Abstract
Among the three human apolipoprotein E (apoE) isoforms, apoE4 increases the risk of Alzheimer’s disease (AD). While transporting cholesterol is a primary function, apoE also regulates amyloid-β (Aβ) metabolism, aggregation and deposition. Although earlier work suggests that different affinities of apoE isoforms to Aβ might account for their effects on Aβ clearance, recent studies indicate that apoE also competes with Aβ for cellular uptake through apoE receptors. Thus, several factors likely determine the variable effects apoE has on Aβ. In this review, we examine biochemical, structural, and functional studies and propose testable models that address the complex mechanisms underlying apoE-Aβ interaction and how apoE4 may increase AD risk and also serve as a target pathway for therapy.
Keywords: Alzheimer’s disease, apolipoprotein E, amyloid-β, aggregation, clearance, cholesterol, endocytosis, lysosome, degradation, LRP1, LDLR, HSPG
Introduction
Alzheimer’s disease (AD) is the most common form of late-life mental failure in humans, which accounts for an estimated 60 to 80% of dementia cases (Thies and Bleiler, 2013). Senile plaques and intracellular neurofibrillary tangles are hallmarks of AD pathology. The generation of amyloid-β (Aβ) peptides of either 40 or 42 amino acids in length, from amyloid precursor protein (APP), and their subsequent accumulation, aggregation and deposition in brain parenchyma, as senile plaques, and in perivascular regions, as cerebral amyloid angiopathy (CAA), are central and perhaps defining events in the pathogenesis of AD (Hardy and Selkoe, 2002). In addition to Aβ deposits, soluble Aβ oligomers are shown to injure synapses by disrupting normal synaptic functions and triggering downstream toxic pathways leading to eventual neurodegeneration and cognitive deficits (Mucke and Selkoe, 2012). Thus, the “amyloid cascade hypothesis” is strongly supported by evidence of Aβ-related pathology early in the disease process and the specific roles of different forms of Aβ in neurotoxicity.
Fewer than 1% of AD cases are caused by dominantly inherited genetic mutations in genes including APP, PSEN1 and PSEN2 (Thies and Bleiler, 2013). Inheriting any of these genetic mutations acceleratesA β production resulting in the development of AD usually before the age of 60, whichis commonly referred to as early-onset familial AD(FAD). Late-onset AD (LOAD), or the sporadic occurrence of the disease later in life, represents the majority of AD cases (Hardy and Selkoe, 2002). In LOAD, the disturbance of Aβ clearance machinery appears to be a leading cause of Aβ accumulation in the brain (Mawuenyega et al., 2010). Indisputable evidence showing the ε4 allele of the APOE gene, which encodes a lipid/cholesterol carrier apolipoprotein E (apoE), is a stronger genetic risk factor for AD than the more common ε3 allele, whereas the presence of the ε2 allele is protective (Bu, 2009; Liu et al., 2013). The presence of the ε4 allele of the APOE gene not only dose-dependently increases the risk for AD but also lowers the age of onset (Bu, 2009; Corder et al., 1993; Liu et al., 2013). ApoE4 contributes to the pathogenesis of AD likely by both loss-of-function in neuroprotection and gain-of-function in neurotoxicity compared to apoE3 (Bu, 2009; Huang and Mucke, 2012). Although several pathways underlying the risk associated with apoE4-linkedAD have been defined through in vitro and in vivo studies, the exact mechanisms are still not completely understood and those that have been proposed remain controversial. Nonetheless, the differential effects of apoE isoforms on amyloid pathology and Aβ metabolism have been confirmed in humans, animal models and cellular studies. In this review, we will discuss how apoE isoforms differentially regulate AD pathogenic pathways with particular focus on Aβ-dependent pathways.
Back to basics: Biochemical and structural features of apoE
ApoE is a glycoprotein of 299 amino acids with a molecular mass of ~34 kDa. It was originally identified as an apolipoprotein enriched on cholesterol- and triglyceride-rich plasma lipoproteins synthesized by the liver in humans and animals (Mahley, 1988; Mahley and Rall, 2000). ApoE mediates the transport and delivery of cholesterol and other lipids through cell surface apoE receptors (Mahley, 1988; Mahley and Rall, 2000). The mRNA profile has shown that the liver is the major tissue in which apoE is synthesized accounting for >75% of total apoE, followed by the brain (Elshourbagy et al., 1985). The concentrations of apoE in plasma and cerebrospinal fluid (CSF) are estimated to be ~ 40–70 μg/ml and ~ 3–5 μg/ml, respectively (Mahley et al., 2009; Pitas et al., 1987b). ApoE reporter mice with EGFP insertion into the Apoe gene locus exhibit highly expressed apoE in hepatocytes and peritoneal macrophages. In the brain, astrocytes, microglia, vascular smooth muscle cells and choroid plexus constitutively express apoE, whereas neurons predominantly synthesize apoE under stress conditions (Xu et al., 2006). The multiple isoforms of human apoE were first identified when human apoE polymorphisms were characterized using isoelectric focusing with plasma samples from patients with familial lipoprotein disorder type III hyperlipoproteinemia (Utermann et al., 1977). Because of their different charges, human apoE has mainly three patterns of band designated with each isoelectric point as E2 (pH 5.4), E3 (pH 5.55) and E4 (pH 6.1) (Mahley and Rall, 2000; Utermann et al., 1977). The heterogeneity of major isoforms apoE2, apoE3 and apoE4 is due to genetic polymorphisms (Zannis and Breslow, 1981). Complete amino acid sequencing revealed that apoE2 has Cys residues at positions 112 and 158, apoE3 has a Cys residue at 112 and an Arg residue at 158 and apoE4 has Arg residues at both positions (Rall et al., 1982; Weisgraber et al., 1981) (Figure 1). These differences at amino acid residues 112 and 158 among apoE isoforms likely induce significant, perhaps profound, changes in their structures and associated biological functions. The minor isoforms result from variable posttranslational O-linked glycosylation/sialylation of major apoE isoforms at Thr 194, resulting in further negative charges (Figure 1) (Wernette-Hammond et al., 1989; Zannis and Breslow, 1981). Posttranslational modification and subsequent deglycosylation of apoE likely lead to its heterogeneous sialylation. Although newly secreted apoE is highly sialylated (Wernette-Hammond et al., 1989), 80–85% of human plasma apoE is in the asialo form, with monosialo and disialo isoforms as the minor species (Zannis and Breslow, 1981). Interestingly, apoE in the brain has higher sialylation than that in plasma (Pitas et al., 1987a). Further studies are needed to understand how glycosylation of apoE affects its metabolism and functions in an isoform-dependent and/or tissue-dependent manner.
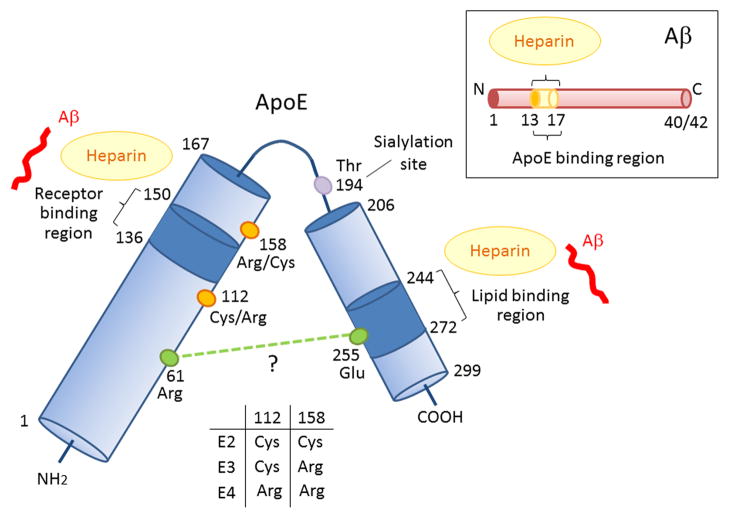
Figure 1. Schematic illustration of structural and functional regions of apoE and Aβ.
Human apoE is a glycosylated protein of 299 amino acids consisting of a receptor-binding region (residues 136–150) in the N-terminal domain (residues 1–167) and a lipid-binding region (residues 244–272) in the C-terminal domain (residues 206–299) (Chen et al., 2011). ApoE also has two heparin-binding sites, each within the N-terminal and the C-terminal domains (Ji et al., 1993; Saito et al., 2003). The residues that distinguish the apoE isoforms are located at residues 112 and 158, where apoE2 has Cys residues at both positions, apoE3 has a Cys residue at 112 and an Arg residue at 158, and apoE4 has Arg residues at both positions (Rall et al., 1982; Weisgraber et al., 1981). The domain interaction between Arg 61 and Glu 255 in apoE4 is also indicated (Mahley and Rall, 2000; Mahley et al., 2009; Wilson et al., 1991), although other structural studies have not confirmed this (Chen et al., 2011). Aβ can interact with both the receptor-binding region and lipid-binding region of apoE, as well as with heparin through its residues 13–17 (Strittmatter et al., 1993; Winkler et al., 1999).
The nuclear magnetic resonance (NMR) of a monomeric mutant form of apoE3, which has mutations in the C-terminal domain to prevent aggregation, revealed for the first time the full-length structure of apoE in which the N-terminal domain (residues 1–167) and C-terminal domain (residues 206–299) are separated by a hinge region (residues 168–205) (Chen et al., 2011). The N-terminal domain contains a four-helix-bundle, which was detected by both X-ray crystallography (Wilson et al., 1991) and NMR (Sivashanmugam and Wang, 2009). In monomeric mutant apoE3, Arg 61 forms a hydrogen bond with Thr 194 and Glu 255 forming a salt-bridge with Lys 95 (Chen et al., 2011). In contrast to this, X-ray crystallographic analyses of the N-terminal domain structure of apoE revealed that Arg 112 forms a salt bridge with Glu 109 resulting inthe exposure of Arg 61 away from the four-helix bundle in apoE4, whereas this side chain in apoE3 is buried (Dong et al., 1994). As a result, Arg 61 in apoE4 is predicted to interact with Glu 255 leading to interactions between the N- and C-terminal domains (Mahley and Rall, 2000; Mahley et al., 2009; Wilson et al., 1991). Fluorescence resonance energy transfer (FRET) and electron paramagnetic resonance (EPR) revealed that the distance between Arg 61 and Glu 255 is closer in both lipid-free and phospholipid-bound apoE4 than in apoE3 (Hatters et al., 2005). This domain-domain interaction was also observed in apoE4-expression neuronal cells by live cell imaging (Xu et al., 2004). Although the NMR structure does not support the domain-domain interaction in apoE4, it is likely that the single amino acid difference at position 112 between apoE3 (Cys) and apoE4 (Arg) has an influence on its structure. Future technology that allows for determination of full-length wild-type apoE3 and apoE4, either in lipid-free or lipid-bound state, should teach us how a single amino acid difference can have a profound impact on structure and functions, in particular those related to AD pathways.
The major functional regions of apoE are the receptor-binding site (residues 136–150) in the N-terminal domain and the lipid-binding site (residues 244–272) in the C-terminal domain (Figure 1) (Hatters et al., 2006a). NMR analysis of apoE3 demonstrated that several hydrophilic residues in the C-terminal domain are buried within the domain interface. The exposed hydrophobic residues destabilize the C-terminal domain and form a large exposed hydrophobic surface to attract lipids for initial binding (Chen et al., 2011). ApoE3 and apoE2 display a preference for high-density lipoproteins (HDL), whereas apoE4 is frequently associated with very low-density lipoproteins (VLDL) and low-density lipoproteins (LDL) (Dong et al., 1994; Hatters et al., 2006a). In apoE4, replacement of Arg 61 with Thr, or Glu 255 with Ala leads to apoE3-like HDL preference (Dong et al., 1994; Hatters et al., 2006a). Therefore, Arg 61, Glu 255 or their interaction is likely necessary for VLDL preference in apoE4. One consequence of different lipoprotein preference by apoE4 is the increased plasma cholesterol levels, which in turn increases the risk for cardiovascular diseases, in particular atherosclerosis(Davignon et al., 1988).
Major apoE receptors belong to the LDL receptor (LDLR) family, including LDLR (Innerarity and Mahley, 1978), LDLR-related protein 1 (LRP1) (Beisiegel et al., 1989; Kowal et al., 1989), VLDL receptor (VLDLR) and apoE receptor 2 (apoER2) (Kim et al., 1996). LDLR interacts with both apoB- LDL and apoE-HDL particles (Innerarity et al., 1979); however, apoE-HDL particles have a 10- to 100-fold higher binding affinity to the cell surface receptor than the apoB-LDL particles (Innerarity and Mahley, 1978). LRP1 recognizes more than 30 structurally and functionally distinct ligands, including apoE, α2-macroglobulin, tissue-type plasminogen activator, APP and Aβ (Herz and Strickland, 2001). Although both LRP1 and LDLR are major apoE metabolic receptors, they differ in ligand recognition. While LRP1 prefers apoE-enriched particles or apoE aggregates, LDLR binds naturedly secreted or circulating apoE particles (Bu, 2009). Thus, the receptor-binding specificity of apoE is influenced by its lipidation state and the density of apoE epitopes. For binding to LDLR, the positively charged residues within the N-terminal receptor-binding region of apoE are likely buried in a lipid-free state but are exposed upon lipidation (Chen et al., 2011). LRP1 binding, which appears to require either apoE enrichment or aggregation, is less understood but may require simultaneous exposure of multiple receptor-binding epitopes. In addition to the LDLR family, apoE also binds to cell surface heparan sulfate proteoglycans (HSPG) (Ji et al., 1993), either in lipid-free or lipidated forms (Saito et al., 2003), through both the receptor-binding region in the N-terminal domain and the basic residues around Lys 233 in the C-terminal domain (Saito et al., 2003) (Figure 1). The C-terminal domain of recombinant apoE protein displays a stronger affinity and faster kinetics in terms of binding to heparin. Thus, HSPG-binding through basic residues around Lys 233 is likely to be the first step, which makes the receptor-binding region in N-terminal domain available for the second-step of HSPG-binding (Futamura et al., 2005). Furthermore, LRP1 can form a complex with HSPG (Ji et al., 1993). In the HSPG/LRP1 uptake pathway, either apoE first binds to HSPG and is then transferred to LRP1 for subsequent uptake, or it can bind directly to the HSPG/LRP1 complex (Ji et al., 1993). The receptor-binding function of apoE has been shown to be isoform-dependent. While apoE3 and apoE4 bind with similar affinity to LDLR, apoE2 has less binding affinity (Schneider et al., 1981). The unique Cys 158 in apoE2 eliminates a salt bridge between Arg 158 and Asp 154 with the formation of a new salt bridge between Arg 150 and Asp 154, thus reducing its affinity to the LDLR (Dong et al., 1994). Due to this reduced binding of apoE2 to LDLR, apoE2 homozygous individuals are at higher risk for a genetic disorder termed type III hyperlipoproteinemia, which is characterized by increased plasma levels of cholesterol and triglycerides (Hatters et al., 2006a). The reduced affinity of apoE2 to LRP1 compared to apoE3 and apoE4 appears to be less severe (Kowal et al., 1990) and there seem to be no major differences amongst the apoE isoforms in their ability to bind to HSPG (Mahley and Rall, 2000).
Although we still do not know the full-length structures of wild-type apoE due to its highly aggregative nature, numerous biochemical, structural and functional studies in lipid transport have provided important clues as to why subtle changes in sequence among the three apoE isoforms can generate sufficient differences in risk for cardiovascular diseases and AD. As we further explore the mechanisms of apoE isoforms in AD pathogenesis, in particular those related to Aβ pathways, it is important to consider all available knowledge, some of which was generated decades ago (Mahley, 1988), as we design further studies to advance our understanding of apoE physiological and pathophysiological functions.
ApoE and Alzheimer’s disease: Amyloid is the clue
In humans, the APOE gene exists as three polymorphic alleles (ε2, ε3 and ε4) with the APOE ε3 allele being the most common (77%) and ε2 allele the least common (8%) (Mahley, 1988). Much evidence has confirmed that APOE4 is the strongest genetic risk factor for AD (Harold et al., 2009; Lambert et al., 2009), as APOE4 significantly increases the risk for both early-onset AD and late-onset AD (Chartier-Harlin et al., 1994; Houlden et al., 1998). The ε4 allele frequency is estimated to be about 15% in the general population, but is ~40% in AD patients (Farrer et al., 1997). In addition to increasing the prevalence of AD, the presence of the APOE4 allele also lowers the age of onset for AD in a gene dose-dependent manner (Corder et al., 1993; Farrer et al., 1997). Among ε4 homozygotes, the frequency of AD and mean age at clinical onset are 91% and 68 years of age respectively, compared to, 47% and 76 years of age in ε4 heterozygotes, and 20% and 84 years in ε4 non-carriers (Corder et al., 1993). Importantly, Aβ deposition as senile plaques is more abundant in APOE4 carriers compared with non-carriers (Kok et al., 2009; Polvikoski et al., 1995), and APOE4 increases Aβ deposition in the brains of elderly subjects with normal cognitive function, although the association is weaker than that in AD patients (Reiman et al., 2009).
A large scale study of CSF samples from both normal individuals and AD patients demonstrated that apoE protein levels in CSF positively associated with CSF Aβ42 levels independent of APOE4 genotype, although there was no significant difference in CSF apoE levels between AD patients and normal individuals (Cruchaga et al., 2012). Because there is a strong correlation between low CSF Aβ42 and high Aβ brain deposition detected by Pittsburgh compound B (PiB)-positron emission tomography (PET) imaging, lower CSF apoE levels in effect correlate with increased Aβ deposition in the brain (Fagan et al., 2006). Thus, CSF apoE protein levels appear to hold potential as an endophenotype marker for AD pathology. Interestingly, CSF and plasma apoE protein levels are reduced in APOE4 carriers (Cruchaga et al., 2012). As there was no association between apoE genotype and apoE mRNA levels (Cruchaga et al., 2012), the effect of apoE isoforms on CSF apoE levels is likely due to the different conformational stability which affects their proteolysis. The apoE isoforms differ in the stability of their N-terminal domains with apoE4 being the least resistant to thermal and chemical denaturation, apoE2 the most, and apoE3 showing intermediate resistance (Hatters et al., 2006a). These studies suggest that reduced apoE levels in APOE4 carriers and in AD patients can facilitate the accumulation of Aβ in the brain. Supporting this, when Aβ and apoE levels were analyzed in multiple brain areas of non-demented individuals, apoE levels negatively correlated with Aβ levels both in APOE4 carriers and non-carriers, indicating that apoE affects region-specific Aβ levels by preventing Aβ accumulation (Shinohara et al., 2013). Taken together, these clinical studies using human samples suggest that apoE isoforms impact AD pathogenesis by driving Aβ pathology. The following sections in this review discuss preclinical studies highlighting potential pathways by which apoE isoforms affect Aβ metabolism and aggregation.
ApoE/Aβ complexes: How we assess them makes a world of difference
Histological analyses of AD brains reveal that apoE is co-deposited with Aβ in amyloid plaques (Namba et al., 1991), indicating a direct association between apoE and Aβ in AD pathogenesis. Several in vitro studies have shown that synthetic Aβ peptides can bind to secreted apoE (cell lines) (LaDu et al., 1994) or purified apoE from human CSF (Wisniewski et al., 1993) and plasma (Strittmatter et al., 1993). Human recombinant apoE binds to immobilized Aβ40 with a high affinity (KD = ~20 nM). Its interaction is greatly influenced by the conformational state of Aβ, where apoE shows stronger preference for Aβ peptides with higher β-sheet structure (Golabek et al., 1996). Epitope mapping reveals that residues 13–17 in Aβ and residues 144–148 in the apoE N-terminal region, as part of the receptor-binding domain (Winkler et al., 1999), are common sites that interact with each other. In addition, residues 244–272 in the apoE C-terminal region also appear to be critical for the formation of apoE/Aβ complexes (Strittmatter et al., 1993). Interestingly, heparin interacts with both Aβ-binding sites in apoE and apoE binding site in Aβ (Figure 1) (Brunden et al., 1993; Saito et al., 2003). Thus, the heparin binding motifs in both apoE and Aβ are critical for their interaction. HSPG is a major cell surface receptor for both apoE and Aβ; thus, improved understanding of the interactions between apoE, Aβ and cell surface HSPG, as discussed later, should provide further clues as to how these interactions influence AD pathogenesis.
The notion that an Aβ binding site overlaps with the lipid binding region within the apoE C-terminal domain suggests that Aβ and lipids might compete with one another for apoE binding (Figure 1). In the brain, apoE is mainly secreted by astrocytes, where a plasma membrane ATP binding cassette transporter A1 (ABCA1) loads cholesterol and other lipids onto lipoprotein particles (Koldamova et al., 2010). Astrocyte-secreted apoE particles are small and have different lipid components compared with apoE particles in CSF (LaDu et al., 1998), indicating that newly synthesized apoE particles likely undergo modifications before they are transferred to neurons. Indeed, in vitro experiments have shown that lipid-free recombinant apoE interacts with immobilized Aβ with a higher affinity than lipidated recombinant apoE particles (Verghese et al., 2013), although conflicting results have also been reported (Tokuda et al., 2000). Since Aβ can interact with both the lipid-binding site and the receptor-binding site within apoE, the lipidation condition of apoE may dictate both Aβ binding affinity and the binding site. Further, it has been reported that incubation of recombinant apoE with Aβ oligomers severely impairs its lipid-binding ability (Tamamizu-Kato et al., 2008), suggesting that Aβ oligomers can impair the physiological function of apoE. As such ability of apoE to transport lipids to neurons is critical for synaptic maintenance and repair (Mahley and Rall, 2000), Aβ oligomer-mediated disturbance of lipid binding in AD brains may further compromise synaptic integrity and function.
The interaction between apoE and Aβ appears to be predicated on the isoform being studies, its lipidation status and the cellular compartment generating it, however, the methods of choice for evaluating apoE/Aβ complexes may significantly influence results. For example, initial studies using purified and delipidated apoE suggest that apoE4 binds to Aβ with faster kinetics than apoE3 when detected by Western blotting, and such an interaction is sensitive to both reducing agents and pH (Strittmatter et al., 1993). Subsequent studies using both HEK293 cell-secreted and native plasma apoE particles produced opposite results with apoE3 forming more abundant SDS-stable complexes with Aβ than apoE4 (LaDu et al., 1994; LaDu et al., 1995). By directly comparing purified, delipidated apoE with native apoE, it was shown that apoE purification, which de-lipidates apoE, changes the behaviors of apoE such that purification itself attenuates isoform-specific binding to Aβ (LaDu et al., 1995). Using EPR to assess apoE binding to Aβ oligomers in solution and surface plasmon resonance (SPR) on solid phase, Petrlova et al. also detected stronger interactions for apoE3 than apoE4 (Petrlova et al., 2011), further supporting the notion that apoE3 might be more proficient in carrying Aβ for cellular clearance and/or preventing Aβ from aggregation or neurotoxicity.
Our recent collaborative study led by the LaDu group has shown apoE4/Aβ complexes are less stable than those formed with apoE2 and apoE3 (Tai et al., 2013). Using a novel ELISA quantifying apoE/Aβ complexes, it was shown that despite similar amounts of apoE/Aβ complexes formed between Aβ and apoE2, apoE3, and apoE4, apoE4/Aβ complexes are less stable in the presence of a denaturing agent or lower pH. Interestingly, the amounts of apoE/Aβ complexes are fewer in APOE4 carriers and in CSF from AD patients, which also have higher levels of Aβ oligomers. These results indicate the apoE isoform-dependent function of apoE/Aβ complexes and suggest that the amounts of both apoE/Aβ complexes and Aβ oligomers can serve as potential biomarkers for AD. One possible mechanism responsible for the less stable nature of apoE4/Aβ complexes is the poorer lipidation status of apoE4/lipoprotein particles. However, the pathophysiological factors that influence the stability of apoE/Aβ complexes and how Aβ binding to apoE impacts both the amounts and toxic functions of Aβ oligomers require further investigation.
Despite abundant evidence that apoE exhibits isoform-specific binding to Aβ and that apoE/Aβ complexes might modulate Aβ metabolism, the significance of apoE/Aβ complexes was called into question by a recent study led by the Holtzman group (Verghese et al., 2013). In this study, when astrocyte-secreted or in vitro-reconstituted apoE particles were mixed with cell-derived Aβ using concentrations resembling physiological conditions, i.e., apoE:Aβ ratio at 50–150:1 (Hesse et al., 2000; Wahrle et al., 2007), the authors detected minimal amounts of apoE/Aβ complexes, accounting for ~5% of total Aβ (Verghese et al., 2013). Interestingly in astrocytes, apoE particles compete with, rather than facilitate, cellular Aβ uptake. These results further highlight the importance of the source of apoE and Aβ, as well as experimental conditions, in addressing the roles of apoE/Aβ complexes in AD-related pathways. It is important to note that despite significantly higher concentrations of apoE than Aβ in CSF, the exact concentrations of these molecules in brain parenchyma, in particular around the synapses where Aβ is likely produced and exhibits neurotoxicity, is unclear. Future studies should focus on developing technology, for example real-time in vivo microdialysis, to assess apoE-Aβ interactions and how a perturbation of their interaction affects Aβ metabolism, oligomerizationand toxicity.
ApoE and Aβ aggregation: ApoE is needed to seed amyloid
Several in vivo studies have clearly shown that apoE is essential for Aβ deposition in APP transgenic amyloid model mice. When Apoe knockout (KO) mice were crossed with amyloid model PDAPP or Tg2576 mice, Aβ deposition in the form of amyloid plaques and cerebral amyloid angiopathy (CAA) was dramatically reduced (Bales et al., 1997; Irizarry et al., 2000). In particular, although there is still significant, sometimes increased, Aβ deposition as diffused plaques, thioflavin S-positive fibril plaques were virtually absent. These results clearly demonstrate the essential role of mouse apoE in Aβ fibrillogenesis, stabilization of fibrillar Aβ, and/or maturation of amyloid plaques (Bales et al., 1997; Irizarry et al., 2000). However, the effects of apoE on Aβ fibrillogenesis appear to depend on the origin of apoE. In this respect, it is interesting to note that expression of human apoE3 and apoE4 by astrocytes in the Apoe-KO background decreased early Aβ deposition in PDAPP mice (Holtzman et al., 1999). Human apoE-targeted replacement (TR) mice, in which the mouse Apoe gene was replaced with those encoding human apoE isoforms, had less Aβ deposition in the background of Tg2576 mice compared with the control mice expressing mouse apoE (Fryer et al., 2005b). Thus, human apoE isoforms likely determine the amounts, morphology, localization and fibrillogenesis of Aβ depositsdepending on whether mouse apoE is present or not.
During the Aβ aggregation process, soluble Aβ peptides are known to change their conformation into a β-sheet structure and form nucleuses (lag phase), which further accelerates the process of fibrillogenesis to form insoluble fibrils with enriched β-sheet structures as “seed” (elongation phase) (Harper and Lansbury, 1997). There are numerous studies analyzing the effects of apoE on Aβ aggregation in vitro. However, the conclusions are controversial, where apoE can either facilitate or inhibit Aβ aggregation. It has been shown that high concentrations of apoE form high molecular weight co-aggregates with Aβ (Chan et al., 1996), where apoE4 is likely to promote Aβ aggregation more than apoE3 (Castano et al., 1995; Ma et al., 1994). Moreover, it was shown that apoE increases the level of Aβ oligomers in an isoform-dependent manner (apoE4 > apoE3 > apoE2) (Hashimoto et al., 2012). Furthermore, apoE4 stabilizes Aβ oligomers more than apoE3 (Cerf et al., 2011). These findings imply that apoE4 harmfully accelerates Aβ aggregation in AD.
In contrast, other studies have reported that apoE decreases Aβ fibrillogenesis. ApoE strongly inhibits the initiation of Aβ fibril formation, when analyzed either with or without the addition of pre-formed Aβ aggregates as seeds (Naiki et al., 1997; Wood et al., 1996b). Because apoE prefers to interact with Aβ peptides that are in β-sheet structure (Golabek et al., 1996), apoE likely captures Aβ nuclei and prevents its seeding effects (Figure 2) (Wood et al., 1996a). ApoE3 appears to interact with Aβ more than apoE4 as described; therefore, it is possible that apoE4 is less effective in the inhibition of Aβ fibril formation. In this case, apoE4 may be less effective in supporting the beneficial effects of apoE in preventing Aβ fibrillation in AD. If the amount of apoE/Aβ complex increases as the sole product of the reaction, they may form large co-aggregates (Figure 2) (Wood et al., 1996a). ApoE is also known to aggregate with irregular protofilament-like morphology, where the aggregates form at substantially different rates depending on the isoform (apoE4 > apoE3 > apoE2) (Hatters et al., 2006b). Thus, apoE4 may be able to produce more co-aggregates with Aβ through its self-aggregating propensity. Consistent with these findings, in vivo experiments have also shown that apoE4-TR mice had more Aβ deposition than apoE3-TR mice in the background of amyloid model mice (Bales et al., 2009; Fryer et al., 2005b). In yet a more aggressive amyloid model mice termed 5xFAD (Oakley et al., 2006), amyloid plaque deposition was in general greater in E4FAD mice, E2/E3FAD mice have significantly more diffuse plaques with E4FAD exhibiting more compact plaques (Youmans et al., 2012). Collectively, these studies indicate that compared to apoE2 or apoE3, apoE4 is either more likely to promote Aβ fibrillogenesis or less effective in preventing Aβ aggregation, or both. Again, the specific outcomes can be influenced by apoE isoform, lipidation status, aggregation states, and the time and location of its presence during the disease process. Further studies, perhaps by time-lapsed recording in living mice with exogenously introduced apoE isoforms with different lipidation statuses, might allow us to generate a clearer picture regarding the specific effects of apoE on Aβ and deposition. The effects of diffuse and fibrillar plaques on synaptic functions and behaviors, in the presence of different apoE isoforms, also warrant further investigation.
Figure 2. Aβ aggregation: role of apoE.
During the Aβ aggregation process, Aβ monomers change their conformation to a β-sheet-rich structure and form soluble oligomers or insoluble intermediate aggregates. Such nuclei further accelerate the fibrillogenesis to form large insoluble fibrils as “seeds” (Harper and Lansbury, 1997). The association of apoE with an Aβ nucleus is likely to block its seeding effect which accelerates Aβ fibrillogenesis. Under certain conditions, apoE and Aβ may form large co-aggregates. Newly generated Aβ fibrils can bind to existing aggregates, resulting in the formation of even larger co-aggregates, with or without additional apoE (Wood et al., 1996a). Finally, these aggregates may deposit as amyloid plaques in the brain.
ApoE and Aβ in endocytic trafficking: Common receptors and trafficking to lysosomes
The major apoE receptors, LDLR, LRP1 and HSPG, are abundantly expressed in the brain. These receptors mediate cellular uptake of Aβ as well as apoE (Figure 3). The mRNAs of LDLR and LRP1 are detected in several types of brain cells including neurons, astrocytes, microglia and oligodendrocytes (Fan et al., 2001). While LDLR deficiency in mice leads to apoE accumulation in the brain (Fryer et al., 2005a), overexpression of LDLR decreases apoE levels by 50–90% (Kim et al., 2009), demonstrating the important role of LDLR in brain catabolism of apoE. Since apoE2 has a lower affinity to the LDLR (Dong et al., 1994), apoE2 catabolism is likely slower compared to apoE3 or apoE4. Consistent with this notion, apoE levels in CSF and plasma are higher than those of apoE3 or apoE4 both in human (Cruchaga et al., 2012) and in apoE-TR mice (Bales et al., 2009). The increased absolute levels of apoE in APOE2 individuals might help with preventing Aβ accumulation in the brain and decrease the risk for developing AD. In vitro experiments have revealed that Aβ can bind directly to LDLR (Basak et al., 2012). Overexpression of LDLR significantly increases cellular uptake of Aβ in astrocytes, whereas deletion of LDLR has the opposite effect (Basak et al., 2012). Consistent with these findings, overexpression of LDLR in the brain decreases Aβ deposition in amyloid model mice (Kim et al., 2009); however, the effects of LDLR deletion on Aβ deposition is less clear (Cao et al., 2006; Fryer et al., 2005a), indicating that although LDLR is capable of metabolizing Aβ in the brain, its pathophysiological role requires further investigation.
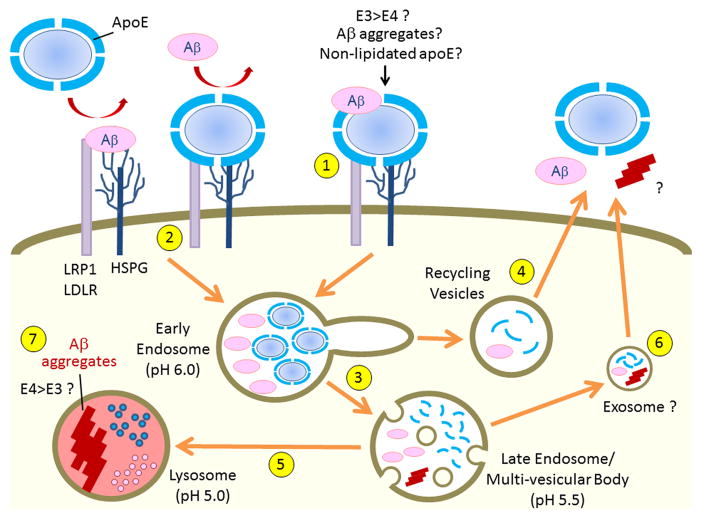
Figure 3. Cell surface binding and endocytic trafficking of apoE and Aβ.
ApoE likely binds to Aβ in an isoform-dependent manner with apoE3 forming more stable apoE/Aβ complexes than apoE4 (LaDu et al., 1994; LaDu et al., 1995). LRP1, LDLR and HSPG are major cell surface receptors that bind apoE, Aβ and apoE/Aβ complexes. In addition to forming a stable complex with Aβ (1), apoE likely competes with Aβ to common cell surface receptors (2) (Verghese et al., 2013). Endocytosed apoE either dissociates from lipid components within the early endosomes due to lower pH (3) and recycles (4), or be transported to lysosomes for degradation (5). Endocytosed Aβ is typically delivered to lysosomes for degradation (5), although a small amount of Aβ can be recycled (4) (Li et al., 2012). In some conditions, apoE and Aβ may be transferred through exosomes from the late endosomes/multi-vesicular body (6). When Aβ accumulation overwhelms the capacity of lysosomes for degradation, the low pH in the lysosomes provide a suitable environment to initiate Aβ aggregation (7) (Hu et al., 2009), which could injure lysosomes and also provide seeding for further Aβ aggregation.
LRP1 is an important apoE metabolic receptor in the brain and also among the most studied Aβ receptors. In vivo experiments show that overexpression of a functional LRP1 minireceptor (mLRP2) in mouse brain significantly decreases apoE levels (Zerbinatti et al., 2006), whereas neuronal-specific LRP1 deletion leads to an increase of apoE levels (Liu et al., 2007). LRP1 has been shown to mediate cellular Aβ uptake in neurons (Kanekiyo et al., 2011), astrocytes (Koistinaho et al., 2004), microglia (Laporte et al., 2004), vascular smooth muscle cells (Bell et al., 2009; Kanekiyo et al., 2012) and endothelial cells (Deane et al., 2004; Yamada et al., 2008). The in vivo experiments have shown that exogenous Aβ application through intracerebral microinjections is rapidly removed from the brain, whereas an LRP1-specific antibody slows this elimination process (Shibata et al., 2000). Consistent with these results, conditional knockout of the Lrp1 gene in mouse forebrain neurons (Kanekiyo et al., 2013) or vascular smooth muscle cells (Kanekiyo et al., 2012) exacerbated amyloid pathology in amyloid model mice by suppressing cellular Aβ uptake and lysosomal degradation. Aβ binds directly to immobilized LRP1 receptor fragments with high affinity (Deane et al., 2004), although an opposing result was also reported (Yamada et al., 2008). LRP1 also indirectly interacts with Aβ through its ligands, including apoE, α2-macroglobulin (Narita et al., 1997) and prion proteins (Rushworth et al., 2013). It was shown that cellular prion protein (PrPc) mediates Aβ oligomer binding to the cell surface (Lauren et al., 2009; Wang et al., 2013), where LRP1 functions as a co-receptor of PrPc (Rushworth et al., 2013). Furthermore, it is important to note that LRP1 and HSPG can form immunoprecipitable complexes at the cell surface, which might further regulate the metabolism of apoE and Aβ (Wilsie and Orlando, 2003). Our work has shown that Aβ appears to initially bind to cell surface HSPG, followed by endocytosis through either LRP1-independent or dependent manners (Kanekiyo et al., 2011). HSPG deficiency significantly decreases cellular Aβ binding and uptake (Kanekiyo et al., 2011). Given several types of HSPG are found to be co-localized with senile plaques and CAA in the brain of AD patients (van Horssen et al., 2003), further supports the important role HSPG plays in apoE and Aβ cellular metabolism. Interestingly, peripheral treatment with a low-molecular-weight heparin inhibits the binding of Aβ to HSPG, and significantly decreases Aβ concentration and deposition in the brain of amyloid model mice (Bergamaschini et al., 2004). The effects observed with heparin treatment are different to the phenotype displayed with LRP1 or LDLR deletion, where Aβ deposition is exacerbated. These observations suggest that the pathways of cellular Aβ and apoE uptake are differently regulated depending on their binding to specific apoE receptor LDLR, LRP1 and/or HSPG. Conditional deletion of HSPG in the brain can help to address the in vivo roles of this sometimes promiscuous molecule in brain Aβ metabolism. In this regard, it is important to note that HSPG also plays important roles in cellular uptake of tau and α-synuclein (Holmes et al., 2013), a critical step for propagation of disease-related pathology (Guo et al., 2013). Thus, further studies to dissect how HSPG regulates the endocytosis and trafficking of a variety of pathogenic proteins including Aβ, apoE, tau and α-synuclein may provide new insights into the pathogenesis of multiple neurodegenerative diseases.
Several studies have demonstrated that apoE enhances cellular Aβ uptake through its ability to form a complex. Soluble Aβ and recombinant apoE form SDS-stable complexes, which enhance Aβ internalization in primary neurons (Gylys et al., 2003). Recombinant apoE accelerates neuronal Aβ uptake in an isoform-dependent manner, with apoE3 more efficiently facilitating Aβ binding to the cell surface than apoE4 (Li et al., 2012). However, other studies have shown that apoE might compete with Aβ for receptor binding and subsequent cellular uptake. Recombinant apoE was shown to reduce the uptake of Aβ oligomers, but not fibrils, in astrocytes (Nielsen et al., 2010). ApoE particles interfere with the cellular uptake of soluble Aβ through an LRP1-dependent pathway in astrocytes (Verghese et al., 2013). These seemingly conflicting results may be induced by specific experimental conditions, including the source and the amounts of apoE and Aβ, the specific cell types tested, and/or the specific detection methods. Because Aβ and heparin are capable of binding to both the lipid-binding region and the receptor-binding region in apoE (Figure 1) and the complex interacting network of apoE receptors, apoE might either facilitate or compete with Aβ for cellular binding and uptake depending on their concentrations, Aβ aggregation state, apoE isoform, apoE lipidation state and expression pattern of the receptors on the cell surface (Figure 3). Further studies, in particular those designed in vivo, arenecessary to clarify apoE isoform-dependent functions in cellular Aβ uptake and metabolism.
It is clear that the majority of cell-internalized Aβ traffics through the early and late endosomes en route to lysosomes for degradation (Hu et al., 2009; Li et al., 2012), although a small portion of cell-internalized Aβ does recycle through Rab11-positive recycling endosomes (Li et al., 2012). Thus, cellular Aβ uptake is in general a beneficial pathway for Aβ clearance. However, when lysosomal degradation is impaired (Li et al., 2012) or when Aβ concentration overwhelms this compartment in AD brains (Hu et al., 2009; Knauer et al., 1992), trafficking to lysosomes can be detrimental as it could lead to Aβ aggregation (Hu et al., 2009), which is highly favourable in the acidic environment found in lysosomes (Peralvarez-Marin et al., 2008) (Figure 3). Aβ aggregates in turn likely induce lysosomal dysfunction and cellular toxicity. More importantly, these lysosomal initiated Aβ aggregates can further accelerate Aβ aggregation, which could eventually contribute to the genesis of extracellular Aβ oligomers and amyloid plaque deposition. Interestingly, apoE also aggregates faster under acidic pH conditions found in lysosomes with apoE4 aggregating more than apoE3 (Garai et al., 2011). Thus, the aggregative nature of both Aβ and apoE4 in the acidic lysosomes demonstrates that these AD pathogenic molecules might co-aggregate in the lysosomes as accidental encounters and/or partners.
ApoE in Aβ clearance: Distinct roles and isoform-specific effects
While Aβ is continuously generated in the brain, it is efficiently eliminated under physiological conditions. In human brains, the Aβ clearance rate is calculated to be 8.3% per hour (Bateman et al., 2006). There are three major pathways by which Aβ is cleared from the brain: 1) through proteolytic degradation, 2) by cellular clearance through lysosomal degradation in brain parenchyma cells (microglia, astrocytes, neurons), and 3) by cerebrovascular system-mediated clearance including the interstitial fluid (ISF) drainage pathway, local cellular clearance and blood-brain barrier (BBB) (Figure 4). In vivo experiments have shown that Aβ clearance is slower in apoE4-TR mice compared with apoE3-TR mice (Castellano et al., 2011). However, the pathways contributing to apoE-regulated Aβ clearance can be complex as discussed above as the clearance of soluble Aβ in brain ISF is increased in apoE-KO mice (DeMattos et al., 2004).
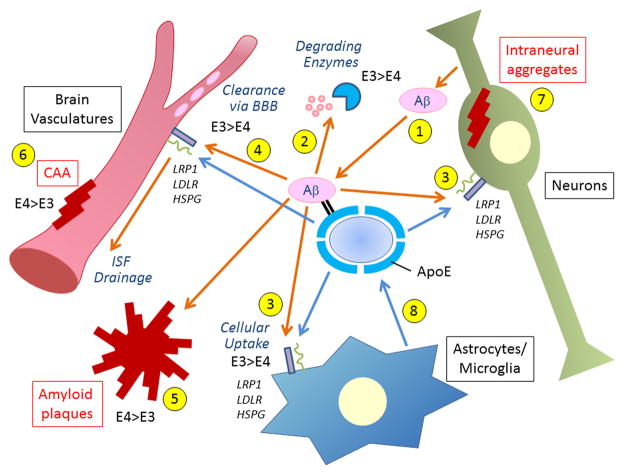
Figure 4. Major Aβ clearance pathways and effects of apoE isoforms.
Aβ is predominantly generated in neurons (1) and eliminated through three major clearance pathways including proteolytic degradation by (2) endopeptidases (e.g., NEP, IDE) (Saido and Leissring, 2012), (3) cellular clearance by cells in the brain parenchyma (neurons, astrocytes and microglia) (Kanekiyo et al., 2013; Koistinaho et al., 2004), and (4) ISF drainage where it is degraded by vascular cells (Bell et al., 2009; Kanekiyo et al., 2012) or transported out of the brain through BBB (Ito et al., 2013). Disturbance of these pathways induce Aβ accumulation and deposition in the brain parenchyma as amyloid plaques (5), in the perivascular region as CAA (6) and sometimes also inside neurons (7). ApoE is generated mainly by the glial cells (8) and encounters Aβ in all these pathways. ApoE likely facilitates Aβ clearance by activating enzymatic degradation (2) and phagocytosis (3) in an isoform-dependent manner (apoE3 > apoE4). However, apoE might also suppress Aβ clearance (apoE4 > apoE3) by either competing with Aβ for receptor binding or by retaining Aβ from it clearance through the BBB (4). ApoE4 exacerbates Aβ deposition as amyloid plaques and CAA formation when compared with apoE3 (5 and 6) (Fryer et al., 2005b). LRP1, LDLR and HSPG, which are expressed in all major cellular Aβ clearance pathways, regulate Aβ clearance either directly or through apoE.
Aβ is degraded by a large set of proteases including neprilysin (NEP) and insulin-degrading enzyme (IDE) in both intracellular and extracellular compartments (Figure 4) (Saido and Leissring, 2012). Deletion of NEP or treatment with a NEP inhibitor leads to increased levels of Aβ (Farris et al., 2007; Iwata et al., 2000). The IDE-KO mice show increased accumulation of endogenous Aβ in the brain (Farris et al., 2003). Consistent with these findings, overexpression of NEP and/or IDE lowers Aβ levels by around 90% and reduces amyloid pathology (Leissring et al., 2003). Soluble apoE/Aβ complexes isolated from human brains were more susceptible to proteolytic degradation than apoE-free Aβ (Russo et al., 1998). Similarly, NEP-mediated Aβ degradation in microglia is enhanced by exogenous apoE, in an isoform-dependent manner (apoE2 > apoE3 > apoE4). Extracellular Aβ degradation by IDE and NEP is also facilitated by apoE. Interestingly, lipidated apoE shows stronger effects on the capacities of NEP and IDE to degrade Aβ than non-lipidated apoE (Jiang et al., 2008). Although further studies are needed, it is tempting to speculate that apoE facilitates Aβ degradation by converting the Aβ structure into one that is more recognizable by NEP and IDE.
Cellular uptake of Aβ by astrocytes and microglia is likely to represent a more functional pathway for Aβ clearance (Figure 4). When brain sections bearing Aβ plaques from amyloid model mice were cultured with adult mouse astrocytes, the astrocytes internalized and degraded Aβ in both apoE and LRP1-dependent manner (Koistinaho et al., 2004). In microglia, soluble Aβ is likely internalized by fluid-phase macropinocytosis into lysosomes for degradation (Mandrekar et al., 2009), whereas Aβ aggregates interact with a multicomponent cell surface receptor complex and are internalized through phagocytosis (Bamberger et al., 2003). Microglia and macrophage are known to have two activation statuses; classically activated (M1) and alternatively activated (M2) stages. M1 cells produce proinflammatory cytokines, which damage neurons; whereas M2 cells trigger anti-inflammatory/neurotrophic pathways and clear Aβ by phagocytosis in AD (Aguzzi et al., 2013). Microglial activation is likely modulated by apoE, as apoE has been shown to convert macrophages from the proinflammatory M1 to the anti-inflammatory M2 phenotype (Baitsch et al., 2011). ApoE4 is less effective in anti-inflammatory functions than apoE3 in microglia (Zhu et al., 2012). Furthermore, apoE isoforms differently regulate microglia migration in response to activation, with microglia from apoE4-TR mice exhibiting slower migration than those from apoE3-TR mice (Cudaback et al., 2011). Thus, apoE promotes Aβ clearance by activating phagocytosis and migration in microglia, where apoE4 has a reduced capacity to induce these phenotypes than apoE3. Recent genetic studies have also identified several new AD risk genes that are potentially involved in regulating neuroinflammation-related functions including TREM2, CLU, CR1, CD33 and ABCA7 (Guerreiro et al., 2013; Harold et al., 2009; Hollingworth et al., 2011; Lambert et al., 2009; Naj et al., 2011). Although the functional relationships between apoE and proteins encoded by these genes are not fully understood, the discovery of these new AD risk genes further implies a critical relationship between neuroinflammation and AD pathogenesis. As Clu and ABCA7 are also related to lipid metabolism, further studies may demonstrate their cooperative roles with apoE in Aβ clearance.
Neurons also have an ability to eliminate Aβ through its uptake and lysosomal degradation (Figure 4) (Li et al., 2012). It should be noted that neurons have the highest risk of encountering Aβ in the brain, because Aβ is predominantly produced in neurons. The functional role of neurons in brain Aβ clearance is clearly shown by our recent work demonstrating that when LRP1 is deleted exclusively in neurons in adult mouse brain, the half-life of ISF Aβ increases, which leads to more abundant Aβ accumulation and pathology (Kanekiyo et al., 2013).
Cerebrovascular systems also play critical roles in Aβ clearance (Figure 4). Pathological studies from AD patients show that Aβ deposits in the vascular smooth muscle cell layer of cerebral blood vessels and capillaries in the form of CAA as well as senile plaques (Rensink et al., 2003; Revesz et al., 2003). ISF flows along periarterial spaces in the brain and joins the CSF to drain into the cervical lymph nodes and/or venous blood flow. Part of ISF can directly enter the arterial blood flow through the BBB (de Boer and Gaillard, 2007; Marques et al., 2013). In the ISF drainage pathway, Aβ is thought to be eliminated through cellular degradation by vascular cells (smooth muscle cells, pericytes, endothelial cells and astrocytes), proteolytic degradation and clearance through the BBB. Remaining Aβ in ISF may be eventually drained into lymph flow and venous blood flow. Thus, the disturbance of ISF drainage pathway likely causes Aβ accumulation in the brain. In fact, the water channel aquaporin-4 has been shown to be a crucial component in this process. Aquaporin-4 null mice exhibit a ~70% reduction in ISF clearance rate and a suppressed clearance of soluble Aβ through reduction of ISF drainage (Iliff et al., 2012), supporting an importance role of this pathway in Aβ clearance. Epidemiological studies have also shown that practically all well-described risk factors for AD, including diabetes mellitus, atherosclerosis, stroke, hypertension, transient ischemic attacks, microvessel pathology and smoking, have a vascular component that disturbs cerebral vascular functions (de la Torre, 2002). Pathological evidence indicates that LRP1 levels are significantly decreased during aging and AD, in particular in the brain vasculature (Bell et al., 2009; Deane et al., 2004). LRP1 in vascular smooth muscle cells regulates Aβ clearance in the ISF drainage pathway. Deletion of LRP1 in vascular mural cells enhances Aβ deposition both in brain parenchyma as amyloid plaques and in vasculature as CAA (Kanekiyo et al., 2012). BBB efflux of Aβ also represents a major pathway for brain Aβ clearance. The overall clearance rate constant of Aβ40 in mouse cerebral cortex is 3.21 × 10−2/min, while the elimination rate for brain-blood clearance is 1.48 × 10−2/min (Ito et al., 2013). Due to technical limitations, it is difficult to precisely distinguish Aβ clearance through the BBB and simultaneous clearance through other pathways. Current research implicates apoE and apoE receptors as major regulators in Aβ clearance through the BBB. One such study demonstrates that Aβ is cleared rapidly across the BBB through LRP1, where apoE significantly disturbs this process (Bell et al., 2007). ApoE2/Aβ and apoE3/Aβ complexes are cleared at the BBB via LRP1 and VLDLR at a substantially faster rate than apoE4/Aβ complexes (Figure 4) (Deane et al., 2008). In addition, LDLR has also been shown to mediate brain-blood Aβ clearance (Castellano et al., 2012). Moreover, apoE4, but not apoE2 or apoE3, leads to the breakdown of the BBB (Bell et al., 2012; Nishitsuji et al., 2011), which causes a reduction in cerebral blood flow (Bell et al., 2012). This harmful effect of apoE4 on BBB integrity might further compromise Aβ clearance. Consistent with these findings, APOE4 carriers have more severe CAA pathology, in particular capillary CAA when compared with APOE4 non-carriers (Richard et al., 2010; Thal et al., 2002). Taken together, apoE and apoE receptors are clearly involved in Aβ clearance through the cerebrovascular systems, likely in ways depending on the presence of specific apoE isoforms and apoE receptors. Future studies should focus on addressing whether apoE isoforms produced in brain parenchyma differ from those by vascular cells in regulating brain Aβ clearance and how apoE receptors expressed in specific vascular cell types regulate these events.
ApoE-targeted therapy for AD: The dilemma when modulating apoE
Because of the importance of apoE in AD pathogenesis, several therapeutic strategies that target apoE for AD have been proposed. Among them, retinoid X receptor (RXR) and liver X receptor (LXR) agonists are promising candidates. RXR forms heterodimers with LXR, peroxisome proliferator-activated receptor γ (PPARγ) or retinoic acid receptor (RAR) to regulate gene networks that control multiple metabolic systems. In particular, these nuclear receptors are known to control the transcription of ABCA1 and apoE (Perez et al., 2012). Oral administration of an RXR agonist, Bexarotene, suppressed Aβ deposition and improved cognitive function in an apoE-dependent manner in amyloid model mice (Cramer et al., 2012), although these results are likely somewhat disputed by several follow up studies. The LXR agonist TO901317 is also shown to increase apoE levels in the brain, facilitate Aβ clearance and reverse the memory deficit in amyloid model mice (Riddell et al., 2007; Terwel et al., 2011). Thus, treatments that result in increased levels of apoE in the brain are likely beneficial in AD therapy. However, this notion has to be carefully dissected as apoE4 clearly has harmful effects (Bu, 2009). Under stress conditions, neuronally expressed apoE often becomes fragmented, which damages mitochondria and cytoskeleton (Huang and Mucke, 2012). In addition, in vitro experiments have shown that apoE4 facilitates Aβ production by promoting amyloidogenic processing of APP as a functional consequence of its interaction with APP (Vincent and Smith, 2001). Thus, it might be particularly detrimental to up-regulate apoE4 production in AD. Rather than focusing on increasing apoE, it has been proposed that increasing apoE lipidation might be the key for apoE-based therapy. ApoE lipidation is mediated by ABCA1 (Koldamova et al., 2010). Importantly, deletion of ABCA1 increases Aβ deposition in amyloid model mice (Wahrle et al., 2005), whereas its overexpression suppresses Aβ deposition (Wahrle et al., 2008). Furthermore, the haploinsufficiency of ABCA1 significantly decreases Aβ clearance in apoE4-TR mice, but not in apoE3-TR mice (Fitz et al., 2012), suggesting that the less efficient lipid transport phenotype associated with apoE4 might allow for greater manipulation at the level of apoE lipidation.
The argument to decrease apoE expression to treat AD is championed by the fact that apoE is essential for Aβ deposition as discussed above. Recent studies have also shown that decreased apoE expression under haploinsufficiency of human apoE also results in less Aβ deposition in amyloid mouse models, which is independent of apoE isoforms (Bien-Ly et al., 2012; Kim et al., 2011). Further, immunotherapy for apoE also reduces Aβ accumulation. When amyloid model mice were intraperitoneally administered with anti-mouse apoE specific antibody for 14 weeks, amyloid deposition was dramatically reduced by 60 – 80% and insoluble Aβ levels were significantly decreased (Kim et al., 2012). These results suggest that decreasing apoE levels has beneficial effects and that anti-apoE immunization can be explored as a novel therapeutic tool, at least from the perspective of Aβ deposition. However, further studies are needed to determine how decreased apoE levels affect Aβ oligomerization, neurotoxicity, synaptic integrity and cognitive function. It is also an undisputed fact that apoE mediates cholesterol homeostasis, particularly in the brain, thus, these approaches to solely decrease apoE levels over extended periods of time may induce harmful, perhaps unforeseen side effects.
The interaction between apoE and Aβ can also be targeted for therapy, although the outcome is less clear. Despite evidence supporting a beneficial role of apoE/Aβ complexes, the presence of these proteins in an environment that favors Aβ aggregation, such as lysosomes and amyloid plaques, could be harmful. Supporting this, a synthetic peptide mimicking the apoE binding sequence of Aβ, Aβ12–28P, reduces Aβ deposition and ameliorates memory deficits in amyloid model mice (Sadowski et al., 2004). Other methods that target apoE for therapy include apoE4 structural correctors (Chen et al., 2012) and increasing the functions of apoE receptors (Shinohara et al., 2010).
Taken together, the choice of increasing or decreasing apoE is an unavoidable dilemma in our efforts to develop apoE-based AD therapies. Despite this dilemma, apoE remains a promising target. Both pharmacological manipulations of apoE expression and lipidation, and potentially new genetic mouse models that allow for testing the effects of increasing or reducing apoE should help to clarify our path to new AD therapy.
Summary and Perspective
APOE4 is the strongest genetic risk factor for late-onset AD. ApoE4 appears to drive amyloid pathology in humans and in animal models; however, it is clear that apoE4 also contributes to AD pathogenesis in a manner that is independent of Aβ. ApoE and Aβ interact with each other and share common receptors including LRP1, LDLR and HSPG. Importantly, the interactions among apoE, Aβ and their receptors likely vary depending on their concentrations, the apoE isoform involved, lipidation status, Aβ aggregation status and receptor distribution patterns. Thus, apoE isoforms likely have different, sometimes seemingly conflicting, roles in Aβ aggregation and clearance. First, apoE reduces Aβ oligomerization and fibril formation but is also essential for amyloid deposition. Second, apoE competes with Aβ for their receptor binding but can also facilitate cellular Aβ uptake by forming apoE/Aβ complexes. Third, apoE facilitates the enzymatic degradation and phagocytosis of Aβ by glial cells, but apoE compromises Aβ elimination through the BBB. As a result, it has been difficult to elucidate the exact pathological mechanism by which apoE regulates Aβ clearance. Nonetheless, it is clear that multiple pathways are involved and apoE and Aβ appear to share several properties suggesting their interaction goes beyond accidental encounters. In fact, apoE-targeted therapy aimed at ameliorating amyloid pathology and improving cognitive function via RXR/LXR agonists or increasing apoE lipidation hold promise and warrant further investigation.
In summary, apoE has multiple functions in regulating Aβ clearance, Aβ aggregation and Aβ-independent pathways in AD pathogenesis. A critical challenge is to determine whether increasing or decreasing apoE, or simply its lipidation is beneficial, and how the presence of apoE4 affects the outcomes. Future studies should also be focused on addressing why a single amino acid change from apoE3 to apoE4 causes profound differences in their properties and functions and how we can utilize our knowledge to design new therapies for AD targeting apoE pathway.
Acknowledgments
This work was supported by NIH grants R01AG035355, R01AG027924, P01AG030128, and P01NS074969 (to G.B.), and Mayo Clinic CRM Career Developmental Award (to T.K.). We thank Caroline T. Stetler, Melissa C. Wren and Caroline S. Casey for careful readings of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguzzi A, Barres BA, Bennett ML. Microglia: scapegoat, saboteur, or something else? Science. 2013;339:156–161. doi: 10.1126/science.1227901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baitsch D, Bock HH, Engel T, Telgmann R, Muller-Tidow C, Varga G, Bot M, Herz J, Robenek H, von Eckardstein A, et al. Apolipoprotein E induces antiinflammatory phenotype in macrophages. Arterioscler Thromb Vasc Biol. 2011;31:1160–1168. doi: 10.1161/ATVBAHA.111.222745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bales KR, Liu F, Wu S, Lin S, Koger D, DeLong C, Hansen JC, Sullivan PM, Paul SM. Human APOE isoform-dependent effects on brain beta-amyloid levels in PDAPP transgenic mice. J Neurosci. 2009;29:6771–6779. doi: 10.1523/JNEUROSCI.0887-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bales KR, Verina T, Dodel RC, Du Y, Altstiel L, Bender M, Hyslop P, Johnstone EM, Little SP, Cummins DJ, et al. Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat Genet. 1997;17:263–264. doi: 10.1038/ng1197-263. [DOI] [PubMed] [Google Scholar]
- Bamberger ME, Harris ME, McDonald DR, Husemann J, Landreth GE. A cell surface receptor complex for fibrillar beta-amyloid mediates microglial activation. J Neurosci. 2003;23:2665–2674. doi: 10.1523/JNEUROSCI.23-07-02665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak JM, Verghese PB, Yoon H, Kim J, Holtzman DM. Low-density lipoprotein receptor represents an apolipoprotein E-independent pathway of Abeta uptake and degradation by astrocytes. J Biol Chem. 2012;287:13959–13971. doi: 10.1074/jbc.M111.288746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman RJ, Munsell LY, Morris JC, Swarm R, Yarasheski KE, Holtzman DM. Human amyloid-beta synthesis and clearance rates as measured in cerebrospinal fluid in vivo. Nat Med. 2006;12:856–861. doi: 10.1038/nm1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisiegel U, Weber W, Ihrke G, Herz J, Stanley KK. The LDL-receptor-related protein, LRP, is an apolipoprotein E-binding protein. Nature. 1989;341:162–164. doi: 10.1038/341162a0. [DOI] [PubMed] [Google Scholar]
- Bell RD, Deane R, Chow N, Long X, Sagare A, Singh I, Streb JW, Guo H, Rubio A, Van Nostrand W, et al. SRF and myocardin regulate LRP-mediated amyloid-beta clearance in brain vascular cells. Nat Cell Biol. 2009;11:143–153. doi: 10.1038/ncb1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RD, Sagare AP, Friedman AE, Bedi GS, Holtzman DM, Deane R, Zlokovic BV. Transport pathways for clearance of human Alzheimer’s amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J Cereb Blood Flow Metab. 2007;27:909–918. doi: 10.1038/sj.jcbfm.9600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RD, Winkler EA, Singh I, Sagare AP, Deane R, Wu Z, Holtzman DM, Betsholtz C, Armulik A, Sallstrom J, et al. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature. 2012;485:512–516. doi: 10.1038/nature11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschini L, Rossi E, Storini C, Pizzimenti S, Distaso M, Perego C, De Luigi A, Vergani C, De Simoni MG. Peripheral treatment with enoxaparin, a low molecular weight heparin, reduces plaques and beta-amyloid accumulation in a mouse model of Alzheimer’s disease. J Neurosci. 2004;24:4181–4186. doi: 10.1523/JNEUROSCI.0550-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien-Ly N, Gillespie AK, Walker D, Yoon SY, Huang Y. Reducing human apolipoprotein E levels attenuates age-dependent Abeta accumulation in mutant human amyloid precursor protein transgenic mice. J Neurosci. 2012;32:4803–4811. doi: 10.1523/JNEUROSCI.0033-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunden KR, Richter-Cook NJ, Chaturvedi N, Frederickson RC. pH-dependent binding of synthetic beta-amyloid peptides to glycosaminoglycans. J Neurochem. 1993;61:2147–2154. doi: 10.1111/j.1471-4159.1993.tb07453.x. [DOI] [PubMed] [Google Scholar]
- Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Fukuchi K, Wan H, Kim H, Li L. Lack of LDL receptor aggravates learning deficits and amyloid deposits in Alzheimer transgenic mice. Neurobiology of aging. 2006;27:1632–1643. doi: 10.1016/j.neurobiolaging.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Castano EM, Prelli F, Wisniewski T, Golabek A, Kumar RA, Soto C, Frangione B. Fibrillogenesis in Alzheimer’s disease of amyloid beta peptides and apolipoprotein E. Biochem J. 1995;306(Pt 2):599–604. doi: 10.1042/bj3060599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JM, Deane R, Gottesdiener AJ, Verghese PB, Stewart FR, West T, Paoletti AC, Kasper TR, DeMattos RB, Zlokovic BV, et al. Low-density lipoprotein receptor overexpression enhances the rate of brain-to-blood Abeta clearance in a mouse model of beta-amyloidosis. Proc Natl Acad Sci U S A. 2012;109:15502–15507. doi: 10.1073/pnas.1206446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, et al. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci Transl Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerf E, Gustot A, Goormaghtigh E, Ruysschaert JM, Raussens V. High ability of apolipoprotein E4 to stabilize amyloid-beta peptide oligomers, the pathological entities responsible for Alzheimer’s disease. FASEB J. 2011;25:1585–1595. doi: 10.1096/fj.10-175976. [DOI] [PubMed] [Google Scholar]
- Chan W, Fornwald J, Brawner M, Wetzel R. Native complex formation between apolipoprotein E isoforms and the Alzheimer’s disease peptide A beta. Biochemistry. 1996;35:7123–7130. doi: 10.1021/bi952852v. [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Parfitt M, Legrain S, Perez-Tur J, Brousseau T, Evans A, Berr C, Vidal O, Roques P, Gourlet V, et al. Apolipoprotein E, epsilon 4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer’s disease: analysis of the 19q13.2 chromosomal region. Hum Mol Genet. 1994;3:569–574. doi: 10.1093/hmg/3.4.569. [DOI] [PubMed] [Google Scholar]
- Chen HK, Liu Z, Meyer-Franke A, Brodbeck J, Miranda RD, McGuire JG, Pleiss MA, Ji ZS, Balestra ME, Walker DW, et al. Small molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons. J Biol Chem. 2012;287:5253–5266. doi: 10.1074/jbc.M111.276162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Q, Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc Natl Acad Sci U S A. 2011;108:14813–14818. doi: 10.1073/pnas.1106420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Cramer PE, Cirrito JR, Wesson DW, Lee CY, Karlo JC, Zinn AE, Casali BT, Restivo JL, Goebel WD, James MJ, et al. ApoE-directed therapeutics rapidly clear beta-amyloid and reverse deficits in AD mouse models. Science. 2012;335:1503–1506. doi: 10.1126/science.1217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C, Kauwe JS, Nowotny P, Bales K, Pickering EH, Mayo K, Bertelsen S, Hinrichs A, Fagan AM, Holtzman DM, et al. Cerebrospinal fluid APOE levels: an endophenotype for genetic studies for Alzheimer’s disease. Hum Mol Genet. 2012;21:4558–4571. doi: 10.1093/hmg/dds296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudaback E, Li X, Montine KS, Montine TJ, Keene CD. Apolipoprotein E isoform-dependent microglia migration. FASEB J. 2011;25:2082–2091. doi: 10.1096/fj.10-176891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davignon J, Gregg RE, Sing CF. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 1988;8:1–21. doi: 10.1161/01.atv.8.1.1. [DOI] [PubMed] [Google Scholar]
- de Boer AG, Gaillard PJ. Drug targeting to the brain. Annu Rev Pharmacol Toxicol. 2007;47:323–355. doi: 10.1146/annurev.pharmtox.47.120505.105237. [DOI] [PubMed] [Google Scholar]
- de la Torre JC. Alzheimer disease as a vascular disorder: nosological evidence. Stroke. 2002;33:1152–1162. doi: 10.1161/01.str.0000014421.15948.67. [DOI] [PubMed] [Google Scholar]
- Deane R, Sagare A, Hamm K, Parisi M, Lane S, Finn MB, Holtzman DM, Zlokovic BV. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J Clin Invest. 2008;118:4002–4013. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane R, Wu Z, Sagare A, Davis J, Du Yan S, Hamm K, Xu F, Parisi M, LaRue B, Hu HW, et al. LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron. 2004;43:333–344. doi: 10.1016/j.neuron.2004.07.017. [DOI] [PubMed] [Google Scholar]
- DeMattos RB, Cirrito JR, Parsadanian M, May PC, O’Dell MA, Taylor JW, Harmony JA, Aronow BJ, Bales KR, Paul SM, et al. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41:193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- Dong LM, Wilson C, Wardell MR, Simmons T, Mahley RW, Weisgraber KH, Agard DA. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J Biol Chem. 1994;269:22358–22365. [PubMed] [Google Scholar]
- Elshourbagy NA, Liao WS, Mahley RW, Taylor JM. Apolipoprotein E mRNA is abundant in the brain and adrenals, as well as in the liver, and is present in other peripheral tissues of rats and marmosets. Proc Natl Acad Sci U S A. 1985;82:203–207. doi: 10.1073/pnas.82.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan AM, Mintun MA, Mach RH, Lee SY, Dence CS, Shah AR, LaRossa GN, Spinner ML, Klunk WE, Mathis CA, et al. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol. 2006;59:512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- Fan QW, Iosbe I, Asou H, Yanagisawa K, Michikawa M. Expression and regulation of apolipoprotein E receptors in the cells of the central nervous system in culture: A review. J Am Aging Assoc. 2001;24:1–10. doi: 10.1007/s11357-001-0001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, Eckman CB, Tanzi RE, Selkoe DJ, Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci U S A. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris W, Schutz SG, Cirrito JR, Shankar GM, Sun X, George A, Leissring MA, Walsh DM, Qiu WQ, Holtzman DM, et al. Loss of neprilysin function promotes amyloid plaque formation and causes cerebral amyloid angiopathy. Am J Pathol. 2007;171:241–251. doi: 10.2353/ajpath.2007.070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz NF, Cronican AA, Saleem M, Fauq AH, Chapman R, Lefterov I, Koldamova R. Abca1 deficiency affects Alzheimer’s disease-like phenotype in human ApoE4 but not in ApoE3-targeted replacement mice. J Neurosci. 2012;32:13125–13136. doi: 10.1523/JNEUROSCI.1937-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer JD, Demattos RB, McCormick LM, O’Dell MA, Spinner ML, Bales KR, Paul SM, Sullivan PM, Parsadanian M, Bu G, et al. The low density lipoprotein receptor regulates the level of central nervous system human and murine apolipoprotein E but does not modify amyloid plaque pathology in PDAPP mice. J Biol Chem. 2005a;280:25754–25759. doi: 10.1074/jbc.M502143200. [DOI] [PubMed] [Google Scholar]
- Fryer JD, Simmons K, Parsadanian M, Bales KR, Paul SM, Sullivan PM, Holtzman DM. Human apolipoprotein E4 alters the amyloid-beta 40:42 ratio and promotes the formation of cerebral amyloid angiopathy in an amyloid precursor protein transgenic model. J Neurosci. 2005b;25:2803–2810. doi: 10.1523/JNEUROSCI.5170-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futamura M, Dhanasekaran P, Handa T, Phillips MC, Lund-Katz S, Saito H. Two-step mechanism of binding of apolipoprotein E to heparin: implications for the kinetics of apolipoprotein E-heparan sulfate proteoglycan complex formation on cell surfaces. J Biol Chem. 2005;280:5414–5422. doi: 10.1074/jbc.M411719200. [DOI] [PubMed] [Google Scholar]
- Garai K, Baban B, Frieden C. Self-association and stability of the ApoE isoforms at low pH: implications for ApoE-lipid interactions. Biochemistry. 2011;50:6356–6364. doi: 10.1021/bi2006702. [DOI] [PubMed] [Google Scholar]
- Golabek AA, Soto C, Vogel T, Wisniewski T. The interaction between apolipoprotein E and Alzheimer’s amyloid beta-peptide is dependent on beta-peptide conformation. J Biol Chem. 1996;271:10602–10606. doi: 10.1074/jbc.271.18.10602. [DOI] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JL, Covell DJ, Daniels JP, Iba M, Stieber A, Zhang B, Riddle DM, Kwong LK, Xu Y, Trojanowski JQ, et al. Distinct alpha-synuclein strains differentially promote tau inclusions in neurons. Cell. 2013;154:103–117. doi: 10.1016/j.cell.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gylys KH, Fein JA, Tan AM, Cole GM. Apolipoprotein E enhances uptake of soluble but not aggregated amyloid-beta protein into synaptic terminals. J Neurochem. 2003;84:1442–1451. doi: 10.1046/j.1471-4159.2003.01643.x. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JD, Lansbury PT., Jr Models of amyloid seeding in Alzheimer’s disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Serrano-Pozo A, Hori Y, Adams KW, Takeda S, Banerji AO, Mitani A, Joyner D, Thyssen DH, Bacskai BJ, et al. Apolipoprotein E, especially apolipoprotein E4, increases the oligomerization of amyloid beta peptide. J Neurosci. 2012;32:15181–15192. doi: 10.1523/JNEUROSCI.1542-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatters DM, Budamagunta MS, Voss JC, Weisgraber KH. Modulation of apolipoprotein E structure by domain interaction: differences in lipid-bound and lipid-free forms. J Biol Chem. 2005;280:34288–34295. doi: 10.1074/jbc.M506044200. [DOI] [PubMed] [Google Scholar]
- Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006a;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Hatters DM, Zhong N, Rutenber E, Weisgraber KH. Amino-terminal domain stability mediates apolipoprotein E aggregation into neurotoxic fibrils. J Mol Biol. 2006b;361:932–944. doi: 10.1016/j.jmb.2006.06.080. [DOI] [PubMed] [Google Scholar]
- Herz J, Strickland DK. LRP: a multifunctional scavenger and signaling receptor. J Clin Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse C, Larsson H, Fredman P, Minthon L, Andreasen N, Davidsson P, Blennow K. Measurement of apolipoprotein E (apoE) in cerebrospinal fluid. Neurochem Res. 2000;25:511–517. doi: 10.1023/a:1007516210548. [DOI] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes BB, DeVos SL, Kfoury N, Li M, Jacks R, Yanamandra K, Ouidja MO, Brodsky FM, Marasa J, Bagchi DP, et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc Natl Acad Sci U S A. 2013;110:E3138–3147. doi: 10.1073/pnas.1301440110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Bales KR, Wu S, Bhat P, Parsadanian M, Fagan AM, Chang LK, Sun Y, Paul SM. Expression of human apolipoprotein E reduces amyloid-beta deposition in a mouse model of Alzheimer’s disease. J Clin Invest. 1999;103:R15–R21. doi: 10.1172/JCI6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlden H, Crook R, Backhovens H, Prihar G, Baker M, Hutton M, Rossor M, Martin JJ, Van Broeckhoven C, Hardy J. ApoE genotype is a risk factor in nonpresenilin early-onset Alzheimer’s disease families. Am J Med Genet. 1998;81:117–121. doi: 10.1002/(sici)1096-8628(19980207)81:1<117::aid-ajmg19>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Hu X, Crick SL, Bu G, Frieden C, Pappu RV, Lee JM. Amyloid seeds formed by cellular uptake, concentration, and aggregation of the amyloid-beta peptide. Proc Natl Acad Sci U S A. 2009;106:20324–20329. doi: 10.1073/pnas.0911281106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliff JJ, Wang M, Liao Y, Plogg BA, Peng W, Gundersen GA, Benveniste H, Vates GE, Deane R, Goldman SA, et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid beta. Sci Transl Med. 2012;4:147ra111. doi: 10.1126/scitranslmed.3003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innerarity TL, Mahley RW. Enhanced binding by cultured human fibroblasts of apo-E-containing lipoproteins as compared with low density lipoproteins. Biochemistry. 1978;17:1440–1447. doi: 10.1021/bi00601a013. [DOI] [PubMed] [Google Scholar]
- Innerarity TL, Pitas RE, Mahley RW. Binding of arginine-rich (E) apoprotein after recombination with phospholipid vesicles to the low density lipoprotein receptors of fibroblasts. J Biol Chem. 1979;254:4186–4190. [PubMed] [Google Scholar]
- Irizarry MC, Rebeck GW, Cheung B, Bales K, Paul SM, Holzman D, Hyman BT. Modulation of A beta deposition in APP transgenic mice by an apolipoprotein E null background. Ann N Y Acad Sci. 2000;920:171–178. doi: 10.1111/j.1749-6632.2000.tb06919.x. [DOI] [PubMed] [Google Scholar]
- Ito S, Matsumiya K, Ohtsuki S, Kamiie J, Terasaki T. Contributions of degradation and brain-to-blood elimination across the blood-brain barrier to cerebral clearance of human amyloid-beta peptide(1–40) in mouse brain. J Cereb Blood Flow Metab. 2013 doi: 10.1038/jcbfm.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata N, Tsubuki S, Takaki Y, Watanabe K, Sekiguchi M, Hosoki E, Kawashima-Morishima M, Lee HJ, Hama E, Sekine-Aizawa Y, et al. Identification of the major Abeta1–42-degrading catabolic pathway in brain parenchyma: suppression leads to biochemical and pathological deposition. Nat Med. 2000;6:143–150. doi: 10.1038/72237. [DOI] [PubMed] [Google Scholar]
- Ji ZS, Brecht WJ, Miranda RD, Hussain MM, Innerarity TL, Mahley RW. Role of heparan sulfate proteoglycans in the binding and uptake of apolipoprotein E-enriched remnant lipoproteins by cultured cells. J Biol Chem. 1993;268:10160–10167. [PubMed] [Google Scholar]
- Jiang Q, Lee CY, Mandrekar S, Wilkinson B, Cramer P, Zelcer N, Mann K, Lamb B, Willson TM, Collins JL, et al. ApoE promotes the proteolytic degradation of Abeta. Neuron. 2008;58:681–693. doi: 10.1016/j.neuron.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo T, Cirrito JR, Liu CC, Shinohara M, Li J, Schuler DR, Holtzman DM, Bu G. Neuronal clearance of amyloid-beta by endocytic receptor LRP1. J Neurosci. 2013 doi: 10.1523/JNEUROSCI.3487-13.2013. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo T, Liu CC, Shinohara M, Li J, Bu G. LRP1 in brain vascular smooth muscle cells mediates local clearance of Alzheimer’s amyloid-beta. J Neurosci. 2012;32:16458–16465. doi: 10.1523/JNEUROSCI.3987-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo T, Zhang J, Liu Q, Liu CC, Zhang L, Bu G. Heparan sulphate proteoglycan and the low-density lipoprotein receptor-related protein 1 constitute major pathways for neuronal amyloid-beta uptake. J Neurosci. 2011;31:1644–1651. doi: 10.1523/JNEUROSCI.5491-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Iijima H, Goto K, Sakai J, Ishii H, Kim HJ, Suzuki H, Kondo H, Saeki S, Yamamoto T. Human apolipoprotein E receptor 2. A novel lipoprotein receptor of the low density lipoprotein receptor family predominantly expressed in brain. J Biol Chem. 1996;271:8373–8380. doi: 10.1074/jbc.271.14.8373. [DOI] [PubMed] [Google Scholar]
- Kim J, Castellano JM, Jiang H, Basak JM, Parsadanian M, Pham V, Mason SM, Paul SM, Holtzman DM. Overexpression of low-density lipoprotein receptor in the brain markedly inhibits amyloid deposition and increases extracellular A beta clearance. Neuron. 2009;64:632–644. doi: 10.1016/j.neuron.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Eltorai AE, Jiang H, Liao F, Verghese PB, Stewart FR, Basak JM, Holtzman DM. Anti-apoE immunotherapy inhibits amyloid accumulation in a transgenic mouse model of Abeta amyloidosis. J Exp Med. 2012;209:2149–2156. doi: 10.1084/jem.20121274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jiang H, Park S, Eltorai AE, Stewart FR, Yoon H, Basak JM, Finn MB, Holtzman DM. Haploinsufficiency of human APOE reduces amyloid deposition in a mouse model of amyloid-beta amyloidosis. J Neurosci. 2011;31:18007–18012. doi: 10.1523/JNEUROSCI.3773-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer MF, Soreghan B, Burdick D, Kosmoski J, Glabe CG. Intracellular accumulation and resistance to degradation of the Alzheimer amyloid A4/beta protein. Proc Natl Acad Sci U S A. 1992;89:7437–7441. doi: 10.1073/pnas.89.16.7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koistinaho M, Lin S, Wu X, Esterman M, Koger D, Hanson J, Higgs R, Liu F, Malkani S, Bales KR, et al. Apolipoprotein E promotes astrocyte colocalization and degradation of deposited amyloid-beta peptides. Nat Med. 2004;10:719–726. doi: 10.1038/nm1058. [DOI] [PubMed] [Google Scholar]
- Kok E, Haikonen S, Luoto T, Huhtala H, Goebeler S, Haapasalo H, Karhunen PJ. Apolipoprotein E–dependent accumulation of Alzheimer disease–related lesions begins in middle age. Annals of Neurology. 2009;65:650–657. doi: 10.1002/ana.21696. [DOI] [PubMed] [Google Scholar]
- Koldamova R, Fitz NF, Lefterov I. The role of ATP-binding cassette transporter A1 in Alzheimer’s disease and neurodegeneration. Biochim Biophys Acta. 2010;1801:824–830. doi: 10.1016/j.bbalip.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal RC, Herz J, Goldstein JL, Esser V, Brown MS. Low density lipoprotein receptor-related protein mediates uptake of cholesteryl esters derived from apoprotein E-enriched lipoproteins. Proc Natl Acad Sci U S A. 1989;86:5810–5814. doi: 10.1073/pnas.86.15.5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal RC, Herz J, Weisgraber KH, Mahley RW, Brown MS, Goldstein JL. Opposing effects of apolipoproteins E and C on lipoprotein binding to low density lipoprotein receptor-related protein. J Biol Chem. 1990;265:10771–10779. [PubMed] [Google Scholar]
- LaDu MJ, Falduto MT, Manelli AM, Reardon CA, Getz GS, Frail DE. Isoform-specific binding of apolipoprotein E to beta-amyloid. J Biol Chem. 1994;269:23403–23406. [PubMed] [Google Scholar]
- LaDu MJ, Gilligan SM, Lukens JR, Cabana VG, Reardon CA, Van Eldik LJ, Holtzman DM. Nascent astrocyte particles differ from lipoproteins in CSF. J Neurochem. 1998;70:2070–2081. doi: 10.1046/j.1471-4159.1998.70052070.x. [DOI] [PubMed] [Google Scholar]
- LaDu MJ, Pederson TM, Frail DE, Reardon CA, Getz GS, Falduto MT. Purification of apolipoprotein E attenuates isoform-specific binding to beta-amyloid. J Biol Chem. 1995;270:9039–9042. doi: 10.1074/jbc.270.16.9039. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Laporte V, Lombard Y, Levy-Benezra R, Tranchant C, Poindron P, Warter JM. Uptake of Abeta 1–40- and Abeta 1–42-coated yeast by microglial cells: a role for LRP. J Leukoc Biol. 2004;76:451–461. doi: 10.1189/jlb.1203620. [DOI] [PubMed] [Google Scholar]
- Lauren J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature. 2009;457:1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leissring MA, Farris W, Chang AY, Walsh DM, Wu X, Sun X, Frosch MP, Selkoe DJ. Enhanced proteolysis of beta-amyloid in APP transgenic mice prevents plaque formation, secondary pathology, and premature death. Neuron. 2003;40:1087–1093. doi: 10.1016/s0896-6273(03)00787-6. [DOI] [PubMed] [Google Scholar]
- Li J, Kanekiyo T, Shinohara M, Zhang Y, LaDu MJ, Xu H, Bu G. Differential regulation of amyloid-beta endocytic trafficking and lysosomal degradation by apolipoprotein E isoforms. J Biol Chem. 2012;287:44593–44601. doi: 10.1074/jbc.M112.420224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zerbinatti CV, Zhang J, Hoe HS, Wang B, Cole SL, Herz J, Muglia L, Bu G. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron. 2007;56:66–78. doi: 10.1016/j.neuron.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Yee A, Brewer HB, Jr, Das S, Potter H. Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature. 1994;372:92–94. doi: 10.1038/372092a0. [DOI] [PubMed] [Google Scholar]
- Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Rall SC., Jr Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J Lipid Res. 2009;50(Suppl):S183–188. doi: 10.1194/jlr.R800069-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrekar S, Jiang Q, Lee CY, Koenigsknecht-Talboo J, Holtzman DM, Landreth GE. Microglia mediate the clearance of soluble Abeta through fluid phase macropinocytosis. J Neurosci. 2009;29:4252–4262. doi: 10.1523/JNEUROSCI.5572-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques F, Sousa JC, Sousa N, Palha JA. Blood-brain-barriers in aging and in Alzheimer’s disease. Mol Neurodegener. 2013;8:38. doi: 10.1186/1750-1326-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC, Yarasheski KE, Bateman RJ. Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science. 2010;330:1774. doi: 10.1126/science.1197623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucke L, Selkoe DJ. Neurotoxicity of amyloid beta-protein: synaptic and network dysfunction. Cold Spring Harb Perspect Med. 2012;2:a006338. doi: 10.1101/cshperspect.a006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki H, Gejyo F, Nakakuki K. Concentration-dependent inhibitory effects of apolipoprotein E on Alzheimer’s beta-amyloid fibril formation in vitro. Biochemistry. 1997;36:6243–6250. doi: 10.1021/bi9624705. [DOI] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba Y, Tomonaga M, Kawasaki H, Otomo E, Ikeda K. Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer’s disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 1991;541:163–166. doi: 10.1016/0006-8993(91)91092-f. [DOI] [PubMed] [Google Scholar]
- Narita M, Holtzman DM, Schwartz AL, Bu G. Alpha2-macroglobulin complexes with and mediates the endocytosis of beta-amyloid peptide via cell surface low-density lipoprotein receptor-related protein. J Neurochem. 1997;69:1904–1911. doi: 10.1046/j.1471-4159.1997.69051904.x. [DOI] [PubMed] [Google Scholar]
- Nielsen HM, Mulder SD, Belien JA, Musters RJ, Eikelenboom P, Veerhuis R. Astrocytic A beta 1–42 uptake is determined by A beta-aggregation state and the presence of amyloid-associated proteins. Glia. 2010;58:1235–1246. doi: 10.1002/glia.21004. [DOI] [PubMed] [Google Scholar]
- Nishitsuji K, Hosono T, Nakamura T, Bu G, Michikawa M. Apolipoprotein E regulates the integrity of tight junctions in an isoform-dependent manner in an in vitro blood-brain barrier model. J Biol Chem. 2011;286:17536–17542. doi: 10.1074/jbc.M111.225532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, Guillozet-Bongaarts A, Ohno M, Disterhoft J, Van Eldik L, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26:10129–10140. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralvarez-Marin A, Barth A, Graslund A. Time-resolved infrared spectroscopy of pH-induced aggregation of the Alzheimer Abeta(1–28) peptide. J Mol Biol. 2008;379:589–596. doi: 10.1016/j.jmb.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Perez E, Bourguet W, Gronemeyer H, de Lera AR. Modulation of RXR function through ligand design. Biochim Biophys Acta. 2012;1821:57–69. doi: 10.1016/j.bbalip.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Petrlova J, Hong HS, Bricarello DA, Harishchandra G, Lorigan GA, Jin LW, Voss JC. A differential association of Apolipoprotein E isoforms with the amyloid-beta oligomer in solution. Proteins. 2011;79:402–416. doi: 10.1002/prot.22891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitas RE, Boyles JK, Lee SH, Foss D, Mahley RW. Astrocytes synthesize apolipoprotein E and metabolize apolipoprotein E-containing lipoproteins. Biochim Biophys Acta. 1987a;917:148–161. doi: 10.1016/0005-2760(87)90295-5. [DOI] [PubMed] [Google Scholar]
- Pitas RE, Boyles JK, Lee SH, Hui D, Weisgraber KH. Lipoproteins and their receptors in the central nervous system. Characterization of the lipoproteins in cerebrospinal fluid and identification of apolipoprotein B,E(LDL) receptors in the brain. J Biol Chem. 1987b;262:14352–14360. [PubMed] [Google Scholar]
- Polvikoski T, Sulkava R, Haltia M, Kainulainen K, Vuorio A, Verkkoniemi A, Niinisto L, Halonen P, Kontula K. Apolipoprotein E, dementia, and cortical deposition of beta-amyloid protein. N Engl J Med. 1995;333:1242–1247. doi: 10.1056/NEJM199511093331902. [DOI] [PubMed] [Google Scholar]
- Rall SC, Jr, Weisgraber KH, Mahley RW. Human apolipoprotein E. The complete amino acid sequence. J Biol Chem. 1982;257:4171–4178. [PubMed] [Google Scholar]
- Reiman EM, Chen K, Liu X, Bandy D, Yu M, Lee W, Ayutyanont N, Keppler J, Reeder SA, Langbaum JBS, et al. Fibrillar amyloid-β burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease. Proceedings of the National Academy of Sciences. 2009;106:6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink AA, de Waal RM, Kremer B, Verbeek MM. Pathogenesis of cerebral amyloid angiopathy. Brain Res Brain Res Rev. 2003;43:207–223. doi: 10.1016/j.brainresrev.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Revesz T, Ghiso J, Lashley T, Plant G, Rostagno A, Frangione B, Holton JL. Cerebral amyloid angiopathies: a pathologic, biochemical, and genetic view. Journal of neuropathology and experimental neurology. 2003;62:885–898. doi: 10.1093/jnen/62.9.885. [DOI] [PubMed] [Google Scholar]
- Richard E, Carrano A, Hoozemans JJ, van Horssen J, van Haastert ES, Eurelings LS, de Vries HE, Thal DR, Eikelenboom P, van Gool WA, et al. Characteristics of dyshoric capillary cerebral amyloid angiopathy. J Neuropathol Exp Neurol. 2010;69:1158–1167. doi: 10.1097/NEN.0b013e3181fab558. [DOI] [PubMed] [Google Scholar]
- Riddell DR, Zhou H, Comery TA, Kouranova E, Lo CF, Warwick HK, Ring RH, Kirksey Y, Aschmies S, Xu J, et al. The LXR agonist TO901317 selectively lowers hippocampal Abeta42 and improves memory in the Tg2576 mouse model of Alzheimer’s disease. Mol Cell Neurosci. 2007;34:621–628. doi: 10.1016/j.mcn.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Rushworth JV, Griffiths HH, Watt NT, Hooper NM. Prion protein-mediated toxicity of amyloid-beta oligomers requires lipid rafts and the transmembrane LRP1. J Biol Chem. 2013;288:8935–8951. doi: 10.1074/jbc.M112.400358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo C, Angelini G, Dapino D, Piccini A, Piombo G, Schettini G, Chen S, Teller JK, Zaccheo D, Gambetti P, et al. Opposite roles of apolipoprotein E in normal brains and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 1998;95:15598–15602. doi: 10.1073/pnas.95.26.15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M, Pankiewicz J, Scholtzova H, Ripellino JA, Li Y, Schmidt SD, Mathews PM, Fryer JD, Holtzman DM, Sigurdsson EM, et al. A synthetic peptide blocking the apolipoprotein E/beta-amyloid binding mitigates beta-amyloid toxicity and fibril formation in vitro and reduces beta-amyloid plaques in transgenic mice. Am J Pathol. 2004;165:937–948. doi: 10.1016/s0002-9440(10)63355-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saido T, Leissring MA. Proteolytic degradation of amyloid beta-protein. Cold Spring Harb Perspect Med. 2012;2:a006379. doi: 10.1101/cshperspect.a006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Dhanasekaran P, Nguyen D, Baldwin F, Weisgraber KH, Wehrli S, Phillips MC, Lund-Katz S. Characterization of the heparin binding sites in human apolipoprotein E. J Biol Chem. 2003;278:14782–14787. doi: 10.1074/jbc.M213207200. [DOI] [PubMed] [Google Scholar]
- Schneider WJ, Kovanen PT, Brown MS, Goldstein JL, Utermann G, Weber W, Havel RJ, Kotite L, Kane JP, Innerarity TL, et al. Familial dysbetalipoproteinemia. Abnormal binding of mutant apoprotein E to low density lipoprotein receptors of human fibroblasts and membranes from liver and adrenal of rats, rabbits, and cows. J Clin Invest. 1981;68:1075–1085. doi: 10.1172/JCI110330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Yamada S, Kumar SR, Calero M, Bading J, Frangione B, Holtzman DM, Miller CA, Strickland DK, Ghiso J, et al. Clearance of Alzheimer’s amyloid-ss(1–40) peptide from brain by LDL receptor-related protein-1 at the blood-brain barrier. J Clin Invest. 2000;106:1489–1499. doi: 10.1172/JCI10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Petersen RC, Dickson DW, Bu G. Brain regional correlation of amyloid-beta with synapses and apolipoprotein E in non-demented individuals: potential mechanisms underlying regional vulnerability to amyloid-beta accumulation. Acta Neuropathol. 2013;125:535–547. doi: 10.1007/s00401-013-1086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Sato N, Kurinami H, Takeuchi D, Takeda S, Shimamura M, Yamashita T, Uchiyama Y, Rakugi H, Morishita R. Reduction of brain beta-amyloid (Abeta) by fluvastatin, a hydroxymethylglutaryl-CoA reductase inhibitor, through increase in degradation of amyloid precursor protein C-terminal fragments (APP-CTFs) and Abeta clearance. J Biol Chem. 2010;285:22091–22102. doi: 10.1074/jbc.M110.102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivashanmugam A, Wang J. A unified scheme for initiation and conformational adaptation of human apolipoprotein E N-terminal domain upon lipoprotein binding and for receptor binding activity. J Biol Chem. 2009;284:14657–14666. doi: 10.1074/jbc.M901012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Weisgraber KH, Huang DY, Dong LM, Salvesen GS, Pericak-Vance M, Schmechel D, Saunders AM, Goldgaber D, Roses AD. Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai LM, Bilousova T, Jungbauer L, Roeske SK, Youmans KL, Yu C, Poon WW, Cornwell LB, Miller CA, Vinters HV, et al. Levels of soluble apolipoprotein E/amyloid-beta (Abeta) complex are reduced and oligomeric Abeta increased with APOE4 and Alzheimer disease in a transgenic mouse model and human samples. J Biol Chem. 2013;288:5914–5926. doi: 10.1074/jbc.M112.442103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamizu-Kato S, Cohen JK, Drake CB, Kosaraju MG, Drury J, Narayanaswami V. Interaction with amyloid beta peptide compromises the lipid binding function of apolipoprotein E. Biochemistry. 2008;47:5225–5234. doi: 10.1021/bi702097s. [DOI] [PubMed] [Google Scholar]
- Terwel D, Steffensen KR, Verghese PB, Kummer MP, Gustafsson JA, Holtzman DM, Heneka MT. Critical role of astroglial apolipoprotein E and liver X receptor-alpha expression for microglial Abeta phagocytosis. J Neurosci. 2011;31:7049–7059. doi: 10.1523/JNEUROSCI.6546-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thal DR, Ghebremedhin E, Rub U, Yamaguchi H, Del Tredici K, Braak H. Two types of sporadic cerebral amyloid angiopathy. J Neuropathol Exp Neurol. 2002;61:282–293. doi: 10.1093/jnen/61.3.282. [DOI] [PubMed] [Google Scholar]
- Thies W, Bleiler L. 2013 Alzheimer’s disease facts and figures. Alzheimers Dement. 2013;9:208–245. doi: 10.1016/j.jalz.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Tokuda T, Calero M, Matsubara E, Vidal R, Kumar A, Permanne B, Zlokovic B, Smith JD, Ladu MJ, Rostagno A, et al. Lipidation of apolipoprotein E influences its isoform-specific interaction with Alzheimer’s amyloid beta peptides. Biochem J. 2000;348(Pt 2):359–365. [PMC free article] [PubMed] [Google Scholar]
- Utermann G, Hees M, Steinmetz A. Polymorphism of apolipoprotein E and occurrence of dysbetalipoproteinaemia in man. Nature. 1977;269:604–607. doi: 10.1038/269604a0. [DOI] [PubMed] [Google Scholar]
- van Horssen J, Wesseling P, van den Heuvel LP, de Waal RM, Verbeek MM. Heparan sulphate proteoglycans in Alzheimer’s disease and amyloid-related disorders. Lancet Neurol. 2003;2:482–492. doi: 10.1016/s1474-4422(03)00484-8. [DOI] [PubMed] [Google Scholar]
- Verghese PB, Castellano JM, Garai K, Wang Y, Jiang H, Shah A, Bu G, Frieden C, Holtzman DM. ApoE influences amyloid-beta (Abeta) clearance despite minimal apoE/Abeta association in physiological conditions. Proc Natl Acad Sci U S A. 2013;110:E1807–1816. doi: 10.1073/pnas.1220484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent B, Smith JD. Astrocytes down-regulate neuronal beta-amyloid precursor protein expression and modify its processing in an apolipoprotein E isoform-specific manner. Eur J Neurosci. 2001;14:256–266. doi: 10.1046/j.0953-816x.2001.01643.x. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Hartman RE, Bales KR, Paul SM, Holtzman DM. Deletion of Abca1 increases Abeta deposition in the PDAPP transgenic mouse model of Alzheimer disease. J Biol Chem. 2005;280:43236–43242. doi: 10.1074/jbc.M508780200. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Kim J, Li A, Knoten A, Jain S, Hirsch-Reinshagen V, Wellington CL, Bales KR, et al. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J Clin Invest. 2008;118:671–682. doi: 10.1172/JCI33622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahrle SE, Shah AR, Fagan AM, Smemo S, Kauwe JS, Grupe A, Hinrichs A, Mayo K, Jiang H, Thal LJ, et al. Apolipoprotein E levels in cerebrospinal fluid and the effects of ABCA1 polymorphisms. Mol Neurodegener. 2007;2:7. doi: 10.1186/1750-1326-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ren CH, Gunawardana CG, Schmitt-Ulms G. Overcoming barriers and thresholds - signaling of oligomeric Abeta through the prion protein to Fyn. Mol Neurodegener. 2013;8:24. doi: 10.1186/1750-1326-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisgraber KH, Rall SC, Jr, Mahley RW. Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J Biol Chem. 1981;256:9077–9083. [PubMed] [Google Scholar]
- Wernette-Hammond ME, Lauer SJ, Corsini A, Walker D, Taylor JM, Rall SC., Jr Glycosylation of human apolipoprotein E. The carbohydrate attachment site is threonine 194. J Biol Chem. 1989;264:9094–9101. [PubMed] [Google Scholar]
- Wilsie LC, Orlando RA. The low density lipoprotein receptor-related protein complexes with cell surface heparan sulfate proteoglycans to regulate proteoglycan-mediated lipoprotein catabolism. The Journal of biological chemistry. 2003;278:15758–15764. doi: 10.1074/jbc.M208786200. [DOI] [PubMed] [Google Scholar]
- Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- Winkler K, Scharnagl H, Tisljar U, Hoschutzky H, Friedrich I, Hoffmann MM, Huttinger M, Wieland H, Marz W. Competition of Abeta amyloid peptide and apolipoprotein E for receptor-mediated endocytosis. J Lipid Res. 1999;40:447–455. [PubMed] [Google Scholar]
- Wisniewski T, Golabek A, Matsubara E, Ghiso J, Frangione B. Apolipoprotein E: binding to soluble Alzheimer’s beta-amyloid. Biochem Biophys Res Commun. 1993;192:359–365. doi: 10.1006/bbrc.1993.1423. [DOI] [PubMed] [Google Scholar]
- Wood SJ, Chan W, Wetzel R. An ApoE-Abeta inhibition complex in Abeta fibril extension. Chem Biol. 1996a;3:949–956. doi: 10.1016/s1074-5521(96)90183-0. [DOI] [PubMed] [Google Scholar]
- Wood SJ, Chan W, Wetzel R. Seeding of A beta fibril formation is inhibited by all three isotypes of apolipoprotein E. Biochemistry. 1996b;35:12623–12628. doi: 10.1021/bi961074j. [DOI] [PubMed] [Google Scholar]
- Xu Q, Bernardo A, Walker D, Kanegawa T, Mahley RW, Huang Y. Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J Neurosci. 2006;26:4985–4994. doi: 10.1523/JNEUROSCI.5476-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Brecht WJ, Weisgraber KH, Mahley RW, Huang Y. Apolipoprotein E4 domain interaction occurs in living neuronal cells as determined by fluorescence resonance energy transfer. J Biol Chem. 2004;279:25511–25516. doi: 10.1074/jbc.M311256200. [DOI] [PubMed] [Google Scholar]
- Yamada K, Hashimoto T, Yabuki C, Nagae Y, Tachikawa M, Strickland DK, Liu Q, Bu G, Basak JM, Holtzman DM, et al. The low density lipoprotein receptor-related protein 1 mediates uptake of amyloid beta peptides in an in vitro model of the blood-brain barrier cells. J Biol Chem. 2008;283:34554–34562. doi: 10.1074/jbc.M801487200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youmans KL, Tai LM, Nwabuisi-Heath E, Jungbauer L, Kanekiyo T, Gan M, Kim J, Eimer WA, Estus S, Rebeck GW, et al. APOE4-specific changes in Abeta accumulation in a new transgenic mouse model of Alzheimer disease. J Biol Chem. 2012;287:41774–41786. doi: 10.1074/jbc.M112.407957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis VI, Breslow JL. Human very low density lipoprotein apolipoprotein E isoprotein polymorphism is explained by genetic variation and posttranslational modification. Biochemistry. 1981;20:1033–1041. doi: 10.1021/bi00507a059. [DOI] [PubMed] [Google Scholar]
- Zerbinatti CV, Wahrle SE, Kim H, Cam JA, Bales K, Paul SM, Holtzman DM, Bu G. Apolipoprotein E and low density lipoprotein receptor-related protein facilitate intraneuronal Abeta42 accumulation in amyloid model mice. J Biol Chem. 2006;281:36180–36186. doi: 10.1074/jbc.M604436200. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Nwabuisi-Heath E, Dumanis SB, Tai LM, Yu C, Rebeck GW, LaDu MJ. APOE genotype alters glial activation and loss of synaptic markers in mice. Glia. 2012;60:559–569. doi: 10.1002/glia.22289. [DOI] [PMC free article] [PubMed] [Google Scholar]