Abstract
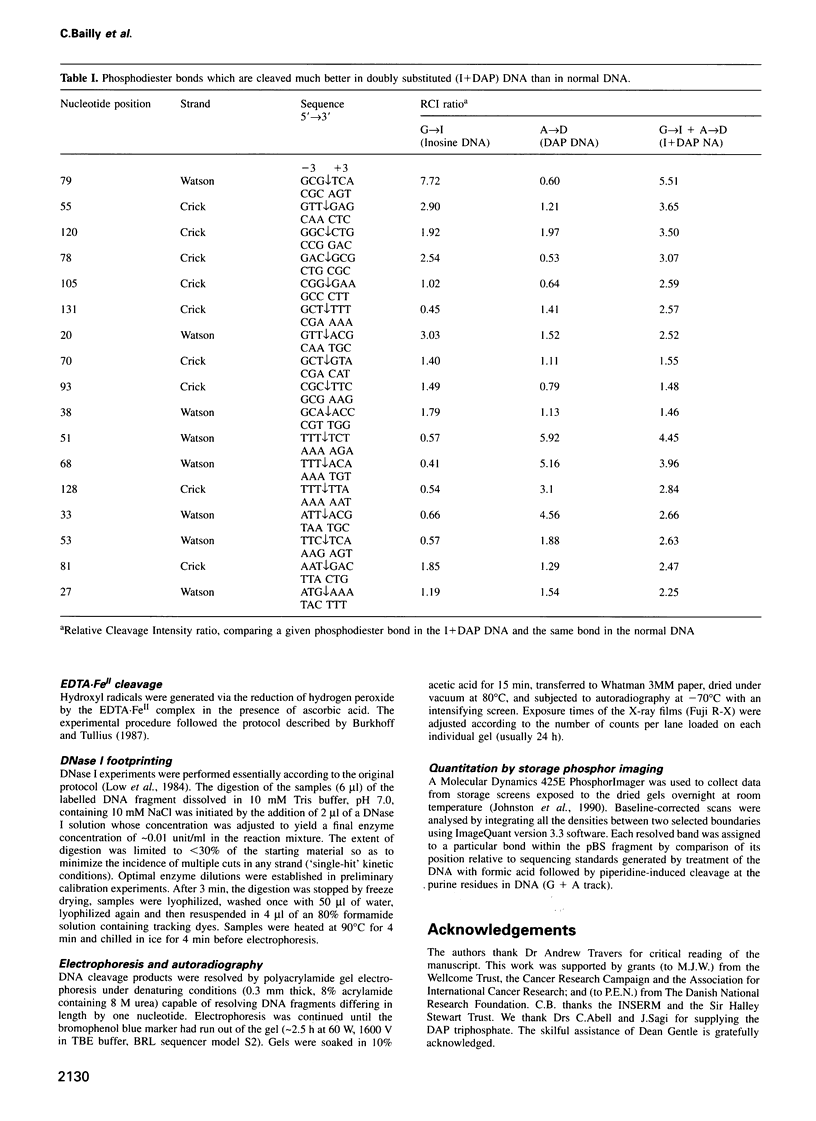
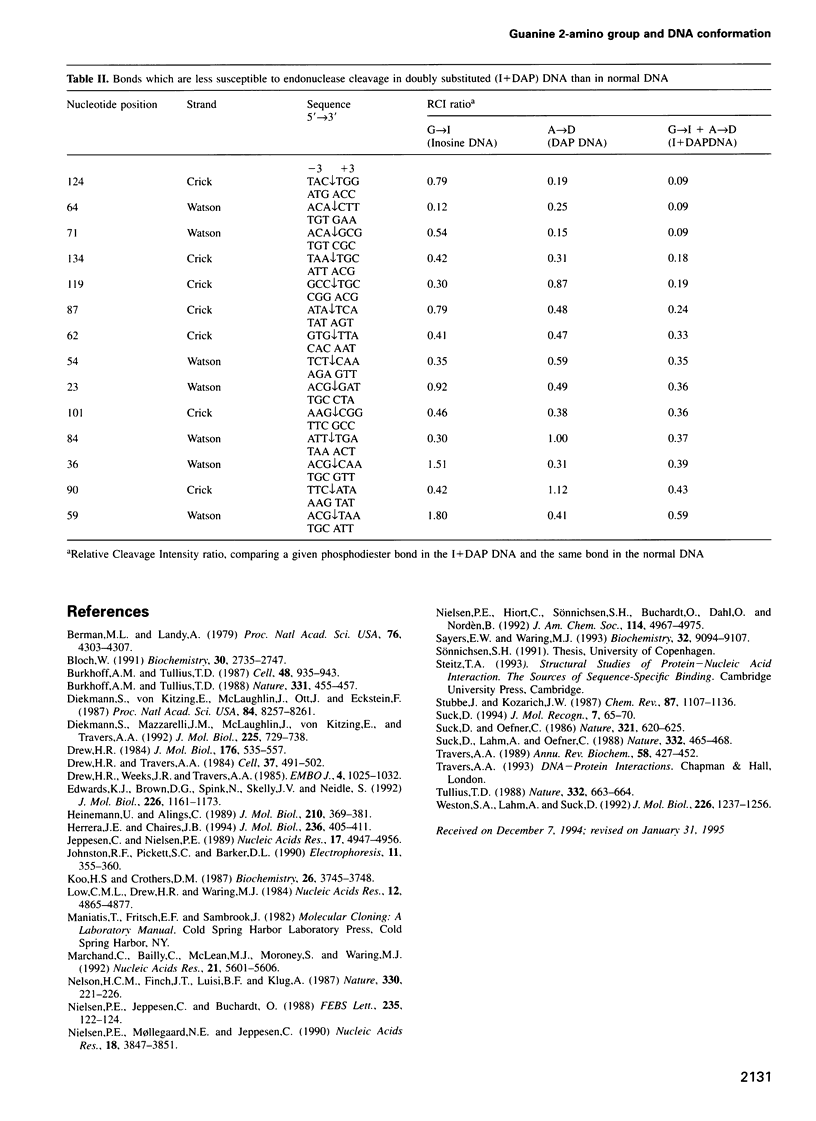
The conformation of the DNA helix is supposed to be a critical element in site-specific recognition by ligands both large and small. Groove width is one important measure of the conformation which varies with the local nucleotide composition, perhaps because of the presence of a purine 2-amino group on G.C base pairs. We have probed DNA with G-->inosine (I) and/or A-->diaminopurine (DAP) substitutions to see whether the location of the purine 2-amino group can indeed affect the minor groove width. At acid pH, the reactivity towards uranyl nitrate is modulated in substituted DNA quite differently from natural DNA, consistent with a marked narrowing of the minor groove at sites of G-->I substitution and widening at sites of A-->DAP replacement. The latter exerts the dominant effect. The expected changes in conformation are equally evident in the patterns of susceptibility to DNase I cleavage, but not to hydroxyl radical attack. Nuclease cleavage is maximal in normal and substituted DNA at regions of inferred moderate groove width which are generally little affected by the nucleotide substitutions. Consistent with models of sequence-dependent cutting by DNase I we find that the presence of a purine 2-amino group on the base pair three places upstream of the cutting site has a profound influence on the rate of reaction.
Full text
PDF
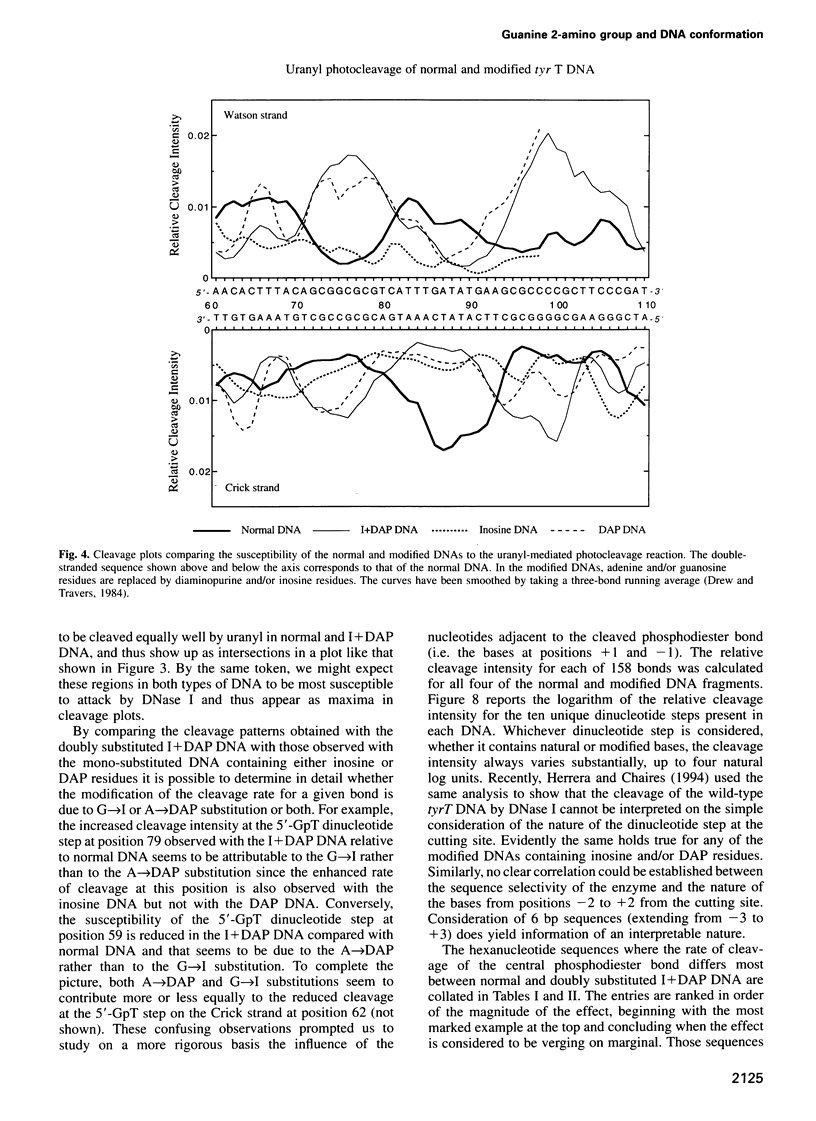
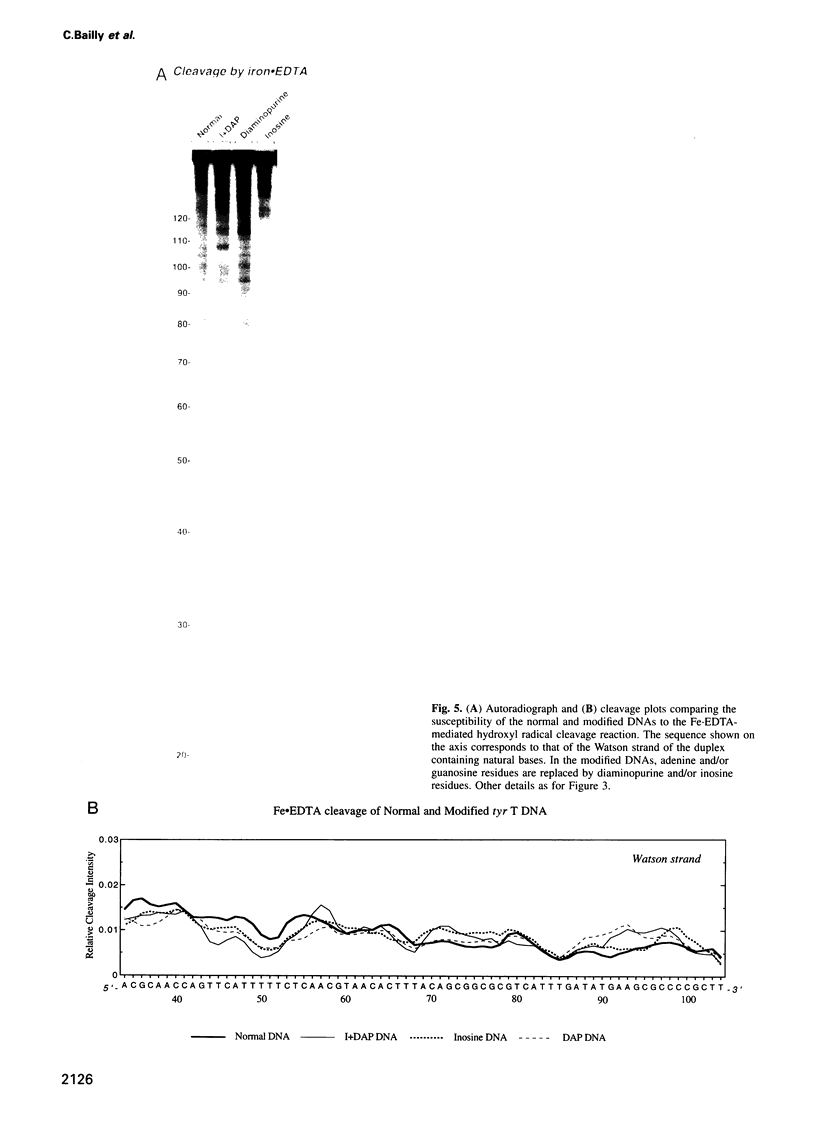

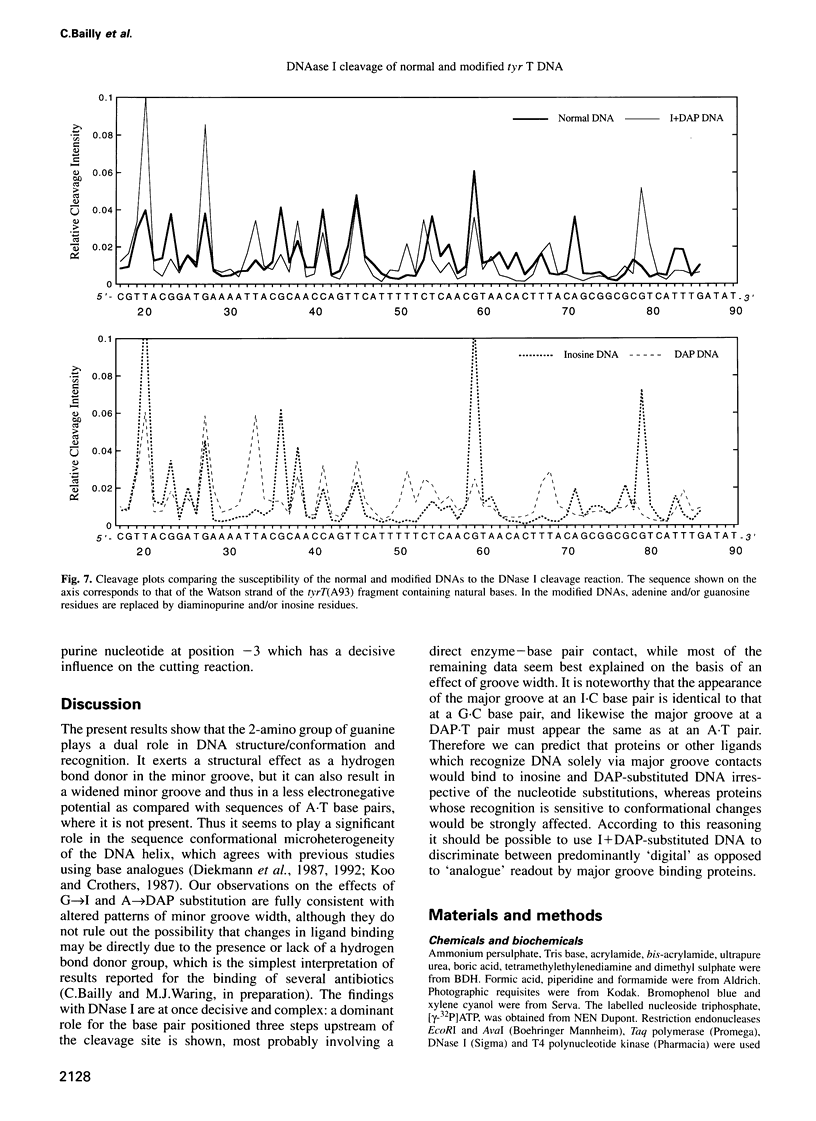
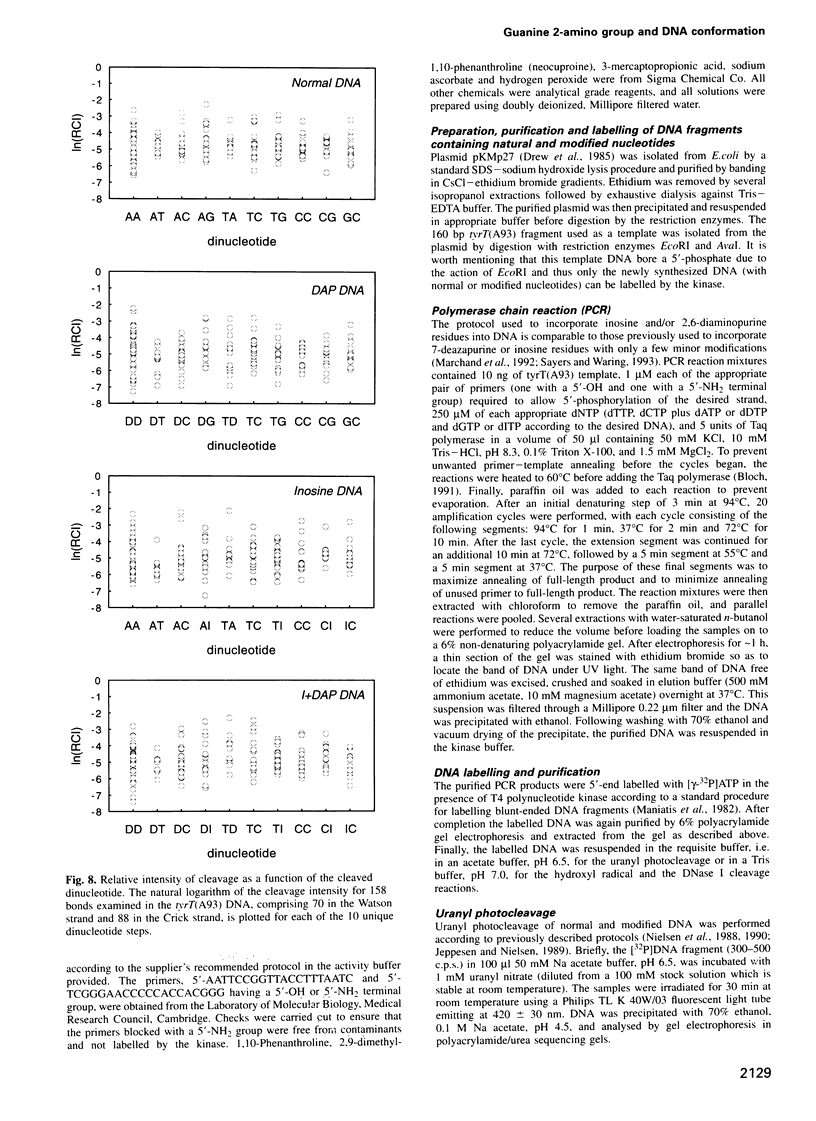
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman M. L., Landy A. Promoter mutations in the transfer RNA gene tyrT of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4303–4307. doi: 10.1073/pnas.76.9.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch W. A biochemical perspective of the polymerase chain reaction. Biochemistry. 1991 Mar 19;30(11):2735–2747. doi: 10.1021/bi00225a001. [DOI] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. Structural details of an adenine tract that does not cause DNA to bend. Nature. 1988 Feb 4;331(6155):455–457. doi: 10.1038/331455a0. [DOI] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Diekmann S., Mazzarelli J. M., McLaughlin L. W., von Kitzing E., Travers A. A. DNA curvature does not require bifurcated hydrogen bonds or pyrimidine methyl groups. J Mol Biol. 1992 Jun 5;225(3):729–738. doi: 10.1016/0022-2836(92)90397-3. [DOI] [PubMed] [Google Scholar]
- Diekmann S., von Kitzing E., McLaughlin L., Ott J., Eckstein F. The influence of exocyclic substituents of purine bases on DNA curvature. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8257–8261. doi: 10.1073/pnas.84.23.8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Weeks J. R., Travers A. A. Negative supercoiling induces spontaneous unwinding of a bacterial promoter. EMBO J. 1985 Apr;4(4):1025–1032. doi: 10.1002/j.1460-2075.1985.tb03734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K. J., Brown D. G., Spink N., Skelly J. V., Neidle S. Molecular structure of the B-DNA dodecamer d(CGCAAATTTGCG)2. An examination of propeller twist and minor-groove water structure at 2.2 A resolution. J Mol Biol. 1992 Aug 20;226(4):1161–1173. doi: 10.1016/0022-2836(92)91059-x. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Alings C. Crystallographic study of one turn of G/C-rich B-DNA. J Mol Biol. 1989 Nov 20;210(2):369–381. doi: 10.1016/0022-2836(89)90337-9. [DOI] [PubMed] [Google Scholar]
- Herrera J. E., Chaires J. B. Characterization of preferred deoxyribonuclease I cleavage sites. J Mol Biol. 1994 Feb 18;236(2):405–411. doi: 10.1006/jmbi.1994.1152. [DOI] [PubMed] [Google Scholar]
- Jeppesen C., Nielsen P. E. Uranyl mediated photofootprinting reveals strong E. coli RNA polymerase--DNA backbone contacts in the +10 region of the DeoP1 promoter open complex. Nucleic Acids Res. 1989 Jul 11;17(13):4947–4956. doi: 10.1093/nar/17.13.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. F., Pickett S. C., Barker D. L. Autoradiography using storage phosphor technology. Electrophoresis. 1990 May;11(5):355–360. doi: 10.1002/elps.1150110503. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Crothers D. M. Chemical determinants of DNA bending at adenine-thymine tracts. Biochemistry. 1987 Jun 16;26(12):3745–3748. doi: 10.1021/bi00386a070. [DOI] [PubMed] [Google Scholar]
- Low C. M., Drew H. R., Waring M. J. Sequence-specific binding of echinomycin to DNA: evidence for conformational changes affecting flanking sequences. Nucleic Acids Res. 1984 Jun 25;12(12):4865–4879. doi: 10.1093/nar/12.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand C., Bailly C., McLean M. J., Moroney S. E., Waring M. J. The 2-amino group of guanine is absolutely required for specific binding of the anti-cancer antibiotic echinomycin to DNA. Nucleic Acids Res. 1992 Nov 11;20(21):5601–5606. doi: 10.1093/nar/20.21.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E., Jeppesen C., Buchardt O. Uranyl salts as photochemical agents for cleavage of DNA and probing of protein-DNA contacts. FEBS Lett. 1988 Aug 1;235(1-2):122–124. doi: 10.1016/0014-5793(88)81245-6. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E., Møllegaard N. E., Jeppesen C. DNA conformational analysis in solution by uranyl mediated photocleavage. Nucleic Acids Res. 1990 Jul 11;18(13):3847–3851. doi: 10.1093/nar/18.13.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E. W., Waring M. J. Footprinting titration studies on the binding of echinomycin to DNA incapable of forming Hoogsteen base pairs. Biochemistry. 1993 Sep 7;32(35):9094–9107. doi: 10.1021/bi00086a014. [DOI] [PubMed] [Google Scholar]
- Suck D. DNA recognition by DNase I. J Mol Recognit. 1994 Jun;7(2):65–70. doi: 10.1002/jmr.300070203. [DOI] [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Suck D., Oefner C. Structure of DNase I at 2.0 A resolution suggests a mechanism for binding to and cutting DNA. Nature. 1986 Jun 5;321(6070):620–625. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Tullius T. D. DNA footprinting with hydroxyl radical. Nature. 1988 Apr 14;332(6165):663–664. doi: 10.1038/332663a0. [DOI] [PubMed] [Google Scholar]
- Weston S. A., Lahm A., Suck D. X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution. J Mol Biol. 1992 Aug 20;226(4):1237–1256. doi: 10.1016/0022-2836(92)91064-v. [DOI] [PubMed] [Google Scholar]