Abstract
Several small-molecule CDK inhibitors have been identified, but none have been approved for clinical use in the past few years. A new series of 4-[(3-hydroxybenzylamino)-methylene]-4H-isoquinoline-1,3-diones were reported as highly potent and selective CDK4 inhibitors. In order to find more potent CDK4 inhibitors, the interactions between these novel isoquinoline-1,3-diones and cyclin-dependent kinase 4 was explored via in silico methodologies such as 3D-QSAR and docking on eighty-one compounds displaying potent selective activities against cyclin-dependent kinase 4. Internal and external cross-validation techniques were investigated as well as region focusing, bootstraping and leave-group-out. A training set of 66 compounds gave the satisfactory CoMFA model (q 2 = 0.695, r 2 = 0.947) and CoMSIA model (q 2 = 0.641, r 2 = 0.933). The remaining 15 compounds as a test set also gave good external predictive abilities with r 2 pred values of 0.875 and 0.769 for CoMFA and CoMSIA, respectively. The 3D-QSAR models generated here predicted that all five parameters are important for activity toward CDK4. Surflex-dock results, coincident with CoMFA/CoMSIA contour maps, gave the path for binding mode exploration between the inhibitors and CDK4 protein. Based on the QSAR and docking models, twenty new potent molecules have been designed and predicted better than the most active compound 12 in the literatures. The QSAR, docking and interactions analysis expand the structure-activity relationships of constrained isoquinoline-1,3-diones and contribute towards the development of more active CDK4 subtype-selective inhibitors.
Introduction
Cyclin-dependent kinases (CDKs), a family of serine/threonine protein kinases, play a central role in the growth, development, proliferation and death of eukaryotic cells [1], [2]. There are more than 13 CDKs of which 12 different cyclin families have been identified up to now, and different CDK/cyclin combinations are active during each phase of the cell cycle [3]–[5]. Among these CDK/cyclin complexes, the D/CDK4 and E/CDK2 complexes have been greatly concerned [6]. In the G1-S phase transition, the retinoblastoma susceptibility gene family of proteins (Rb) was phosphorylated by the D-type cyclins (D1, D2 or D3) in combination with CDK4 and cyclin E/CDK2 complexes. Phosphorylation of the Rb activated the E2F transcription factors and resulted in the transcription of genes required for DNA synthesis. This kind of function exerted by D/CDK4 and E/CDK2 complexes is positively regulated by the mitogenic signaling pathways and negatively regulated by the cyclin-dependent kinase inhibitors (CKIs) [7]–[13]. Inhibition of cyclin-dependent kinases (CDKs) with small molecules has been suggested as a strategy for treatment of cancer, based on deregulation of CDKs commonly found in many types of human tumors. Selective CDK inhibitors such as CYC-202 [14] and BMS-387032 [15] targeting CDK2, and PD0332991 [16] targeting CDK4 have been under clinical evaluations. Recently, a series of novel isoquinoline-1, 3-(2H, 4H)-diones have been found to possess excellent selective inhibitory activity against the CDK4 [7], [8].
The three-dimensional quantitative structure-activity relationship (3D-QSAR) models derived from the most widely used computational methods, CoMFA (comparative molecular field analysis) and CoMSIA (comparative molecular similarity indices analysis), could be used to guide rational synthesis of potent novel inhibitors and now aimed to elucidate the structural features required for CDK4 inhibitors. The best developed models have been duly validated by a systemic external validation, on the basis of which a set of twenty new potent molecules have been designed and predicted stronger activity than 12, the most active compound reported in the literatures[7], [8].
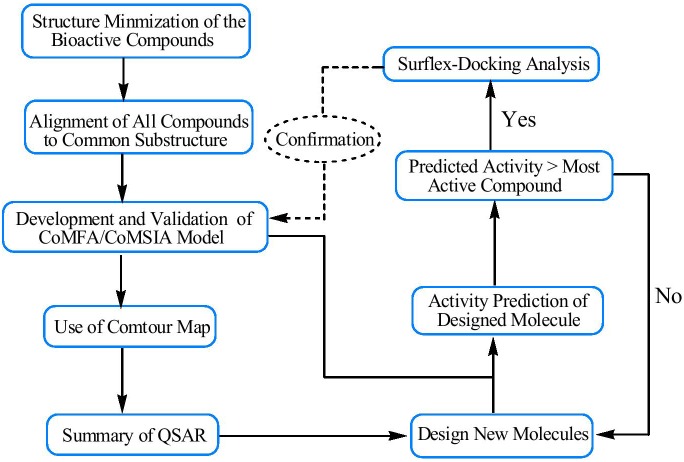
Molecular docking techniques have been extensively used as an important tool in the discovery of new small-molecule drugs for targeting proteins.[17]–[21] Based on an idealized representation that a ligand makes every potential interaction with the binding sites, docking uses an incremental construction algorithm to place flexible ligands into a fully specified active site. Surflex-Dock is particularly successful at eliminating false positive results and therefore used to narrow down the screening pool significantly, while retaining a large number of active compounds. The binding interactions of the isoquinolinedione derivatives within the CDK4 active sites were discussed. The step-wise description of methodology used for 3D-QSAR analysis, molecular docking and designing of new CDK4 inhibitors is as shown in Figure 1.
Figure 1. Step-wise description ofmethodology used for 3D-QSAR analysis, molecular docking and designing of new inhibitors for CDK4.
Materials and Methods
Data Set
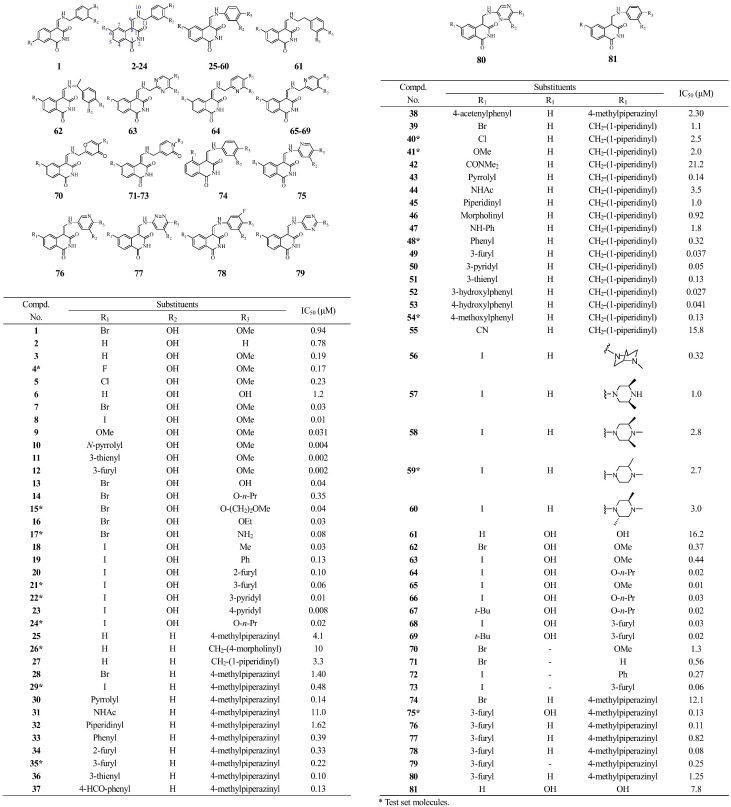
All isoquinoline-1,3-(2H,4H)-dione derivatives and their biological activities were collected from literatures [7], [8] (Figure 2). A total set of 81 molecules were randomly segregated into training and test sets comprising 66 and 15 molecules, respectively, based on the following rules: (i) Diversity of the molecules was very necessary to assess the statistical significance. (ii) To avoid any redundancy or bias in terms of structure features and activity range, the information of the selected compounds must be clear and concise. (iii) The most active and least active compounds should be included in the training set. The activities of the CDK4 inhibitors were reported in IC50 and converted to pIC50 by taking Log (1/IC50) for the convenience. The activity range from 4.6 to 8.6 log units of these compounds provided a broad and homogenous data set for 3D-QSAR study. In general, the spread of activity should cover at least 3 log units for a reliable 3D-QSAR model [22].
Figure 2. Chemical structures and IC50 values of the training and test set molecules.
Molecular Modeling and Alignment
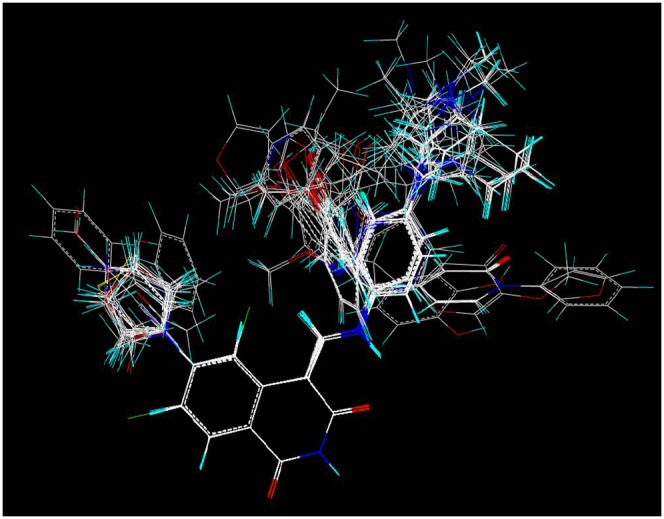
The structures of the derivatives were sketched in SYBYL 8.1 (Tripos, Inc., St. Louis, MO, USA) molecular modeling package and Gasteiger-Hückel charges were assigned to the atoms of all the compounds. A good alignment is the most essential for the quality and the predictive ability of CoMFA and CoMSIA models [23], and common substructure, pharmacophore or docking overlaps can be available to align molecules [24], [25]. The isoquinoline-1, 3-(2H, 4H)-dione ring with structural rigidity was selected as the common substructure and the compound 12 with the strongest inhibitory activity as the template molecule (Figure 3 ). It can be seen that all the compounds studied have similar active conformations.
Figure 3. Alignment of the compounds used in the training set.
CoMFA and CoMSIA Setup
Three-dimensional grid spacing was set at 2 Å in the x, y, and z directions. The steric and electrostatic field energies were calculated using the Lennard-Jones and the Coulomb potentials [26]. For CoMFA method, a sp 3 hybridized carbon atom with a+1 charge was identified as the probe atom to determine the magnitude of the steric and electrostatic field values, whose truncation was set at 30 kcal/mol [27]–[29].
The CoMSIA method, similar to CoMFA in terms of fields around the molecule, was based on the assumption that changes of ligands in binding affinities are associated with changes of molecular properties. Besides steric and electrostatic fields, three other different fields of hydrophobic, hydrogen bond donor and hydrogen bond acceptor are also calculated [30]. Moreover, a Gaussian function introduced in similarity indices makes it be calculated at all grid points, inside and outside different molecular surfaces. Equation used to calculate the similarity indices is as follows:
Where, A is the similarity index at grid point q, summed over all atoms i of the molecule j under investigation. W probe, k is the probe atom with radius 1 Å, charge +1, hydrophobicity +1, hydrogen bond donating +1 and hydrogen bond accepting +1. W ik is the actual value of the physicochemical property k of atom i. r iq is the mutual distance between the probe atom at grid point q and atom i of the test molecule. α is the attenuation factor whose optimal value is normally between 0.2 and 0.4, with a default value of 0.3 [31].
Partial Least Squares (PLS) Analysis
For Partial Least Squares (PLS) analysis [32], the “leave-one-out” cross-validation method was first carried out to generate a cross-validated r 2 (q 2) value and the optimal number of components (ONC), based on the lowest standard error of prediction (SEP) which usually corresponds to the highest cross-validated squared coefficient (q 2). To avoid over-fitting the models, a higher component was accepted only when the q 2 differences between two components was larger than 10% [24]. Region focusing was performed to maximize q 2 value by rotating the extracted principal components [33]. The q 2 is a good indicator of the accuracy of actual predictions and a q 2 value of 0.5 means halfway between no model and a perfect model [34]. Non-cross-validation was then executed to establish the final 3D-QSAR model after the optimal number of components was determined. The consequential final PLS models gave the conventional correlation coefficient (r 2), standard errors of estimate (SEE) and F ratio between the variances of calculated and observed activities. The equation for SEE is given below
Where, n and c is the number of compounds and components, respectively, and PRESS is the sum of squared deviations between predicted and actual activity values for each molecule in the test set.
External Validation
q
2 is often a useful but not sufficient criterion for model validation. In many cases, a model with high r2cv and r
2 values were proved to be inaccurate. Even though a model may exhibit a good predictive ability based on the statistics for the test set, it is not always sure that the model will perform well on a new set of data [35]. Therefore, an external test sets (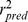 ) [36] was recommended for the estimation of predictive ability. Predictive values
) [36] was recommended for the estimation of predictive ability. Predictive values 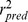 were calculated as follows:
were calculated as follows:
Therein, SD is the sum of squared differences between the measured activities of the test set and the average measured activity of the training set.
Several other statistics such as r 2 m, r 2 0, R and k were calculated using the following equations, and 3D-QSAR models were considered acceptable only if they satisfy the following conditions: r2cv>0.5, r2>0.6, [(r2−r02)/r2] <0.1, 0.85≤k≤1.15 and r 2 m>0.5 [36], [37].
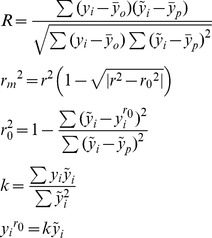 |
Where, yi and 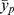 are the actual and predicted activities, respectively.
are the actual and predicted activities, respectively. 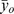 and
and 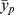 are the average values of the observed and predicted pIC50 values of the test set molecules, respectively. r2 is the non-cross-validated correlation coefficient from PLS process.
are the average values of the observed and predicted pIC50 values of the test set molecules, respectively. r2 is the non-cross-validated correlation coefficient from PLS process.
Molecular Docking
Surflex-Dock in SYBYL 8.1, using a patented search engine and an empirical scoring function to dock ligands into a protein’s binding site [19], was applied to study molecular docking in the present paper. The crystal structure of CDK4 with ligand 1GIH was retrieved from the RCSB Protein Data Bank [38]. A protomol, a computational representation of the receptor’s binding cavity to which putative ligands are aligned, was generated automatically with a threshold parameter of 0.31 and a bloat parameter of 1 Å, and composed of a collection of fragments or probe molecules such as CH4, N-H, and C = O that characterize steric effects in the binding pocket, hydrogen bond donor and acceptor groups, respectively.[39], [40] All the water molecules and sulfate salt in CDK4 1GIH (receptor) were deleted, and hydrogen atoms and Gasteiger charges were added [41], [42]. All of the eighty-one ligands were docked sequentially into the binding pocket of CDK4 using the parameters previously optimized. Surflex-Dock total scores are expressed in log10(Kd) to represent binding affinities. The scores of 10 docked conformers of each isoquinolinedione derivatives were ranked in a molecular spreadsheet, and the highest total score was taken into consideration for ligand-receptor interactions. To visualize the binding mode between the protein and ligands, the MOLCAD (Molecular Computer Aided Design) program was applied to calculate and display the surfaces of channels and cavities, as well as the separating surface between protein subunits. MOLCAD program provides several types to create a molecular surface, in which the Robbin surfaces illustrating the secondary structure elements of the binding structure was applied to build the MOLCAD Robbin and Multi-Channel surfaces displayed with several potentials. Other parameters were established in default.
Results and Discussion
CoMFA and CoMSIA Analysis
The results taken from the PLS analysis were summarized in Table 1 . The actual versus the predicted pIC50 values for the training and the test set molecules were listed in Table 2 and depicted graphically in Figure 4. For the CoMFA model after region focusing, the leave-one-out cross-validated q 2 value was 0.695 (>0.5) and non-cross-validated r 2 value was 0.947 with an optimized component of 6, standard error estimate (SEE) of 0.185 and F value of 139.423. Contributions of steric and electrostatic fields were 0.479 and 0.521, respectively.
Table 1. PLS results of CoMFA and CoMSIA models.
| CoMFA(before region focusing) | CoMFA (after region focusing) | CoMSIA | |
| PLS statistics | |||
| LOO cross q 2/SEP | 0.544/0.727 | 0.695/0.348 | 0.641/0.496 |
| Group cross q 2/SEP | 0.578/0.706 | 0.711/0.334 | 0.638/0.498 |
| Non-validated r 2/SEE | 0.914/0.294 | 0.947/0.185 | 0.933/0.210 |
| F | 104.511 | 139.423 | 121.534 |
| r 2 bootstrap | 0.916±0.023 | 0.965±0.010 | 0.928±0.015 |
| Sbootstrap | 0.285±0.098 | 0.152±0.071 | 0.231±0.135 |
| Optimal components | 6 | 6 | 6 |
| Field distribution% | |||
| Steric | 45.0 | 47.9 | 16.0 |
| Electrostatic | 55.0 | 52.1 | 25.2 |
| Hydrophobic | 18.9 | ||
| H-bond Donor | 10.1 | ||
| H-bond Acceptor | 29.8 |
Table 2. The actual pIC50, predicted pIC50 (Pred.) and their residuals (Res.) of the training and test set molecules.
| Compd. | pIC50 | CoMFA | CoMSIA | ||
| No. | Actual | Pred. | Res. | Pred. | Res. |
| 1 | 6.027 | 5.905 | −0.122 | 6.329 | 0.302 |
| 2 | 6.108 | 6.099 | −0.009 | 6.005 | −0.103 |
| 3 | 6.721 | 6.801 | 0.080 | 6.579 | −0.142 |
| 4 * | 6.770 | 6.568 | −0.102 | 6.556 | −0.114 |
| 5 | 6.638 | 6.715 | 0.077 | 6.734 | 0.096 |
| 6 | 5.921 | 5.893 | −0.028 | 6.210 | 0.289 |
| 7 | 7.523 | 7.482 | −0.041 | 7.601 | 0.078 |
| 8 | 8.000 | 7.907 | −0.093 | 7.693 | −0.307 |
| 9 | 7.509 | 7.605 | 0.096 | 7.309 | −0.200 |
| 10 | 8.398 | 8.474 | 0.076 | 8.511 | 0.113 |
| 11 | 8.699 | 8.759 | 0.060 | 8.559 | −0.140 |
| 12 | 8.699 | 8.736 | 0.037 | 8.368 | −0.331 |
| 13 | 7.398 | 7.411 | 0.013 | 7.223 | −0.175 |
| 14 | 6.456 | 6.806 | 0.350 | 6.931 | 0.475 |
| 15 * | 7.398 | 7.189 | −0.209 | 7.098 | −0.300 |
| 16 | 7.523 | 7.205 | −0.318 | 7.167 | −0.356 |
| 17 * | 7.097 | 7.089 | −0.008 | 7.193 | 0.096 |
| 18 | 7.523 | 7.401 | −0.122 | 7.347 | −0.176 |
| 19 | 6.886 | 7.209 | 0.323 | 6.400 | −0.486 |
| 20 | 7.000 | 6.890 | −0.110 | 7.208 | 0.208 |
| 21 * | 7.222 | 6.800 | −0.422 | 7.377 | 0.155 |
| 22 * | 8.000 | 7.823 | −0.177 | 7.633 | −0.367 |
| 23 | 8.097 | 7.907 | −0.190 | 7.911 | −0.186 |
| 24 * | 7.699 | 7.409 | −0.260 | 7.205 | −0.494 |
| 25 | 5.397 | 5.517 | 0.120 | 5.591 | 0.194 |
| 26 * | 5.000 | 5.299 | 0.299 | 5.394 | 0.394 |
| 27 | 5.482 | 5.478 | −0.004 | 5.389 | −0.093 |
| 28 | 5.854 | 5.726 | −0.128 | 5.898 | 0.044 |
| 29 * | 6.319 | 6.820 | 0.501 | 6.907 | 0.588 |
| 30 | 6.854 | 6.703 | −0.151 | 7.017 | 0.163 |
| 31 | 4.959 | 5.028 | 0.069 | 4.784 | −0.175 |
| 32 | 5.791 | 6.134 | 0.343 | 6.132 | 0.341 |
| 33 | 6.409 | 6.632 | 0.223 | 6.703 | 0.294 |
| 34 | 6.482 | 6.539 | 0.057 | 6.192 | −0.290 |
| 35 * | 6.658 | 6.605 | −0.053 | 6.523 | −0.135 |
| 36 | 7.000 | 6.898 | −0.102 | 6.749 | −0.251 |
| 37 | 6.886 | 6.769 | −0.117 | 7.007 | 0.121 |
| 38 | 5.638 | 5.700 | 0.062 | 6.094 | 0.456 |
| 39 | 5.959 | 5.780 | −0.179 | 6.043 | 0.084 |
| 40 * | 5.602 | 5.460 | −0.142 | 5.950 | 0.348 |
| 41 * | 5.699 | 5.711 | 0.012 | 5.901 | 0.202 |
| 42 | 4.674 | 4.597 | −0.077 | 4.406 | −0.268 |
| 43 | 6.854 | 6.616 | −0.238 | 7.010 | 0.156 |
| 44 | 5.456 | 5.502 | 0.046 | 5.512 | 0.056 |
| 45 | 6.000 | 6.003 | 0.003 | 6.237 | 0.237 |
| 46 | 6.036 | 6.104 | 0.068 | 5.989 | −0.047 |
| 47 | 5.745 | 5.972 | 0.227 | 6.001 | 0.256 |
| 48 * | 6.495 | 6.502 | 0.007 | 6.618 | 0.123 |
| 49 | 7.432 | 7.207 | −0.225 | 7.629 | 0.197 |
| 50 | 7.301 | 7.405 | 0.104 | 7.198 | −0.103 |
| 51 | 6.886 | 6.789 | −0.097 | 7.099 | 0.213 |
| 52 | 7.569 | 7.500 | −0.069 | 7.613 | 0.044 |
| 53 | 7.387 | 7.237 | −0.150 | 7.403 | 0.016 |
| 54 * | 6.886 | 7.066 | 0.180 | 7.100 | 0.214 |
| 55 | 4.801 | 4.834 | 0.033 | 4.748 | −0.053 |
| 56 | 6.495 | 5.909 | −0.586 | 6.001 | −0.494 |
| 57 | 6.000 | 6.079 | 0.079 | 5.867 | −0.133 |
| 58 | 5.553 | 5.604 | 0.051 | 5.713 | 0.160 |
| 59 * | 5.569 | 5.803 | 0.234 | 5.737 | 0.168 |
| 60 | 5.523 | 5.543 | 0.020 | 5.624 | 0.101 |
| 61 | 4.790 | 4.643 | −0.147 | 4.996 | 0.206 |
| 62 | 6.432 | 6.507 | 0.075 | 6.670 | 0.238 |
| 63 | 6.357 | 6.503 | 0.146 | 6.609 | 0.252 |
| 64 | 7.699 | 7.499 | −0.200 | 7.803 | 0.104 |
| 65 | 8.000 | 7.865 | −0.135 | 7.787 | −0.213 |
| 66 | 7.523 | 7.570 | 0.047 | 7.632 | 0.109 |
| 67 | 7.699 | 7.398 | −0.301 | 8.004 | 0.305 |
| 68 | 7.523 | 7.723 | 0.200 | 7.422 | −0.101 |
| 69 | 7.699 | 7.609 | −0.090 | 7.865 | 0.166 |
| 70 | 5.886 | 6.003 | 0.117 | 5.599 | −0.287 |
| 71 | 6.252 | 6.331 | 0.079 | 6.271 | 0.019 |
| 72 | 6.569 | 6.796 | 0.227 | 6.358 | −0.211 |
| 73 | 7.222 | 7.301 | 0.079 | 6.933 | −0.289 |
| 74 | 4.917 | 5.230 | 0.313 | 5.274 | 0.357 |
| 75 * | 6.886 | 6.558 | −0.328 | 6.587 | −0.299 |
| 76 | 6.959 | 6.960 | 0.001 | 6.789 | −0.170 |
| 77 | 6.086 | 5.867 | −0.219 | 6.372 | 0.286 |
| 78 | 7.097 | 7.109 | 0.012 | 6.903 | −0.194 |
| 79 | 6.602 | 6.604 | 0.002 | 6.517 | −0.085 |
| 80 | 5.903 | 5.913 | 0.010 | 6.043 | 0.140 |
| 81 | 5.108 | 5.140 | 0.032 | 5.006 | −0.102 |
*Test set molecules.
Figure 4. Graph of actual versus predicted pIC50 of the training set and the test set using CoMFA (Left) and CoMSIA (Right).
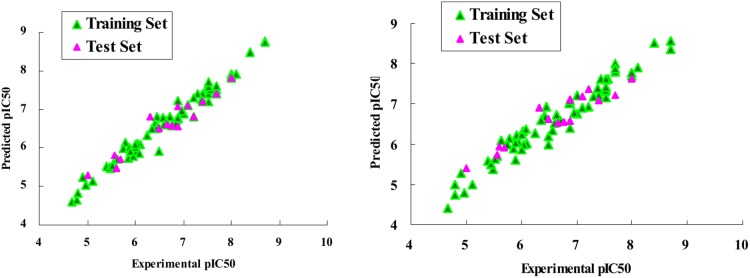
The CoMSIA model comprising all five descriptors gave a q2 value of 0.641 and r 2 value of 0.933 with an optimized component of 6, standard error estimate (SEE) of 0.210 and F value of 121.534. Contributions of steric, electrostatic, hydrophobic, hydrogen bond donor and acceptor fields were 0.160, 0.252, 0.189, 0.101 and 0.298, correspondingly.
External Validation Results
The calculated results of the external validation were listed in Table 3
. For CoMFA and CoMSIA models, the calculated r
2
pred values were 0.875 and 0.769, with the slope ( ) values of 1.021 and 1.201 (close to 1), intercept (
) values of 1.021 and 1.201 (close to 1), intercept (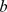 ) values of −0.025 and −0.039 (close to 0) and the correlation coefficient (R) values of 0.950 and 0.943 (close to 1), respectively. The valid r
2
m values of 0.669 and 0.631 (>0.5) as well as high slope of regression lines through the origin (
) values of −0.025 and −0.039 (close to 0) and the correlation coefficient (R) values of 0.950 and 0.943 (close to 1), respectively. The valid r
2
m values of 0.669 and 0.631 (>0.5) as well as high slope of regression lines through the origin (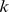 ) values of 0.986 and 0.991 (0.85≤k≤1.15) and the calculated [(r2−r02)/r2] values of −0.079 and −0.100 (<0.1) were also obtained respectively. These external validation statistics revealed that both the CoMFA and CoMSIA models possessed high accommodating capacities and they would be reliable for predicting the pIC50 values of new derivatives.
) values of 0.986 and 0.991 (0.85≤k≤1.15) and the calculated [(r2−r02)/r2] values of −0.079 and −0.100 (<0.1) were also obtained respectively. These external validation statistics revealed that both the CoMFA and CoMSIA models possessed high accommodating capacities and they would be reliable for predicting the pIC50 values of new derivatives.
Table 3. Results of the external validation for CoMFA and CoMSIA models.
| Parameters | r 2 pred |
 slope slope |
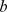 intercept intercept |
correlationcoefficient R |
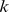 slope slope |
r 2 m | [(r2−r02)/r2] |
| CoMFA | 0.875 | 1.021 | −0.025 | 0.950 | 0.986 | 0.669 | −0.079 |
| CoMSIA | 0.769 | 1.201 | −1.319 | 0.943 | 0.991 | 0.631 | −0.100 |
CoMFA Contour Maps
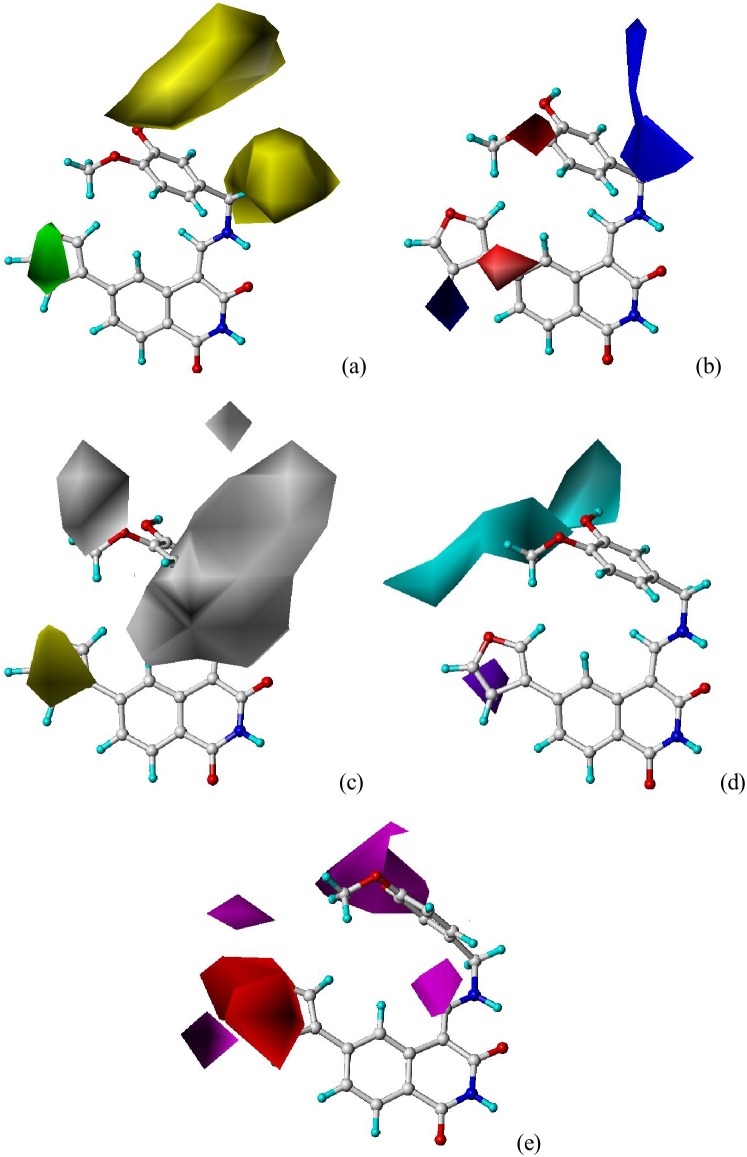
Figure 5 . depicted the CoMFA steric and electrostatic contour plots for the most active compound 12. For the steric field, the green contours represent regions of high steric tolerance (80% contribution) and the yellow contours (20% contribution) for unfavorable steric effect. The electrostatic field defined blue contours (80%) and red contours (20%) for electron-donating and -withdrawing substituents, respectively.
Figure 5. Std* coeff contour maps of CoMFA analysis in combination with compound 12.
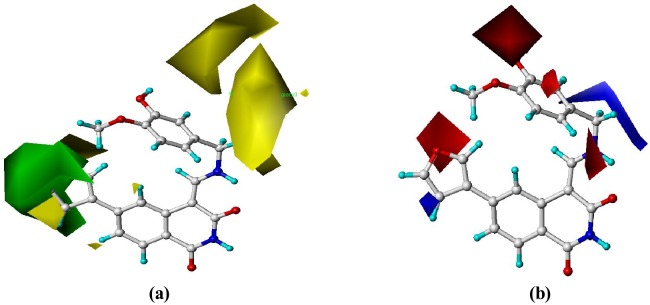
(a) Steric fields: green contours suggest regions where bulky groups increase activity, while yellow contours indicate regions where bulky groups decrease activity, and (b) Electrostatic fields: blue contours represent regions where electron-donating groups increase activity, while red contours highlight regions where electron-withdrawing groups increase activity.
In Figure 5a , one huge green contour around the R1 position revealed that bulky substituents at this site would benefit the activity, and two huge yellow contours near the R2 and R3 positions suggested bulky groups at these sites unfavorable. This may explain the facts that derivatives 10–12 with relative bulkier groups (e.g. N-pyrrolyl, 3-furyl and 3-thienyl) at R1 displayed the strongest activity, while derivatives 25–28, 31–32, 38–42, 44–45, 47, 55, 57–60, 74 and 80 bearing a relative bulkier substituents (e.g. 4-methylpiperazinyl, -CH2-(1-piperidinyl)) at R3 position showed a weak activity. Especially, derivatives 5, 7 and 8 with the corresponding substituent of chloro, bromo and iodo showed their activities in the following order of 5<7<8.
One red contour near the R1 and one red around the R2 and R3 in Figure 5b indicated an electron-withdrawing group favorable. The most potential derivatives 10–12 possessed corresponding electron-withdrawing aromatic groups (e.g. N-pyrrolyl, 3-furyl and 3-thienyl) at R1, while the activities of compounds 31–32, 42 and 44–46 bearing electron-donating substituents (e.g. piperidinyl, -NHAc, morpholinyl, N,N-dimethylformamido-) decreased significantly. This also explained why compounds 1, 3–5, 7–16 and 19–24 with the corresponding methoxyl, hydroxyl, 2-furyl, 3-furyl, 3-pyridyl or 4-pyridyl group at R3 showed much more active than derivatives 25–60 and 74–75 with electron-donating substituent (1-piperidinylmethyl, substituted piperazinyl). A blue contour around the N-10 and C-11 positions emphasized the extreme importance of the electron-donating aminomethyl group.
CoMSIA Contour Maps
The CoMSIA steric, electrostatic, hydrophobic, hydrogen bond donor and acceptor contours plots for the compound 12 were shown in Figure 6 . The CoMSIA steric and electrostatic contour maps (Figs. 6a and 6b ) were similar as the CoMFA steric and electrostatic contour maps (Figure 5 ). For hydrophobic field, white (20% contribution) and yellow (80% contribution) contours highlighted hydrophilic and hydrophobic properties, respectively. Hydrogen bond donor and acceptor fields take the cyan (80%) and purple (20%) contours for hydrogen bond donor, and the magenta (80%) and red (20%) contours as favor and unfavor for hydrogen bond acceptor, correspondingly.
Figure 6. Std* coeff contour maps of CoMSIA in combination with compound 12.
(a) Steric contour map. Green and yellow contours refer to sterically favored and unfavored regions. (b) Electrostatic contour map. Blue and red contours refer to regions where electron-donating and electron withdrawing groups are favored. (c) Hydrophobic contour map. White and yellow contours represent regions where hydrophilic and hydrophobic substituent are favored. (d) Hydrogen bond donor contour map. The cyan and purple contours indicate favorable and unfavorable hydrogen bond donor groups. (e) Hydrogen bond acceptor contour map. The magenta and red contours demonstrated favorable and unfavorable hydrogen bond acceptor groups.
In Figure 6c , one yellow contour near the R1 region demonstrated that a hydrophobic substituent at this site would benefit the activity. Most of the active derivatives involved in present study possessed a hydrophobic group (e.g. 3-furyl, N-pyrrolyl, 3-thienyl, bromo-, chloro-, iodo-, phenyl) at R1, while those with only a hydrogen atom (e.g. 2, 3, 6 and 25–27) exhibited significantly decreased potencies. Three pieces of white contour around the R2 position highlighted the hydrophilic properties of compounds 1–25 and 62–69. One purple contour near the R1 position in Figure 6d suggested a hydrogen bond donor group unfavorable. Therefore, the compounds 10–12, 49, 50 and 52–53 with hydrogen bond acceptor oxygen or nitrogen atoms at R1 site exhibited better potencies. Both one red and two magenta contours located near the R1 position in Figure 6e revealed that the hydrogen bond acceptor field was not very important for this site. The magenta contour near the R2 and R3 sites indicated hydrogen bond acceptor properties favorable. The hydroxyl groups at R2 could act as hydrogen bond acceptor at the same time. Therefore, the magenta contour confirmed the importance of the hydroxyl group at this region. Compounds 7–12 and 23 bearing a hydrogen bond acceptor substituent (methoxyl, 4-pyridyl) at R3 showed the most activities.
Molecular Docking Analysis
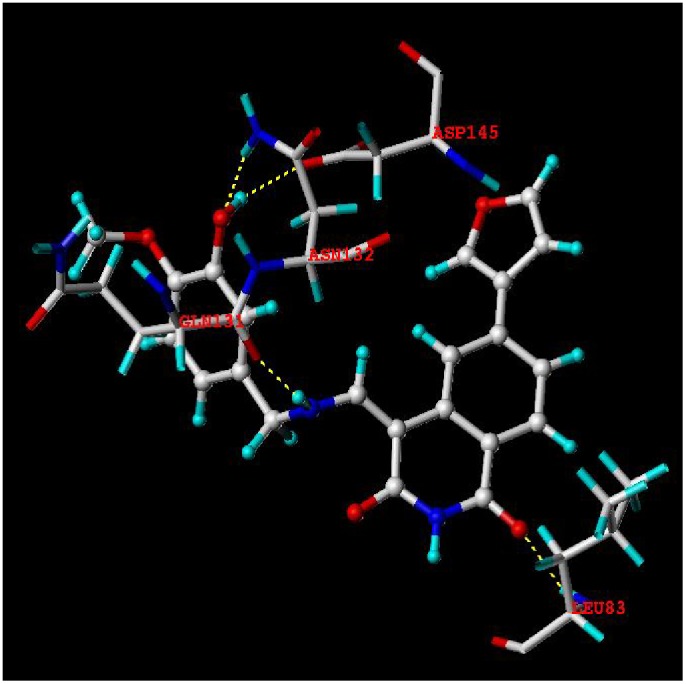
Figure 7 . illustrated the binding modes between compound 12 and the ATP pocket. The carbonyl group at C-3 position acted as a hydrogen bond acceptor by forming a H-bond with the -NH group of Leu83 residue; the imino group at N-10 position served as a hydrogen bond donor and formed H-bond with the carbonyl group of Gln131 residue; the hydroxyl group at R2 site acted as both hydrogen bond donor and acceptor and formed two H-bonds with the carbonyl group of Asp145 and the -NH group of Asn132 residues, respectively. The observations taken from Figure 7 satisfactorily matched the corresponding CoMSIA hydrogen bond donor and acceptor contour maps.
Figure 7. The binding mode between selected compound 12 and the ATP pocket of CDK4 (PDB code: 1GIH).
Summary of Structure-activity Relationship
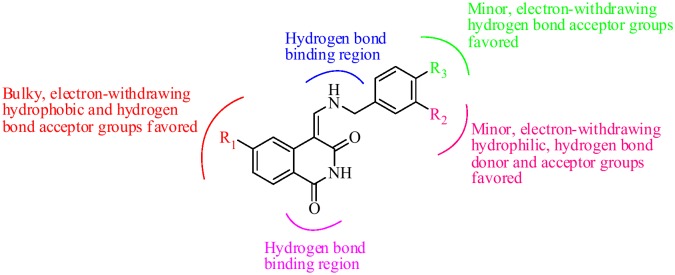
The structure-activity relationship revealed by 3D-QSAR and docking studies was illustrated in Figure 8 . In short, the bulky, electron-withdrawing, hydrophobic and hydrogen bond acceptor groups at R1 position are favorable; the minor, electron-withdrawing, hydrophilic, hydrogen bond donor and acceptor groups at R2 position may benefit the potency; the minor, electron-withdrawing and hydrogen bond acceptor substituent at R3 position would increase the activity. The carbonyl group at C-3 and the imino group at N-10 site were essential for binding to the ATP pocket of CDK4.
Figure 8. Structure-activity relationship revealed by 3D-QSAR and docking studies.
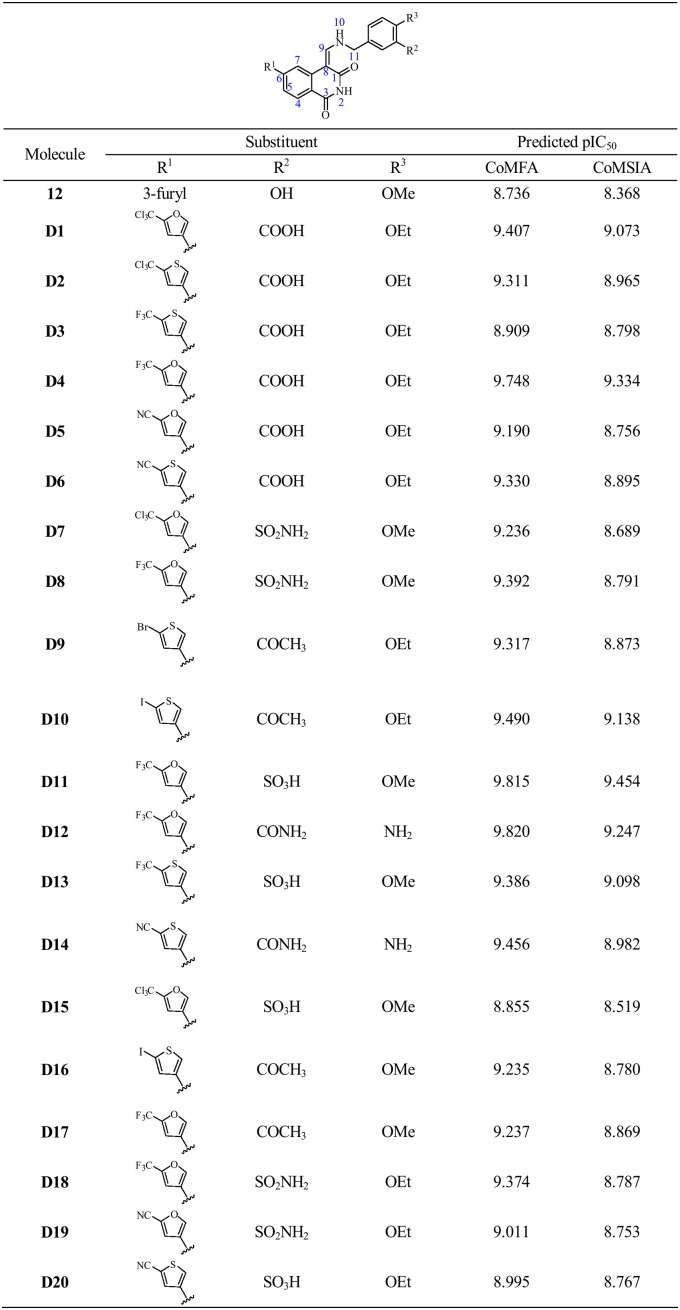
Designing of Potent Derivatives
Based on the structure-activity relationship revealed by the present study, twenty novel isoquinoline- 1, 3-(2H,4H)-dione derivatives were designed. These molecules were aligned to the database and their activities were predicted better than compound 12 by the best CoMFA and CoMSIA models established previously, especially D4, D11 and D12 showed 10 folds more active than compound 12. The chemical structures and predicted pIC50 values of these compounds were shown in Figure 9 . The results validated the structure-activity relationship in this present work.
Figure 9. Structures and predicted pIC50 values of newly designed molecules.
Conclusions
In this frame-work, a combined docking and 3D-QSAR analysis was performed to explore the interactions between isoquinoline-1,3-diones and CDK4 protein. The satisfactory CoMFA model (q 2 = 0.695, r 2 = 0.947) and CoMSIA model (q 2 = 0.641, r 2 = 0.933) showing good correlative and predictive abilities were obtained via internal and external cross-validation techniques, region focusing, bootstraping and leave-group-out. Our analyses found that all five parameters (steric, electrostatic, hydrophobic, hydrogen bond donor and acceptor properties) are highly desirable for potent inhibitory activity. The contour maps and the docking binding structures showed that the carbonyl group at C-3 and the imino group at N-10 site were necessary for binding to the ATP pocket of CDK4. Based on the interactions, twenty new designed molecules predicted higher activities than compound 12, confirming that the models could provide a valuable clue for the development of more active CDK4 subtype-selective inhibitors.
Funding Statement
This work was supported by the grant from National Nature Science Foundation of China (81202889). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chu XJ, DePinto W, Bartkovitz D, So SS, Vu BT, et al. (2006) Discovery of [4-Amino-2-(1-methanesulfonyl-piperidin-4-ylamino)pyrimidin-5-yl] (2,3-difluoro-6-methoxyphenyl) methanone (R547), a potent and selective cyclin-dependent kinase inhibitor with significant in vivo antitumor activity. J Med Chem 49: 6549–6560. [DOI] [PubMed] [Google Scholar]
- 2. Kuo GH, Deangelis A, Emanuel S, Wang A, Zhang Y, et al. (2005) Synthesis and identification of [1,3,5]triazine-pyridine biheteroaryl as a novel series of potent cyclin-dependent kinase inhibitors. J Med Chem 48: 4535–4546. [DOI] [PubMed] [Google Scholar]
- 3. Shimamura T, Shibata J, Kurihara H, Mita T, Otsuki S, et al. (2006) Identification of potent 5-pyrimidinyl-2-aminothiazole CDK4, 6 inhibitors with significant selectivity over CDK1, 2, 5, 7, and 9. Bioorg Med Chem Lett 16: 3751–3754. [DOI] [PubMed] [Google Scholar]
- 4. Jones CD, Andrews DM, Barker AJ, Blades K, Daunt P, et al. (2008) The discovery of AZD5597, a potent imidazole pyrimidine amide CDK inhibitor suitable for intravenous dosing. Bioorg Med Chem Lett 18: 6369–6373. [DOI] [PubMed] [Google Scholar]
- 5. Power DP, Lozach O, Meijer L, Grayson DH, Connon SJ (2010) Concise synthesis and CDK/GSK inhibitory activity of the missing 9-azapaullones. Bioorg Med Chem Lett 20: 4940–4944. [DOI] [PubMed] [Google Scholar]
- 6. McInnes C, Wang S, Anderson S, O’Boyle J, Jackson W, et al. (2004) Structural determinants of CDK4 inhibition and design of selective ATP competitive inhibitors. Chem Biol 11: 525–534. [DOI] [PubMed] [Google Scholar]
- 7. Tsou HR, Liu X, Birnberg G, Kaplan J, Otteng M, et al. (2009) Discovery of 4-(benzylaminomethylene)isoquinoline-1,3-(2H,4H)-diones and 4-[(pyridylmethyl)aminomethylene] isoquinoline-1,3-(2H,4H)-diones as potent and selective inhibitors of the cyclin-dependent kinase 4. J Med Chem 52 2289–2310. [DOI] [PubMed] [Google Scholar]
- 8. Tsou HR, Otteng M, Tran T, Floyd MB Jr, Reich M, et al. (2008) 4-(Phenylaminomethy- lene)isoquinoline-1,3(2H,4H)-diones as potent and selective inhibitors of the cyclin-dependent kinase 4 (CDK4). J Med Chem 51: 3507–3525. [DOI] [PubMed] [Google Scholar]
- 9. Amendola D, DeSalvo M, Marchese R, Verga FC, Stigliano A, et al. (2009) Myc down-regulation affects cyclin D1/cdk4 activity and induces apoptosis via Smac/Diablo pathway in an astrocytoma cell line. Cell Prolif 42: 94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pratt DJ, Bentley J, Jewsbury P, Boyle FT, Endicott JA, et al. (2006) Dissecting the determinants of cyclin-dependent kinase 2 and cyclin-dependent kinase 4 inhibitor selectivity. J Med Chem 49: 5470–5477. [DOI] [PubMed] [Google Scholar]
- 11. Heady L, Fernandez-Serra M, Mancera RL, Joyce S, Venkitaraman AR, et al. (2006) Novel structural features of CDK inhibition revealed by an ab initio computational method combined with dynamic simulations. J Med Chem 49: 5141–5153. [DOI] [PubMed] [Google Scholar]
- 12. Lu H, Schulze-Gahmen U (2006) Toward understanding the structural basis of cyclin-dependent kinase 6 specific inhibition. J Med Chem 49: 3826–3831. [DOI] [PubMed] [Google Scholar]
- 13. Horiuchi T, Chiba J, Uoto K, Soga T (2009) Discovery of novel thieno[2,3-d]pyrimidin-4-yl hydrazone-based inhibitors of Cyclin D1-CDK4: synthesis, biological evaluation, and structure-activity relationships. Bioorg Med Chem Lett 19: 305–308. [DOI] [PubMed] [Google Scholar]
- 14. McClue SJ, Blake D, Clarke R, Cowan A, Cummings L, et al. (2002) In vitro and in vivo antitumor properties of the cyclin dependent kinase inhibitor CYC202 (R-roscovitine). Int J Cancer 102: 463–468. [DOI] [PubMed] [Google Scholar]
- 15. Misra RN, Xiao HY, Kim KS, Lu S, Han WC, et al. (2004) N-(cycloalkylamino)acyl-2-aminothia- zole inhibitors of cyclin-dependent kinase 2. N-[5-[[[5-(1,1-dimethylethyl)-2–oxazolyl]methyl]thio]- 2-thiazolyl]-4-piperidinecarboxamide (BMS-387032), a highly efficacious and selective antitumor agent. J Med Chem 47: 1719–1728. [DOI] [PubMed] [Google Scholar]
- 16. Toogood PL, Harvey PJ, Repine JT, Sheehan DJ, VanderWel SN, et al. (2005) Discovery of a potent and selective inhibitor of cyclin-dependent kinase 4/6. J Med Chem 48: 2388–2406. [DOI] [PubMed] [Google Scholar]
- 17. Wang J, Kollman PA, Kuntz ID (1999) Flexible ligand docking: a multistep strategy approach. Proteins 36: 1–19. [PubMed] [Google Scholar]
- 18. Spitzer GM, Wellenzohn B, Laggner C, Langer T, Liedl KR (2007) DNA minor groove pharmacophores describing sequence specific properties. J Chem Inf Model 47: 1580–1589. [DOI] [PubMed] [Google Scholar]
- 19. Jain AN (2003) Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem 46: 499–511. [DOI] [PubMed] [Google Scholar]
- 20. Jain AN (2007) Surflex-Dock 2.1: robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J Comput Aided Mol Des 21: 281–306. [DOI] [PubMed] [Google Scholar]
- 21. Ambure PS, Gangwal RP, Sangamwar AT (2012) 3D-QSAR and molecular docking analysis of biphenyl amide derivatives as p38 alpha mitogen-activated protein kinase inhibitors. Molec Div 16: 377–388. [DOI] [PubMed] [Google Scholar]
- 22. Cramer RD 3rd, Patterson DE, Bunce JD (1989) Recent advances in comparative molecular field analysis (CoMFA). Prog Clin Biol Res 291: 161–165. [PubMed] [Google Scholar]
- 23. Cho SJ, Tropsha A (1995) Cross-validated R2-guided region selection for comparative molecular field analysis: a simple method to achieve consistent results. J Med Chem 38: 1060–1066. [DOI] [PubMed] [Google Scholar]
- 24. Zheng J, Xiao G, Guo J, Zheng Y, Gao H, et al. (2011) Exploring QSARs for 5-lipoxygenase (5-LO) inhibitory activity of 2-substituted 5-hydroxyindole-3-carboxylates by CoMFA and CoMSIA. Chem Biol Drug Des 78: 314–321. [DOI] [PubMed] [Google Scholar]
- 25. Afantitis A, Melagraki G, Sarimveis H, Koutentis PA, Markopoulos J, et al. (2006) A novel simple QSAR model for the prediction of anti-HIV activity using multiple linear regression analysis. Molec Div 10: 405–414. [DOI] [PubMed] [Google Scholar]
- 26. Murumkar PR, Giridhar R, Yadav MR (2008) 3D-quantitative structure-activity relationship studies on benzothiadiazepine hydroxamates as inhibitors of tumor necrosis factor-alpha converting enzyme. Chem Biol Drug Des 71: 363–373. [DOI] [PubMed] [Google Scholar]
- 27. Lan P, Huang ZJ, Sun JR, Chen WM (2010) 3D-QSAR and Molecular Docking Studies on Fused Pyrazoles as p38alpha Mitogen-Activated Protein Kinase Inhibitors. Int J Mol Sci 11: 3357–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu R, Barbault F, Delamar M, Zhang R (2009) Receptor- and ligand-based 3D-QSAR study for a series of non-nucleoside HIV-1 reverse transcriptase inhibitors. Bioorg Med Chem 17: 2400–2409. [DOI] [PubMed] [Google Scholar]
- 29. Reddy RN, Mutyala RR, Aparoy P, Reddanna P, Reddy MR (2010) An analysis of hydrophobic interactions of thymidylate synthase with methotrexate: free energy calculations involving mutant and native structures bound to methotrexate. J Mol Model 16: 203–209. [DOI] [PubMed] [Google Scholar]
- 30. Klebe G, Abraham U (1999) Comparative molecular similarity index analysis (CoMSIA) to study hydrogen-bonding properties and to score combinatorial libraries. J Comput Aided Mol Des 13: 1–10. [DOI] [PubMed] [Google Scholar]
- 31. Sivan SK, Manga V (2010) Molecular docking and 3D-QSAR studies on triazolinone and pyridazinone, non-nucleoside inhibitor of HIV-1 reverse transcriptase. J Mol Model 16: 1169–1178. [DOI] [PubMed] [Google Scholar]
- 32. Zhang N, Zhong R (2010) Docking and 3D-QSAR studies of 7-hydroxycoumarin derivatives as CK2 inhibitors. Eur J Med Chem 45: 292–297. [DOI] [PubMed] [Google Scholar]
- 33. Lindgren F, Geladi P, Rännar S, Wold S (1994) Interactive variable selection (IVS) for PLS. Part 1: Theory and algorithms. J Chemometrics 8: 349–363. [Google Scholar]
- 34. Clark M, Cramer RD 3rd (1993) The probability of chance correlation using partial least squares (PLS). QSAR 12: 137–145. [Google Scholar]
- 35. Roy K, Paul S (2010) Docking and 3D-QSAR studies of acetohydroxy acid synthase inhibitor sulfonylurea derivatives. J Mol Model 16: 951–964. [DOI] [PubMed] [Google Scholar]
- 36. Golbraikh A, Tropsha A (2002) Beware of q2! J Mol Graph Model. 20: 269–276. [DOI] [PubMed] [Google Scholar]
- 37. Roy PP, Roy K (2008) On some aspects of variable selection for partial least squares regression models. Qsar Comb Sci 27: 302–313. [Google Scholar]
- 38. Ikuta M, Kamata K, Fukasawa K, Honma T, Machida T, et al. (2001) Crystallographic approach to identification of cyclin-dependent kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein. J Biol Chem 276: 27548–27554. [DOI] [PubMed] [Google Scholar]
- 39. Ruppert J, Welch W, Jain AN (1997) Automatic identification and representation of protein binding sites for molecular docking. Protein Sci 6: 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Holt PA, Chaires JB, Trent JO (2008) Molecular docking of intercalators and groove-binders to nucleic acids using Autodock and Surflex. J Chem Inf Model 48: 1602–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Muthas D, Sabnis YA, Lundborg M, Karlen A (2008) Is it possible to increase hit rates in structure-based virtual screening by pharmacophore filtering? An investigation of the advantages and pitfalls of post-filtering. J Mol Graph Model 26: 1237–1251. [DOI] [PubMed] [Google Scholar]
- 42. Clark RD (2008) A ligand’s-eye view of protein binding. J Comput Aid Mol Des 22: 507–521. [DOI] [PubMed] [Google Scholar]