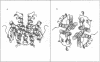Abstract
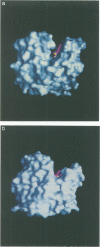
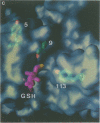
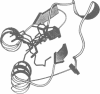
Glutathione S-transferases (GSTs) are a family of enzymes involved in the cellular detoxification of xenotoxins. Cytosolic GSTs have been grouped into four evolutionary classes for which there are representative crystal structures of three of them. Here we report the first crystal structure of a theta-class GST. So far, all available GST crystal structures suggest that a strictly conserved tyrosine near the N-terminus plays a critical role in the reaction mechanism and such a role has been convincingly demonstrated by site-directed mutagenesis. Surprisingly, the equivalent residue in the theta-class structure is not in the active site, but its role appears to have been replaced by either a nearby serine or by another tyrosine residue located in the C-terminal domain of the enzyme.
Full text
PDF
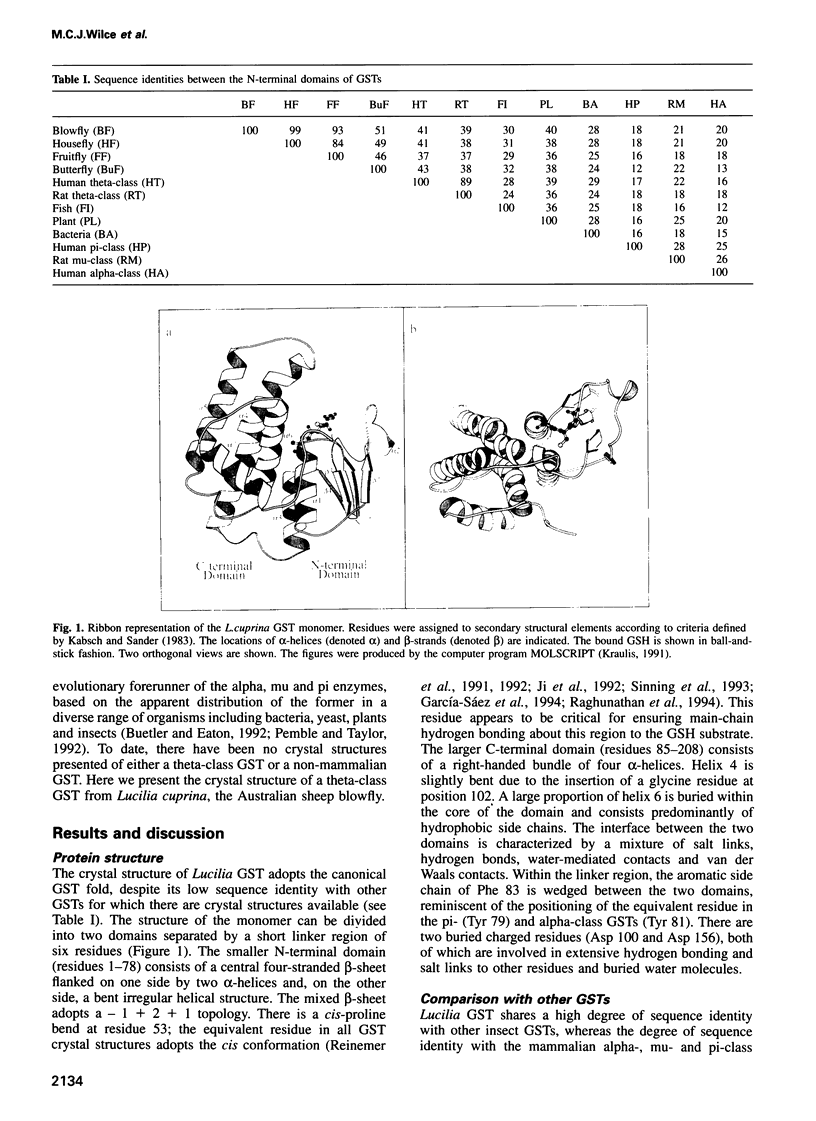
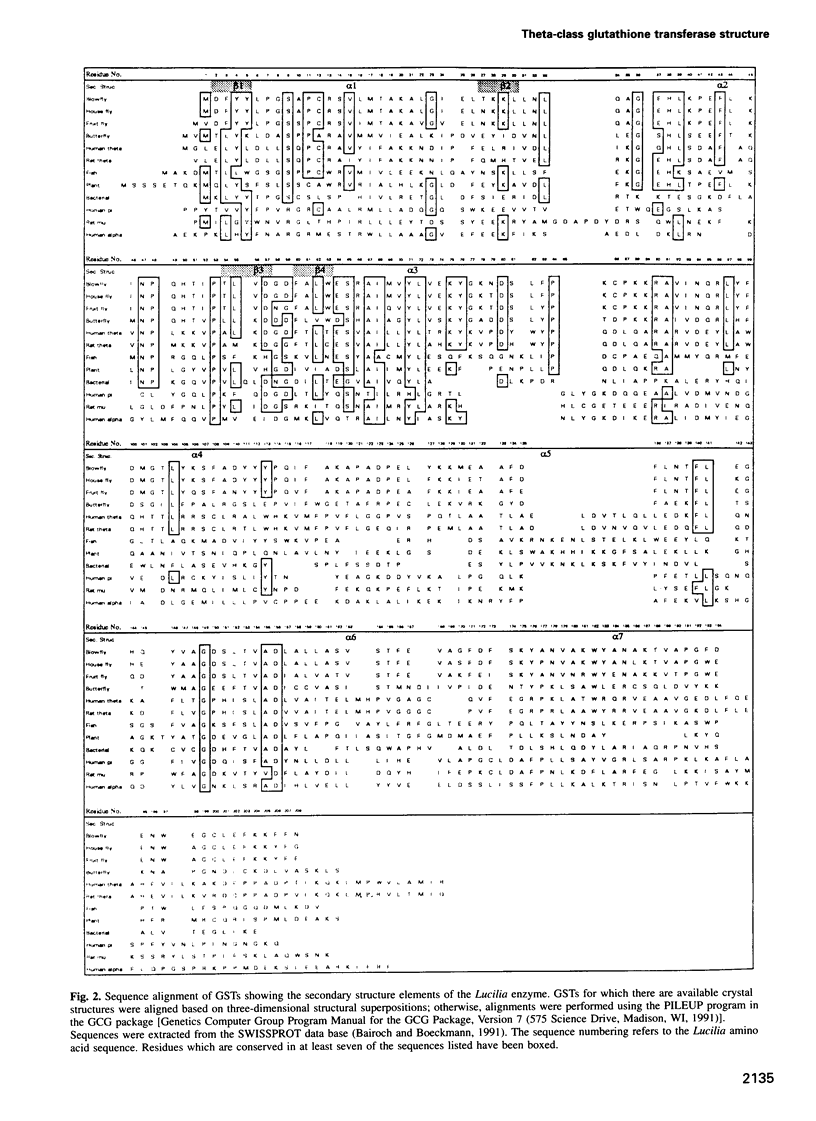
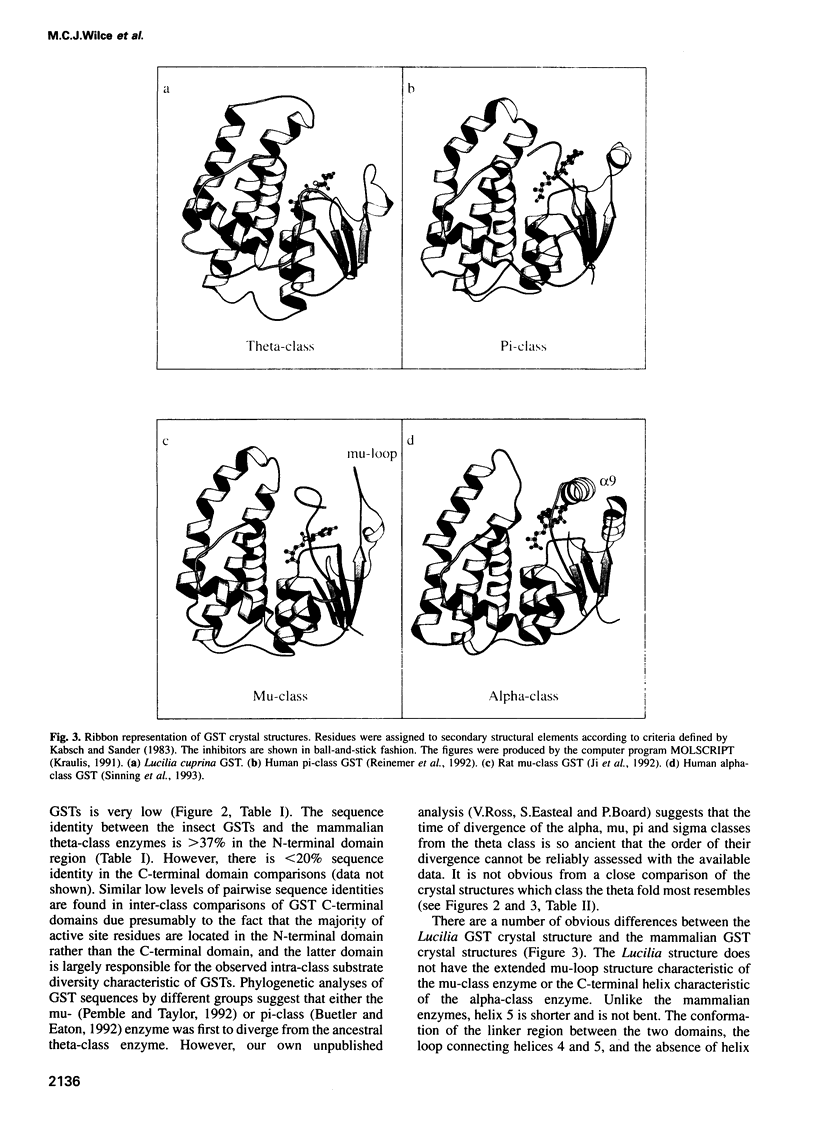

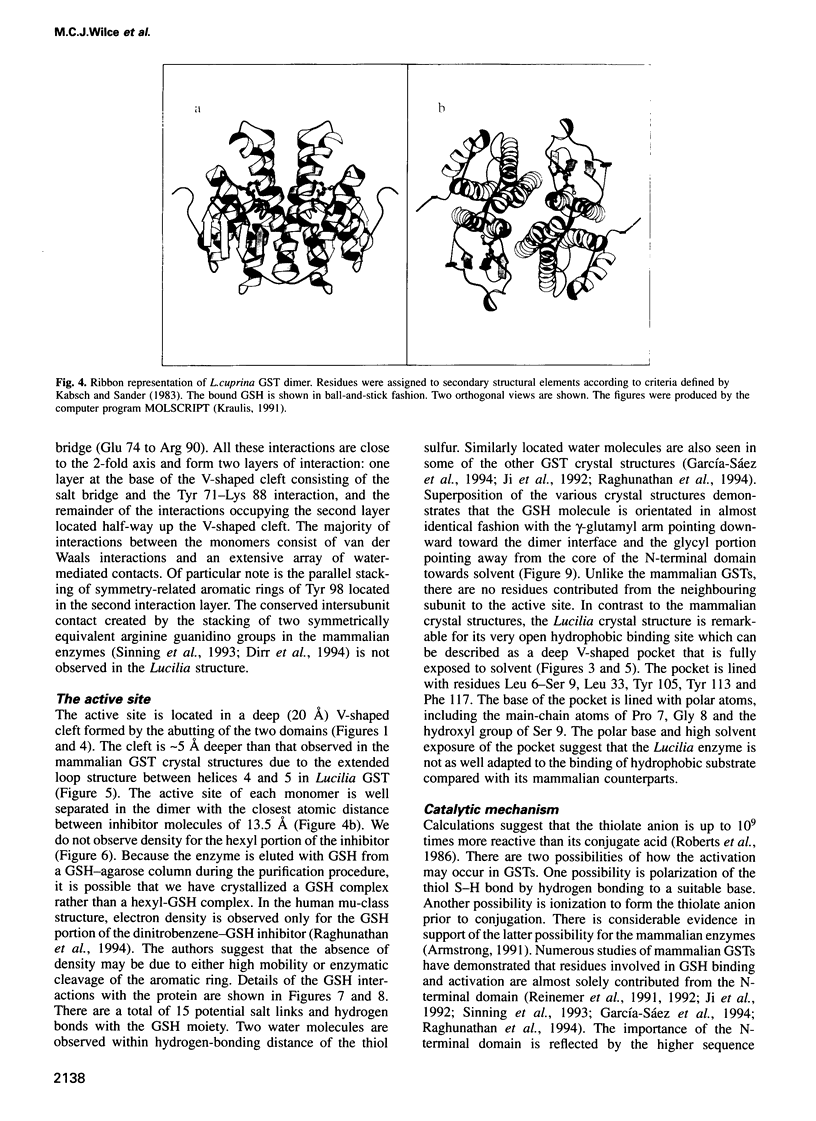




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong R. N. Glutathione S-transferases: reaction mechanism, structure, and function. Chem Res Toxicol. 1991 Mar-Apr;4(2):131–140. doi: 10.1021/tx00020a001. [DOI] [PubMed] [Google Scholar]
- Atkins W. M., Wang R. W., Bird A. W., Newton D. J., Lu A. Y. The catalytic mechanism of glutathione S-transferase (GST). Spectroscopic determination of the pKa of Tyr-9 in rat alpha 1-1 GST. J Biol Chem. 1993 Sep 15;268(26):19188–19191. [PubMed] [Google Scholar]
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2247–2249. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barycki J. J., Colman R. F. Affinity labeling of glutathione S-transferase, isozyme 4-4, by 4-(fluorosulfonyl)benzoic acid reveals Tyr115 to be an important determinant of xenobiotic substrate specificity. Biochemistry. 1993 Dec 7;32(48):13002–13011. doi: 10.1021/bi00211a008. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Board P., Coggan M., Johnston P., Ross V., Suzuki T., Webb G. Genetic heterogeneity of the human glutathione transferases: a complex of gene families. Pharmacol Ther. 1990;48(3):357–369. doi: 10.1016/0163-7258(90)90054-6. [DOI] [PubMed] [Google Scholar]
- Board P., Russell R. J., Marano R. J., Oakeshott J. G. Purification, molecular cloning and heterologous expression of a glutathione S-transferase from the Australian sheep blowfly (Lucilia cuprina). Biochem J. 1994 Apr 15;299(Pt 2):425–430. doi: 10.1042/bj2990425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Clark A. G. The comparative enzymology of the glutathione S-transferases from non-vertebrate organisms. Comp Biochem Physiol B. 1989;92(3):419–446. doi: 10.1016/0305-0491(89)90114-4. [DOI] [PubMed] [Google Scholar]
- Cowtan K. D., Main P. Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta Crystallogr D Biol Crystallogr. 1993 Jan 1;49(Pt 1):148–157. doi: 10.1107/S0907444992007698. [DOI] [PubMed] [Google Scholar]
- Dirr H., Reinemer P., Huber R. X-ray crystal structures of cytosolic glutathione S-transferases. Implications for protein architecture, substrate recognition and catalytic function. Eur J Biochem. 1994 Mar 15;220(3):645–661. doi: 10.1111/j.1432-1033.1994.tb18666.x. [DOI] [PubMed] [Google Scholar]
- Fournier D., Bride J. M., Poirie M., Bergé J. B., Plapp F. W., Jr Insect glutathione S-transferases. Biochemical characteristics of the major forms from houseflies susceptible and resistant to insecticides. J Biol Chem. 1992 Jan 25;267(3):1840–1845. [PubMed] [Google Scholar]
- García-Sáez I., Párraga A., Phillips M. F., Mantle T. J., Coll M. Molecular structure at 1.8 A of mouse liver class pi glutathione S-transferase complexed with S-(p-nitrobenzyl)glutathione and other inhibitors. J Mol Biol. 1994 Apr 1;237(3):298–314. doi: 10.1006/jmbi.1994.1232. [DOI] [PubMed] [Google Scholar]
- Hiratsuka A., Sebata N., Kawashima K., Okuda H., Ogura K., Watabe T., Satoh K., Hatayama I., Tsuchida S., Ishikawa T. A new class of rat glutathione S-transferase Yrs-Yrs inactivating reactive sulfate esters as metabolites of carcinogenic arylmethanols. J Biol Chem. 1990 Jul 15;265(20):11973–11981. [PubMed] [Google Scholar]
- Ji X., Armstrong R. N., Gilliland G. L. Snapshots along the reaction coordinate of an SNAr reaction catalyzed by glutathione transferase. Biochemistry. 1993 Dec 7;32(48):12949–12954. doi: 10.1021/bi00211a001. [DOI] [PubMed] [Google Scholar]
- Ji X., Johnson W. W., Sesay M. A., Dickert L., Prasad S. M., Ammon H. L., Armstrong R. N., Gilliland G. L. Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene. Biochemistry. 1994 Feb 8;33(5):1043–1052. doi: 10.1021/bi00171a002. [DOI] [PubMed] [Google Scholar]
- Ji X., Zhang P., Armstrong R. N., Gilliland G. L. The three-dimensional structure of a glutathione S-transferase from the mu gene class. Structural analysis of the binary complex of isoenzyme 3-3 and glutathione at 2.2-A resolution. Biochemistry. 1992 Oct 27;31(42):10169–10184. doi: 10.1021/bi00157a004. [DOI] [PubMed] [Google Scholar]
- Johnson W. W., Liu S., Ji X., Gilliland G. L., Armstrong R. N. Tyrosine 115 participates both in chemical and physical steps of the catalytic mechanism of a glutathione S-transferase. J Biol Chem. 1993 Jun 5;268(16):11508–11511. [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kolm R. H., Sroga G. E., Mannervik B. Participation of the phenolic hydroxyl group of Tyr-8 in the catalytic mechanism of human glutathione transferase P1-1. Biochem J. 1992 Jul 15;285(Pt 2):537–540. doi: 10.1042/bj2850537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong K. H., Nishida M., Inoue H., Takahashi K. Tyrosine-7 is an essential residue for the catalytic activity of human class PI glutathione S-transferase: chemical modification and site-directed mutagenesis studies. Biochem Biophys Res Commun. 1992 Feb 14;182(3):1122–1129. doi: 10.1016/0006-291x(92)91848-k. [DOI] [PubMed] [Google Scholar]
- Kong K. H., Takasu K., Inoue H., Takahashi K. Tyrosine-7 in human class Pi glutathione S-transferase is important for lowering the pKa of the thiol group of glutathione in the enzyme-glutathione complex. Biochem Biophys Res Commun. 1992 Apr 15;184(1):194–197. doi: 10.1016/0006-291x(92)91177-r. [DOI] [PubMed] [Google Scholar]
- Liu S., Zhang P., Ji X., Johnson W. W., Gilliland G. L., Armstrong R. N. Contribution of tyrosine 6 to the catalytic mechanism of isoenzyme 3-3 of glutathione S-transferase. J Biol Chem. 1992 Mar 5;267(7):4296–4299. [PubMed] [Google Scholar]
- Lopez M. F., Patton W. F., Sawlivich W. B., Erdjument-Bromage H., Barry P., Gmyrek K., Hines T., Tempst P., Skea W. M. A glutathione S-transferase (GST) isozyme from broccoli with significant sequence homology to the mammalian theta-class of GSTs. Biochim Biophys Acta. 1994 Mar 16;1205(1):29–38. doi: 10.1016/0167-4838(94)90088-4. [DOI] [PubMed] [Google Scholar]
- Mannervik B., Danielson U. H. Glutathione transferases--structure and catalytic activity. CRC Crit Rev Biochem. 1988;23(3):283–337. doi: 10.3109/10409238809088226. [DOI] [PubMed] [Google Scholar]
- Manoharan T. H., Gulick A. M., Puchalski R. B., Servais A. L., Fahl W. E. Structural studies on human glutathione S-transferase pi. Substitution mutations to determine amino acids necessary for binding glutathione. J Biol Chem. 1992 Sep 15;267(26):18940–18945. [PubMed] [Google Scholar]
- Manoharan T. H., Gulick A. M., Reinemer P., Dirr H. W., Huber R., Fahl W. E. Mutational substitution of residues implicated by crystal structure in binding the substrate glutathione to human glutathione S-transferase pi. J Mol Biol. 1992 Jul 20;226(2):319–322. doi: 10.1016/0022-2836(92)90949-k. [DOI] [PubMed] [Google Scholar]
- Meyer D. J., Coles B., Pemble S. E., Gilmore K. S., Fraser G. M., Ketterer B. Theta, a new class of glutathione transferases purified from rat and man. Biochem J. 1991 Mar 1;274(Pt 2):409–414. doi: 10.1042/bj2740409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D. J., Xia C., Coles B., Chen H., Reinemer P., Huber R., Ketterer B. Unusual reactivity of Tyr-7 of GSH transferase P1-1. Biochem J. 1993 Jul 15;293(Pt 2):351–356. doi: 10.1042/bj2930351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp K. A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Pemble S. E., Taylor J. B. An evolutionary perspective on glutathione transferases inferred from class-theta glutathione transferase cDNA sequences. Biochem J. 1992 Nov 1;287(Pt 3):957–963. doi: 10.1042/bj2870957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penington C. J., Rule G. S. Mapping the substrate-binding site of a human class mu glutathione transferase using nuclear magnetic resonance spectroscopy. Biochemistry. 1992 Mar 24;31(11):2912–2920. doi: 10.1021/bi00126a010. [DOI] [PubMed] [Google Scholar]
- Raghunathan S., Chandross R. J., Kretsinger R. H., Allison T. J., Penington C. J., Rule G. S. Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity. J Mol Biol. 1994 May 20;238(5):815–832. doi: 10.1006/jmbi.1994.1336. [DOI] [PubMed] [Google Scholar]
- Reinemer P., Dirr H. W., Ladenstein R., Huber R., Lo Bello M., Federici G., Parker M. W. Three-dimensional structure of class pi glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 A resolution. J Mol Biol. 1992 Sep 5;227(1):214–226. doi: 10.1016/0022-2836(92)90692-d. [DOI] [PubMed] [Google Scholar]
- Reinemer P., Dirr H. W., Ladenstein R., Schäffer J., Gallay O., Huber R. The three-dimensional structure of class pi glutathione S-transferase in complex with glutathione sulfonate at 2.3 A resolution. EMBO J. 1991 Aug;10(8):1997–2005. doi: 10.1002/j.1460-2075.1991.tb07729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. D., Lewis S. D., Ballou D. P., Olson S. T., Shafer J. A. Reactivity of small thiolate anions and cysteine-25 in papain toward methyl methanethiosulfonate. Biochemistry. 1986 Sep 23;25(19):5595–5601. doi: 10.1021/bi00367a038. [DOI] [PubMed] [Google Scholar]
- Sinning I., Kleywegt G. J., Cowan S. W., Reinemer P., Dirr H. W., Huber R., Gilliland G. L., Armstrong R. N., Ji X., Board P. G. Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the Mu and Pi class enzymes. J Mol Biol. 1993 Jul 5;232(1):192–212. doi: 10.1006/jmbi.1993.1376. [DOI] [PubMed] [Google Scholar]
- Stenberg G., Board P. G., Mannervik B. Mutation of an evolutionarily conserved tyrosine residue in the active site of a human class Alpha glutathione transferase. FEBS Lett. 1991 Nov 18;293(1-2):153–155. doi: 10.1016/0014-5793(91)81174-7. [DOI] [PubMed] [Google Scholar]
- Wang R. W., Newton D. J., Huskey S. E., McKeever B. M., Pickett C. B., Lu A. Y. Site-directed mutagenesis of glutathione S-transferase YaYa. Important roles of tyrosine 9 and aspartic acid 101 in catalysis. J Biol Chem. 1992 Oct 5;267(28):19866–19871. [PubMed] [Google Scholar]
- Wilce M. C., Feil S. C., Board P. G., Parker M. W. Crystallization and preliminary X-ray diffraction studies of a glutathione S-transferase from the Australian sheep blowfly, Lucilia cuprina. J Mol Biol. 1994 Mar 11;236(5):1407–1409. doi: 10.1016/0022-2836(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Wilce M. C., Parker M. W. Structure and function of glutathione S-transferases. Biochim Biophys Acta. 1994 Mar 16;1205(1):1–18. doi: 10.1016/0167-4838(94)90086-8. [DOI] [PubMed] [Google Scholar]