving Frz proteins, governs the localization of soluble RomRMx, MglAMx and MglBMx proteins to alternately activate or deactivate each cell pole for surface-motility directionality [23], [24]. In B. bacteriovorus, we detected an interaction between RomRBd and the CdgA GVNEF domain c-di-GMP binding protein (Figure S3) which has been shown to affect prey entry [25]. There is no Frz system in Bdellovibrio [23] but our finding that CdgA binds RomRBd (Figure S3) suggests that this c-di-GMP signalling pathway could contribute to RomRBd localization in the control of the prey-invasive pole. Further work is underway to define any signalling-link to RomRBd and CdgA from our previous observations that c-di-GMP synthases control gliding motility, predation and the switch from predatory to host-independent growth [25].
The data we present here show how the “phenotype space” and function of B. bacteriovorus MglA has diverged from that in M. xanthus. MglABd functions in the control of unipolar prey-invasion: a critical process in the predatory lifecycle of B. bacteriovorus. Our present observations indicate (Figure 7) that MglABd, RomRBd and the interacting TPR-domain protein TPRBd and TamABBd complex act at a single pole in B. bacteriovorus to facilitate prey-invasion via a mechanism that has diverged from that which controls M. xanthus S-motility.
Materials and Methods
Bacteria, plasmids and primers
Bacterial strains and plasmids used are listed in Table S1. Primers used for gene manipulation or PCR amplification are listed in Table S2.
Deletion construction
Markerless deletion strains of mglABd and bd2492 (encoding TPRBd) were generated using a modified technique of that of the Pineiro lab [49], and as described previously [25]. Construction of each mutant is described in full in Text S1.
Fluorescent protein tagging
Fluorescent protein tags were generated as described previously [25] by cloning of a whole gene fused to mCherry at the 3′ end. Construction of each tag is described fully in Text S1.
Fluorescent microscopy
To observe the fluorescence of B. bacteriovorus mCherry-tagged strains during attachment to E. coli prey cells, 1 ml of a B. bacteriovorus predatory culture (containing 2.5×108 pfu ml−1) was concentrated 20-fold and added to a microcentrifuge tube containing 30 µl CaHEPES and 40 µl E. coli S17-1 pZMR100 (from a culture grown for 16 hours at 37°C 200 rpm in YT broth supplemented with Km50) diluted to OD600 2.0 in CaHEPES, before incubating at 29°C for 5 minutes to allow attachment to occur. Cells were immobilised on a 1% agarose/CaHEPES pad and images were taken on using a Nikon Eclipse E600 epifluorescence microscope with a 100× objective lens and an hcRED filter (excitation 550 to 600 nm; emission 610 to 665 nm) with a Hamamatsu Orca ER camera. Images were analysed using Simple PCI software (version 5.3.1 Hamamatsu).
Host-independent predation, invasion and attachment assays
Procedures for attachment, invasion and predation assays of HI Bdellovibrio cells on E. coli prey are described in Text S1. 3 biological replicates were performed.
Gliding motility assay
B. bacteriovorus gliding motility was observed on 1% agarose/CaHEPES by timelapse microscopy as previously described [3]. Briefly, 1 ml of an predatory culture (containing 2.5×108 pfu ml−1) was concentrated 10-fold (HI cultures were not concentrated) and 8 µl was spotted onto the agarose pad. Measurements of gliding reversals were calculated after cells had been gliding for >1 hr.
Electron microscopy
To analyse percentages of piliated cells, each HI strain was back-diluted and grown to OD600 0.1–0.5 in PY broth at 29°C 200 rpm. Cells were then stained with 2.0% phosphotungstic acid (PTA) on carbon formvar copper grids (Agar Scientific) and analysed for the presence/absence of a pilus structure, as described previously [26].
Bacterial two-hybrid and protein co-purification
Procedures for bacterial two-hybrid and protein co-purification are described in Text S1.
Supporting Information
Tree showing co-evolution of G21-encoding mglA with mglB versus lone mglA S21 in deltaproteobacteria. (A) A Maximum Likelihood phylogenetic tree of deltaproteobacteria small subunit rRNA gene sequences: the majority of these bacteria encode an MglA with a G21 residue - these also encode an MglB homologue. Bdellovibrio bacteriovorus and Bacteriovorax marinus diverge separately from these mglB-encoding deltaproteobacteria, including Myxococcus xanthus. The B. marinus genome encodes MglA G21 and a degenerate MglB; the B. bacteriovorus genome encodes MglA with an S21 residue, but no MglB homologue. Tree generated using Phylogeny.fr [50] and rooted with Shewanella onidensis; confidence values represent approximate likelihood-ratio (aLRT) values. (B) The mglB-like gene of B. marinus (BMS_00553) is found at the same location as the mglB gene in M. xanthus (MXAN_1926; accession: YP_630170.1) (upstream of mglA). (C) B. marinus BMS_0053 has only limited sequence similarity to M. xanthus MglB (MXAN_1926).
(TIF)
MglA co-purified with hypothetical protein Bd2492 (TPRBd). SDS-PAGE on 10–20% Tris-Tricine gel with protein molecular weights (left), HID13 control (left lane) and HI MglA His8 (right lane). Differential bands are indicated by arrows A and B. Each differential band was excised and analysed by LC-MS/MS. The lower band (A, 22.2 kDa) was identified as Bd3734 (the protein bait) and the upper band (B, 40.5 kDa) was identified as Bd2492.
(TIF)
Bacterial two-hybrid shows MglA and RomR interact with Bd2492; RomR and Bd2492 interact with CdgA. A bacterial two-hybrid (BTH) assay between Bd2492 and MglA produces a positive signal on spot tests (A); the interaction between pUT18C-MglA and pKT25-Bd2492 was confirmed by beta-galactosidase assay (C). A positive result was also obtained for a BTH interaction between RomR homologue Bd2761 and Bd2492 on spot tests (A); the interaction between pUT18C-RomR pKT25-Bd2492 was confirmed by beta-galactosidase assay (C). Both RomR and Bd2492 were found to interact with CdgA (Bd3125) by BTH (A). The interactions between pKT25 Bd3125 and pUT18C-RomR or pUT18C-Bd2492 were confirmed by beta-galactosidase assay (C). When MglA and RomR interactions were assayed with tags at either end of the proteins, one combination (pUT18C-RomR and pKNT25-MglA), indicated by an asterisk (2 independent transformants) reproducibly produced a positive result on spot tests suggesting these two proteins interact (B). This interaction could not be confirmed as significant by beta-galactosidase assay, suggesting there is no interaction (as detected by BTH) between RomR and MglA. Positive control (+) = pUT18-zip and pKT25-zip and negative control (−) = pUT18C and pKT25. Error bars represent 1 SD from the mean.
(TIF)
Purification of the MglA-Bd2492-His6 complex. SDS-PAGE of fractions collected during nickel purification of the MglA-Bd2492-His6 complex expressed in E. coli cells harbouring plasmid pD2492N/3734. Soluble E. coli lysate (lane 1); insoluble material (lane 2); flow-through from nickel agarose column (lane 3); proteins eluted from column in the presence of 40 mM imidazole (lanes 4–6) and proteins eluted in the presence of 200 mM imidazole (lane 7). The positions of MglA and Bd2492-His6 on the gel are marked with arrows.
(TIF)
Predation of B. bacteriovorus Δbd2492 HI strains assayed against predatory wild-type controls. (A) Predation efficiency of the Δbd2492 HI strain was assayed against predatory and non-predatory controls by the reduction of E. coli numbers over 48 hours. Wild-type HI strain HID26 reduced E. coli numbers in liquid cultures by over four logs. The Δbd2492 HI strain showed no reduction in E. coli numbers, comparable to a known non-predatory ΔpilA HI strain, and to E. coli with no added B. bacteriovorus. (B) Reintroduction of the bd2492 ORF in cis to the Δbd2492 HI strain in plasmid pK18::bd2492 restored predatory growth. Error bars represent 1 SD from the mean Error bars represent 1 SD from the mean.
(TIF)
B. bacteriovorus genes bd2492-2495 are co-transcribed. RT-PCR on B. bacteriovorus HD100 attack-phase RNA showed that bd2492 and bd2494 (left) are co-transcribed, as are bd2494 and bd2495 (right). This suggests that the three genes are all co-transcribed in the same operon. Bd = attack-phase B. bacteriovorus RNA; Ec = E.coli S17-1 RNA; (−) no template; (+) B. bacteriovorus genomic DNA.
(TIF)
ClustalW protein alignment of M. xanthus RomR (MXAN_4461) and B. bacteriovorus putative RomR homologue Bd2761. The N-terminal REC domain and the C-terminal C-domain are highly conserved between the two proteins, whilst the Pro-rich linker region of M. xanthus RomR (MXAN_4461; accession: YP_632632.1) is not well conserved in Bd2761. A phosphorylatable aspartic acid at residue D53 of M. xanthus (red arrow) is conserved between the two proteins.
(TIF)
Plasmids and strains used in this study.
(DOCX)
Primers used in this study.
(DOCX)
Supplemental Materials and Methods.
(DOCX)
Acknowledgments
We thank members of the Sockett lab for helpful comments, Trevor Lithgow (Monash University) for helpful comments on BAM proteins and Andrew K. Fenton (Harvard University) for advice on bacterial two-hybrid.
Funding Statement
DSM was funded by a quota PhD studentship BB/F016999/1 to RES from BBSRC UK, SMB was funded by a quota PhD studentship NE/I528469/1 to RES from NERC UK. EBS was an undergraduate student at University of Nottingham. IC was funded by BBSRC grant BB/J015229/1 to ALL and RES. LEW was funded by the USDA. RES, ALL and RT were funded by the Higher Education Funding Council of England which funds UK university staff salaries. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Iida Y, Hobley L, Lambert C, Fenton AK, Sockett RE, et al. (2009) Roles of multiple flagellins in flagellar formation and flagellar growth post bdelloplast lysis in Bdellovibrio bacteriovorus . J Mol Biol 394: 1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lambert C, Evans KJ, Till R, Hobley L, Capeness M, et al. (2006) Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus . Mol Microbiol 60: 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lambert C, Fenton AK, Hobley L, Sockett RE (2011) Predatory Bdellovibrio bacteria use gliding motility to scout for prey on surfaces. J Bacteriol 193: 3139–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Evans KJ, Lambert C, Sockett RE (2007) Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J Bacteriol 189: 4850–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mahmoud KK, Koval SF (2010) Characterization of type IV pili in the life cycle of the predator bacterium Bdellovibrio . Microbiology 156: 1040–1051. [DOI] [PubMed] [Google Scholar]
- 6. Leonardy S, Miertzschke M, Bulyha I, Sperling E, Wittinghofer A, et al. (2010) Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. Embo J 29: 2276–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y, Franco M, Ducret A, Mignot T (2010) A bacterial Ras-like small GTP-binding protein and its cognate GAP establish a dynamic spatial polarity axis to control directed motility. PLoS Biol 8: e1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mauriello EM, Nan B, Zusman DR (2009) AglZ regulates adventurous (A-) motility in Myxococcus xanthus through its interaction with the cytoplasmic receptor, FrzCD. Mol Microbiol 72: 964–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang R, Bartle S, Otto R, Stassinopoulos A, Rogers M, et al. (2004) AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus . J Bacteriol 186: 6168–6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaiser D (1979) Social Gliding Is Correlated with the Presence of Pili in Myxococcus xanthus . Proc Natl Acad Sci U S A 76: 5952–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nan B, Mauriello EM, Sun IH, Wong A, Zusman DR (2010) A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol Microbiol 76: 1539–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mauriello EM, Mouhamar F, Nan B, Ducret A, Dai D, et al. (2010) Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. Embo J 29: 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun MZ, Wartel M, Cascales E, Shaevitz JW, Mignot T (2011) Motor-driven intracellular transport powers bacterial gliding motility. P Natl Acad Sci USA 108: 7559–7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ducret A, Valignat MP, Mouhamar F, Mignot T, Theodoly O (2012) Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. P Natl Acad Sci USA 109: 10036–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wartel M, Ducret A, Thutupalli S, Czerwinski F, Le Gall AV, et al. (2013) A Versatile Class of Cell Surface Directional Motors Gives Rise to Gliding Motility and Sporulation in Myxococcus xanthus . PLoS Biol 11: e1001728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bulyha I, Schmidt C, Lenz P, Jakovljevic V, Hone A, et al. (2009) Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol Microbiol 74: 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luciano J, Agrebi R, Le Gall AV, Wartel M, Fiegna F, et al. (2011) Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet 7: e1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Medina AA, Shanks RM, Kadouri DE (2008) Development of a novel system for isolating genes involved in predator-prey interactions using host independent derivatives of Bdellovibrio bacteriovorus 109J. BMC Microbiol 8: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cotter TW, Thomashow MF (1992) Identification of a Bdellovibrio bacteriovorus genetic locus, hit, associated with the host-independent phenotype. J Bacteriol 174: 6018–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horowitz AT, Kessel M, Shilo M (1974) Growth cycle of predacious Bdellovibrios in a host-free extract system and some properties of the host extract. Journal of Bacteriology 117: 270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaiser D (2003) Coupling cell movement to multicellular development in myxobacteria. Nat Rev Microbiol 1: 45–54. [DOI] [PubMed] [Google Scholar]
- 22. Blackhart BD, Zusman DR (1985) “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci U S A 82: 8767–8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keilberg D, Wuichet K, Drescher F, Sogaard-Andersen L (2012) A response regulator interfaces between the Frz chemosensory system and the MglA/MglB GTPase/GAP module to regulate polarity in Myxococcus xanthus . PLoS Genet 8: e1002951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Y, Guzzo M, Ducret A, Li YZ, Mignot T (2012) A dynamic response regulator protein modulates G-protein-dependent polarity in the bacterium Myxococcus xanthus . PLoS Genet 8: e1002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hobley L, Fung RK, Lambert C, Harris MA, Dabhi JM, et al. (2012) Discrete Cyclic di-GMP-Dependent Control of Bacterial Predation versus Axenic Growth in Bdellovibrio bacteriovorus . PLoS Pathog 8: e1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Capeness MJ, Lambert C, Lovering AL, Till R, Uchida K, et al. (2013) Activity of Bdellovibrio Hit Locus Proteins, Bd0108 and Bd0109, Links Type IVa Pilus Extrusion/Retraction Status to Prey-Independent Growth Signalling. PLoS One 8: e79759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fenton AK, Kanna M, Woods RD, Aizawa SI, Sockett RE (2010) Shadowing the Actions of a Predator: Backlit Fluorescent Microscopy Reveals Synchronous Nonbinary Septation of Predatory Bdellovibrio inside Prey and Exit through Discrete Bdelloplast Pores. J Bacteriol 192: 6329–6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fremgen SA, Burke NS, Hartzell PL (2010) Effects of site-directed mutagenesis of mglA on motility and swarming of Myxococcus xanthus . BMC Microbiol 10: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miertzschke M, Koerner C, Vetter IR, Keilberg D, Hot E, et al. (2011) Structural analysis of the Ras-like G protein MglA and its cognate GAP MglB and implications for bacterial polarity. Embo J 30: 4185–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sei S, Mussio JK, Yang QE, Nagashima K, Parchment RE, et al. (2009) Synergistic antitumor activity of oncolytic reovirus and chemotherapeutic agents in non-small cell lung cancer cells. Mol Cancer 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hartzell P, Kaiser D (1991) Upstream gene of the mgl operon controls the level of MglA protein in Myxococcus xanthus . J Bacteriol 173: 7625–7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hobley L, Lerner TR, Williams LE, Lambert C, Till R, et al. (2012) Genome analysis of a simultaneously predatory and prey-independent, novel Bdellovibrio bacteriovorus from the River Tiber, supports in silico predictions of both ancient and recent lateral gene transfer from diverse bacteria. BMC Genomics 13: 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Snyder AR, Williams HN, Baer ML, Walker KE, Stine OC (2002) 16S rDNA sequence analysis of environmental Bdellovibrio-and-like organisms (BALO) reveals extensive diversity. Int J Syst Evol Microbiol 52: 2089–2094. [DOI] [PubMed] [Google Scholar]
- 34. Karimova G, Pidoux J, Ullmann A, Ladant D (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95: 5752–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karpenahalli MR, Lupas AN, Soding J (2007) TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 37. Leonardy S, Freymark G, Hebener S, Ellehauge E, Sogaard-Andersen L (2007) Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus . Embo J 26: 4433–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seidler RJ, Starr MP (1969) Isolation and characterisation of host-independent Bdellovibrios. J Bacteriol 100: 769–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Selkrig J, Mosbahi K, Webb CT, Belousoff MJ, Perry AJ, et al. (2012) Discovery of an archetypal protein transport system in bacterial outer membranes. Nat Struct Mol Biol 19: 506–510, S501. [DOI] [PubMed] [Google Scholar]
- 40. Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, et al. (2006) YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli . Mol Microbiol 61: 151–164. [DOI] [PubMed] [Google Scholar]
- 41. Fan E, Fiedler S, Jacob-Dubuisson F, Muller M (2012) Two-partner secretion of gram-negative bacteria: a single beta-barrel protein enables transport across the outer membrane. J Biol Chem 287: 2591–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Youderian P, Hartzell PL (2006) Transposon insertions of magellan-4 that impair social gliding motility in Myxococcus xanthus . Genetics 172: 1397–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Macara IG, Lounsbury KM, Richards SA, McKiernan C, Bar-Sagi D (1996) The Ras superfamily of GTPases. FASEB Journal 10: 625–630. [DOI] [PubMed] [Google Scholar]
- 44. Rosenberg E, Keller KH, Dworkin M (1977) Cell density-dependent growth of Myxococcus xanthus on casein. J Bacteriol 129: 770–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Evans AG, Davey HM, Cookson A, Currinn H, Cooke-Fox G, et al. (2012) Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology 158: 2742–2752. [DOI] [PubMed] [Google Scholar]
- 46. Cowles KN, Moser TS, Siryaporn A, Nyakudarika N, Dixon W, et al. (2013) The putative Poc complex controls two distinct Pseudomonas aeruginosa polar motility mechanisms. Mol Microbiol 90: 923–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wolfgang M, van Putten JP, Hayes SF, Dorward D, Koomey M (2000) Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. Embo J 19: 6408–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Y, Sun H, Ma X, Lu A, Lux R, et al. (2003) Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus . Proc Natl Acad Sci U S A 100: 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Steyert SR, Pineiro SA (2007) Development of a novel genetic system to create markerless deletion mutants of Bdellovibrio bacteriovorus . Appl Environ Microbiol 73: 4717–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tree showing co-evolution of G21-encoding mglA with mglB versus lone mglA S21 in deltaproteobacteria. (A) A Maximum Likelihood phylogenetic tree of deltaproteobacteria small subunit rRNA gene sequences: the majority of these bacteria encode an MglA with a G21 residue - these also encode an MglB homologue. Bdellovibrio bacteriovorus and Bacteriovorax marinus diverge separately from these mglB-encoding deltaproteobacteria, including Myxococcus xanthus. The B. marinus genome encodes MglA G21 and a degenerate MglB; the B. bacteriovorus genome encodes MglA with an S21 residue, but no MglB homologue. Tree generated using Phylogeny.fr [50] and rooted with Shewanella onidensis; confidence values represent approximate likelihood-ratio (aLRT) values. (B) The mglB-like gene of B. marinus (BMS_00553) is found at the same location as the mglB gene in M. xanthus (MXAN_1926; accession: YP_630170.1) (upstream of mglA). (C) B. marinus BMS_0053 has only limited sequence similarity to M. xanthus MglB (MXAN_1926).
(TIF)
MglA co-purified with hypothetical protein Bd2492 (TPRBd). SDS-PAGE on 10–20% Tris-Tricine gel with protein molecular weights (left), HID13 control (left lane) and HI MglA His8 (right lane). Differential bands are indicated by arrows A and B. Each differential band was excised and analysed by LC-MS/MS. The lower band (A, 22.2 kDa) was identified as Bd3734 (the protein bait) and the upper band (B, 40.5 kDa) was identified as Bd2492.
(TIF)
Bacterial two-hybrid shows MglA and RomR interact with Bd2492; RomR and Bd2492 interact with CdgA. A bacterial two-hybrid (BTH) assay between Bd2492 and MglA produces a positive signal on spot tests (A); the interaction between pUT18C-MglA and pKT25-Bd2492 was confirmed by beta-galactosidase assay (C). A positive result was also obtained for a BTH interaction between RomR homologue Bd2761 and Bd2492 on spot tests (A); the interaction between pUT18C-RomR pKT25-Bd2492 was confirmed by beta-galactosidase assay (C). Both RomR and Bd2492 were found to interact with CdgA (Bd3125) by BTH (A). The interactions between pKT25 Bd3125 and pUT18C-RomR or pUT18C-Bd2492 were confirmed by beta-galactosidase assay (C). When MglA and RomR interactions were assayed with tags at either end of the proteins, one combination (pUT18C-RomR and pKNT25-MglA), indicated by an asterisk (2 independent transformants) reproducibly produced a positive result on spot tests suggesting these two proteins interact (B). This interaction could not be confirmed as significant by beta-galactosidase assay, suggesting there is no interaction (as detected by BTH) between RomR and MglA. Positive control (+) = pUT18-zip and pKT25-zip and negative control (−) = pUT18C and pKT25. Error bars represent 1 SD from the mean.
(TIF)
Purification of the MglA-Bd2492-His6 complex. SDS-PAGE of fractions collected during nickel purification of the MglA-Bd2492-His6 complex expressed in E. coli cells harbouring plasmid pD2492N/3734. Soluble E. coli lysate (lane 1); insoluble material (lane 2); flow-through from nickel agarose column (lane 3); proteins eluted from column in the presence of 40 mM imidazole (lanes 4–6) and proteins eluted in the presence of 200 mM imidazole (lane 7). The positions of MglA and Bd2492-His6 on the gel are marked with arrows.
(TIF)
Predation of B. bacteriovorus Δbd2492 HI strains assayed against predatory wild-type controls. (A) Predation efficiency of the Δbd2492 HI strain was assayed against predatory and non-predatory controls by the reduction of E. coli numbers over 48 hours. Wild-type HI strain HID26 reduced E. coli numbers in liquid cultures by over four logs. The Δbd2492 HI strain showed no reduction in E. coli numbers, comparable to a known non-predatory ΔpilA HI strain, and to E. coli with no added B. bacteriovorus. (B) Reintroduction of the bd2492 ORF in cis to the Δbd2492 HI strain in plasmid pK18::bd2492 restored predatory growth. Error bars represent 1 SD from the mean Error bars represent 1 SD from the mean.
(TIF)
B. bacteriovorus genes bd2492-2495 are co-transcribed. RT-PCR on B. bacteriovorus HD100 attack-phase RNA showed that bd2492 and bd2494 (left) are co-transcribed, as are bd2494 and bd2495 (right). This suggests that the three genes are all co-transcribed in the same operon. Bd = attack-phase B. bacteriovorus RNA; Ec = E.coli S17-1 RNA; (−) no template; (+) B. bacteriovorus genomic DNA.
(TIF)
ClustalW protein alignment of M. xanthus RomR (MXAN_4461) and B. bacteriovorus putative RomR homologue Bd2761. The N-terminal REC domain and the C-terminal C-domain are highly conserved between the two proteins, whilst the Pro-rich linker region of M. xanthus RomR (MXAN_4461; accession: YP_632632.1) is not well conserved in Bd2761. A phosphorylatable aspartic acid at residue D53 of M. xanthus (red arrow) is conserved between the two proteins.
(TIF)
Plasmids and strains used in this study.
(DOCX)
Primers used in this study.
(DOCX)
Supplemental Materials and Methods.
(DOCX)