Abstract
An abundant literature dealing with the population genetics and taxonomy of Giardia duodenalis, Cryptosporidium spp., Pneumocystis spp., and Cryptococcus spp., pathogens of high medical and veterinary relevance, has been produced in recent years. We have analyzed these data in the light of new population genetic concepts dealing with predominant clonal evolution (PCE) recently proposed by us. In spite of the considerable phylogenetic diversity that exists among these pathogens, we have found striking similarities among them. The two main PCE features described by us, namely highly significant linkage disequilibrium and near-clading (stable phylogenetic clustering clouded by occasional recombination), are clearly observed in Cryptococcus and Giardia, and more limited indication of them is also present in Cryptosporidium and Pneumocystis. Moreover, in several cases, these features still obtain when the near-clades that subdivide the species are analyzed separately (“Russian doll pattern”). Lastly, several sets of data undermine the notion that certain microbes form clonal lineages simply owing to a lack of opportunity to outcross due to low transmission rates leading to lack of multiclonal infections (“starving sex hypothesis”). We propose that the divergent taxonomic and population genetic inferences advanced by various authors about these pathogens may not correspond to true evolutionary differences and could be, rather, the reflection of idiosyncratic practices among compartmentalized scientific communities. The PCE model provides an opportunity to revise the taxonomy and applied research dealing with these pathogens and others, such as viruses, bacteria, parasitic protozoa, and fungi.
Author Summary
Micropathogen species definition is extremely difficult, since concepts applied to higher organisms (the biological species concept) are inadequate. In particular, the pathogens here surveyed have given rise to long-lasting controversies about their species status and that of the genotypes that subdivide them. The population genetic approach based on the predominant clonal evolution (PCE) concept proposed by us could bring simple solutions to these controversies, since it permits the description of clearly defined evolutionary entities (clonal multilocus genotypes and near-clades [incompletely isolated clades]) that could be the basis for species description, if the concerned specialists find it justified for applied research. The PCE model also provides a convenient framework for applied studies (molecular epidemiology, vaccine and drug design, clinical research) dealing with these pathogens and others.
Introduction: The Model of Predominant Clonal Evolution (PCE)
The PCE model [1], [2] defines clonal evolution as scarcity or absence of genetic recombination, a definition that is accepted by most authors working on pathogen population genetics [3], including the species here surveyed [4]–[9]. The PCE model [3], [10], [11] (i) does not presume that recombination is absent [12], [13] or plays a minor evolutionary role, but that it is too rare to break the prevalent pattern of clonality; (ii) addresses each species as a whole, and not their genetic subdivisions considered individually [14]; and (iii) definitely includes selfing/inbreeding/homogamy (which lead to restrained recombination) as particular cases of PCE, rather than as distinct evolutionary models [1]–[3], [10], [11], [15]. This view is shared by many authors working on the pathogens here analyzed [12], [16]–[19] and by others [20]. A few authors [21], [22] prefer to limit the concept of clonality to “strict” clonality (i.e., mitotic propagation) and consider that it should be distinguished from selfing/inbreeding/“unisex.” This is a matter of definition. It is nevertheless worth noting that in the examples cited in [21], differently from the authors of the article, all scientists working on parthenogenesis in insects, amphibians, fishes, and reptiles definitely include parthenogenesis in clonality.
As we have exposed extensively [1]–[3], biases that could lead to wrong conclusions of restrained recombination (mainly isolation by distance and/or time or Wahlund effect) should be carefully considered before concluding a PCE pattern.
Lastly, as we have insisted in [3], the PCE model states that restrained recombination is mainly due to built-in properties of microbes, rather than to the downstream elimination of most possible recombinants by natural selection and epistasis phenomena. If natural selection were the main factor that would maintain clonality, it would be at unacceptable costs for the organisms considered, because this would mean that most of the offspring is eliminated at each generation. Natural selection certainly acts on microbes, as it does on any organism. However, our proposal is that it cannot be the main factor responsible for PCE in organisms that would be otherwise potentially panmictic.
Recent Developments
We have recently proposed new insights about PCE, applicable to all kinds of micropathogens (including viruses, bacteria, parasites, and fungi) [3] and, more specifically, to Trypanosoma and Leishmania [10] and to Plasmodium and Toxoplasma [11]. We have proposed replacing subjective and imprecise assertions such as “recombination at a high rate” [14] or “gross incongruences” [23] with a clear-cut PCE definition relying on two complementary criteria: (i) statistically significant linkage disequilibrium (LD), or nonrandom association of genotypes occurring at different loci, and (ii) growing phylogenetic signal when more reliable data are added. Lastly, we have discussed the possibility of distinguishing PCE from cryptic biological speciation. We have also distinguished clonality by lack of available mating partners (due to scarcity of multiclonal infections) from built-in clonality.
LD is the very statistic that permits one to evidence lack of recombination, the basic definition of PCE. Contrary to segregation tests, LD analysis does not require that the organism under survey is diploid, nor does it require knowledge of ploidy [3]. This is highly relevant when micropathogens are concerned [3] since widespread aneuploidy seems to be very frequent in them, including in fungi, Trypanosoma, and Leishmania [12], which renders tests based on diploidy invalid. When a sufficient set of loci is analyzed, LD is a very powerful statistic [1].
One has to ascertain that LD cannot be explained by trivial physical obstacles (isolation by space or time: the Wahlund effect) [2]. It is widely used as circumstantial evidence for PCE by authors working on the pathogens here considered [7], [24]–[26] and by others [27], [28]. A telling consequence of LD is the spread of stable multilocus genotypes (MLGs) over vast time and space scales [3]. However, this pattern depends on the rate of evolution (molecular clock) of the marker considered and might not be observed with fast-evolving markers such as microsatellites, even in the case of strong linkage disequilibrium [3].
The criterion of a growing phylogenetic signal when more adequate data are added relies on the congruence principle [29], which states that if the working hypothesis is correct, evidence increases as more data are considered. For example, when a set of Multilocus Sequence Typing (MLST) data are considered, although some discrepancies can be observed between individual gene trees, the phylogenetic signal gets stronger and stronger when more loci are included in the combined tree. Or, the genetic distances calculated from different molecular markers are strongly correlated (the “g” test [1]). If the impact of recombination were stronger than clonal propagation in the long run, the contrary would obtain. This approach, relying on congruence, may not be verified when inadequate data are compared, such as, for example, markers with different molecular clocks or undergoing different selective pressures or different evolutionary tendencies. This could lead to wrong assertions of recombination [10]. The main manifestation of this growing phylogenetic signal is the existence of genetic subdivisions that are stable in space and time (“near-clades” [3]). The term “clade” [26], [30], [31] is not adequate when micropathogens are concerned, because even when PCE obtains, some residual recombination can always occur [3].
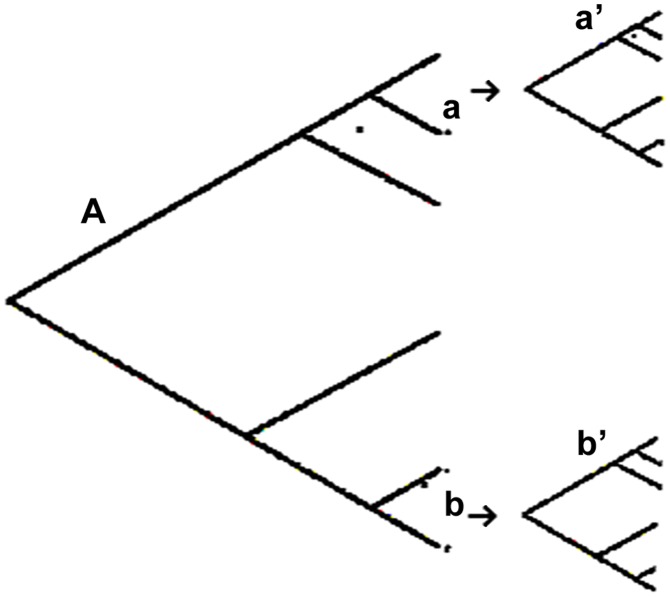
We have differentiated PCE from cryptic speciation. It has been inferred that apparent clonality could be explained by the fact that the species under study is subdivided into discrete genetic clusters, among which recombination is inhibited while it is not within them [32]. Such a model amounts to equating these genetic subdivisions to cryptic biological species. To distinguish this case from PCE, we have proposed [10] the “Russian doll model.” If the PCE criteria are uncovered, not only at the level of the whole species but also within its genetic subdivisions, it favors PCE rather than cryptic speciation. In this case, the genetic subdivisions of the species show a miniature picture of the whole species, with LD and lesser near-clades (Figure 1). However, this approach should be conveniently applied by selecting markers with an adequate resolution power (molecular clock). As a matter of fact, when addressing lesser genetic subdivisions rather than the whole species, one changes evolutionary scales. If the resolution of the markers is not consequently adapted, lack of PCE signal could be due to a statistical type II error (lack of resolution). For the same reason, the sampling size should not become too small.
Figure 1. “Russian doll” model [10].
When population genetic tests are performed with appropriate markers (of sufficient resolution) within each of the near-clades, a and b, that subdivide the species, A, under study (large tree, left part of the figure), they reveal within these near-clades a miniature picture of the whole species, with the two main PCE features, namely linkage LD and lesser near-clades (two small trees, a′ and b′, right part of the figure). This shows that PCE obtains also within the near-clades, and that these do not correspond to cryptic, potentially panmictic, biological species.
We have also discussed apparent clonality by lack of available mating partners in low transmission cycles. To explain apparent manifestations of clonality in Plasmodium falciparum [1], [2], it has been proposed that selfing/inbreeding occurred “mechanically” in low transmission areas because mixed infections of different genotypes are rare, which makes outcrossing impossible [33]. We have called this model the “starving sex hypothesis” and have shown that it was frequently at odds with the available data in P. falciparum as well as in P. vivax [11]. The alternative hypothesis [11] is that restrained recombination by selfing, inbreeding, or any other mechanism, is a built-in evolutionary strategy used by the pathogen to avoid the “recombinational load” (break-up of favorable MLGs by recombination [34]), even when different MLGs are available for mating. Inbreeding/selfing, unisexual reproduction can be considered as a way to add limited phenotypic and genotypic diversity in a clonal population without breaking favorable multilocus combinations [12], [18]. Cryptococcus and Giardia possess meiosis genes [17], [35]. However, these genes could be associated with other functions than meiosis: “Evolution is constantly re-using old genes for new purposes” [16]. We have proposed [3] that many micropathogens could possess a “clonality/sexuality machinery” rather than meiosis genes for switching between clonal evolution and recombination to face various evolutionary challenges. Selfing could be used by them instead of outcrossing, even when mating partners are available.
PCE Manifestations in the Pathogens under Survey
We have proposed [1], [2] that Giardia duodenalis and Cryptococcus neoformans undergo PCE. Contrary to Plasmodium [1], [11], this proposal did not lead to hot controversy. That clonality is strong or preponderant is accepted in Cryptococcus [5], [17], [32] and G. duodenalis [4], [36], [37] and has been proposed for Cryptosporidium hominis [7]. As a matter of fact, the main PCE manifestations are easily observable in these pathogens. A few examples among the many available include the following:
LD: It has been recorded in C. gattii [30], [38]–[40], C. neoformans [26], [30], [38], [40], [41], Pneumocystis jirovecii [25], Cr. hominis [7] and G. duodenalis [13], [37].
Widespread, stable MLGs: In C. gatti, the MLG responsible for the “Vancouver epidemics,” sequence type (ST) 39 has been isolated in Vancouver, the United States Pacific Coast, and Korea, in humans and in animals [39], [42]. It is identical to the NIH 444 strain, isolated in 1970 [43]. In C. neoformans var. grubii, the MLG ST4 has been isolated from 1996 to 2007 in six different countries in Africa and Asia. ST5 has been isolated from 1983 to 2009 in four countries in North and South America, Europe, and Asia [26]. The MLG M5 is distributed in North and South America, Asia, Europe, and Africa [44]. In Pn. jirovecii, identical MLGs have been isolated in ten different European hospitals over 9 years, and in the same patients over 8 weeks [45].
Near-clading: Near-clades are clearly identifiable in G. duodenalis [24], [36], [46], [47]. As a matter of fact, the Giardia “assemblages” are perfectly equivalent to near-clades. They are stable, widespread, and occur in sympatry, including in the same host [36]. As we have stated [3], [10], [11], the near-clades are not defined by strict phylogenetic congruence among loci, but rather, by a clear increasing phylogenetic signal when more loci are added. This is the case for Giardia assemblages, even if some discrepancies are observed among loci [48]. We have already called attention [3] to the fact that the many terms used by various authors to designate pathogen subspecific genetic subdivisions do not correspond to true different evolutionary entities and are rather a manifestation of the compartmentalization in this scientific milieu. We propose that the “assemblages,” “clusters,” “clonal groups,” and many other terms (see Table 1) correspond to a unique evolutionary entity, the near-clade. Using this only term instead of the many other ones that are now used in this field (see Table 1) has two main advantages: (i) the term near-clading has a clear evolutionary definition and (ii) the same evolutionary entity should not de designated by a wealth of different, imprecise terms. Obviously, this field of research calls for urgent semantic simplification. Near-clades are identified in Cr. hominis [7]. In the “C. neoformans complex of species” (CNC), the “molecular types” in C. neoformans VN I–IV and C. gattii VG I–IV [30]–[32] correspond to clearly delimited near-clades. The former species Pn. carinii proved to be subdivided into clearly-differentiated genotypes with strong host specificity [49], [50]. These host-specific genotypes have been given the species status, although (i) host specificity is far from absolute and (ii) indications of hybridization are recorded among them [50]. Since some indications for clonality are recorded within these genotypes [25], [45], they might be as well considered as mere near-clades.
Russian doll patterns: In C. gattii, within the cluster (near-clade) VGI, clonality obtains, and four lesser subdivisions, namely C1–4, are observed [32]. In VG II, three “clonal groups,” a, b, and c, are evidenced [31], [42]. In G. duodenalis, assemblage A shows clear subdivisions (“subassemblages”); assemblage B and other assemblages may also exhibit subdivisions, although they are less ascertained [13], [24], [47], [51], [52].
Data congruence: In the CNC, the near-clades are corroborated by Amplified Fragment Length Polymorphism (AFLP), MLST, PCR fingerprinting, Random Amplified Polymorphic DNA (RAPD), and Restriction Fragment Length Polymorphism (RFLP) [5], [8], [38]. The Giardia assemblages and their subdivisions (Russian dolls) are corroborated by Multilocus Enzyme Electrophoresis (MLEE) and sequence data [36], [47].
Table 1. The many different terms used in the pathogen population genetic literature to designate the same evolutionary entity (near-clade).
| Viruses | Bacteria | Parasitic protozoa | Fungi |
| clades | clades | assemblages | AFLP groups |
| clusters | clonal complexes | clades | clades |
| genogroups | clonal lineages | clonal lineages | clonal lineages |
| genotypes | clusters | clones | clusters |
| major genotypes | genetic groups | clusters | clonal groups |
| major lineages | genoclouds | core subgroups | genetically distinct subgroups |
| phylogenetic groups | groups | discrete typing units (DTUs) | genotypes |
| lineages | genetic groups | genotypic groups | |
| genotypes | groups | ||
| groups | lineages | ||
| haplotypes | molecular genotypes | ||
| lesser subgroups | molecular types | ||
| populations | phylogenetic species | ||
| subassemblages | subclusters | ||
| subgroups | subgenotypes | ||
| subpopulations | subgroups | ||
| subtypes | subpopulations | ||
| subtype groups | varieties | ||
| types |
Starving sex versus built-in restrained recombination
Clonality in Cryptosporidium, whose cycle includes meiosis, is generally considered explainable by lack of outcrossing opportunity due to low transmission, or starving sex [53]. However, some data do not rule out the alternative hypothesis of built-in restrained recombination, even if the data are less conclusive than for Plasmodium [11]. In Ireland, Cr. parvum is considered panmictic due to high transmission rates. However, the percentage of multiclonal infections is lower in Ireland than in other European countries such as Italy, where Cr. parvum is not panmictic [54]. In the US Midwest, Cr. parvum is overall panmictic. However, it is “epidemic” (unstable clonality [27]) in Minnesota, where the transmission is high [55]. The C. gatti widespread genotype responsible for the Vancouver epidemics is supposed to be the result of “same sex mating” between identical MLGs [38]. This results in “meiotically-derived clones undetectable by molecular approaches” [43]. However, it cannot be inferred from the data whether same-sex mating is the result of starving sex or of built-in restrained recombination.
In summary, evidence that the main PCE signs obtain is strong in G. duodenalis and the CNC. Both present striking similarities with many other pathogens, for example, Trypanosoma cruzi [10] and Toxoplasma gondii [11], [56], with significant LD; clearly delimited near-clades; ubiquitous, stable MLGs; and “Russian doll” patterns within the near-clades. Both Giardia and the CNC also present indications for limited recombination or hybridization, both within and between near-clades [36], [47], [57], and even between species in the case of the CNC [41]. As is the case for T. cruzi [58] and Toxoplasma [56], patterns of hybridization might be complex [41]. The case of Cryptosporidium is less clear. This apicomplexa genus is known to undergo a sexual phase during transmission cycles, as do Plasmodium and Toxoplasma. Indications for clonal evolution are present in some populations. One Cr. hominis MLG is dominant and widespread in the UK [59]. Some Cr. andersoni MLGs are widespread in North America and the Czech Republic [60] and in several Chinese regions [61]. LD evidence is strong in Cr. hominis [7], [59], [62] and Cr. parvum [9], [59]. However, the impact of the Wahlund effect was not taken into account in [7], [62]. Near-clading can be suspected in Cr. hominis [7], Cr. parvum [13], and Cr. muris [61], although the evidence is less clear than for Giardia and the CNC. Lastly, panmixia was inferred in some populations of Cr. parvum [54], [55]. It is possible that Cryptosporidium population structure is similar to that of P. falciparum and P. vivax [11], with a continuum between panmixia and clonality and the existence of unstable near-clades. As for Plasmodium, whether clonality is due to starving sex or in-built genetic properties should be explored in depth. Obviously, the issue of Cryptosporidium population structure deserves further investigation.
Lastly, some indications for clonality were found in Pn. jirovecii [45]. However, evidence is far too limited to reach any firm conclusions.
Implications for Molecular Epidemiology and Experimental Evolution
LD permits indirect typing; that is to say, the characterization of whole genotypes with only one gene, or a few genes. When LD is doubtful, indirect typing can be grossly misleading. This could be the case for Cryptosporidium subtyping with the unique gp60 gene [63]. If recombination is frequent, multilocus typing [64] is not a solution since frequent recombination makes the MLGs ephemeral. Still, the fact remains that the population structure of Cryptosporidium is far from being panmictic. Even if it is not strong enough to lead to stable near-clades, restrained recombination in these parasites constitutes a major stratification factor that should be taken into account in molecular epidemiology and all applied studies, as it should in Plasmodium [11].
When the evidence for PCE is clear, clonal MLGs and near-clades are convenient units of analysis for both molecular epidemiology and experimental evolution [3], thanks to their stability in space and time. Near-clades can be characterized by specific markers [13].
Taxonomical Implications
We have called attention to the fact that radically dissimilar taxonomical inferences could be drawn from similar sets of data [65]. Scientists working on the pathogens here surveyed have granted considerable attention to taxonomical problems and species definition and delimitation. The conclusions they have reached vary considerably. The PCE model allows reconsidering these questions.
Two main species concepts are involved in these debates: the biological species concept (BSC) [66] and the phylogenetic species concept (PSC) [67]. The BSC demands two criteria: (i) within the species, genetic flow should have no other limitations than physical obstacles (potential panmixia) and (ii) it should be inhibited between species by built-in biological mechanisms. The PSC stipulates that species should correspond to clades, between which, by definition, gene flow is interrupted. Generally, authors propose a mix of genetic and biological characteristics to define species [68]. Some attempts have been made to apply the BSC concept to the CNC: experiments have shown that crosses within C. gattii VG II are easy, while they are difficult between II and III [31]. The authors have proposed that II and III deserve the status of biological species. This is debatable for two reasons: (i) experiments tell nothing about the frequency of recombination in nature [3] and (ii) the presence of stable genetic subdivisions (Russian doll near-clades) in VG II [31], [42] clearly shows that VG II is not a potentially panmictic entity. Also, by the survey of natural populations, it has been proposed [5] to equate the CNC “genotypic groups” to biological species. Nevertheless, as shown above, many PCE manifestations are observed within these groups.
The BSC has been proposed for the Cryptosporidum species [64], although, as we have seen above, recombination is restrained in some populations of this parasite.
Attributing the species status to the Giardia assemblages still is a matter of debate [4], [6], [16], [69], [70].
Lastly, as we have seen, the host-specific Pneumocystis genotypes are now considered as distinct species, although they could be equated, as well, to near-clades.
We propose that the BSC is not applicable to most, if not all, micropathogens. First, even between different species, very often, some genetic exchange occurs. Second, more importantly, clonality occurring in many populations of micropathogens makes it impossible to consider them as potentially panmictic units.
The PCE concept, and more specifically, the near-clade and Russian doll models, give an opportunity to apply the PSC to most pathogen species. The flexible phylogenetic approach based on the congruence principle relaxes the demands of a strict cladistic approach. The near-clades can be the starting units (necessary, but not sufficient) for species description based on the PSC adapted to the special case of micropathogens (lack of strictly separated intraspecific clades). It would then be the decision of specialists working on the considered pathogen to decide whether the specific biological properties and medical relevance of the near-clades (host specificity, pathogenicity, and drug resistance) justify that they be described as new species.
Conclusion
We have provided clear evidence that the PCE model as it is formulated in the present study is verified in many pathogens, including viruses, bacteria, parasitic protozoa, and fungi [1]–[3], [10], [11]. The PCE model provides a convenient population genetics framework for all applied studies (strain typing, vaccine and drug design, and molecular and immunological diagnosis) dealing with the pathogens here surveyed and for experimental evolution. As a matter of fact, it provides these studies with stable, clearly defined units of analysis (clonal MLGs, near-clades). Moreover, it might bring a renewal of the long-lasting controversies concerning the species status of Cryptosporidium, Giardia, Cryptococcus, and Pneumocystis.
Funding Statement
This study was supported by the ANR Clonix project (ANR-11-BSV7-007). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tibayrenc M, Kjellberg F, Ayala FJ (1990) A clonal theory of parasitic protozoa: the population structure of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas and Trypanosoma, and its medical and taxonomical consequences. Proc Nat Acad Sci U S A 87: 2414–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tibayrenc M, Kjellberg F, Arnaud J, Oury B, Brenière SF, et al. (1991) Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci U S A 88: 5129–5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tibayrenc M, Ayala FJ (2012) Reproductive clonality of pathogens: A perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc Nat Acad Sci U S A 109: E3305–E3313 doi:10.1073/pnas.1212452109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersson JO (2012) Double peaks reveal rare diplomonad sex. Trends Parasitol 28: 46–52. [DOI] [PubMed] [Google Scholar]
- 5. Bovers M, Hagen F, Kuramae EE, Boekhout T (2008) Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet Biol 45: 400–421. [DOI] [PubMed] [Google Scholar]
- 6. Cooper MA, Adam RD, Worobey M, Sterling CR (2007) Population Genetics Provides Evidence for Recombination in Giardia . Curr Biol 17: 1984–1988. [DOI] [PubMed] [Google Scholar]
- 7. Gatei W, Das P, Dutta P, Sen A, Cama V, et al. (2007) Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect Genet Evol 7: 197–205. [DOI] [PubMed] [Google Scholar]
- 8. Lin X, Heitman J (2006) The Biology of the Cryptococcus neoformans Species Complex. Ann Rev Microbiol 60: 69–105. [DOI] [PubMed] [Google Scholar]
- 9. Morrison LJ, Mallon ME, Smith HV, MacLeod A, Xiao L, et al. (2008) The population structure of the Cryptosporidium parvum population in Scotland: A complex picture. Infect Genet Evol 8: 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tibayrenc M, Ayala FJ (2013) How clonal are Trypanosoma and Leishmania? Trends Parasitol 29: 264–269. [DOI] [PubMed] [Google Scholar]
- 11. Tibayrenc M, Ayala FJ (2014) New insights into Clonality and Panmixia in Plasmodium and Toxoplasma . Adv Parasitol 84: 253–268. [DOI] [PubMed] [Google Scholar]
- 12. Calo S, Billmyre BB, Heitman J (2013) Generators of Phenotypic Diversity in the Evolution of Pathogenic Microorganisms. PLoS Pathog 9: e1003181 doi:10.1371/journal.ppat.1003181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ortega-Pierres G, Smith HV, Caccio SM, Thompson RC (2009) New tools provide further insights into Giardia and Cryptosporidium biology. Trends Parasitol 25: 410–416. [DOI] [PubMed] [Google Scholar]
- 14. Ramírez JD, Tapia-Calle G, Guhl F (2013) Genetic structure of Trypanosoma cruzi in Colombia revealed by a High-throughput Nuclear Multilocus Sequence Typing (nMLST) approach. BMC Genet 14: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tibayrenc M, Ayala FJ (2013) Unisexual reproduction is a particular case of clonality. Comment on: Feretzaki F, Heitman J. (2013) Unisexual Reproduction Drives Evolution of Eukaryotic Microbial Pathogens. PLoS Pathog 9: e1003674 doi:10.1371/journal.ppat.1003674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Birky CW Jr (2009) Giardia Sex? Yes, but how and how much? Trends Parasitol 26: 70–74. [DOI] [PubMed] [Google Scholar]
- 17. Heitman J (2006) Sexual reproduction and the evolution of microbial pathogens. Curr Biol 16: R711–R725. [DOI] [PubMed] [Google Scholar]
- 18. Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, et al. (2013) Unisexual and Heterosexual Meiotic Reproduction Generate Aneuploidy and Phenotypic Diversity De Novo in the Yeast Cryptococcus neoformans . PLoS Biol 11: e1001653 doi:10.1371/journal.pbio.1001653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu J (2006) Fundamentals of Fungal Molecular Population Genetic Analyses. Curr Issues Mol Biol 8: 75–80. [PubMed] [Google Scholar]
- 20. Lehtonen J, Schmidt DJ, Heubel K, Kokko H (2013) Evolutionary and ecological implications of sexual parasitism. Trends Ecol Evol 28: 297–306. [DOI] [PubMed] [Google Scholar]
- 21. Feretzaki F, Heitman J (2013) Unisexual Reproduction Drives Evolution of Eukaryotic Microbial Pathogens. PLoS Pathog 9: e1003674 doi:10.1371/journal.ppat.1003674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rougeron V, De Meeûs T, Kako Ouraga S, Hide M, Bañuls AL (2010) “Everything You Always Wanted to Know about Sex (but Were Afraid to Ask)” in Leishmania after Two Decades of Laboratory and Field Analyses. PLoS Pathog 6: e1001004 doi:10.1371/journal.ppat.1001004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Messenger LA, Llewellyn MS, Bhattacharyya T, Franzén O, Lewis MD, et al. (2012) Multiple Mitochondrial Introgression Events and Heteroplasmy in Trypanosoma cruzi Revealed by Maxicircle MLST and Next Generation Sequencing. PLoS Negl Trop Dis 6: e1584 doi:10.1371/journal.pntd.0001584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cacciò SM, Ryan U (2008) Molecular epidemiology of giardiasis. Molec Biochem Parasitol 160: 75–80. [DOI] [PubMed] [Google Scholar]
- 25. Esteves F, Gaspar J, Tavares A, Moser I, Antunes F, et al. (2010) Population structure of Pneumocystis jirovecii isolated from immunodeficiency virus-positive patients. Infect Genet Evol 10: 192–199. [DOI] [PubMed] [Google Scholar]
- 26. Khayhan K, Hagen F, Pan W, Simwami S, Fisher MC, et al. (2013) Geographically Structured Populations of Cryptococcus neoformans Variety grubii in Asia Correlate with HIV Status and Show a Clonal Population Structure. PLoS ONE 8: e72222 doi:10.1371/journal.pone.0072222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maynard Smith J, Smith NH, O'Rourke M, Spratt BG (1993) How clonal are bacteria? Proc Natl Acad Sci U S A 90: 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schurko AM, Neiman M, Logsdon JM Jr (2008) Signs of sex: what we know and how we know it. Trends Ecol Evol 2: 208–217. [DOI] [PubMed] [Google Scholar]
- 29.Avise JC (2004) Molecular markers, Natural History and Evolution. 2nd ed. New York, London: Chapman & Hall. [Google Scholar]
- 30. Ngamskulrungroj P, Gilgado F, Faganello J, Litvintseva AP, Leal AL, et al. (2009) Genetic Diversity of the Cryptococcus Species Complex Suggests that Cryptococcus gattii Deserves to Have Varieties. PLoS ONE 4: e5862 doi:10.1371/journal.pone.0005862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Voelz K, Ma H, Phadke S, Byrnes EJ, Zhu P, et al. (2013) Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen Cryptococcus gattii . PLoS Genet 9: e1003771 doi:10.1371/journal.pgen.1003771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Campbell LT, Currie BJ, Krockenberger M, Malik R, Meyer W, et al. (2005) Clonality and Recombination in Genetically Differentiated Subgroups of Cryptococcus gattii . Eukaryotic Cell 4: 1403–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, et al. (2000) Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum . Mol Biol Evol 17: 1467–1482. [DOI] [PubMed] [Google Scholar]
- 34. Agrawal AF (2006) Evolution of Sex: Why Do Organisms Shuffle Their Genotypes? Curr Biol 16: R696–R704. [DOI] [PubMed] [Google Scholar]
- 35. Birky CW (2005) Sex: is Giardia doing it in the dark? Curr Biol 15: R56–R58. [DOI] [PubMed] [Google Scholar]
- 36. Monis PT, Caccio SM, Thompson RCA (2009) Variation in Giardia: towards a taxonomic revision of the genus. Trends Parasitol 25: 93–100. [DOI] [PubMed] [Google Scholar]
- 37. Takumi K, Swart A, Mank T, Lasek-Nesselquist E, Lebbad M, et al. (2012) Population-based analyses of Giardia duodenalis is consistent with the clonal assemblage structure. Parasit Vectors 5: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Campbell LT, Carter DE (2006) Looking for sex in the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii . FEMS Yeast Res 6: 588–598. [DOI] [PubMed] [Google Scholar]
- 39. Carriconde F, Gilgado F, Arthur I, Ellis D, Malik R, et al. (2011) Clonality and α-a Recombination in the Australian Cryptococcus gattii VGII Population - An Emerging Outbreak in Australia. PLoS ONE 6: e16936 doi:10.1371/journal.pone.0016936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chowdhary A, Hiremath SS, Sun S, Kowshik T, Randhawa HS, et al. (2011) Genetic differentiation, recombination and clonal expansion in environmental populations of Cryptococcus gattii in India. Environm Microbiol 13: 1875–1888. [DOI] [PubMed] [Google Scholar]
- 41. Li W, Averette AF, Desnos-Ollivier M, Ni M, Dromer F, et al. (2012) Genetic Diversity and Genomic Plasticity of Cryptococcus neoformans AD Hybrid Strains. G3 (Bethesda) 2: 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chaturvedi V, Chaturvedi S (2011) Cryptococcus gattii: a resurgent fungal pathogen. Trends Microbiol 19: 564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, et al. (2005) Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437: 1360–1364. [DOI] [PubMed] [Google Scholar]
- 44. Litvintseva AP, Mitchell TG (2012) Population Genetic Analyses Reveal the African Origin and Strain Variation of Cryptococcus neoformans var. grubii . PLoS Pathog 8: e1002495 doi:10.1371/journal.ppat.1002495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matos O, Esteves F (2010) Pneumocystis jirovecii multilocus gene sequencing: findings and implications. Future Microbiol 5: 1257–1267. [DOI] [PubMed] [Google Scholar]
- 46. Cacciò SM, Sprong H (2010) Giardia duodenalis: Genetic recombination and its implications for taxonomy and molecular epidemiology. Exp Parasitol 124: 107–112. [DOI] [PubMed] [Google Scholar]
- 47. Feng Y, Xiao L (2011) Zoonotic Potential and Molecular Epidemiology of Giardia Species and Giardiasis. Clin Microbiol Rev 24: 110 doi:10.1128/CMR.00033-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ryan A, Cacciò SM (2013) Zoonotic potential of Giardia . Int J Parasitol 43: 943–956. [DOI] [PubMed] [Google Scholar]
- 49. Mazars E, Guyot K, Durand I, Dei-Cas E, Boucher S, et al. (1997) Isoenzyme Diversity in Pneumocystis carinii from Rats, Mice, and Rabbits. J Infect Dis 175: 655–660. [DOI] [PubMed] [Google Scholar]
- 50. Aliouat-Denis CM, Chabé M, Demanche C, Aliouat EM, Viscogliosi E, et al. (2008) Pneumocystis species, co-evolution and pathogenic power. Infect Genet Evol 8: 708–726. [DOI] [PubMed] [Google Scholar]
- 51. Plutzer J, Ongerth J, Karanis P (2010) Giardia taxonomy, phylogeny and epidemiology: Facts and open questions. Int J Hyg Environm Health 213: 321–333. [DOI] [PubMed] [Google Scholar]
- 52. Wielinga CM, Thompson RCA (2007) Comparative evaluation of Giardia duodenalis sequence data. Parasitol 134: 1795–1821. [DOI] [PubMed] [Google Scholar]
- 53. Widmer G, Sullivan S (2012) Genomics and population biology of Cryptosporidium species. Parasite Immunol 34: 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. De Waele V, Van den Broeck F, Huyse T, McGrath G, Higgins I, et al. (2013) Panmictic Structure of the Cryptosporidium parvum Population in Irish Calves: Influence of Prevalence and Host Movement. Appl Environ Microbiol 79: 2534–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Herges GR, Widmer G, Clark ME, Khan E, Giddings CW, et al. (2012) Evidence that Cryptosporidium parvum Populations Are Panmictic and Unstructured in the Upper Midwest of the United States. Appl Environ Microbiol 78: 8096–8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Su C, Khan A, Zhou P, Majumdara D, Ajzenberg D, et al. (2012) Globally diverse Toxoplasmagondii isolates comprise six major clades originating from a small number of distinct ancestral lineages. Proc Nat Acad Sci U S A 109: 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu J (2006) Microbial ecology in the age of genomics and metagenomics: concepts, tools, and recent advances. Molec Ecol 15: 1713–1731. [DOI] [PubMed] [Google Scholar]
- 58. Zingales B, Miles MA, Campbell D, Tibayrenc M, Macedo AM, et al. (2012) The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol 12: 240–253. [DOI] [PubMed] [Google Scholar]
- 59. Tanrıverdi S, Grinberg A, Chalmers RM, Hunter PR, Petrovic Z, et al. (2008) Inferences about the Global Population Structures of Cryptosporidium parvum and Cryptosporidium hominis . Appl Env Microbiol 74: 7227–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Feng Y, Yang W, Ryan U Zhang L, Kvác M, et al. (2011) Development of a Multilocus Sequence Tool for Typing Cryptosporidium muris and Cryptosporidium andersoni . J Clin Microbiol 49: 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang R, Jian F, Zhang L, Ning C, Liu A, et al. (2012) Multilocus Sequence Subtyping and Genetic Structure of Cryptosporidium muris and Cryptosporidium andersoni . PLoS ONE 7: e43782 doi:10.1371/journal.pone.0043782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ngouanesavanh T, Guyot K, Certad G, Le Fichoux Y, Chartier C, et al. (2006) Cryptosporidium Population Genetics: Evidence of Clonality in Isolates from France and Haiti. J Euk Microbiol 53: S33–S36. [DOI] [PubMed] [Google Scholar]
- 63. Xiao L (2010) Molecular epidemiology of cryptosporidiosis: An update. Exp Parasitol 124: 80–89. [DOI] [PubMed] [Google Scholar]
- 64. Widmer G, Lee Y (2010) Comparison of Single- and Multilocus Genetic Diversity in the Protozoan Parasites Cryptosporidium parvum and C. hominis . Appl Environm Microbiol 76: 6639–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tibayrenc M (1993) Entameba, Giardia and Toxoplasma: clones or cryptic species? Parasitol Today 9: 102–105. [DOI] [PubMed] [Google Scholar]
- 66.Dobzhansky T (1937) Genetics and the origin of species. New York: Columbia University Press. [Google Scholar]
- 67.Cracraft J (1983) Species concept and speciation analysis. In: Johnson RF, editor. Current ornithology. New York: Plenum Press. pp. 159–187 [Google Scholar]
- 68. Fayer R (2010) Taxonomy and species delimitation in Cryptosporidium . Exp Parasitol 124: 90–97. [DOI] [PubMed] [Google Scholar]
- 69. Lasek-Nesselquist E, Welch DM, Thompston RCA, Steuart RF, Sogin ML (2009) Genetic Exchange Within and Between Assemblages of Giardia duodenalis . J Euk Microbiol 56: 504–518. [DOI] [PubMed] [Google Scholar]
- 70. Xu F, Jerlström-Hultqvist J, Andersson JO (2012) Genome-Wide Analyses of Recombination Suggest That Giardia intestinalis Assemblages Represent Different Species. Mol Biol Evol 29: 2895–2898. [DOI] [PubMed] [Google Scholar]