Abstract
In this study, we investigated the molecular adhesion between the major constituents of cartilage extracellular matrix, namely, the highly negatively charged proteoglycan aggrecan and the type II/IX/XI fibrillar collagen network, in simulated physiological conditions. Colloidal force spectroscopy was applied to measure the maximum adhesion force and total adhesion energy between aggrecan end-attached spherical tips (end radius R ≈ 2.5 μm) and trypsin-treated cartilage disks with undamaged collagen networks. Studies were carried out in various aqueous solutions to reveal the physical factors that govern aggrecan–collagen adhesion. Increasing both ionic strength and [Ca2+] significantly increased adhesion, highlighting the importance of electrostatic repulsion and Ca2+-mediated ion bridging effects. In addition, we probed how partial enzymatic degradation of the collagen network, which simulates osteoarthritic conditions, affects the aggrecan–collagen interactions. Interestingly, we found a significant increase in aggrecan–collagen adhesion even when there were no detectable changes at the macro- or microscales. It is hypothesized that the aggrecan–collagen adhesion, together with aggrecan–aggrecan self-adhesion, works synergistically to determine the local molecular deformability and energy dissipation of the cartilage matrix, in turn, affecting its macroscopic tissue properties.
Introduction
The unique biomechanical properties of articular cartilage, including compressive and shear resistance as well as shock absorption, are directly governed by collective intra- and intermolecular interactions between its extracellular matrix (ECM) molecules. These interactions include electrostatics, steric and entropic repulsion, and water–proteoglycan molecular friction between the type II/IX/XI fibrillar collagen network and the enmeshed large proteoglycan aggrecan (Figure 1).1−4 In addition, it was reported that binding activities between ECM molecules, while not directly contributing to cartilage biomechanics, are critical in governing chondrocyte activities and ECM assembly.5,6 For example, aggrecan monomers bind to hyaluronan via its G1 domain to form aggregates,7 stabilized by link proteins (Figure 1a),8 which prevents aggrecan loss from cartilage ECM. Bindings between aggrecan keratan sulfate glycosaminoglycan (KS-GAG) side chains and collagen have been suggested to affect the aggrecan spatial distribution in vivo,9 and to protect collagen fibrils from proteolytic degradation.10 Specific bindings involving quantitatively minor matrix proteins and collagen, including decorin-type II collagen, biglycan-type II collagen,11 cartilage oligometric matrix protein (COMP)-type II collagen,12 and biglycan-type VI collagen,13 regulate the fibrillogenesis and cross-linking of the collagen network. Bindings between these proteins and cytokines, such as decorin and transforming growth factor (TGF)-β, regulate cell signaling and mechanotransduction.14,15 Besides these specific bindings, at physiological density and molecular strain, aggrecan can undergo nonspecific self-adhesion,16 despite the presence of strong electrostatic repulsion between GAG side chains. This aggrecan–aggrecan self-adhesion was suggested to be an important factor contributing to the self-assembled hierarchical architecture of cartilage ECM.16
Figure 1.
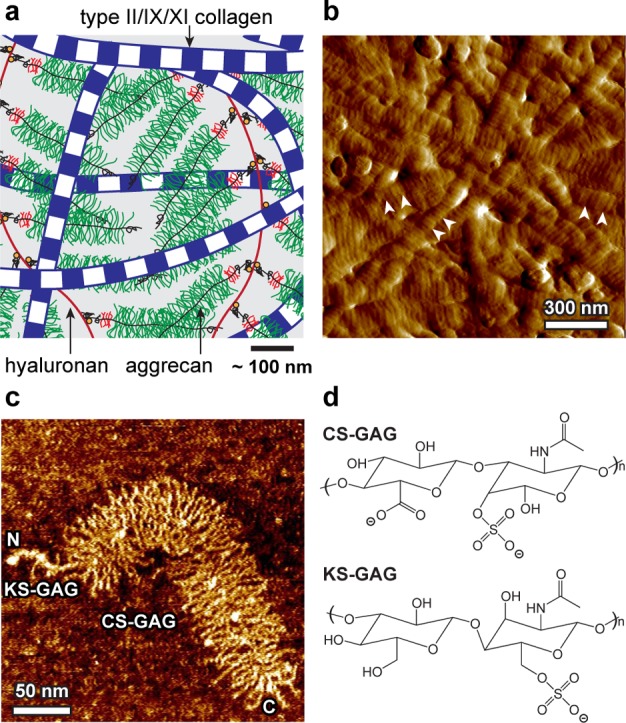
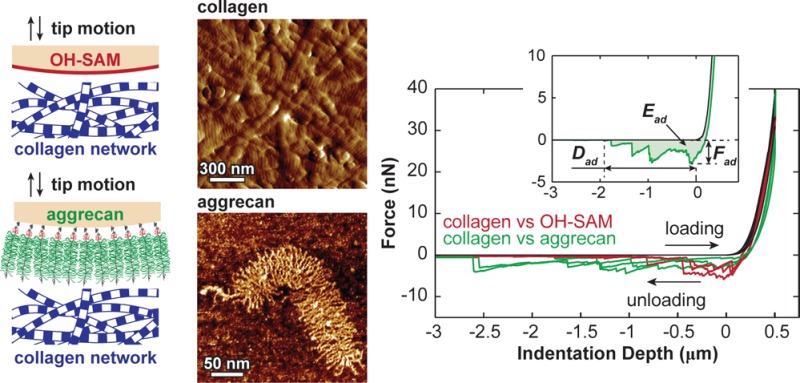
(a) Schematic of the structure and major molecular constituents of the articular cartilage extracellular matrix (ECM), including the type II/IX/XI fibrillar collagen network, aggrecan moiety, and hyaluronan that aggrecan binds to,7 which is stabilized by the link protein.8 The scale bar is an estimate based on the dimensions of aggrecan and collagen fibrils. Molecular density is reduced to increase clarity. (b) Tapping mode atomic force microscopy (AFM) amplitude image of air-dried, proteoglycan-depleted calf knee cartilage surface, which displays the transversely isotropically aligned collagen fibrils and the nanoscale d-banding patterns (arrows). (c) Tapping mode AFM height image of individual fetal epiphyseal aggrecan monomer (adapted with permission from ref (27)), illustrating the N- and C-termini of the core protein, chondroitin sulfate glycosaminoglycan (CS-GAG), and keratan sulfate (KS)-GAG side chains. (d) Schematics of the dissacharide constituents of the CS-GAG (chondroitin-4-sulfate GAG) and KS-GAG.3,4
While aggrecan self-adhesion has been investigated in detail,16,17 there is a lack of understanding of direct, nonspecific interactions between the two primary ECM constituents, aggrecan and the fibrillar collagen network. Knowledge of aggrecan–collagen interactions could provide a critical step forward in our understanding of the molecular basis of cartilage tissue function and the origins and characteristics of osteoarthritis. Toward this end, the objective of this study is to investigate the mechanisms of aggrecan–collagen molecular adhesion under simulated physiological conditions. We utilized atomic force microscope (AFM)-based colloidal force spectroscopy to measure the adhesion between gold-coated spherical colloid tips (R ≈ 2.5 μm) functionalized with end-grafted aggrecan and the transversely isotropically aligned collagen fibrils of native bovine superficial zone cartilage surfaces in 0.15 M phosphate buffered saline (PBS; Figure 2a). We studied the molecular origins of adhesion by comparing the adhesions measured on the collagen specimen using the aggrecan tip to those measured by a hydroxyl-terminated spherical tip (Figure 2b,c), in which effects like electrostatic repulsion, hydrophobicity, and macromolecular entanglements are eliminated. We quantified the influences of electrostatic repulsion by changing the bath solution conditions, including ionic strength (IS) and concentration of Ca2+. Furthermore, we probed how osteoarthritic-like enzymatic degradation of collagen affects aggrecan–collagen adhesion in PBS (Figure 2a). These observations were interpreted in the context of cartilage ECM macromolecular composition and structure to provide insights into the tissue integrity of cartilage and the characteristics of osteoarthritic degradation.
Figure 2.
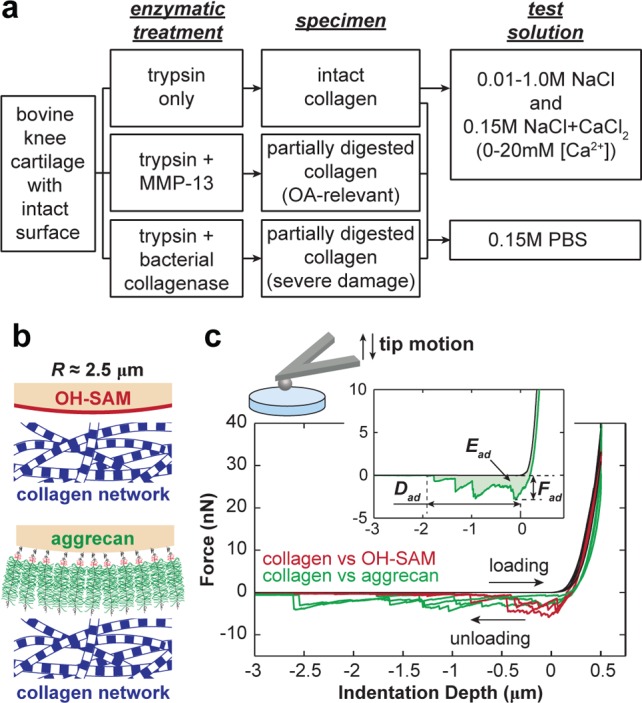
(a) Flowchart of types of collagen networks specimens as a result of different enzymatic treatments and the types of aqueous solutions for aggrecan–collagen adhesion test. (b) Schematics of colloidal force spectroscopy using microspherical tips (end radius R ≈ 2.5 μm) functionalized with hydroxyl-terminated self-assembled monolayer (OH-SAM) and aggrecan. (c) Typical force vs depth (F–D) curves measured via OH-SAM and aggrecan tips (PBS, [Ca2+] = 0 mM, surface dwell time td = 30 s). Curves shown are from three different locations for each tip. (Inset) Definitions of the maximum adhesion force, Fad, total adhesion energy, Ead, and maximum adhesion interaction distance, Dad, for each F–D curve.
Materials and Methods
Sample Preparation
Cartilage plugs were harvested from the femoropatellar grooves of 1–2 week old bovine calves (Research ’87, Hopkinton, MA) using a 6 mm dermal punch. Cartilage disks of ≈1.0 mm thickness were extracted from the plugs with intact superficial zone and surface. Disks were incubated for 12 h in phosphate buffered saline (PBS, IS = 0.15 M, pH = 7.4) at 37 °C in the presence of 0.1 mg/mL bovine pancreatic trypsin (Sigma-Aldrich, St. Louis, MO) to remove proteoglycans (PGs)18 without interrupting the macroscopic19 and microscopic20 structure or static tensile properties21 of the collagen network. Tapping mode AFM images of trypsin-treated (PG-depleted) disks showed the collagen network maintains its nanostructure integrity after the trypsin treatment (Figures 1b and S1). Following the trypsin digestion, disks were separated into three groups (Figure 2a). The first group was directly used for nanomechanical tests, discussed in detail in the next section. The second group was further treated with 10 μg/mL human recombinant matrix metalloprotease-13 (rhMMP-13, gift from Ivan Otterness, Pfizer) for 24 h in 37 °C water bath. MMP-13, or collagenase-3, cleaves type II collagen molecules and is highly overexpressed in osteoarthritis, resulting in degradation of collagen fibrils.22 The third group was further treated with 0.1 mg/mL bacterial collagenase (BC) from Clostridium histolyticum (Worthington Biochemical Corporation, Lakewood, NJ) for 10 min at 37 °C to induce more severe partial degradation of the collagen network. The BC was added to PBS with Ca2+ for activation and preheated in 37 °C water bath for 30 min before the treatment.23
Histology and Structural Characterization
Histology was carried out on disks from untreated, trypsin-only treated, and disks treated with both trypsin and BC to analyze the gross level morphology. Aggrecan/proteoglycan and collagen were visualized using Safranin-O and Masson’s Trichrome, respectively.24 To characterize the collagen network structure, additional disks from all three trypsin-treated groups were fixed via the Ohtani’s procedure to retain its three-dimensional architecture and subsequently imaged using scanning electron microscopy (SEM).25,26 Briefly, disks were fixed in 10% formalin for 1 day and then immersed in 10% NaOH for 6 days. Specimens were subsequently washed with Milli-Q filtered water for 1 day and then immersed in 1–2% tannic acid for 5 h. A second one-day water rinse was followed by ascending alcohol series dehydration and counterfixing in 1% OsO4 (Sigma-Aldrich, St. Louis, MO) for 2 h. Specimens were then lyophilized (FreeZone Freeze-Dry System, Labconco, Kansas City, MO) and Au–Pd sputter-coated (≈ 8 nm thickness; Quorum Technologies, Guelph, Ontario, Canada) prior to imaging. These disks were then imaged via SEM (Helios 600 Dual Beam FIB/SEM, FEI, Hillsboro, OR). The nanostructure of the trypsin only treated collagen network was characterized via tapping mode AFM imaging on overnight air-dried disks, using a Multimode IIIA AFM (Veeco, Santa Barbara, CA) and Olympus AC240TS-2 rectangular Si cantilevers (nominal tip radius R < 10 nm, spring constant k ∼ 2 N/m, Asylum Research, Santa Barbara, CA).
Colloidal Atomic Force Microscope Tip Preparation
Purified fetal bovine epiphyseal A1A1D1D1 aggrecan, MW ≈ 3 MDa,27 was chemically functionalized with thiol groups at the N-terminal, as described previously.28 Gold-coated borosilicate colloidal AFM spherical tips (end radius R ≈ 2.5 μm, nominal spring constant k = 0.58 N/m, Novascan, Ames, IA) were chemically end-attached with aggrecan by immersion in 100 μL of 1 mg/mL thiol-functionalized aggrecan solution in a humidity chamber for 48 h.16,29 The thiol-gold bonding between aggrecan and the colloid resulted in an aggrecan packing density of ≈50 mg/mL (one monomer per ≈25 nm × 25 nm square),16,29,30 which is within the physiological range of aggrecan in cartilage (20–80 mg/mL).31 As a control, identical colloidal probe tips with the same specifications were functionalized with hydroxyl-terminated self-assembled monolayer (OH-SAM) by immersion in 3 mM 11-mercaptoundecanol (HS(CH2)11OH, Sigma-Aldrich, St. Louis, MO) ethanol solution for 24 h. With this neutral, hydrophilic, hard-wall tip, the effects of electrostatic repulsion, hydrophobicity, and macromolecular entanglement are minimized. Results measured from the OH-SAM tip can thus be used to elucidate the origins of the ionic strength and [Ca2+] dependence of aggrecan–collagen adhesion. Both the aggrecan and OH-SAM functionalized tips have been shown to have surface roughness less than 5 nm (≪ than the tip radius of curvature),29 suggesting that the surface roughness had negligible impact on the outcomes.
Nanomechanical Experiments
Colloidal force spectroscopy was performed using a 3D Molecular Force Probe (MFP-3D, Asylum Research, Santa Barbara, CA) to quantify the adhesion between the proteoglycan-removed cartilage disks with undamaged collagen network and the aggrecan-functionalized tip. The experiment was carried out on the disk surface away from the cutting edges in several aqueous solutions (Figure 2a): (1) physiological-like solution of PBS (IS = 0.15 M, pH ≈ 7.4), (2) 0.01–1.0 M NaCl solutions (pH ≈ 5.6), and (3) 0.15 M IS NaCl + CaCl2 solutions with varying [Ca2+] = 0–20 mM (pH ≈ 5.6). Within this pH range, the GAG chains of aggrecan maintain constant negative charge density and compressive nanomechanical behaviors.28 The tip was programmed to indent into the disk for a maximum depth d ≈ 500 nm at 0.5 μm/s indentation depth rate and then to retract from the sample at the same rate after holding at the constant depth for a given surface dwell time td (0–60 s). The test was carried out in the indenter mode, in which the z-piezo displacement was continuously adjusted to compensate for cantilever bending and to maintain constant indentation rate and maximum indentation depth. Additional control experiments were carried out on the same disks using the hard-wall OH-SAM tips under the same conditions (Figure 2b).
To quantify the aggrecan–collagen adhesion force and energy, we calibrated the cantilever deflection sensitivity (nm/V) on a hard silica surface in 1.0 M NaCl solution to minimize the electrostatic repulsion. At this ionic strength, the aggrecan monolayer can be approximated as incompressible at forces >40 nN, as shown in our previous aggrecan compression studies using the same tips.29 The spring constant was then determined via the thermal oscillation method.32 Proper functionalization of aggrecan tips was verified on mica by confirming the >300 nm long-range repulsion at low IS (0.001 M) and its absence at high IS (1.0 M).28,29 It is unlikely that this repulsion is due to electrical double layer repulsion arising from surface charges, given the interaction distance (>300 nm) is substantially greater than the Debye length κ–1 ≈ 10 nm at IS = 0.001 M. The effective tip–sample contact point was determined by the Golden Section-based algorithm described previously.33,34 The maximum adhesion force, Fad, and total adhesion energy, Ead, were calculated on each of the indentation force-depth (F–D) retract curves (Figure 2c). For each experimental condition, the measurement was repeated for n ≥ 10 locations on each of the disks from the joints of at least three different calves. One measurement was carried out at each location, except for the test of the surface dwell time td dependence, where a total of nine repeats were conducted at the same location, one repeat for each td from 0–60 s.
Statistical Analysis
To avoid the assumptions of data normal distribution and homoscedasticity, nonparametric statistical tests (e.g., Mann–Whitney test, Kruskal–Wallis analysis of variance test, and Friedman repeated-measure analysis of variance test) were performed to examine the overall significance of various test conditions, including the surface dwell time td, ionic strength IS, [Ca2+], and enzymatic treatments of the collagen network. Mann–Whitney test was carried out to compare the data between each pair of ionic strength, [Ca2+] or enzymatic treatments. Data from different calves under the same experimental conditions were pooled, as no statistical differences in Fad or Ead were found between collagen specimens from different animals via Mann–Whitney test (p > 0.05).
Results
When using both the OH-SAM and aggrecan tips to indent onto the trypsin-treated (PG-depleted, collagen network only) cartilage disks, we observed characteristic long-range force–indentation depth curves at 500 nm indentation depth, td = 30 s in PBS (Figure 2c). Increasing the surface dwell time td resulted in significant, nonlinear increase in both the maximum adhesion force, Fad, and the total adhesion energy, Ead (Figure 3, Friedman test, p < 0.001). In comparison, changing td had no appreciable effects on the maximum distance of the adhesion interactions, Dad (Figure 2c). For the control experiment (OH-SAM tip), Fad varied from 1.2 ± 0.1 to 5.3 ± 0.4 nN, and Ead varied from 0.7 ± 0.1 to 4.2 ± 0.4 fJ, when changing td from 0 to 60 s, respectively. The aggrecan probe tip showed significantly lower Fad of 0.8 ± 0.1 nN at td = 0 s and 3.1 ± 0.2 nN at td = 60 s (Figure 3a, Friedman test, p < 0.01), and similar adhesion energy, Ead = 1.3 ± 0.2 fJ at td = 0 s to 3.9 ± 0.3 fJ at td = 60 s (Figure 3b, Friedman test, p > 0.05).
Figure 3.
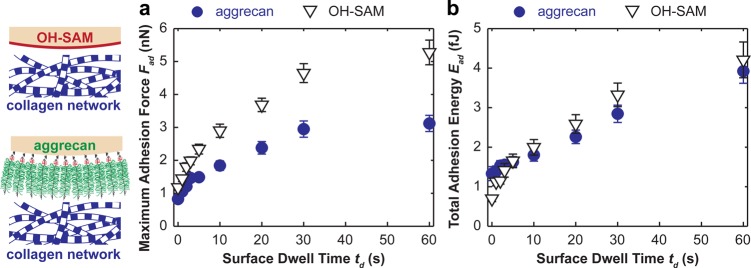
(a) Maximum adhesion force, Fad, and (b) the total adhesion energy, Ead, for the indentation of proteoglycan-depleted cartilage with OH-SAM and aggrecan functionalized microspherical tips (R ≈ 2.5 μm) in 0.15 M PBS (mean ± SEM, n ≥ 50 locations from more than four cartilage disks).
At the same indentation depth (500 nm) and surface dwell time (td = 30 s), increasing ionic strength (IS) from 0.01 to 1.0 M significantly increased both Fad and Ead for the aggrecan tip (Figure 4, Kruskal–Wallis test, p < 0.001), with the exception that Ead, which was similar for 0.01 and 0.15 M IS (p > 0.05). Similarly, at IS = 0.15 M, increasing [Ca2+] from 0 to 20 mM also markedly increased Fad and Ead for the aggrecan tip (Figure 5, Kruskal–Wallis test, p < 0.001). In comparison, the effects of both IS and [Ca2+] are absent for the adhesion between collagen (PG-depleted cartilage disk) and the hard wall, neutral, hydrophilic OH-SAM tip (Figures 4 and 5, Kruskal–Wallis test, p > 0.05).
Figure 4.
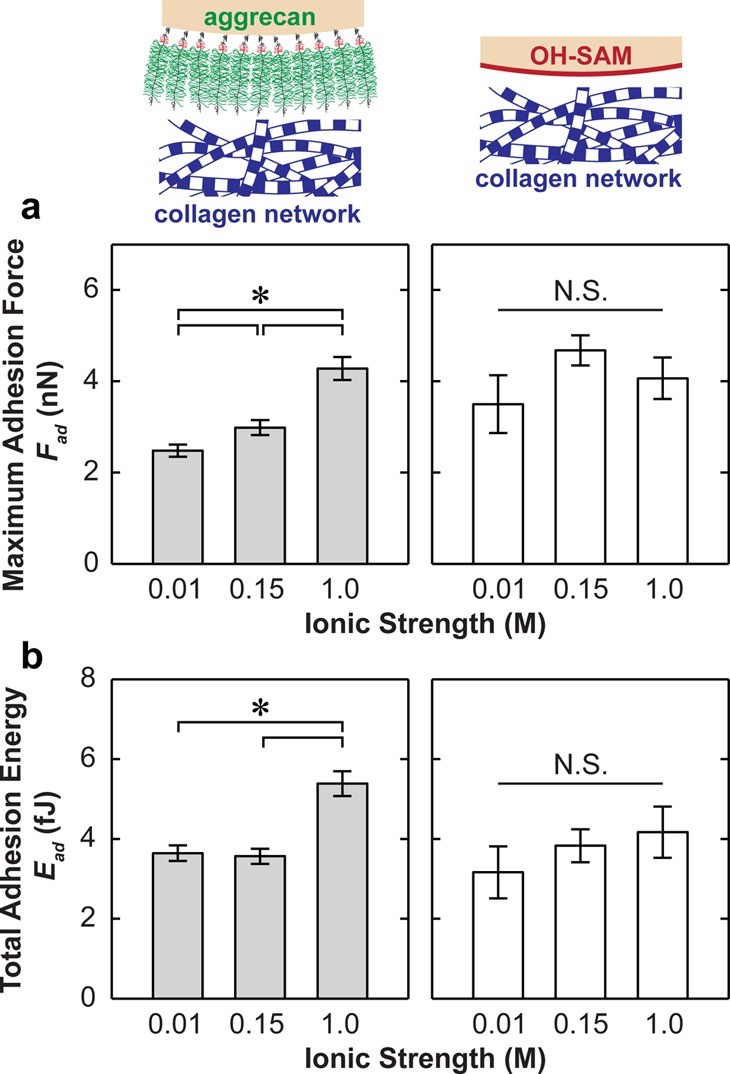
(a) Maximum adhesion force, Fad, and (b) the total adhesion energy, Ead, for the indentation of proteoglycan-depleted cartilage with OH-SAM and aggrecan functionalized microspherical tips (R ≈ 2.5 μm) in NaCl solutions at different ionic strengths, td = 30 s (mean ± SEM, n ≥ 50 locations on more than four cartilage disks, *p < 0.001 via Mann–Whitney test).
Figure 5.
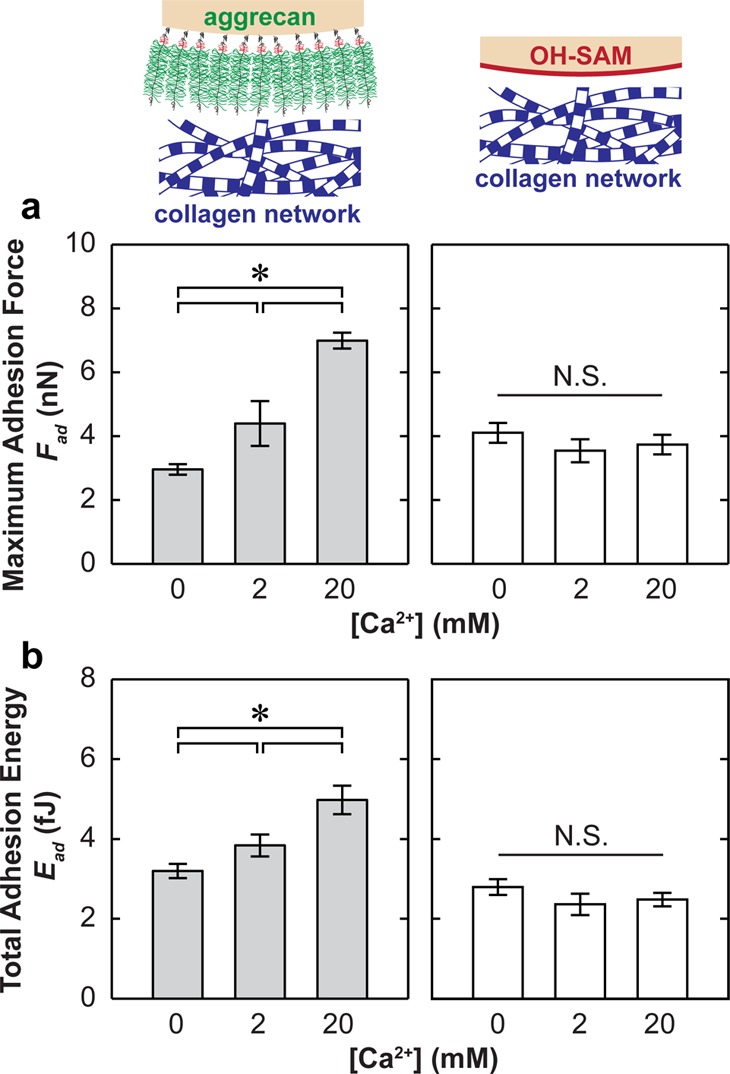
(a) Maximum adhesion force, Fad, and (b) the total adhesion energy, Ead, for the indentation of proteoglycan-depleted cartilage with OH-SAM and aggrecan functionalized microspherical tips (R ≈ 2.5 μm) in NaCl + CaCl2 solutions at [Cl–] = 0.15 M and different [Ca2+], td = 30 s (mean ± SEM, n ≥ 50 locations from more than four cartilage disks, *p < 0.001 via Mann–Whitney test).
Removal of proteoglycans by trypsin digestion revealed the transversely isotropic collagen fibril alignment in the 2D surface plane of the cartilage surface (Figures 1b and 6).35 For the trypsin-only treated collagen network, the fibril diameter was found to be 40.5 ± 4.7 nm (mean ± STD, n ≥ 100 fibrils), with a packing density of ≈50 fibrils per μm2 area (Figure 6). At this length scale, effects of either MMP-13 or bacterial collagenase treatments were not noticeable (Figure 6a,b). In addition, at the tissue level, the more severe collagen digestion by 10 min in bacterial collagenase introduced no changes in collagen concentration or structure (Figure 7). We thus expected similar results from the milder MMP-13 digestion. While we did not observe any appreciable effects via fibril-level imaging (Figure 6) or tissue-level histology (Figure 7), severe damage, and disassembly of the molecular level structure of the fibrillar collagen network were expected.36 For the less severe, more physiological-like MMP-13 digestion, immunohistochemistry of untreated and MMP-13 treated cartilage disks using monoclonal neo-epitope antibody 9A4 to reveal collagenase cleavage of collagen showed that 24 h MMP-13 treatment introduced partial degradation of the collagen network in the upper 30 μm of the superficial zone (Figure S2).37 As a result, the maximum adhesion force, Fad, between aggrecan and collagen significantly increased by a factor of ≈2.5× for both MMP-13 and BC treatments at the same indentation depth (≈500 nm) and td = 30 s in PBS (Figure 8a, Mann–Whitney test, p < 0.001). The total adhesion energy, Ead, also increased substantially for both treatments, where the BC treatment (≈3× increase) had even greater effects than the MMP-13 treatment (≈1.5× increase; Figure 8b, Mann–Whitney test, p < 0.001).
Figure 6.
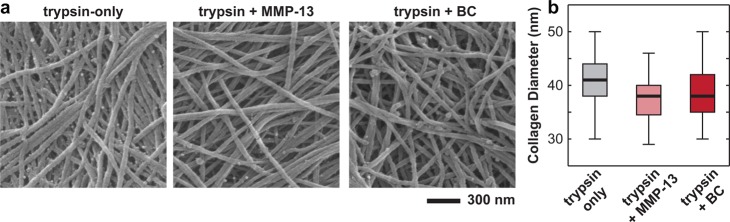
(a) Scanning electron microscopy images of trypsin-treated cartilage disk surfaces, prepared via Ohtani’s procedure25 to retain its 3D architecture, including trypsin only, trypsin + MMP-13, and trypsin + bacterial collagenase treated disks. (b) Box-and-whisker plot of the distribution of collagen fibril diameters measured for the three types of disks (n ≥ 100 fibrils for each treatment).
Figure 7.
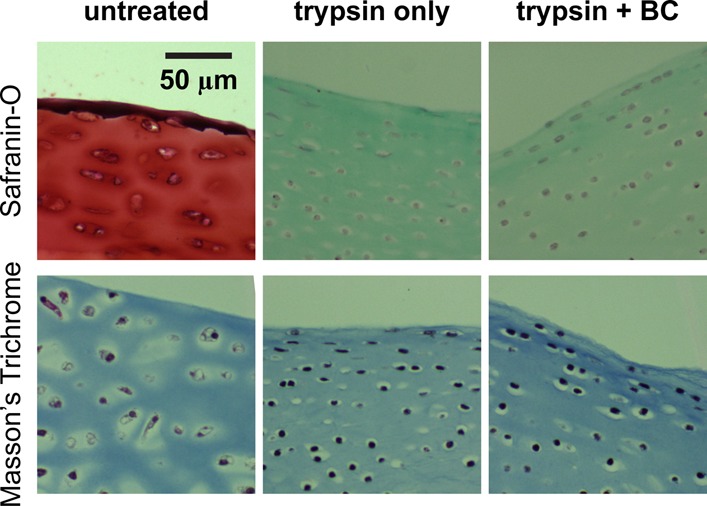
Histology images of the cross sections of untreated (normal), trypsin-treated, and trypsin + bacterial collagenase (BC) treated cartilage disks, stained with Safranin-O (for aggrecan) and Masson’s Trichrome (for collagen).
Figure 8.
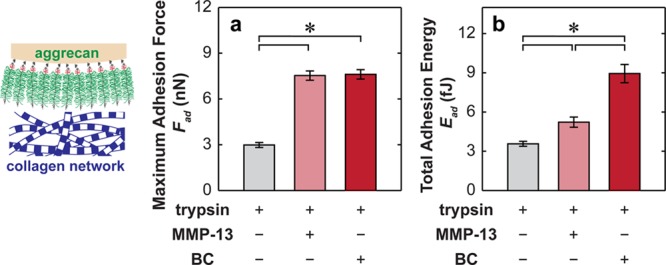
(a) Maximum adhesion force, Fad, and (b) the total adhesion energy, Ead, for the indentation of proteoglycan-depleted cartilage with aggrecan functionalized microspherical tips (R ≈ 2.5 μm) in PBS, td = 30 s. The disks were treated with 0.1 mg/mL trypsin only (intact collagen network), trypsin + 10 μg/mL human recombinant matrix metalloprotease-13 (MMP-13), and trypsin + 0.1 mg/mL Clostridium histolyticum bacterial collagenase (BC; mean ± SEM, n ≥ 50 locations from more than four cartilage disks, *p < 0.001 via Mann–Whitney test).
Discussion
In this study, we investigated the origins and governing factors of the aggrecan–collagen molecular adhesion. In particular, we studied the roles of aggrecan GAG–GAG electrostatic repulsion and Ca2+-mediated ion bridging effects. These interactions were studied in the context of the cartilage extracellular matrix environments to elucidate their contributions to cartilage tissue assembly and biomechanical properties. Furthermore, using partially digested collagen networks, we explored how osteoarthritis-relevant degradation alters the aggrecan–collagen molecular interactions, which in turn, affects cartilage tissue properties at the early stages of OA, when OA-induced matrix changes are indistinguishable via either macroscopic or microscopic analyses.
Relevance to In Vivo Aggrecan–Collagen Adhesion
On native cartilage surfaces, the ECM is dominated by transversely aligned collagen fibrils,35 hyaluronan, proteoglycans, and glycoproteins such as lubricin (proteoglycan 4 or PRG4),38 covered by a physically adsorbed phospholipid layer.39−41 Aggrecan concentration is much lower in the superficial zone compared to the middle and deep zones.31,42,43 In this study, 6 mm diameter bovine cartilage disks with intact superficial zone were treated with bovine trypsin to remove proteoglycans and expose individual collagen fibrils (Figures 1b and S1). While explants diced into small pieces accentuate cell apoptosis and matrix degradation,44 the large explants used here minimize cell death and enable maintenance of normal matrix metabolism;45,46 thus, native collagen architecture is retained away from the cut edges of the disks. The presence of nondegraded collagen is further supported by the absence of collagenase cleavage sites in the superficial zone, which can be detected via immunohistochemistry (Figure S2).37
The fibrillar collagen network is expected to be the major constituent of the trypsin-treated disks, as the physically adsorbed surface-active phospholipid layer39−41 is expected to be removed via rinsing in PBS,47 and proteoglycans such as aggrecan and lubricin were removed by trypsin digestion (Figure 7). While some hyaluronan may remain on the surface, recent studies on cartilage surface lubrication have shown that a 12 h incubation with trypsin likely removed most hyaluronan constituents48 due to the loss of its anchorage with aggrecan7 and lubricin.49 In addition, given that the total hyaluronan content in cartilage is small (<0.3% wet wt)50 and its concentration on the surface is even lower than that in the bulk,51 hyaluronan is expected to have minimal direct contribution to cartilage mechanical behavior. Digestion by hyaluronidase did not significantly impact cartilage surface roughness, modulus, or friction coefficient, as measured by AFM.52 We therefore expect the net adhesion to be dominated by aggrecan–collagen interactions.
Molecular Origins of Aggrecan–Collagen Adhesion
Increasing surface dwell time td allows longer equilibration between the compressed aggrecan and collagen and, therefore, increases the number of effective molecular contacts. As a result, both the maximum adhesion force, Fad, and the total adhesion energy, Ead, significantly increased with td (Figure 3). The measured adhesion is expected to be a complex balance of various attractive and repulsive molecular interaction mechanisms, as in most biomacromolecular systems. The repulsive mechanisms include electrostatic repulsion between the chondroitin sulfate (CS)-GAGs and the negatively charged amino acids on collagen molecules, excluded volume effects, hydration, as well as conformational, translational, and rotational entropic penalties. The attractive mechanisms include van der Waals contacts, hydrophobicity, hydrogen bonding, physical entanglements, and electrostatic attraction between GAGs and the positively charged amino acids on collagen. Hydrogen bonding can take place between the −OH, −COOH, and −SO3– groups (pKa of GAG carboxyl is ≈3 in aqueous solutions53) on aggrecan and collagen.
The cartilage fibrillar collagen network is a type II/IX/XI collagen heteropolymer.54,55 Type II collagen is the major constituent (≈80–90% molar ratio, increase with age).55 Type IX collagen molecules (≈1–10% molar ratio, decrease with age) are attached on the fibril surfaces and provide covalent cross-links between type II and other type IX collagens.56 Type XI collagen (≈ 3 – 10% molar ratio, decreasing with age) forms the fibril nucleation cores that allow self-assembly of type II fibrils.57,58 Aggrecan–collagen adhesion thus mainly takes place between aggrecan and type II collagen fibrils and, to a lesser extent, between aggrecan and the triple helical domain of the surface type IX collagen. Trypsin digestion using bovine pancreatic trypsin has been shown not to affect the type II/XI collagen fibril structure or the triple helical domains of the type IX collagen.20,59,60 However, the positively charged, heparin-binding NC4 domain and the CS-GAG attached on the NC3 domain of type IX collagen were most likely removed by trypsin, as previously reported for full length recombinant collagen IX.61 While removal of these domains could result in underestimation of the aggrecan–collagen adhesion, we expect this effect to be minimal given the relative low concentration of collagen IX.
Each type II collagen triple helix consists of three colIIαI polypeptide chains. One calf colIIα1 molecule contains 1487 amino acids,62 including 995 hydrophobic, 211 hydrophilic (neutral), 140 positively charged, and 141 negatively charged amino acids (Figure S3).62 While each colIIα1 molecule is net neutral at physiological pH, local positive and negative charges could be present along the fibrils. Interactions between aggrecan and the triple helical region of type IX collagens (with three different polypeptide chains, colIXαI, colIXαII, and colIXαIII) also contribute to the net adhesion, albeit to a much lesser extent, due to its low concentration.55 For aggrecan, each CS-GAG chain contains ≈40–50 disaccharide units, in which both polar and nonpolar groups are present1,63 (Figure 1c,d). Nonpolar patches along the aggrecan CS-GAGs and hydrophobic amino acids on collagen (e.g., 995 hydrophobic amino acids on each colIIα1) can lead to hydrophobic interactions.64 At the maximum indentation force ≈40 nN, aggrecan is compressed at ≈50% molecular strain.29 CS-GAGs can also undergo conformational changes and form physical entanglements with relatively stiff collagen fibrils. Given the configuration of aggrecan attachment onto the spherical tip (Figure 2b), it is less likely that the shorter KS-GAG chains play an important role in this measured adhesion. In addition, if multivalent ions such as Ca2+ are present, additional charge redistribution,65 and ion-bridging effects66 will also contribute to the net aggrecan–collagen adhesion. In this study, however, like our previous aggrecan–aggrecan adhesion work,16 we were unable to distinguish a single dominating molecular mechanism due to the complexity of biological macromolecules, i.e., aggrecan and collagen. According to their molecular composition and structure, we expect that all the proposed mechanisms are synergistically involved in the net adhesion.
Effects of Ionic Strength
Variations of Fad and Ead with ionic strength (IS) provide insights into how electrostatic interactions govern aggrecan–collagen adhesion in vivo. Increasing IS effectively shields the GAG–GAG electrostatic repulsion and, consequently, alters the conformation and compressibility of aggrecan monomers. Increasing IS from 0.01 M (Debye length κ–1 ≈ 3 nm) to physiological-like 0.15 M (κ–1 ≈ 1 nm) did not completely screen the GAG–GAG repulsion, given the intra- and inter-GAG charge distance is ≈1–2 nm within aggrecan.27 Aggrecan monomers partially retain the long-range repulsive, more elongated conformation. Increasing IS from 0.01 to 0.15 M thus has only marginal (Fad) or nonsignificant (Ead) effects on the aggrecan–collagen adhesion (Figure 4). Further increase of IS to 1 M (κ–1 ≈ 0.3 nm) completely shields the GAG–GAG electrostatic repulsion, and aggrecan monomers behave similar to neutral brush-like polymers.29,67 Removal of aggrecan electrostatic repulsion significantly increases both the deformability of aggrecan and effective aggrecan–collagen molecular contacts, and therefore, aggrecan–collagen molecular adhesions (≈2× in Fad, ≈1.5× in Ead compared to 0.15 M, Figure 4). Since the collagen network is net neutral, changing ionic strength has negligible effects on the molecular deformability and surface properties of the collagen fibrillar network. From the F–D loading curves (data not shown) measured by the OH-SAM tips, we did not detect significant IS dependence on the indentation resistance, consistent with previous reports showing that the collagen network nanostiffness is independent of bath IS.68 Similarly, increasing IS from 0.01 to 1.0 M had no significant effect on the either Fad or Ead between the OH-SAM tip and the collagen network (Figure 4).
Effects of Divalent Ca2+
When increasing [Ca2+] from 0 to 20 mM at 0.15 M IS, we observed an ≈3× increase in Fad and an ≈1.5× increase in Ead of aggrecan–collagen adhesion (Figure 5). The presence of divalent Ca2+ ions alters the free counterion distribution65 and introduces ion bridging between multiple negative charges.66 Previously, this redistribution effect on aggrecan compressibility was observed to saturate at [Ca2+] ≥ 2 mM.69 Thus, the monolithic increase in aggrecan–collagen adhesion with [Ca2+] from 0 to 20 mM suggests the likely dominant role of the ion bridging effect.66 It is known that one Ca2+ can bind electrostatically between two monovalent negative charges on the GAG side chain70,71 and between the GAG chain and the aggrecan core protein.72 It is also possible that Ca2+ can bind between GAGs and the local negative charges on collagen. Due to the local rigidity of the fibrillar collagen network, it is less likely for Ca2+ ions to simultaneously act on multiple negative charges on colIIα1 molecules, as demonstrated by the negligible [Ca2+] dependence measured by the OH-SAM tip (Figure 5). The physiological concentration of [Ca2+] is ≈2–4 mM,31 and within this range, variations in Ca2+ concentration can strongly affect the aggrecan–collagen adhesion (Figure 5), as well as aggrecan–aggrecan adhesion.16
Comparison to Molecular Adhesion of Other Cartilage Matrix Proteoglycans
At ≈500 nm indentation depth, there are ≈1 × 104 aggrecan monomers and ≈340 collagen fibrils simultaneously in direct molecular contact underneath the ≈7 μm2 contact area (tip radius R ≈ 2.5 μm). In PBS, the average nonspecific binding force is thus ≈0.3 pN per aggrecan monomer and ≈9 pN per collagen fibril. However, since adhesion may occur within only a fraction of these molecules, this is an estimate of the lower limit of the aggrecan–collagen binding strength. This value is comparable to the estimated per pair aggrecan–aggrecan binding strength (≈1 pN) between two opposing end-attached aggrecan layers.16 Thus, in vivo, aggrecan may have no strong preference in binding to adjacent aggrecan monomers or collagen fibrils, despite their drastically dissimilar charged nature and molecular stiffness. These nonspecific bindings are orders of magnitude weaker than the binding strength measured on individual pairs of other cartilage ECM proteoglycans via single molecule force spectroscopy, such as hyaluronan/aggrecan G1 core protein (40 ± 11 pN),73 decorin/decorin (16.5 ± 5.1 pN),73 type IX collagen/biglycan (≈15 pN),73 type I collagen/decorin (core protein; 54.5 ± 20 pN),74 and type I collagen/decorin (GAG side chain; 31.9 ± 12.4 pN).74 They are also much weaker than the adhesion measured between individual pairs of aggrecan molecules (≈150–250 pN).17 These differences can be mainly attributed to two reasons. First, for each aggrecan–collagen pair, the aggrecan–aggrecan and aggrecan–collagen intermolecular interactions from the surrounding environment can affect its local binding interactions. Second, the maximum compressive stresses applied on each molecule were lower than those between the single molecular pairs in other studies. Those experiments were measuring the upper limits of molecular adhesions between each pair of single molecules, while our experiment was designed to estimate the molecular interactions in vivo by more closely simulating the physiological ionic environment, loading conditions, molecular strains, and molecular packing density.
Implications Regarding Cartilage Tissue Assembly and Properties
In articular cartilage, aggrecan is entrapped in the 3D randomly aligned collagen network with ≈30–50% molecular strain at 20–80 mg/mL concentration.31 Cartilage tissue mechanical function is determined by the hierarchy of structure and collagen/aggrecan mechanical properties arising from the nanoscale.2,68,75 In the present study that focuses on the aggrecan–collagen adhesion, aggrecan monomers were end-grafted at a packing density (≈50 mg/mL) within this physiological concentration, and collagen fibrils are transversely randomly aligned on the cartilage surface (Figures 1b and 6). At the maximum compressive force (≈40 nN), aggrecan macromolecules on the tip were also at ≈50% molecular strain.29 This experiment thus provided a two-dimensional analog of the three-dimensional aggrecan–collagen interaction in vivo. In both cases, it is the CS-GAG versus collagen molecular contacts dominating the aggrecan–collagen interactions, as the aggrecan core proteins are buried within the densely packed CS-GAG side chains. Although previous studies have suggested high binding affinity of KS-GAGs to type II collagen,9 our experiment did not investigate the KS-GAG and collagen adhesion due to the aggrecan attachment configuration (Figure 2b). In addition, this experiment most likely excluded the molecular interactions between aggrecan versus the positively charged NC-4 domain and negatively charged CS-GAG attached to the NC-3 domain of the type IX collagen.61 As discussed before, it is also likely that some residual hyaluronan molecules may contribute to the net adhesion. However, again, contributions of these interactions to the net adhesion are believed to be minor given their low concentrations compared to the type II collagen molecules and CS-GAG side chains.
In articular cartilage, the magnitude of the aggrecan–collagen adhesion per pair molecules is much weaker compared to other specific molecular interactions that directly involve the ECM matrix assembly. For example, interactions directly involved in cartilage matrix assembly include the binding of aggrecan versus hyaluronan at the G1 domain (facilitated by the link protein),7 COMP versus type II collagen,12 and fibromodulin/decorin versus collagen fibrils.76 Aggrecan–collagen adhesion therefore is not directly involved in regulating the cartilage matrix assembly. However, given the abundance of aggrecan and collagen in cartilage ECM, aggrecan–collagen adhesion could work synergistically with the aggrecan–aggrecan self-adhesion16 as physical cross-links to affect the local conformation and deformability of aggrecan monomers, and in turn, nanoscale charge distribution heterogeneity, aggrecan entropic elasticity and effective hydraulic permeability in the ECM. Upon external loading, breaking of these physical cross-links can also provide additional energy dissipative mechanisms to enhance the shock absorption. We thus expect that these interactions play essential roles in the organization and mechanical function of cartilage, including electrostatic repulsion-driven elasticity, osmotic swelling, and fluid flow-independent viscoelasticity, as well as the fluid flow-induced poroelasticity.
Implications Regarding Osteoarthritis-Induced Cartilage Degradation
At early stage osteoarthritis, aggrecan is the first major constituent that undergoes fragmentation and depletion,77,78 followed by the disruption of the collagen network.79,80 In severe osteoarthritis, damage of both aggrecan and collagen fibrils take place simultaneously, which eventually leads to the loss of cartilage.81 In order to provide molecular insights into the progression of OA, we previously investigated the OA-induced changes in local compressive and energy-dissipative mechanical properties of cartilage tissue.34,43 Here, we focused on one particular aspect of the OA-related cartilage degradation, that is, the molecular adhesion between intact aggrecan and partially degraded collagen fibrils. Human recombinant matrix metalloprotease-13 (MMP-13) is a typical enzyme up-regulated in osteoarthritic cartilage, which contributes to the collagen fibril degradation in vivo.82 A previous immunohistochemistry study showed that 24 h MMP-13 digestion introduces partial defibrillization of collagen within the top ≈30 μm surface of cartilage (Figure S2),37 although these changes are not visible at the microscale via SEM imaging (Figure 6). As a result, we observed an ≈2.5× increase in Fad and an ≈1.5× increase in Ead (Figure 8). MMP-13 cleaves the colIIα1 molecule amino acid sequence at the locations of PQG775–776LAG and LAG778–779QRG and results in disorganized collagen fibrils.22 This defibrillization could increase the effective molecular surface contacts and deformability of type II collagen molecules and may also expose the type XI collagen molecules wrapped at the core of these fibrils.54 All these effects can increase the effective molecular contacts and, thus, the aggrecan–collagen adhesion. This deviation from normal aggrecan–collagen interactions in healthy cartilage contributes to the changes in both cartilage mechanics and chondrocyte responses. For example, an increase in aggrecan–collagen association could alter the local deformability of aggrecan, and in turn affect the visco/poroelastic energy dissipation directly linked to aggrecan/collagen, aggrecan/aggrecan, and aggrecan/water molecular friction. In addition, altered aggrecan–collagen molecular interactions upon partial degradation of collagen are also expected to affect specific interactions with other chondrocyte-binding growth factors or cytokines and, in turn, lead to altered chondrocyte signaling.83
The C. histolyticum bacterial collagenase enzyme is not directly related to physiological conditions. It cleaves the collagen fibrils at six locations at a much faster rate than MMP-1336 and represents a model of more severely damaged collagen networks. Under this scenario, we observed similar values of Fad but much greater Ead (Figure 8), suggesting more severe damage of collagen could further increase aggrecan–collagen adhesion, deviating from the normal aggrecan–collagen interactions. Interestingly, both histological and SEM studies showed that even under this more severely damaged scenario by C. histolyticum, negligible differences were observed between the undamaged (trypsin-only treated cartilage disk) and partially damaged collagen network at the tissue and fibril levels (Figures 6 and 7). This is because, at this early stage of OA-like degradation, these changes take place at the length scales beyond the resolution of these techniques. This observation thus also indicates that severe OA-induced cartilage degradation may take place at a stage much earlier than the level at which conventional techniques like histology or radiology are able to detect (Figure 7). Interestingly, previous studies have shown that whereas healthy to grade 3 osteoarthritic human cartilage exhibited no significant differences in effective indentation modulus using a microspherical tip, their moduli decreased for a factor ≈95% if measured by a softer, nanosized pyramidal tip.75 Our observation further elucidates the importance and sensitivity of molecular level phenomena taking place at the nanometer scale to osteoarthritic degradation.
Conclusions
In this study, we quantified the molecular adhesion between the two major cartilage extracellular matrix constituents, that is, aggrecan and the type II/IX/XI fibrillar collagen network, in physiological-like aqueous solutions. This aggrecan–collagen adhesion is nonspecific and governed by both GAG–GAG electrostatic repulsion and Ca2+-induced ion bridging effects in vivo. Aggrecan–collagen adhesion, similar to aggrecan–aggrecan self-adhesion, could be an important factor that determines the local assembly and molecular deformability of the cartilage matrix. By introducing osteoarthritis-like degradation via MMP-13, we found partial disruption of collagen structure leads to significant increase in aggrecan–collagen adhesion. This study provides further molecular-level insights into the assembly and degradation of cartilage tissue, as well as disease induced tissue degradation. Information obtained here contributes to the molecular-level knowledge of cartilage and osteoarthritic degradation, which can be used for designing and optimizing the early stage OA-diagnostics tools and tissue-engineering strategies for cartilage repair.
Acknowledgments
This work was supported by the National Science Foundation (Grant CMMI-0758651), the National Institutes of Health (Grant AR60331), the National Security Science and Engineering Faculty Fellowship (Grant N00244-09-1-0064), the Shriners of North America, and the Faculty Start-up Grant at Drexel University (L.H.). The authors thank the Institute for Soldier Nanotechnologies at MIT, funded through the U.S. Army Research Office, for the use of instruments.
Supporting Information Available
(1) Figure S1: Tapping mode AFM amplitude images of air dried bovine articular cartilage surface of untreated and trypsin-treated disks. (2) Figure S2: Immunohistochemistry of the untreated and MMP-13 digested cartilage disks.37 (3) Figure S3: Amino acid sequence of calf type II collagen (colIIα1).62 This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Muir I. H. M. In Adult Articular Cartilage; Freeman M. A. R., Ed.; Pitman Medical: Kent, 1979; pp 145–214. [Google Scholar]
- Han L.; Grodzinsky A. J.; Ortiz C. Annu. Rev. Mater. Res. 2011, 41, 133–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham T. E.; Fosang A. J. FASEB J. 1992, 6, 861–870. [PubMed] [Google Scholar]
- Lee H.-Y.; Han L.; Roughley P. J.; Grodzinsky A. J.; Ortiz C. J. Struct. Biol. 2013, 181, 264–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinegård D. Int. J. Exp. Pathol. 2009, 90, 575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozzo R. V.; Goldoni S.; Berendsen A. D.; Young M. F. In The Extracellular Matrix: An Overview; Mecham R. F., Ed.; Springer-Verlag: Berlin, 2011; pp 197–231. [Google Scholar]
- Hardingham T. E.; Muir H. Biochim. Biophys. Acta 1972, 279, 401–405. [DOI] [PubMed] [Google Scholar]
- Buckwalter J. A.; Rosenberg L. C.; Tang L.-H. J. Biol. Chem. 1984, 259, 5361–5363. [PubMed] [Google Scholar]
- Hedlund H.; Hedbom E.; Heinegård D.; Mengarelli-Widholm S.; Reinholt F. P.; Svensson O. J. Biol. Chem. 1999, 274, 5777–5781. [DOI] [PubMed] [Google Scholar]
- Pratta M. A.; Yao W.; Decicco C.; Tortorella M. D.; Liu R.-Q.; Copeland R. A.; Magolda R.; Newton R. C.; Trzaskos J. M.; Arner E. C. J. Biol. Chem. 2003, 278, 45539–45545. [DOI] [PubMed] [Google Scholar]
- Douglas T.; Heinemann S.; Bierbaum S.; Scharnweber D.; Worch H. Biomacromolecules 2006, 7, 2388–2393. [DOI] [PubMed] [Google Scholar]
- Hálasz K.; Kassner A.; Mörgelin M.; Heinegård D. J. Biol. Chem. 2007, 282, 31166–31173. [DOI] [PubMed] [Google Scholar]
- Wiberg C.; Heinegård D.; Wenglén C.; Timpl R.; Mörgelin M. J. Biol. Chem. 2002, 277, 49120–49126. [DOI] [PubMed] [Google Scholar]
- Hildebrand A.; Romarís M.; Rasmussen L. M.; Heinegård D.; Twardzik D. R.; Border W. A.; Ruoslahti E. Biochem. J. 1994, 302, 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakao K.; Takahashi K. A.; Arai Y.; Saito M.; Honjyo K.; Hiraoka N.; Kishida T.; Mazda O.; Imanishi J.; Kubo T. J. Orthop. Sci. 2009, 14, 738–747. [DOI] [PubMed] [Google Scholar]
- Han L.; Dean D.; Daher L. A.; Grodzinsky A. J.; Ortiz C. Biophys. J. 2008, 95, 4862–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder A.; Walhorn V.; Dierks T.; Fernàndez-Busquets X.; Anselmetti D. Biophys. J. 2010, 99, 3498–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter K.; Kidder L. H.; Levin I. W.; Lewis E. N.; Spencer R. G. S. Arthritis Rheum. 2001, 44, 846–855. [DOI] [PubMed] [Google Scholar]
- Bonassar L. J.; Frank E. H.; Murray J. C.; Paguio C. G.; Moore V. L.; Lark M. W.; Sandy J. D.; Wu J.-J.; Eyre D. R.; Grodzinsky A. J. Arthritis Rheum. 1995, 38, 173–183. [DOI] [PubMed] [Google Scholar]
- Lewis J. L.; Johnson S. L. J. Anat. 2001, 199, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. B.; Mow V. C.; Chun L. E.; Eyre D. R. J. Orthop. Res. 1990, 8, 353–363. [DOI] [PubMed] [Google Scholar]
- Billinghurst R. C.; Dahlberg L.; Ionescu M.; Reiner A.; Bourne R.; Rorabeck C.; Mitchell P.; Hambor J.; Diekmann O.; Tschesche H.; Chen J.; VanWart H.; Poole A. R. J. Clin. Invest. 1997, 99, 1534–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zareian R.; Church K. P.; Saeidi N.; Flynn B. P.; Beale J. W.; Ruberti J. W. Langmuir 2010, 26, 9917–9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farndale R. W.; Buttle D. J.; Barrett A. J. Biochim. Biophys. Acta 1986, 883, 173–177. [DOI] [PubMed] [Google Scholar]
- Ohtani O. Arch. Histol. Jpn. 1987, 50, 557–566. [DOI] [PubMed] [Google Scholar]
- Petersen W.; Tillmann B. Anat. Embryol. 1998, 197, 317–324. [DOI] [PubMed] [Google Scholar]
- Ng L.; Grodzinsky A. J.; Patwari P.; Sandy J.; Plaas A.; Ortiz C. J. Struct. Biol. 2003, 143, 242–257. [DOI] [PubMed] [Google Scholar]
- Dean D.; Han L.; Ortiz C.; Grodzinsky A. J. Macromolecules 2005, 38, 4047–4049. [Google Scholar]
- Dean D.; Han L.; Grodzinsky A. J.; Ortiz C. J. Biomech. 2006, 39, 2555–2565. [DOI] [PubMed] [Google Scholar]
- Han L.; Dean D.; Ortiz C.; Grodzinsky A. J. Biophys. J. 2007, 92, 1384–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroudas A. In Adult Articular Cartilage; Freeman M. A. R., Ed.; Pitman: England, 1979; pp 215–290. [Google Scholar]
- Hutter J. L.; Bechhoefer J. Rev. Sci. Instrum. 1993, 64, 1868–1873. [Google Scholar]
- Lin D. C.; Dimitriadis E. K.; Horkay F. J. Biomech. Eng. 2007, 129, 430–440. [DOI] [PubMed] [Google Scholar]
- Han L.; Frank E. H.; Greene J. J.; Lee H.-Y.; Hung H.-H. K.; Grodzinsky A. J.; Ortiz C. Biophys. J. 2011, 100, 1846–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. M. J. Anat. 1990, 171, 117–130. [PMC free article] [PubMed] [Google Scholar]
- French M. F.; Bhown A.; Vanwart H. E. J. Protein Chem. 1992, 11, 83–97. [DOI] [PubMed] [Google Scholar]
- Treppo S.Physical Diagnostics of Cartilage Degeneration. Massachusetts Institute of Technology, Ph.D. Thesis, 1999. [Google Scholar]
- Swann D. A.; Slayter H. S.; Silver F. H. J. Biol. Chem. 1981, 256, 5921–5925. [PubMed] [Google Scholar]
- Schwarz I. M.; Hills B. A. Br. J. Rheumatol. 1998, 37, 21–26. [DOI] [PubMed] [Google Scholar]
- Schmidt T. A.; Gastelum N. S.; Nguyen Q. T.; Schumacher B. L.; Sah R. L. Arthritis Rheum. 2007, 56, 882–891. [DOI] [PubMed] [Google Scholar]
- Crockett R.; Grubelnik A.; Roos S.; Dora C.; Born W.; Troxler H. J. Biomed. Mater. Res. A 2007, 82, 958–964. [DOI] [PubMed] [Google Scholar]
- Xia Y.; Zheng S.; Bidthanapally A. J. Magn. Reson. Imaging 2008, 28, 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nia H. T.; Bozchalooi I. S.; Li Y.; Han L.; Hung H.-H.; Frank E. H.; Youcef-Toumi K.; Ortiz C.; Grodzinsky A. J. Biophys. J. 2013, 104, 1529–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F. E.; Ismail H. M.; Didangelos A.; Peirce M.; Vincent T. L.; Wait R.; Saklatvala J. Arthritis Rheum. 2013, 65, 397–407. [DOI] [PubMed] [Google Scholar]
- Hascall V. C.; Handley C. J.; McQuillan D. J.; Hascall G. K.; Robinson H. C.; Lowther D. A. Arch. Biochem. Biophys. 1983, 224, 206–223. [DOI] [PubMed] [Google Scholar]
- Li Y.; Frank E. H.; Wang Y.; Chubinskaya S.; Huang H.-H.; Grodzinsky A. J. Osteoarthritis Cartilage 2013, 21, 1933–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crockett R.; Roos S.; Rossbach P.; Dora C.; Born W.; Troxler H. Tribol. Lett. 2005, 19, 311–317. [Google Scholar]
- Sun Y.; Chen M.-Y.; Zhao C.; An K.-N.; Amadio P. C. J. Orthop. Res. 2008, 26, 1225–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.; Homandberg G.; Madsen L.; Su J.; Kuettner K. Trans. Orthop. Res. Soc. 2002, 48, 359. [Google Scholar]
- Holmes M. W. A.; Bayliss M. T.; Muir H. Biochem. J. 1988, 250, 435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkkinen J. J.; Häkkinen T. P.; Savolainen S.; Wang C.; Tammi R.; Ågren U. M.; Lammi M. J.; Arokoski J.; Helminen H. J.; Tammi M. I. Histochem. Cell Biol. 1996, 105, 187–194. [DOI] [PubMed] [Google Scholar]
- Chan S. M. T.; Neu C. P.; DuRaine G.; Komvopoulos K.; Reddi A. H. Osteoarthritis Cartilage 2010, 18, 956–963. [DOI] [PubMed] [Google Scholar]
- Cleland R. L.; Wang J. L.; Detweiler D. M. Macromolecules 1982, 15, 386–395. [Google Scholar]
- Eyre D. Arthritis Res. 2002, 4, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre D. R.; Weis M. A.; Wu J.-J. Eur. Cell. Mater. 2006, 12, 57–63. [DOI] [PubMed] [Google Scholar]
- Ichimura S.; Wu J. J.; Eyre D. R. Arch. Biochem. Biophys. 2000, 378, 33–39. [DOI] [PubMed] [Google Scholar]
- Eyre D. R.; Apon S.; Wu J.-J.; Ericsson L. H.; Walsh K. A. FEBS Lett. 1987, 220, 337–341. [DOI] [PubMed] [Google Scholar]
- Blaschke U. K.; Eikenberry E. F.; Hulmes D. J. S.; Galla H.-J.; Bruckner P. J. Biol. Chem. 2000, 275, 10370–10378. [DOI] [PubMed] [Google Scholar]
- Vaughan L.; Mendler M.; Huber S.; Bruckner P.; Winterhalter K. H.; Irwin M. I.; Mayne R. J. Cell Biol. 1988, 106, 991–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman M.; Ainola M.; Valmu L.; Bjartell A.; Ma G.; Stenman U.-H.; Sorsa T.; Luukkainen R.; Konttinen Y. T. Am. J. Pathol. 2005, 167, 1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihlajamaa T.; Lankinen H.; Ylöstalo J.; Valmu L.; Jäälinoja J.; Zaucke F.; Spitznagel L.; Gösling S.; Puustinen A.; Mörgelin M.; Peränen J.; Maurer P.; Ala-Kokko L.; Kilpeläinen I. J. Biol. Chem. 2004, 279, 24265–24273. [DOI] [PubMed] [Google Scholar]
- CO2A1_BOVIN (P02459). http://www.uniprot.org/uniprot/P02459. In UnitProtKB.
- Spillmann D.; Burger M. M. In Carbohydrates in Chemistry and Biology; Ernst B., Hart G. W., Sinaÿ P., Eds.; Wiley-VCH: Weinheim, 2000; Vol. 2, Chap. 38, pp 1061–1091. [Google Scholar]
- Scott J. E. FASEB J. 1992, 6, 2639–2645. [PubMed] [Google Scholar]
- Parker K. H.; Winlove C. P.; Maroudas A. Biophys. Chem. 1988, 32, 271–282. [DOI] [PubMed] [Google Scholar]
- de la Cruz M. O.; Belloni L.; Delsanti M.; Dalbiez J. P.; Spalla O.; Drifford M. J. Chem. Phys. 1995, 103, 5781–5791. [Google Scholar]
- Stan G.; DelRio F. W.; MacCuspie R. I.; Cook R. F. J. Phys. Chem. B 2012, 116, 3138–3147. [DOI] [PubMed] [Google Scholar]
- Laric M.; Wirz D.; Daniels A. U.; Raiteri R.; VanLandingham M. R.; Guex G.; Martin I.; Aebi U.; Stolz M. Biophys. J. 2010, 98, 2731–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L.; Dean D.; Mao P.; Ortiz C.; Grodzinsky A. J. Biophys. J. 2007, 93, L23–L25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor E. A.; Bowness J. M. Can. J. Biochem. 1971, 49, 417–425. [DOI] [PubMed] [Google Scholar]
- Hunter G. K.; Wong K. S.; Kim J. J. Arch. Biochem. Biophys. 1988, 260, 161–167. [DOI] [PubMed] [Google Scholar]
- Saleque S.; Ruiz N.; Drickamer K. Glycobiology 1993, 3, 185–190. [DOI] [PubMed] [Google Scholar]
- Chen C.-H.; Yeh M.-L.; Geyer M.; Wang G.-J.; Huang M.-H.; Heggeness M. H.; Höök M.; Luo Z.-P. Biochem. Biophys. Res. Commun. 2006, 339, 204–208. [DOI] [PubMed] [Google Scholar]
- Yeh M.-L.; Luo Z.-P. Scanning 2004, 26, 273–276. [DOI] [PubMed] [Google Scholar]
- Stolz M.; Gottardi R.; Raiteri R.; Miot S.; Martin I.; Imer R.; Staufer U.; Raducanu A.; Düggelin M.; Baschong W.; Daniels A. U.; Friederich N. F.; Aszodi A.; Aebi U. Nat. Nanotechnol. 2009, 4, 186–192. [DOI] [PubMed] [Google Scholar]
- Hedbom E.; Heinegård D. J. Biol. Chem. 1993, 268, 27307–27312. [PubMed] [Google Scholar]
- Sandy J. D.; Neame P. J.; Boynton R. E.; Flannery C. R. J. Biol. Chem. 1991, 266, 8683–8685. [PubMed] [Google Scholar]
- Lark M. W.; Bayne E. K.; Lohmander L. S. Acta Orthop. Scand. Suppl. 1995, 266, 92–7. [PubMed] [Google Scholar]
- Poole A. R.; Kobayashi M.; Yasuda T.; Laverty S.; Mwale F.; Kojima T.; Sakai T.; Wahl C.; El-Maadawy S.; Webb G.; Tchetina E.; Wu W. Ann. Rheum. Dis. 2002, 61, ii78–ii81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinegård D.; Saxne T. Nat. Rev. Rheumatol. 2011, 7, 50–56. [DOI] [PubMed] [Google Scholar]
- Shakoor N.; Block J. A.; Shott S.; Case J. P. Arthritis Rheum. 2002, 46, 3185–3189. [DOI] [PubMed] [Google Scholar]
- Goldring M. B.; Otero M.; Plumb D. A.; Dragomir C.; Favero M.; El Hachem K.; Hashimoto K.; Roach H. I.; Olivotto E.; Borzì R. M.; Marcu K. B. Eur. Cells Mater. 2011, 21, 202–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen S. M.; Lammi M. J.; Saarakkala S.; Han S.-K.; Herzog W.; Tanska P.; Korhonen R. K. Biomech. Model. Mechanobiol. 2013, 12, 417–429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


