Abstract

Heat shock protein 70 (Hsp70) is an important emerging cancer target whose inhibition may affect multiple cancer-associated signaling pathways and, moreover, result in significant cancer cell apoptosis. Despite considerable interest from both academia and pharmaceutical companies in the discovery and development of druglike Hsp70 inhibitors, little success has been reported so far. Here we describe structure–activity relationship studies in the first rationally designed Hsp70 inhibitor class that binds to a novel allosteric pocket located in the N-terminal domain of the protein. These 2,5′-thiodipyrimidine and 5-(phenylthio)pyrimidine acrylamides take advantage of an active cysteine embedded in the allosteric pocket to act as covalent protein modifiers upon binding. The study identifies derivatives 17a and 20a, which selectively bind to Hsp70 in cancer cells. Addition of high nanomolar to low micromolar concentrations of these inhibitors to cancer cells leads to a reduction in the steady-state levels of Hsp70-sheltered oncoproteins, an effect associated with inhibition of cancer cell growth and apoptosis. In summary, the described scaffolds represent a viable starting point for the development of druglike Hsp70 inhibitors as novel anticancer therapeutics.
Introduction
The heat shock protein 70 (Hsp70) family members are powerful proteins with major roles in malignancy, such as inhibition of apoptosis, induction of resistance to chemotherapy, and regulation of the stability of oncoproteins.1−3 Specifically, Hsp70 expression blocks apoptosis at several levels, and in this respect the chaperone inhibits key effectors of the apoptotic machinery, and also facilitates proteasome-mediated degradation of apoptosis-regulatory proteins. The contribution of Hsp70 isoforms to tumorigenesis is mainly through their role as cochaperones of heat shock protein 90 (Hsp90), a heat shock protein known to regulate the transforming activities of several kinases and transcription factors. In this process, Hsp70 initiates the association of the client protein with Hsp90 through a bridging protein called HSP-organizing protein (HOP). These biological functions propose Hsp70 as an important target whose inhibition or downregulation may result in significant apoptosis in a wide range of cancer cells and also in inhibition of signaling pathways involved in tumorigenesis and metastasis. Indeed, simultaneous silencing of Hsc70 or Hsp70 expression in human colon cancer cell lines induced proteasome-dependent degradation of Hsp90 onco-client proteins, cell-cycle arrest, and tumor-specific apoptosis.4 Importantly, silencing of Hsp70 isoforms in nontumorigenic cell lines did not result in comparable growth arrest or induction of apoptosis, indicating a potential therapeutic window for Hsp70 targeted therapies.
The Hsp70’s are a family of highly homologous proteins composed of two functional domains: the N-terminal ATPase domain and the C-terminal client protein-binding domain.5,6 The unique interplay between the two domains creates a ligand-activated, bidirectional molecular switch. For example, ATP binding to the ATPase domain induces a conformational change that is rapidly propagated to the C-terminal and that results in accelerated client protein dissociation. Conversely, client protein binding to the C-terminal domain of ATP-bound Hsp70 induces a conformational change that is propagated to the ATPase domain and that results in a stimulation of the ATP hydrolysis rate. The chaperoning activity of Hsp70 is further regulated by cochaperones (e.g., Hsp40s, BAG, and Hsp110) that catalyze the interconversion between the ATP- and ADP-bound states and thus regulate chaperone function. Such structural regulation suggests that Hsp70 may be vulnerable to most strategies that interfere with its flexibility.
Much effort has recently been dedicated toward the discovery of Hsp70 inhibitors, and unsurprisingly, molecules from a number of chemical classes have been reported to interact with Hsp70 through a variety of modes (Figure 1).7,8 A few, such as 15-deoxyspergualin (1) and pifithrin-μ (2-phenylethynesulfonamide) (2), are believed to target the C-terminal of Hsp70,9,10 whereas others, such as dihydropyrimidines (i.e., 3 (MAL3-101)),11 are thought to block J-domain-stimulated ATPase activity of Hsp70. Compounds such as myricetin (4)12 and 5 (MKT-077)13 are proposed to interact with a pocket outside the nucleotide-binding domain, whereas apoptozole (6) may bind to the ATP-binding pocket of Hsp70.14
Figure 1.
Chemical structure of reported potential Hsp70 inhibitors.
The majority of these compounds were discovered in library screens that aimed to identify inhibitors of either the ATPase or the folding capacity of yeast or bacterial Hsp702,7,8 or in the case of 6 a cell-based screen of compounds capable of inducing apoptosis.155 was discovered following optimization efforts16 that had previously identified such rhodacyanine dyes as possessing anticancer activity.17 In the only reported rational design approach to develop Hsp70 inhibitors, nucleotide mimetics such as the dibenzyl-8-aminoadenosine analogue 7 (VER-155008) were developed to bind into the N-terminal ATP pocket of Hsp70.18 While these molecules are reported to elicit their effects through an Hsp70 mechanism, it is likely that they also act on multiple other unrelated and as yet unspecified mechanisms. Furthermore, these molecules have been hindered by a nontractable structure–activity relationship (SAR), with subtle changes resulting in drastic changes in activity. While these molecules have been of some value as tool molecules to offer insight into the consequences of pharmacological modulation of Hsp70, they have limited potential to become useful drugs.
At this point in time it is fair to say that Hsp70 has proven to be a very difficult target to drug. In contrast, Hsp90 has proven highly amenable with numerous small-molecule ATP-competitive inhibitors entering into the clinic.19 In the case of Hsp90, potent small-molecule inhibitors such as geldanamycin and radicicol were known even before their precise mode of action was determined. When X-ray crystal structures showed that they bound to a clearly specified pocket (i.e., ATP pocket) and behaved as ATP-competitive inhibitors, structure-based drug design became possible. Unfortunately, no such molecules that could potentially guide and truly inspire the development of Hsp70 inhibitors exist for Hsp70.
The experiences of scientists at Vernalis offer some further insight into the difficulty of targeting Hsp70 and perhaps as to why no natural product ATP-competitive inhibitor is known.18,20 In the only rational approach reported to date, they designed a series of adenosine analogues to act as direct ATP-competitive inhibitors. However, despite obtaining molecules that bound Hsp70 with high affinity (Kd = 50 nM), their cellular potency was disappointing, and a number of reasons were given for this poor correlation. Hsp70 has a high affinity for ADP (Kd = 0.11–0.5 μM) and coupled with high intracellular ATP concentrations makes the prospect of competitive inhibition a daunting task. Furthermore, the binding mode of ATP is such that important polar contacts are made with its β- and γ-phosphate groups that lie buried within a polar cavity. Attempts to mimic these interactions have resulted in highly polar nondruglike molecules such as 7, which despite potent affinity (Kd = 0.3 μM) possess weak cellular activity. Combined, these factors make the prospect of obtaining compounds with potent in vivo activity low and make a strong case against the development of reversible ATP-competitive inhibitors of Hsp70 as a viable therapeutic strategy.21
We had sought alternative strategies toward inhibiting Hsp70 which would have the potential for potent in vivo activity. Specifically, we had sought and subsequently identified another pocket, outside of the active site.22 This allosteric site, located in the N-terminal domain, was not evident nor entirely predicted by the available crystal structures of Hsp70 and has been recently discovered by us through computational analyses.22 We used the homology model to design ligands that could bind to this allosteric pocket.22 Here we describe structure–activity investigations in this first reported allosteric pocket inhibitor class. These ligands are 2,5′-thiodipyrimidine and 5-(phenylthio)pyrimidine scaffold compounds that we show here and in the accompanying paper23 to be amenable to extensive medicinal chemistry and to act on cancer cells through an Hsp70-mediated mechanism.
Ligand Design and Computational Analysis of Ligand–Hsp70 Interactions
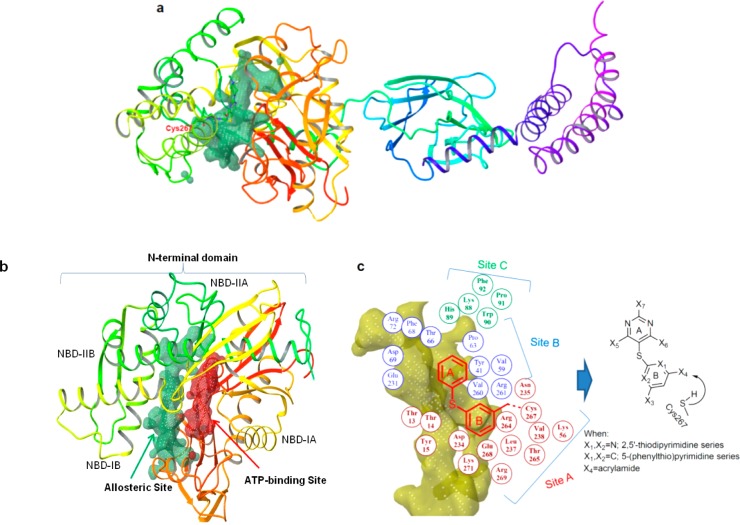
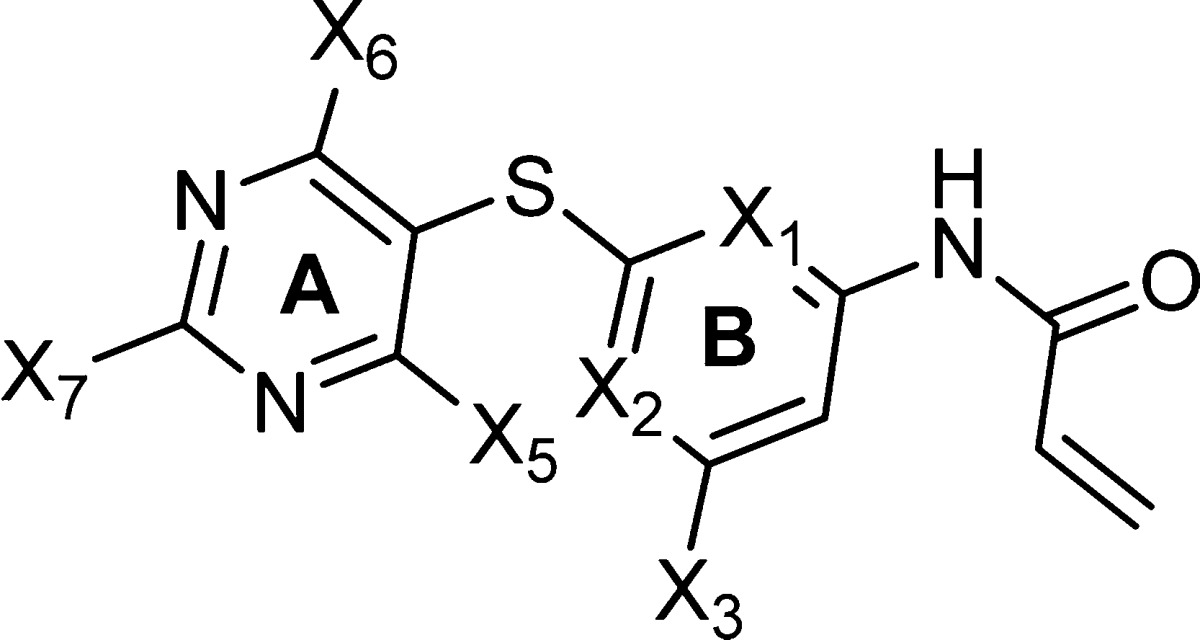
There are several available crystal structures of Hsp70, but most capture a relatively closed nucleotide-binding domain.24−27 In contrast, recent NMR techniques and molecular dynamics studies suggest a model in which subdomains of the N-terminus are bridged and in a close proximity only in the ATP-binding conformation, with residues of these subdomains rearranging and the cleft opening upon ADP binding.28,29 To overcome these limitations of the currently available crystal structures and to investigate pockets other than the ATP-binding site, we recently developed a theoretical model of human Hsp70 (hHsp70).22 This nucleotide-free structure of hHsp70 both captured the N-terminal binding cleft in a conformation that is more open than that captured by X-ray and unveiled a pocket that contains a potentially reactive cysteine residue (Cys267 in hHsp70) embedded inside the cavity (Figure 2a).22 The newly identified allosteric pocket (depicted by the green surface in Figure 2a,b) is located outside the nucleotide-binding domain and is flanked by subregions IB and IIB of the nucleotide-binding domain (NBD) (Figure 2b).
Figure 2.
Rational design of the allosteric pocket ligands. (a,b) The homology-modeled hHsp70 structure led to the identification of a novel allosteric pocket (shown in green) located in the N-terminal domain of human Hsp70.22 This pocket contains a potentially reactive cysteine residue (position shown in red lettering in panel a) and is located outside the ATP-binding site (shown in red in panel b). (c) The geometry of the N-terminal allosteric pocket of hHsp70 as predicted by SiteMap (shown in yellow) and residues lining the pocket and their relative location (depicted as colored circles) are presented. Ligands based on the 2,5′-thiopyrimidine and 5-(phenylthio)pyrimidine chemical scaffolds were designed to fit into the allosteric pocket. They adopt the conformation required for a proper fit and position functionalities toward the shown pocket residues. An acrylamide positioned toward Cys267 was also incorporated into these scaffolds.
This cavity is larger and potentially more druglike than the nucleotide-binding pocket (depicted by the red surface in Figure 2b), because it contains a balanced number of hydrophilic and hydrophobic residues (Figure 2c). At the lower part of the allosteric site lies mainly a hydrophilic cavity containing Lys271 and Glu268 and further apart lined by Thr13, Thr14, Tyr15, Lys56, Asp234, Asn235, Arg264, Thr265, and Arg269 (numbering as in hHsp70) (site A, Figure 2c). Hydrophobic amino acids such as Leu237 and Val238 also line site A. This subpocket also contains the potentially reactive cysteine residue, Cys267, that could covalently link to a ligand containing the appropriate Cys-reactive functionality, such as an acrylamide.30 Adjacent to site A, and placed in the middle, is a larger cavity comprised of both nonpolar and polar amino acid residues, such as Tyr41, Val59, Pro63, Thr66, Asp69, Phe68, Arg72, Glu231, Val260, and Arg261 (site B, Figure 2c). Placed at the upper part of the binding site, and also potentially providing interactions to a small-molecule ligand, are Lys88, His89, Trp90, Pro91, and Phe92 (site C, Figure 2c).
We analyzed the geometry and the environment of the computationally identified allosteric pocket to provide a starting point for the rational design of ligands (Figure 2c). We concluded that ligands built around the 2,5′-thiodipyrimidine and 5-(phenylthio)pyrimidine scaffolds, both little explored chemical spaces, would adapt the necessary conformation and provide the required attachment sites for ligand functionalization (Figure 2c). Each designed ligand used this pharmacophore template to attach X3–7 functionalities pointed toward several amino acids lining the Hsp70 pocket (Figure 2c).
Because our homology model may not entirely mimic the native protein structure, we included an additional binding hook, i.e., an acrylamide functionality, to probe possible covalent bond formation between the inhibitor and Cys267 upon protein binding (Figure 2c). Due to the location of Cys267 deep inside the cavity, such bond formation would be possible only after the ligand was inserted into the pocket and has achieved a proper fit. By gaining affinity through a covalent linkage in addition to enthalpy, we hoped to increase the ligand’s apparent affinity for the protein in the event the fit would be less than optimal. We reasoned that if the initial ligand would be of sufficiently good fit, additional enthalpy could be gained by properly modifying the appended functionalities, such as we proceed to do here and in the adjoining paper.23
Chemistry
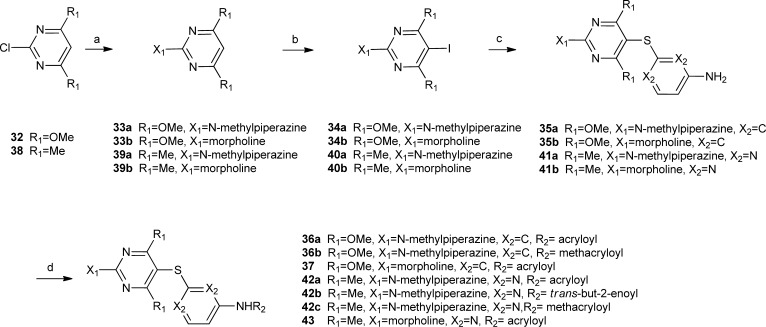
The synthesis of all designed compounds evaluated in this study is shown in Schemes 1–3 and described below. Reaction of 2-amino-4,6-dimethoxypyrimidine (8a) with p-methoxybenzyl chloride ((PMB)Cl) resulted in PMB-protected pyrimidine 9a in 95% yield (Scheme 1). This was iodinated with N-iodosuccinimide (NIS) to give 10a in 98%, which was further coupled to 4,6-diamino-2-mercaptopyrimidine using CuI/neocuproine to give 2,5′-thiodipyrimidine 11a in 65% yield. Acetylation of the amino groups was accomplished with Ac2O/DMAP at 110 °C for 2 h to give 12a in 91% yield. Selective removal of the PMB groups occurred by heating 12a in a 1:1 mixture of TFA/CHCl3 at 62 °C for 24 h to give 13a in 95%. Fluorodediazoniation was accomplished with NaNO2/HF/pyridine to give 14a in 54% yield, which was then reacted with a variety of amines to yield 15a–c. Deacetylation of these intermediates followed by reaction with acryloyl chloride resulted in target compounds 17a–c. Derivatives 20a–c were prepared following a similar route from 2-amino-4,6-diethoxypyrimidine (8b), which was prepared by refluxing a mixture of 2-amino-4,6-dichloropyrimidine and NaH in ethanol in 89% yield (Scheme 1).
Scheme 1.
Scheme 3.
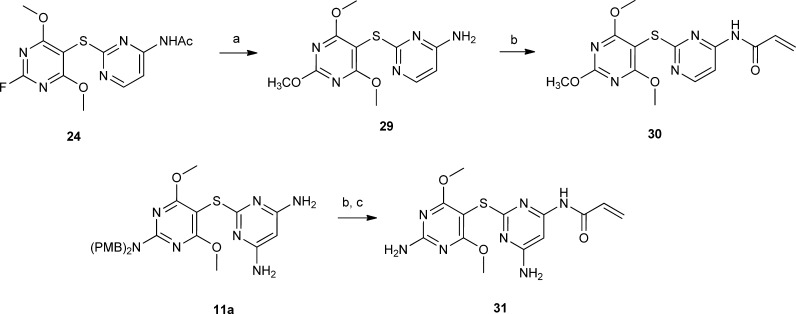
The chemistry used to prepare 27a–d is similar and shown in Scheme 1. Derivative 10a was coupled with 4-amino-2-mercaptopyrimidine to give 21 in 92% yield. Acetylation followed by PMB removal resulted in 23. Reaction with NaNO2/HF/pyridine gave fluoro derivative 24, which was reacted with a variety of amines to give 25a–d. Deacetylation followed by reaction with acryloyl chloride resulted in target compounds 27a–d. Reaction of 27a with m-CPBA at −78 °C resulted in N-oxide 28. Reaction of 24 with NaOMe/MeOH followed by deacetylation resulted in 29, which was reacted with acryloyl chloride to give 30 (Scheme 2). Reaction of 11a with acryloyl chloride followed by treatment with TFA resulted in 31 (Scheme 2).
Scheme 2.
Thioethers shown in Scheme 3 were prepared from chloropyrimidine 32 or 38. Reaction of 32 with N-methylpiperazine or morpholine resulted in 33a and 33b, respectively. Iodination of these derivatives, followed by coupling with 3-aminothiophenol resulted in 35a and 35b, respectively. These compounds were then reacted with a variety of unsaturated acid chlorides to give target compounds 36a,b and 37. Target compounds 42a–c and 43 were prepared similarly starting from chloropyrimidine 38 (Scheme 3).
Biological Evaluation in the Hsp70 Inhibitor Series
Recent studies suggest that, unlike normal cells, cancer cells express modified heat shock protein species characterized by both enhanced cochaperone recruitment and distinct post-translational modifications.31−33 Evidence also indicates that certain small molecules show preferential binding to these tumor-modified heat shock proteins.32 Our goal being to develop Hsp70 inhibitors for cancer treatment, we accordingly employed here a testing strategy that used phenotypic assays to read fingerprints of Hsp70 inhibition in tumor cells rather than a classical recombinant protein-based approach (Figure 3).
Figure 3.
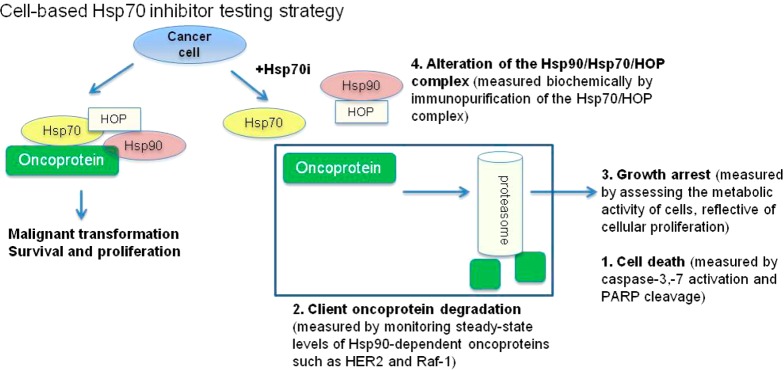
Known Hsp70 functions used to design the biological testing of the Hsp70 inhibitors in cancer cells. Hsp90 in concert with Hsp70 maintains the transforming capacity of several oncoproteins whose aberrant activity leads to increased cell proliferation and survival. The function of the Hsp90 protein complex requires the HSP-organizing protein (HOP), involved in the formation of the active chaperone complex. Altering the formation of the Hsp90–HOP–Hsp70 complex leads to degradation of the oncoprotein via a proteasome-mediated pathway and is associated with cell growth inhibition and/or cell death. To confirm an Hsp70-mediated mechanism, it is expected that compounds will be active at similar concentrations in this battery of assays.
To design a tumor–Hsp70 tailored testing module, we took advantage of known biological activities mediated by Hsp70 in cancer. As mentioned, Hsp70 is both an antiapoptotic molecule4 and a transforming protein that acts together with Hsp90 to regulate the altered activity of several cancer kinases.1,2,34 Thus, our testing strategy incorporated assays that measured induction of apoptosis (i.e., the ability to activate caspase-3,7, Tables 1 and 2 and induce PARP cleavage, Table 3) and those that read a phenotypic outcome similar to that of Hsp90 inhibitors (i.e., degradation of Hsp90/Hsp70-sheltered oncoproteins such as HER2 and Raf-1 in the proper genetic background, Table 3) (see tests 1 and 2 in Figure 3). Because a compound that interferes with nucleotide binding to the ATP-binding pocket of Hsp90, either competitively or allosterically, could result in a phenotype closely resembling Hsp70 inhibition, we also included an assay to test for possible direct binding to Hsp90 (Table 3).
Table 1.
| compd | X1 = X2 | X3 | X5, X6 | X7 | IC50 (μM) growth inhibitiona | IC50 (μM) caspase-3,7 activationb |
|---|---|---|---|---|---|---|
| 17a | N | –NH2 | 2 × −OCH3 | methylpiperazine | 0.90 ± 0.2 | 1.2 ± 0.4 |
| 17b | N | –NH2 | 2 × −OCH3 | morpholine | 12.5 ± 0.7 | 12.1 ± 2.5 |
| 17c | N | –NH2 | 2 × −OCH3 | piperidine | 17.1 ± 2.2 | 16.2 ± 0.5 |
| 20a | N | –NH2 | 2 × −OC2H5 | methylpiperazine | 0.89 ± 0.2 | 4.0 ± 0.0 |
| 20b | N | –NH2 | 2 × −OC2H5 | morpholine | 2.2 ± 1.2 | 10.5 ± 3.5 |
| 20c | N | –NH2 | 2 × −OC2H5 | piperidine | 4.5 ± 0.2 | 4.0 ± 2.0 |
| 27a | N | –H | 2 × −OCH3 | methylpiperazine | 1.25 ± 0.7 | 4.0 ± 0.5 |
| 27b | N | –H | 2 × −OCH3 | morpholine | 2.0 ± 0.5 | 7.2 ± 0.7 |
| 27c | N | –H | 2 × −OCH3 | piperidine | 3.15 ± 1.5 | 12.4 ± 1.5 |
| 27d | N | –H | 2 × −OCH3 | pyrrolidine | 46.1 ± 15.5 | >100 |
| 28 | N | –H | 2 × −OCH3 | 1-methylpiperazine 1-oxide | >100 | >100 |
| 30 | N | –H | 2 × −OCH3 | –OCH3 | ND | 7.5 ± 0.3 |
| 31 | N | –NH2 | 2 × −OCH3 | –NH2 | 3.9 ± 1.4 | 4.5 ± 0.5 |
| 36a | C | –H | 2 × −OCH3 | methylpiperazine | 7.8 ± 1.1 | ND |
| 37 | C | –H | 2 × −OCH3 | morpholine | 46.5 ± 3.4 | 27.5 ± 2.5 |
| 42a | N | –H | 2 × −CH3 | methylpiperazine | ND | 6.0 ± 2.1 |
| 43 | N | –H | 2 × −CH3 | morpholine | 14.4 ± 2.7 | 15.4 ± 0.7 |
| 44a | N | –NH2 | 2 × −OCH3 | 2-(2-(2-(2-(piperazin-1-yl)ethoxy)ethoxy)ethoxy) ethanol | 3.8 ± 1.2 | 14.3 ± 1.5 |
| 45a | N | –NH2 | –OCH3, –(OC2H4O)4H | methylpiperazine | 76.5 ± 21.5 | 100 ± 10.5 |
Inhibition of growth measured in Kasumi-1 acute myeloid leukemia cells. Values are the mean ± SEM.
Caspase-3,7 activation measured in MOLM13 acute myeloid leukemia cells. Values are the mean ± SEM.
Table 2.


Inhibition of growth measured in Kasumi-1 acute myeloid leukemia cells. Values are the mean ± SEM.
Caspase-3,7 activation measured in MOLM13 acute myeloid leukemia cells. Values are the mean ± SEM.
Table 3.
| compd | HER2b | Raf-1b | cPARPb | growth inhibitionb | Hsp90 bindingc |
|---|---|---|---|---|---|
| 4 | >100 | >100 | >100 | NA | 13.5 |
| PU24FCld | 2 | 2.5 | NA | 2.9 | 0.66 |
| 17a | 0.7 | 1.7 | 2.0 | 0.8 | >500 |
| 17c | 2.5 | 7.5 | 5 | 9 | >250 |
| 20a | 1 | 2.5 | 1.5 | 1.1 | >500 |
| 20c | 2 | 3.1 | 10 | 7 | >500 |
| 27a | 7.5 | 5 | 7.5 | 1.2 | >500 |
| 27d | 20 | 17 | 25 | 5 | >500 |
| 31 | 5 | 3.5 | 7.5 | 3.2 | >500 |
| 37 | 15 | 20 | 10 | 16.5 | >500 |
| 42a | 4 | 5 | 5 | 1.8 | >500 |
| 42c | 30 | 50 | 40 | 16.7 | >500 |
| 44a | 2.5 | 7.5 | 10 | 7.5 | >500 |
| 45a | 75 | 60 | 100 | 50 | >500 |
All values are in micromolar units.
HER2 and Raf-1 steady-state levels, PARP cleavage and inhibition of growth measured in SKBr3 breast cancer cells.
Binding to SKBr3 cell extracts.
Structure and activity reported in ref (38).
HER2 is a key transforming protein in the HER2-overexpressing breast cancer cells SKBr3, and its degradation or inhibition is sufficient to inhibit the growth of these cells. Thus, in addition to testing the effect of these derivatives on HER2 expression, we also measured their activity on cell growth (Table 3).35,36 In addition, the compounds were further tested for their ability to inhibit the growth of cancer cells of distinct genetic origin, including the P-gp/MDR-1-expressing Kasumi-1 acute myeloid leukemia cells (Table 1) (see test 3 in Figure 3).
For select derivatives we probed their ability to bind Hsp70 from tumor extracts (Figure 4a) and from live cancer cells (Figure 4b,c) and investigated their ability to alter the Hsp70–HOP complex in cancer cells (Figure 4d; see test 4 in Figure 3).5,6,32
Figure 4.
Designed ligands interact specifically with Hsp70 in cancer cells. (a) Streptavidin beads were incubated with the indicated concentrations of 44b, 45b, and d-biotin, the unbound agent was washed off, and the resulting beads carrying 44b, 45b, or d-biotin were probed with SKBr3 cell extracts (500 μg). Hsp70 isolated on the beads was probed by Western blot (WB). A representative blot is presented (top). Blots were quantified by densitometry and values, in relative luminescence units, graphed against the concentration of added biotinylated Hsp70 inhibitor (bottom). Results from three independent experiments were graphed to determine the relative binding affinity (Kd) of 44b and 45b using equations as implemented in the GraphPad Prism software. Key: points, mean; bars, SD. (b) Cells were treated with the indicated concentrations of 44b for 6 h prior to lysing and precipitation of protein complexes on streptavidin beads. Beads were washed with high-salt (1 M NaCl) buffer before elution of proteins on a denaturing gel and silver staining. BB70 is an antibody specific for Hsp70. This antibody also recognizes Grp78 and Grp75, the endoplasmic reticulum and the mitochondrial Hsp70 paralogues, respectively. (c) Protein complexes were isolated as indicated in (b) in cells pretreated with 17a. CP = chemical precipitation. (d) Analysis of the Hsp70–HOP complex. SKBr3 cells were treated for 24 h with vehicle or indicated concentrations of 20a. Upon cell lysing, Hsp70 complexes were isolated with an anti-Hsp70 antibody (IP BB70) and analyzed by WB. Specificity of binding was tested with a control IgG. Gels were quantified by densitometry, values normalized to the control (vehicle only treated cells), and data graphed against the concentration of 20a. Error bars represent the SEM (n = 2).
For a specific Hsp70-mediated biological effect it is expected that compounds should act with a similar potency in the above-described battery of assays (for example, the IC50 measured in the client degradation assay should be near the IC50 measured in Hsp70–HOP complex alteration and so on).2,35,36 Inclusion in the testing module of three different Hsp70-addicted cell lines, including one P-gp/MDR-1 expressing, increased our likelihood to eliminate early on poorly permeable compounds and those marred by potential for P-gp/MDR-1-induced resistance.
Structure–Activity Relationship in the Hsp70 Inhibitor Series
In addition to ligand design, we used the homology model to investigate structure–activity relationships in the designed Hsp70 inhibitor series (Figure 5). To study potential ligand–protein interactions, we docked each derivative into the allosteric site. This strategy, combined with the phenotypic, target-derived biological investigation (Figure 3), as described above, was used to understand differences in tumor Hsp70 inhibitory activity among the designed derivatives.
Figure 5.
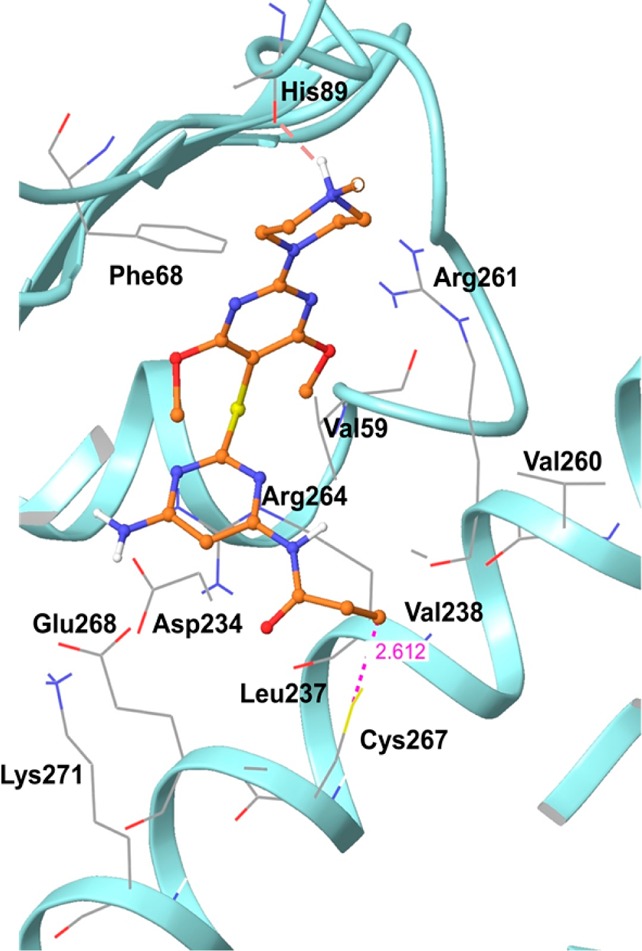
Binding interactions of hHsp70 with derivative 17a, as predicted by Glide (Schrodinger LLC, New York). Hydrogen bonds are shown as dotted red lines, and the interaction distance is shown by dotted purple lines.
Site C
Attached to ring A is substituent X7 pointing toward the exit of the binding site (Figure 2c). At this position we could accommodate a variety of substituents such as piperazine, morpholine, piperidine, pyrrolidine, methoxy, and amino (Table 1). Methylpiperazine, however, was preferred over all others (17a versus 17b, 17c, and 31; 20a versus 20b and 20c; 27a versus 27b, 27c, 27d, and 30; 36a versus 37). At physiological pH, methylpiperazine is likely to be protonated, and this H is predicted by docking to form a hydrogen bond interaction with the backbone carbonyl of His89 (Figure 5). Indeed, an approximately 10-fold drop in activity was observed when the N-Me of methylpiperazine was substituted with O (as in morpholine) or C (as in piperidine). Additionally, when this amine was oxidized to the corresponding N-oxide, as in derivative 28, activity was almost abolished, potentially due to the unfavorable interaction of the N-oxide oxygen with the backbone carbonyl of His89. The pyrrolidine-containing derivative 27d had a poor aqueous solubility, which likely accounted for the erratic cellular activity we observed for this compound.
Site B
Substituents X5 and X6 of ring A are placed in the middle cavity of the binding site comprised of both nonpolar and polar amino acid residues, as described above (Figure 2c). Substituents at these positions, such as methoxy, ethoxy, and methyl, were well tolerated (compare 17a (methoxy), 20a (ethoxy), and 42a (methyl)). Modeling predicts for such substituents a potential for hydrophobic (alkyl portion of these substituents with Val59) and electrostatic (oxygen of methoxy and ethoxy with Arg261) interactions (Figure 5), and all derivatives containing these substituents had favorable biological activity. Modeling also indicated the potential for steric clash for X5 and X6 substituents of larger size, such as in 45a, and indeed, we observed a substantial loss of activity for this derivative (Table 1).
Site A
At the bottom of the allosteric pocket lies Cys267, which is predicted to form a covalent bond with the acrylamide positioned at X4 on ring B (Figure 5). The contribution of the acrylamide moiety toward covalent binding with the protein can be seen by comparing compounds in which it is attached to either a pyrimidine or phenyl ring. Since the two nitrogen atoms of the pyrimidine ring render the ring more electron deficient, acrylamide functions attached to it would be expected to be more active Michael acceptors than in the corresponding phenyl compounds. Indeed, a trend for increased activity was seen for the pyrimidine-based compounds when compared to the similar phenyl derivatives (Table 1, 27a and 27b versus 36a and 37, respectively).
In addition to covalent Cys modification, the acrylamide substituent at position X4 is poised to form several interactions with the adjacent protein residues (Figure 5). Specifically, the alkene group is positioned to establish hydrophobic interactions with Leu237 and Val238. The carbonyl group is poised to form electrostatic interactions with the guanidine group of Arg264, ε-NH2 of Lys271, and backbone −NH of Glu268 and could potentially also form hydrogen bonds with the surrounding residues.
To further understand the contribution of the acrylamide to binding, we prepared several methyl-substituted acrylamide derivatives (Table 2). Each of these derivatives was found to be less active than the unsubstituted acrylamide derivatives (36a versus 36b; 42a versus 42b and 42c). This decrease in reactivity can be explained by both steric and electronic effects. The methyl group could act to sterically hinder nucleophilic attack by the reactive cysteine residue (Cys267). Furthermore, the methyl group is a mild electron-donating group and would decrease the electrophilicity of the Michael acceptor.
Ring B occupies the lower part of the binding site which also contains Arg264. Thus, the aromatic rings that were designed to compose ring B, such as phenyl and pyrimidine, are predicted to be stabilized by cation−π interactions with the guanidine group of this arginine residue (Figure 5). Ring B also orients substituents toward several important pocket residues. Specifically, at the lower part of the pocket are Asp234 and Glu268, which have the potential to interact with substituents on ring B, such as those located at position X3 (Figure 5). Accordingly, derivatives for which X3 was NH2 (i.e., 17a) were more potent than similar derivatives having a hydrogen at this position (i.e., 27a) potentially because NH2 could form hydrogen bonds with the carboxylate groups of Asp234 and Glu268.
Evaluation of Selectivity and Target-Mediated Cellular activity
From these studies, derivatives 17a and 20a emerged as most active (Tables 1 and 3). Next, we attached 17a to biotin with two aims in mind, first, to demonstrate effective and selective isolation of Hsp70 from cancer cells by 17a and, second, to further validate that the acrylamide was not the sole contributor to ligand–protein binding. Specifically, we have created two derivatives of 17a by attaching to it linkers that allow for biotin modification of 17a (Figure 4a). In derivative 44a, the linker was attached from the piperazine ring and thus predicted to point outside the allosteric pocket. Indeed, 44a retained biological activity similar to that of 17a (Tables 1 and 3). In derivative 45a, the linker was attached on ring A through substituent X5, thus at a position that would create steric clashes between the substituent and Hsp70 (Figure 4a). Indeed, 45a was of much reduced activity when compared to 17a (Tables 1 and 3). We next attached 44a and 45a to biotin and then immobilized, dose-dependently, the resulting 44b and 45b biotin-labeled ligands onto streptavidin-containing beads. We then used these beads to isolate Hsp70 from cancer cell extracts.
As expected from a mode of binding in which enthalpy substantially contributes to binding, efficient isolation of Hsp70 from a cancer cell extract was observed for the beads containing derivative 44b but not 45b (Figure 4a). If binding were solely driven by covalent modification through the acrylamide functionality, both 44b and 45b would interact equally well with Hsp70 and, thus, isolate similar amounts of Hsp70 from the cell extract. In conclusion, a proper fit into the protein pocket in addition to covalent modification was important for the ability of these ligands to bind to and inhibit Hsp70.
To demonstrate the selectivity of interaction, we incubated cancer cells with 44b and then proceeded to isolate the captured proteins on streptavidin beads. Upon washing the affinity-purified complexes and running them on a denaturing gel, the selectivity of 44b toward Hsp70 was demonstrated by the presence of a single band upon silver staining (Figure 4b). This band as demonstrated by its size of 70 kDa, its ability to run at a position identical to that of the protein isolated by an anti-Hsp70 antibody (Figure 4b, BB70), and its recognition by an Hsp70-specific antibody (Figure 4a) and mass spectrum analysis22 corresponds to Hsp70. Furthermore, preincubation of cells with soluble 17a prior to affinity purification led to a dose-dependent reduction of immobilized Hsp70, demonstrating the specificity of the interaction (Figure 4c). Altogether, these findings confirm that the designed ligands act in cells by specifically binding to Hsp70.
When cancer cells were incubated with select derivatives (i.e., 20a), we observed a dose-dependent alteration in the formation of Hsp70–HOP complexes in cancer cells (Figure 4d). As mentioned above, HOP is a cochaperone that bridges Hsp70 and Hsp90 to form a megachaperone complex (see test 4 in Figure 3). This chaperone machinery regulates the stability of several onco-client proteins, such as HER2 and Raf-1, whose degradation, as caused by chaperone inhibition, was tested here (Table 3). Of importance, alteration in the Hsp70–HOP complex formation occurred at the same low concentrations (IC50 = 1.73 μM, Figure 4d) where we also observed degradation of HER2 and Raf-1 by this inhibitor (1 and 2.5 μM, respectively, Table 3), further supporting the Hsp70-mediated mechanism of action of these ligands.
We noted no effect on Hsp90 for the ligands reported here at concentrations as high as 500 μM (Table 3). Specifically, unlike the direct Hsp90 inhibitor PU24FCl,38 a compound with cellular activity comparable to that of the most active Hsp70 inhibitors described here, such as 17a and 20a (Table 3, HER2, Raf-1), the Hsp70 inhibitors failed to compete with a fluorescently labeled geldanamycin derivative, GM-Cy3B39 for Hsp90 binding (Table 3). Geldanamycin is an Hsp90 inhibitor that binds to the Hsp90 regulatory pocket37 located in the N-terminal domain of Hsp90. The fluorescence polarization based Hsp90 assay that incorporates GM-Cy3B is designed in such a way that it probes not only for direct binders to the ATP pocket but also for those compounds that may allosterically interfere with the conformation of Hsp90 to block compound access to the ATP pocket.
We also tested myricetin (4; Figure 1) in these assays (Table 3). Using NMR techniques,12 this compound was proposed to interact with the bacterial Hsp70 homologue, DnaK, at a site potentially close to that occupied by our designed Hsp70 inhibitors. 4 was also recently reported to inhibit the DnaK–DnaJ (bacterial Hsp70–Hsp40) complex formation (IC50 = 14.5 μM).12 We could however not measure Hsp70-mediated biological effects for 4 in the SKBr3 cancer cells at concentrations as high as 100 μM, possibly due to the poor stability of 4 in the cells. Indeed, LC/MS–MS analyses of cellular extracts demonstrated rapid myricetin degradation with the agent virtually undetectable after 10 min of incubation (not shown). Interestingly, when tested for its potential to bind Hsp90, we found 4 to alter geldanamycin’s binding to Hsp90 (Table 3). Polyphenols such as 4 are recognized for their high propensity to bind nonspecifically to proteins. As such, caution must be taken when interpreting biological and biochemical results derived through the use of such compounds.
Next, we tested select lead compounds in vitro for potential to interfere with other proteins. At the physiologically relevant concentration of 10 μM, derivative 17a was inert when tested against the scanMAX 402 kinase panel (Figure 6). This panel contains a set of kinases covering the AGC, CAMK, CMGC, CK1, STE, TK, TKL, lipid, and atypical kinase families, plus important mutant forms. Developed by Ambit Biosciences, it employs proprietary active-site-dependent competition binding assays to determine how compounds bind to kinases. It is based on a competition binding assay that quantitatively measures the ability of a compound to compete with an immobilized, active-site-directed ligand and can be used in detection of multiple inhibitor types (e.g., types I and II and non-ATP-competitive).40,41
Figure 6.
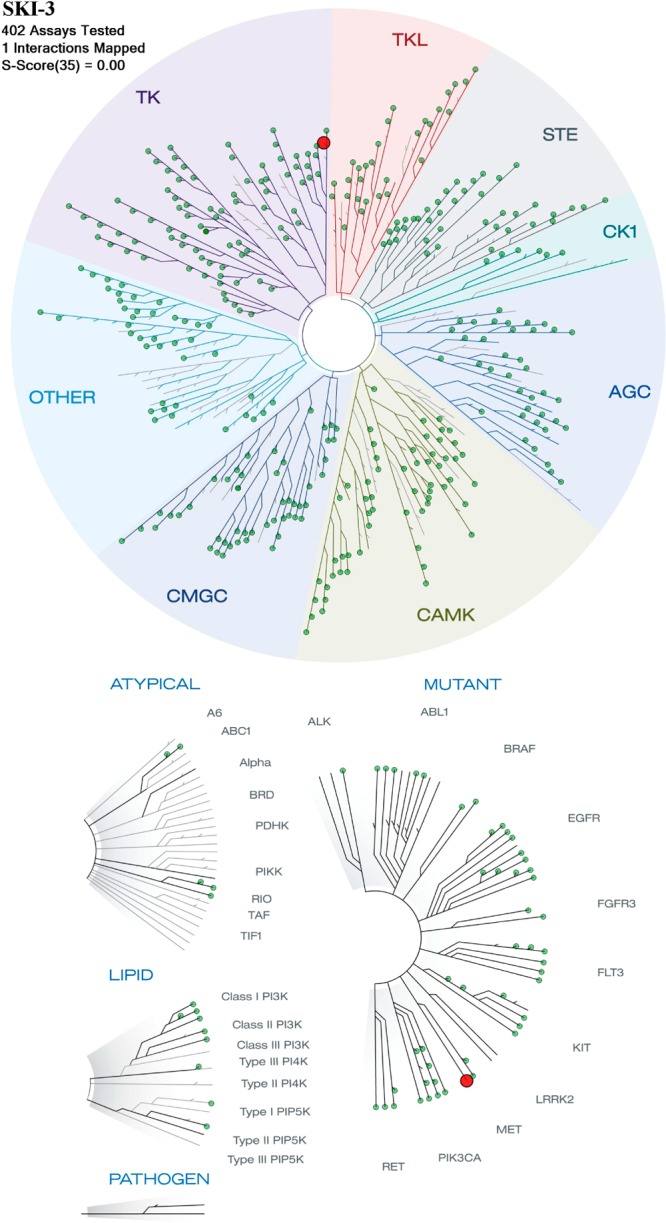
Derivative 17a (at 10 μM) was tested in the scanMAX screen (Ambit) against 402 kinases. The TREEspot interaction map for 17a is presented. Only c-Met (red dot on the kinase tree) appears as a potential low-affinity kinase hit of 17a. KINOMEscan’s selectivity score (S) is a quantitative measure of compound selectivity. It is calculated by dividing the number of kinases that bind to the compound by the total number of distinct kinases tested, excluding mutant variants. S(35) = (number of nonmutant kinases with %Ctrl < 35)/(number of nonmutant kinases tested).
Conclusions
In summary, we describe here SAR studies in the first rationally designed scaffolds with binding potential to a novel allosteric pocket in human Hsp70. This site is located in the N-terminal domain of the chaperone and was recently identified by our homology modeling studies.22 It is not revealed by reported crystal structures of Hsp70, as these capture the protein in a closed conformation in which the reactive Cys267 is unavailable for ligand binding and the pocket is narrow.24−27 The designed ligands are of a yet unexplored chemical space based on the 2,5′-thiodipyrimidine and 5-(phenylthio)pyrimidine scaffolds, and thus, we are also first to report here a chemical strategy that allows for the assembly of such compounds.
The weight of the evidence in this paper and in our previous report22 indicates that these compounds are acting on Hsp70 through binding to the allosteric pocket. Only if reasonably correct could the homology model enable the rational design of a ligand that, when incubated with the thousands of proteins expressed in a cancer cell, substantially affinity purifies one, Hsp70. Seconded by a biological investigation of these ligands based on specific modulation of the target, Hsp70, in the context of a cancer cell, our data clearly define the mode of action of these ligands, in the cancer cell, through inhibition of Hsp70. From these studies, we identify derivatives 17a and 20a as being the most active of the series. Addition of high nanomolar to low micromolar concentrations of these agents to several cancer cells led to a reduction in the steady-state levels of Hsp70–HOP–Hsp90 complex chaperoned oncoproteins, an effect associated with inhibition of cell growth and apoptosis.
Because Hsp70 contains an active cysteine embedded in the allosteric pocket, the derivatives presented herein contain an acrylamide functionality that may create a covalent bond with such a residue upon Hsp70 protein binding. We have used several techniques to demonstrate the potential formation of a covalent bond between the inhibitor and the protein.22 First, when the biotinylated analogue of 17a, 44b, was incubated with cancer cells, we noted a time-dependent progressive increase in the amount of immobilized Hsp70. This profile is indicative of time-dependent covalent modification of the protein and is specific for compounds such as 17a, where irreversible binding plays a role. Second, mass spectrum analysis of the trypsin digest of the 44b–Hsp70 complex identified a major m/z peak at 1867.915.22 This corresponds to 44b bound to LRTAC267ERAK and confirms that the site of interaction of 17a with Hsp70 is within site 1, where Cys267 is located and, furthermore, that 17a forms an irreversible bond with the protein upon binding. Third, when we probed binding of 44b to both the WT Hsp70 protein and the C267S mutant, we noted that high-salt, high-detergent conditions eluted preferentially the Hsp70-C267S mutant protein over the WT form from preformed Hsp70–44b complexes.
There is precedent for the use of an acrylamide “warhead” in the development of several irreversible kinase inhibitors currently in clinical trials for cancer.42−46 There is, of course, a concern that such an entity could indiscriminately react with non-target-related proteins, resulting in unwanted biological effects. However, in spite of the presence of an acrylamide, derivatives described here are not excessively reactive and, in addition, have an appropriate fit in the active site of the target, anticipating a favorable enthalpic effect. It is also important to note that, for proteins with long half-lives, such as Hsp70,47 irreversible inhibition is mechanistically advantageous as it confers complete target inhibition until resynthesis of the target protein48 and thus allows for less frequent dosing, which may also tip the balance toward a better therapeutic index for such compounds.
In addition to providing both a novel pharmacophore and medicinal chemistry for its assembly, in this paper we describe a testing battery for assessing Hsp70-mediated mechanisms in cancer cells and for evaluating specific ligand action in cells through Hsp70 inhibition. Altogether, these findings provide a novel blueprint for a cancer-oriented development of Hsp70-directed ligands.
To conclude, the series of novel chemical entities reported herein interact with a yet unexplored pocket in Hsp70. These compounds of tractable SAR favorably permeate cancer cells and exhibit their biological activity through inhibition of the tumor Hsp70 species, validating the scaffold as an important starting point for the development of Hsp70 inhibitors with potential therapeutic applications. The accompanying paper in this issue describes the synthesis and structure–activity evaluation of inhibitors of this series that target the same allosteric pocket of Hsp70 described herein but act through a reversible mechanism of action.23
Experimental Section
Chemistry
All reagents were purchased from either Aldrich or Acros Organics and used without purification. All reactions were performed under argon protection. NMR spectra were recorded on a Bruker AV-III-500 MHz NMR spectrometer. Chemical shifts are reported in δ values in parts per million downfield from TMS as the internal standard. 1H data are reported as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, br = broad, m = multiplet), coupling constant (Hz), integration. 13C chemical shifts are reported in δ values in parts per million downfield from TMS as the internal standard. High-resolution mass spectra were recorded on a Waters LCT Premier system. Low-resolution mass spectra were obtained on a Waters Acquity Ultra Performance LC instrument with electrospray ionization and an SQ detector. Analytical HPLC was performed on a Waters Autopurification system with PDA, MicroMass ZQ, and ELSD detectors. The purity of the title compounds used in pharmacology testing was determined by HPLC–MS using the following method: 10–12 min gradient on a Waters2525 binary gradient pump of increasing concentrations of acetonitrile in water (5% → 95%) containing 0.1% formic acid with a flow rate of 1.2 mL/min and UV detection at λ = 220 and 254 nm on an XBridge C18 150 mm × 4.6 mm, 5 μm column. Title compounds used in pharmacology testing were >95% pure. Analytical thin-layer chromatography was performed on 250 μM silica gel F254 plates. Preparative thin-layer chromatography was performed on 1000 μM silica gel F254 plates. Flash column chromatography was performed employing 230–400 mesh silica gel. Solvents were HPLC grade. Myricetin was purchased from Indofine Chemical Co. (Hillsborough, NJ). The synthesis of compounds 44a,b and 45a,b is described in detail in the Supporting Information.
4,6-Dimethoxy-N,N-bis(4-methoxybenzyl)pyrimidin-2-amine (9a)
To a solution of 2-amino-4,6-dimethoxypyrimidine (2.0 g, 12.9 mmol) (8a) in 20 mL of DMF at 0 °C was added NaH (1.24 g, 51.5 mmol), and the mixture was stirred at rt for 10 min. 4-Methoxybenzyl chloride (4.03 g, 25.7 mmol) was added, and the mixture was stirred at rt overnight. The reaction was quenched with methanol and solvent removed under reduced pressure. The residue was dissolved in EtOAc, washed with brine, and dried over MgSO4. Solvent was evaporated under reduced pressure, and the residue was purified by column chromatography (hexane/EtOAc, 4:1) to afford 4.8 g (95%) of 9a. 1H NMR (500 MHz, CDCl3): δ 7.26 (d, J = 8.4 Hz, 4H), 6.89 (d, J = 8.4 Hz, 4H), 5.47 (s, 1H), 4.78 (s, 4H), 3.85 (s, 6H), 3.80 (s, 6H). 13C NMR (125 MHz, CDCl3): δ 171.9, 161.5, 158.7, 130.9, 129.1, 113.8, 78.7, 55.2, 53.4, 48.3. MS (m/z): [M + H]+ 396.3.
5-Iodo-4,6-dimethoxy-N,N-bis(4-methoxybenzyl)pyrimidin-2-amine (10a)
To a solution of 9a (4.8 g, 12.3 mmol) in 50 mL of acetonitrile was added NIS (4.13 g, 18.4 mmol), and the resulting solution was stirred at rt for 1 h. Solvent was evaporated, and the residue was purified by column chromatography (hexane/EtOAc, 4:1) to afford 6.3 g (98%) of 10a. 1H NMR (500 MHz, CDCl3): δ 7.18 (d, J = 8.6 Hz, 4H), 6.83 (d, J = 8.6 Hz, 4H), 4.71 (s, 4H), 3.89 (s, 6H), 3.79 (s, 6H). 13C NMR (125 MHz, CDCl3): δ 169.1, 160.9, 158.8, 130.5, 129.0, 113.9, 55.3, 54.7, 48.6, 43.9. MS (m/z): [M + H]+ 522.4.
2-((2-(Bis(4-methoxybenzyl)amino)-4,6-dimethoxypyrimidin-5-yl)thio)pyrimidine-4,6-diamine (11a)
A mixture of 10a (6.2 g, 11.9 mmol), 4,6-diamino-2-mercaptopyrimidine (1.7 g, 11.9 mmol), neocuproine (0.538 g, 2.38 mmol), CuI (0.452 g, 2.38 mmol), and K2CO3 (3.3 g, 33.8 mmol) in 100 mL of DMSO was stirred at 120 °C for 16 h. Solvent was removed under reduced pressure, and the residue was purified by column chromatography (CH2Cl2/MeOH–NH3 (7 N), 20:1) to afford 4.2 g (65%) of 11a. 1H NMR (500 MHz, DMSO-d6): δ 7.18 (d, J = 8.6 Hz, 4H), 6.80 (d, J = 8.6 Hz, 4H), 5.09 (s, 1H), 4.68 (s, 4H), 4.40 (s, 4H), 3.79 (s, 6H), 3.74 (s, 6H). MS (m/z): [M + H]+ 536.5.
N,N′-(2-((2-(Bis(4-methoxybenzyl)amino)-4,6-dimethoxypyrimidin-5-yl)thio)pyrimidine-4,6-diyl)diacetamide (12a)
A solution of 11a (3.2 g, 6.0 mmol) and DMAP (0.037 g, 0.3 mmol) in 20 mL of acetic anhydride was stirred at 110 °C for 2 h. Solvent was removed under reduced pressure, and the residue was purified by column chromatography (hexane/EtOAc, 1:1) to afford 3.4 g (91%) of 12a. 1H NMR (500 MHz, CDCl3/DMSO-d6): δ 8.25 (br s, 1H), 7.70 (br s, 2H), 7.18 (d, J = 10.0 Hz, 4H), 6.81 (d, J = 10.0 Hz, 4H), 4.70 (s, 4H), 3.78 (s, 6H), 3.74 (s, 6H), 2.10 (s, 6H). MS (m/z): [M + H]+ 620.4.
N,N′-(2-((2-Amino-4,6-dimethoxypyrimidin-5-yl)thio)pyrimidine-4,6-diyl)diacetamide (13a)
A solution of 12a (0.950 g, 1.5 mmol) in 20 mL of TFA/CHCl3 (1:1) was heated at 62 °C for 24 h. Excess TFA and solvent were removed under reduced pressure, and the residue was purified by column chromatography (CH2Cl2/MeOH, 20:1) to afford 0.550 g (95%) of 13a. 1H NMR (500 MHz, DMSO-d6): δ 10.51 (br s, 2H), 8.37 (br s, 1H), 6.98 (s, 2H), 3.78 (s, 6H), 2.06 (s, 6H). 13C NMR (125 MHz, DMSO-d6): δ 170.9, 170.1, 169.2, 162.5, 158.9, 92.8, 78.5, 53.9, 24.1. MS (m/z): [M + H]+ 380.2.
N,N′-(2-((2-Fluoro-4,6-dimethoxypyrimidin-5-yl)thio)pyrimidine-4,6-diyl)diacetamide (14a)
13a (2.0 g, 5.3 mmol) was added to a plastic tube fitted with a stir bar and cooled to 0 °C followed by addition of a solution of HF/pyridine (3.6 mL, 144 mmol). NaNO2 (0.545 g, 7.9 mmol) was added in portions over a period of 20 min with stirring. The resulting solution was vigorously stirred for an additional 50 min at 0 °C and 2 h at rt. CaCO3 (14.4 g, 144 mmol) was added to quench excess HF. The mixture was extracted with CH2Cl2 and purified by column chromatography (CH2Cl2/MeOH–NH3(7N), 20:1) to afford 1.1 g (54%) of 14a. 1H NMR (500 MHz, CDCl3): δ 8.45 (s, 1H), 7.61 (s, 2H), 4.00 (s, 6H), 2.18 (s, 6H). MS (m/z): [M + H]+ 383.2.
N,N′-(2-((4,6-Dimethoxy-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidine-4,6-diyl)diacetamide (15a)
To a solution of 14a (30 mg, 0.078 mmol) in 2 mL of DMF was added 1-methylpiperazine (31 mg, 0.31 mmol), and the resulting solution was heated at 90 °C for 1 h. Solvent and excess reagent were removed under reduced pressure, and the residue was purified by column chromatography (CHCl3/MeOH–NH3 (7 N), 10:1) to yield 32 mg (90%) of 15a. 1H NMR (500 MHz, CDCl3): δ 8.36 (br s, 1H), 8.13 (br s, 2H), 3.88 (s, 6H), 3.87 (m, 4H), 2.46 (m, 4H), 2.35 (s, 3H), 1.99 (s, 6H). 13C NMR (125 MHz, CDCl3): δ 172.2, 169.4, 168.9, 160.4, 158.9, 96.1, 95.9, 54.7, 54.3, 46.1, 43.8, 24.7. MS (m/z): [M + H]+ 463.2.
2-((4,6-Dimethoxy-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidine-4,6-diamine (16a)
A mixture of 15a (50 mg, 0.108 mmol) and 1 N NaOH(aq) (2 mL) in 4 mL of methanol was stirred at 60 °C for 1 h. Solvents were removed under reduced pressure, and the residue was purified by preparatory TLC to afford 39 mg (95%) of 16a. 1H NMR (500 MHz, CDCl3): δ 5.19 (s, 1H), 4.48 (s, 4H), 3.88 (s, 6H), 3.87 (m, 4H), 2.48 (m, 4H), 2.35 (s, 3H). MS (m/z): [M + H]+ 378.9.
N-(6-Amino-2-((4,6-dimethoxy-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (17a)
To a solution of 16a (0.370 g, 0.977 mmol) and Et3N (0.988 g, 9.77 mmol) in 10 mL of anhydrous dioxane was added acryloyl chloride (0.855 g, 9.77 mmol) dropwise under a water bath. The resulting mixture was stirred at rt for 24 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to afford 0.211 g (50%) of 17a. 1H NMR (500 MHz, CDCl3): δ 7.96 (br s, 1H), 7.04 (s, 1H), 6.41 (d, J = 16.8 Hz, 1H), 6.17 (dd, J = 16.8, 10.3 Hz, 1H), 5.78 (d, J = 10.3 Hz, 1H), 4.83 (br s, 2H), 3.88 (s, 6H), 3.87 (m, 4H), 2.47 (m, 4H), 2.35 (s, 3H). 13C NMR (125 MHz, CDCl3): δ 169.3, 168.8, 162.6, 162.5, 158.3, 154.9, 128.9, 127.1, 86.7, 78.2, 53.1, 52.4, 44.5, 41.9. HRMS (m/z): [M + H]+ calcd for C18H25N8O3S, 433.1770; found, 433.1750. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 6.28 min.
N,N′-(2-((4,6-Dimethoxy-2-morpholinopyrimidin-5-yl)thio)pyrimidine-4,6-diyl)diacetamide (15b)
To a solution of 14a (60 mg, 0.157 mmol) in 2 mL of DMF was added morpholine (54 mg, 0.620 mmol), and the resulting solution was heated at 90 °C for 1 h. Solvent and excess reagent were removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to yield 56 mg (79%) of 15b. 1H NMR (500 MHz, CDCl3): δ 8.41 (br s, 1H), 7.82 (br s, 2H), 3.83 (s, 6H), 3.77 (m, 4H), 3.68 (m, 4H), 2.16 (s, 6H). HRMS (m/z): [M + H]+ calcd for C18H24N7O5S, 450.1560; found, 450.1548. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 8.47 min.
2-((4,6-Dimethoxy-2-morpholinopyrimidin-5-yl)thio)pyrimidine-4,6-diamine (16b)
A mixture of 15b (45 mg, 0.100 mmol) and 1 N NaOH(aq) (2 mL) in 4 mL of methanol was stirred at 60 °C for 1 h. Solvents were removed under reduced pressure, and the residue was purified by preparatory TLC to afford 35 mg (95%) of 16b. 1H NMR (500 MHz, CDCl3): δ 5.19 (s, 1H), 4.44 (s, 4H), 3.87 (s, 6H), 3.83 (m, 4H), 3.77 (m, 4H), 2.16 (s, 6H). HRMS (m/z): [M + H]+ calcd for C14H20N7O3S, 366.1348; found, 366.1361. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 6.45 min.
N-(6-Amino-2-((4,6-dimethoxy-2-morpholinopyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (17b)
To a solution of 16b (50 mg, 0.111 mmol) and Et3N (56 mg, 0.555 mmol) in 2 mL of anhydrous dioxane was added acryloyl chloride (50 mg, 0.555 mmol) under a water bath. The resulting mixture was stirred at rt for 24 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to afford 26 mg (55%) of 17b. 1H NMR (500 MHz, CDCl3): δ 7.73 (br s, 1H), 7.05 (s, 1H), 6.42 (d, J = 16.9 Hz, 1H), 6.17 (dd, J = 16.7, 10.4 Hz, 1H), 5.80 (d, J = 10.3 Hz, 1H), 4.78 (br s, 2H), 3.89 (s, 6H), 3.81 (m, 4H), 3.75 (m, 4H). HRMS (m/z): [M + H]+ calcd for C17H22N7O4S, 420.1454; found, 420.1470.
N,N′-(2-((4,6-Dimethoxy-2-(piperidin-1-yl)pyrimidin-5-yl)thio)pyrimidine-4,6-diyl)diacetamide (15c)
To a solution of 14a (60 mg, 0.157 mmol) in 2 mL of DMF was added piperidine (52 mg, 0.611 mmol), and the resulting solution was heated at 90 °C for 1 h. Solvent and excess reagent were removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to yield 55 mg (78%) of 15c. 1H NMR (500 MHz, CDCl3): δ 8.32 (br s, 1H), 7.77 (br s, 2H), 3.88 (s, 6H), 2.16 (m, 6H), 1.54–1.70 (m, 6H). HRMS (m/z): [M + H]+ calcd for C19H26N7O4S, 448.1767; found, 448.1766. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 10.13 min.
2-((4,6-Dimethoxy-2-(piperidin-1-yl)pyrimidin-5-yl)thio)pyrimidine-4,6-diamine (16c)
A mixture of 15c (55 mg, 0.123 mmol) and 1 N NaOH(aq) (2 mL) in 4 mL of methanol was stirred at 60 °C for 1 h. Solvents were removed under reduced pressure, and the residue was purified by preparatory TLC to afford 43 mg (96%) of 16c. 1H NMR (500 MHz, CDCl3): δ 5.18 (s, 1H), 4.42 (s, 4H), 3.88 (s, 6H), 3.80 (m, 4H), 1.60–1.71 (m, 6H). HRMS (m/z): [M + H]+ calcd for C15H22N7O2S, 364.1556; found, 364.1544. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 7.43 min.
N-(6-Amino-2-((4,6-dimethoxy-2-(piperidin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (17c)
To a solution of 16c (46 mg, 0.127 mmol) and Et3N (130 mg, 1.30 mmol) in 4 mL of anhydrous dioxane was added acryloyl chloride (119 mg, 1.30 mmol) dropwise under a water bath. The resulting mixture was stirred at rt for 12 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to afford 28 mg (52%) of 17c. 1H NMR (500 MHz, CDCl3/DMSO-d6): δ 9.33 (s, 1H), 7.09 (s, 1H), 6.47–6.35 (m, 2H), 5.71 (d, J = 9.3 Hz, 1H), 5.31 (br s, 2H), 3.88 (s, 6H), 3.81 (m, 4H), 1.69 (m, 2H), 1.62 (m, 4H). HRMS (m/z): [M + H]+ calcd for C18H24N7O3S, 418.1661; found, 418.1660. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 8.06 min.
2-Amino-4,6-diethoxypyrimidine (8b)
To a solution of 2-amino-4,6-dichloropyrimidine (1.0 g, 6.09 mmol) in 20 mL of absolute ethanol was added NaH (0.585 g, 24.39 mmol) at rt. The mixture was stirred under reflux for 12 h. Solvent was removed under reduced pressure, and the residue was dissolved in CH2Cl2 and washed with brine. Solvent was evaporated, and the resulting solid was purified by column chromatography (hexane/EtOAc, 4:1) to afford 1.0 g (89%) of 8b. 1H NMR (500 MHz, CDCl3): δ 5.42 (s, 1H), 4.78 (br s, 2H), 4.24 (q, J = 7.1 Hz, 4H), 1.34 (t, J = 7.1 Hz, 6H). MS (m/z): [M + H]+ 183.9.
4,6-Diethoxy-N,N-bis(4-methoxybenzyl)pyrimidin-2-amine (9b)
To a solution of 8b (1.00 g, 5.46 mmol) in 20 mL of DMF at 0 °C was added NaH (0.524 g, 21.83 mmol), and the resulting solution was stirred at rt for 10 min. 4-Methoxybenzyl chloride (1.88 g, 12.0 mmol) was added, and the mixture was stirred at rt overnight. The reaction was quenched with ethanol, and solvent was removed under reduced pressure. The residue was dissolved in EtOAc, washed with brine, dried over MgSO4, and concentrated to give a residue that was purified by column chromatography (hexane/EtOAc, 4:1) to afford 2.25 g (97%) of 9b. 1H NMR (500 MHz, CDCl3): δ 7.18 (d, J = 8.1, 4H), 6.84 (d, J = 8.1, 4H), 5.38 (s, 1H), 4.70 (s, 4H), 4.27 (q, J = 7.1 Hz, 4H), 3.79 (s, 6H), 1.29 (t, J = 7.1 Hz, 6H). 13C NMR (125 MHz, CDCl3): δ 171.5, 161.4, 158.6, 130.9, 129.0, 113.7, 78.6, 61.8, 55.2, 48.1, 14.6. MS (m/z): [M + H]+ 424.2.
5-Iodo-4,6-diethoxy-N,N-bis(4-methoxybenzyl)pyrimidin-2-amine (10b)
To a solution of 9b (2.2 g, 5.2 mmol) in 50 mL of acetonitrile was added NIS (1.7 g, 8 mmol), and the resulting solution was stirred at rt for 1 h. Solvent was removed under reduced pressure, and the residue was purified by column chromatography (hexane/EtOAc, 4:1) to afford 2.75 g (96%) of 10b. 1H NMR (500 MHz, CDCl3): δ 7.17 (m, 4H), 6.84 (m, 4H), 4.68 (s, 4H), 4.34 (q, J = 7.1 Hz, 4H), 3.80 (s, 6H), 1.32 (t, J = 7.1 Hz, 6H). 13C NMR (125 MHz, CDCl3/DMSO-d6): δ 168.3, 160.4, 158.2, 130.1, 128.4, 113.2, 62.6, 54.8, 48.1, 44.2, 14.1. MS (m/z): [M + H]+ 550.1.
2-((2-(Bis(4-methoxybenzyl)amino)-4,6-diethoxypyrimidin-5-yl)thio)pyrimidine-4,6-diamine (11b)
A mixture of 10b (2.75 g, 5.0 mmol), 4,6-diamino-2-mercaptopyrimidine (0.71 g, 5.0 mmol), neocuproine (0.226 g, 1.0 mmol), CuI (0.190 g, 1.0 mmol), and K2CO3 (1.38 g, 10.0 mmol) in 60 mL of DMSO was stirred at 120 °C for 16 h. Solvent was removed under reduced pressure, and the residue was partially purified by column chromatography (CH2Cl2/MeOH–NH3 (7 N), 20:1) to afford 2.2 g (80%) of impure 11b [MS (m/z): [M + H]+ 564.2], which was used without further purification in the next step.
N,N′-(2-((2-(Bis(4-methoxybenzyl)amino)-4,6-diethoxypyrimidin-5-yl)thio)pyrimidine-4,6-diyl)diacetamide (12b)
A solution of 11b (1.2 g, 2.19 mmol) and DMAP (0.013 g, 0.11 mmol) in 20 mL of acetic anhydride was stirred at 110 °C for 2 h. Solvent was removed under reduced pressure, and the residue was purified by column chromatography (hexane/EtOAc, 1:1) to afford 1.2 g (89%) of 12b. 1H NMR (500 MHz, CDCl3): δ 8.22 (s, 2H), 7.21 (d, J = 8.5 Hz, 4H), 6.86 (d, J = 8.5 Hz, 4H), 4.70 (s, 4H) 4.32 (q, J = 7.1 Hz, 4H), 3.80 (s, 6H), 2.16 (s, 6H), 1.20 (t, J = 7.1 Hz, 6H). MS (m/z): [M + H]+ 648.1.
N,N′-(2-((2-Amino-4,6-diethoxypyrimidin-5-yl)thio)pyrimidine-4,6-diyl)diacetamide (13b)
A solution of 12b (2.00 g, 3.09 mmol) in 20 mL of TFA/CHCl3 (1:1) was heated at 62 °C for 24 h. Excess TFA and solvent were removed under reduced pressure, and the residue was purified by column chromatography (CH2Cl2/MeOH, 20:1) to afford 1.15 g (92%) of 13b. MS (m/z): [M + H]+ 407.8.
N,N′-(2-((2-Fluoro-4,6-diethoxypyrimidin-5-yl)thio)pyrimidine-4,6-diyl)diacetamide (14b)
13b (1.5 g, 3.68 mmol) was added to a plastic tube fitted with a stir bar and cooled to 0 °C followed by addition of a solution of HF/pyridine (3.0 mL, 120 mmol). After several minutes NaNO2 (0.380 g, 5.52 mmol) was added in portions over a period of 20 min with stirring. The resulting solution was vigorously stirred for an additional 50 min at 0 °C. CaCO3 (12.0 g, 120 mmol) was added to quench excess HF. The mixture was extracted with CH2Cl2 and purified by column chromatography (CH2Cl2/MeOH–NH3 (7 N), 20:1) to afford 0.76 g (46%) of 14b. 1H NMR (500 MHz, CDCl3): δ 8.47 (br s, 1H), 7.85 (br s, 2H), 4.44 (q, J = 7.1 Hz, 4H), 2.17 (s, 6H), 1.31 (t, J = 7.1 Hz, 6H). MS (m/z): [M + H]+ 411.3.
N,N′-(2-((4,6-Diethoxy-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidine-4,6-diyl)diacetamide (18a)
To a solution of 14b (0.165 g, 0.402 mmol) in 3 mL of DMF was added 1-methylpiperazine (400 mg, 4.4 mmol), and the resulting solution was heated to 90 °C for 1 h. Solvent was removed under reduced pressure, and the residue was purified by column chromatography (CH2Cl2/MeOH–NH3 (7 N), 10:1) to yield 0.180 g (91%) of 18a. 1H NMR (500 MHz, CDCl3): δ 8.35 (br s, 1H), 8.06 (br s, 2H), 4.35 (q, J = 7.1 Hz, 4H), 3.82 (m, 4H), 2.43 (m, 4H), 2.36 (s, 3H), 2.15 (s, 6H), 1.26 (t, J = 7.1 Hz, 6H). MS (m/z): [M + H]+ 491.2.
2-((4,6-Diethoxy-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidine-4,6-diamine (19a)
A mixture of 18a (0.130 g, 0.265 mmol) and 1 N NaOH(aq) (2 mL) in 7 mL of methanol was stirred at 60 °C for 1 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CH2Cl2/MeOH, 10:1) to afford 0.100 g (93%) of 19a. 1H NMR (500 MHz, CDCl3): δ 5.17 (br s, 1H), 4.48 (s, 4H), 4.34 (q, J = 7.1 Hz, 4H), 3.83 (m, 4H), 2.47 (m, 4H), 2.35 (s, 3H), 1.27 (t, J = 7.1 Hz, 6H). MS (m/z): [M + H]+ 407.1.
N-(6-Amino-2-((4,6-diethoxy-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (20a)
To a solution of 19a (20 mg, 0.049 mmol) and Et3N (49 mg, 0.49 mmol) in 1 mL of anhydrous dioxane was added acryloyl chloride (44 mg, 0.49 mmol). The resulting mixture was stirred at rt for 12 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to afford 9 mg (40%) of 20a. 1H NMR (500 MHz, CDCl3): δ 8.14 (br s, 1H), 7.04 (s, 1H), 6.40 (d, J = 16.9 Hz, 1H), 6.19 (dd, J = 16.9, 10.4 Hz, 1H), 5.77 (d, J = 10.4 Hz, 1H), 4.83 (br s, 2H), 4.35 (q, J = 7.0 Hz, 4H), 3.83 (m, 4H), 2.46 (m, 4H), 2.35 (s, 3H), 1.28 (t, J = 7.0 Hz, 6H). 13C NMR (125 MHz, DMSO-d6): δ 170.1, 169.2, 164.8, 164.2, 159.4, 156.4, 131.3, 128.1, 87.5, 79.9, 62.0, 54.3, 46.6, 43.2, 14.4. HRMS (m/z): [M + H]+ calcd for C20H29N8O3S, 461.2083; found, 461.2096. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 5.57 min.
N,N′-(2-((4,6-Diethoxy-2-morpholinopyrimidin-5-yl)thio)pyrimidine-4,6-diyl)diacetamide (18b)
To a solution of 14b (165 mg, 0.402 mmol) in 3 mL of DMF was added morpholine (350 mg, 4.02 mmol), and the resulting solution was heated at 90 °C for 1 h. Solvent and excess reagent were removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to yield 192 mg (81%) of 18b. MS (m/z): [M + H]+ 478.1. 1H NMR (500 MHz, CDCl3): δ 8.35 (br s, 1H), 8.06 (br s, 2H), 4.34 (q, J = 7.0 Hz, 4H), 3.82–3.84 (m, 4H), 2.43–2.45 (m, 4H), 2.15 (s, 6H), 1.26 (t, J = 7.0 Hz, 6H).
2-((4,6-Diethoxy-2-morpholinopyrimidin-5-yl)thio)pyrimidine-4,6-diamine (19b)
A mixture of 18b (0.200 g, 0.42 mmol) and 1 N NaOH(aq) (2 mL) in 5 mL of methanol was stirred at 60 °C for 1 h. Solvent was removed under reduced pressure, and the residue was purified by chromatography (CH2Cl2/MeOH–NH3 (7 N), 10:1) to afford 0.153 g (93%) of 19b. 1H NMR (500 MHz, CDCl3): δ 6.39 (br s, 1H), 4.31 (q, J = 6.2 Hz, 4H), 3.51–3.83 (m, 4H), 1.90–1.95 (m, 4H), 1.19 (t, J = 6.2 Hz, 6H). MS (m/z): [M + H]+ 394.1.
N-(6-Amino-2-((4,6-diethoxy-2-morpholinopyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (20b)
To a solution of 19b (20 mg, 0.051 mmol) and Et3N (51 mg, 0.51 mmol) in 1 mL of anhydrous dioxane was added acryloyl chloride (46 mg, 0.51 mmol). The resulting mixture was stirred at rt for 12 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to afford 10 mg (42%) of 20b. 1H NMR (500 MHz, CDCl3): δ 8.14 (br s, 1H), 7.04 (s, 1H), 6.40 (d, J = 16.9 Hz, 1H), 6.18 (dd, J = 16.9, 10.4 Hz, 1H), 5.78 (d, J = 10.4 Hz, 1H), 4.82 (s, 2H), 4.35 (q, J = 7.1 Hz, 4H), 3.79 (m, 4H), 3.76 (m, 4H), 1.27 (t, J = 7.2 Hz, 6H). HRMS (m/z): [M + H]+ calcd for C19H26N7O4S, 448.1767; found, 448.1754. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 7.97 min.
N,N′-(2-((4,6-Diethoxy-2-(piperidin-1-yl)pyrimidin-5-yl)thio)pyrimidine-4,6-diyl)diacetamide (18c)
To a solution of 14b (165 mg, 0.402 mmol) in 3 mL of DMF was added piperidine (342 mg, 4.02 mmol), and the resulting solution was heated at 90 °C for 1 h. Solvent and excess reagent were removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to yield 167 mg (87%) of 18c. 1H NMR (500 MHz, CDCl3): δ 8.60 (br s, 2H), 8.35 (br s, 1H), 4.34 (q, J = 7.1 Hz, 4H), 3.75 (m, 4H), 2.17 (s, 6H), 1.67 (m, 2H), 1.58 (m, 4H), 1.27 (t, J = 7.1 Hz, 6H). MS (m/z): [M + H]+ 476.2.
2-((4,6-Diethoxy-2-(piperidin-1-yl)pyrimidin-5-yl)thio)pyrimidine-4,6-diamine (19c)
A mixture of 18c (0.200 g, 0.42 mmol) and 1 N NaOH(aq) (2 mL) in 5 mL of methanol was stirred at 60 °C for 1 h. Solvent was removed under reduced pressure, and the residue was purified by chromatography (CH2Cl2/MeOH–NH3 (7 N), 10:1) to afford 0.156 g (95%) of 19c. MS (m/z): [M + H]+ 392.2.
N-(6-Amino-2-((4,6-diethoxy-2-(piperidin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (20c)
To a solution of 19c (30 mg, 0.076 mmol) and Et3N (76 mg, 0.76 mmol) in 1 mL of anhydrous dioxane was added acryloyl chloride (69 mg, 0.76 mmol) dropwise. The resulting mixture was stirred at rt for 12 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to afford 16 mg (48%) of 20c. 1H NMR (500 MHz, CDCl3): δ 8.09 (br s, 1H), 7.03 (s, 1H), 6.40 (dd, J = 16.9, 1.1 Hz, 1H), 6.19 (dd, J = 16.9, 10.3 Hz, 1H), 5.77 (dd, J = 10.3, 1.1 Hz, 1H), 4.82 (s, 2H), 4.35 (q, J = 7.1 Hz, 4H), 3.76 (m, 4H), 1.65–1.69 (m, 2H), 1.57–1.61 (m, 4H), 1.26 (t, J = 7.1 Hz, 6H). HRMS (m/z): [M + H]+ calcd for C20H28N7O3S, 446.1974; found, 446.1974. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 10.67 min.
5-((4-Aminopyrimidin-2-yl)thio)-4,6-dimethoxy-N,N-bis(4-methoxybenzyl)pyrimidin-2-amine (21)
A mixture of 10a (8.2 g, 15.8 mmol), 4-amino-2-mercaptopyrimidine (2.2 g, 17.3 mmol), neocuproine (0.711 g, 3.16 mmol), CuI (0.616 g, 3.16 mmol), and K2CO3 (4.36 g, 31.6 mmol) in 100 mL of DMSO was stirred at 130 °C for 16 h. Solvent was removed under reduced pressure, and the residue was purified by column chromatography (CH2Cl2/MeOH, 100:0 to 90:10) to afford 7.6 g (92%) of 21. 1H NMR (500 MHz, CDCl3): δ 7.97 (d, J = 5.7 Hz, 1H), 7.23 (d, J = 8.3 Hz, 4H), 6.86 (d, J = 8.3 Hz, 4H), 6.05 (d, J = 5.7 Hz, 1H), 4.94 (br s, 2H), 4.75 (s, 4H), 3.87 (s, 6H), 3.81 (s, 6H). MS (m/z): [M + H]+ 521.1.
N-(2-((2-(Bis(4-methoxybenzyl)amino)-4,6-dimethoxypyrimidin-5-yl)thio)pyrimidin-4-yl)acetamide (22)
A solution of 21 (7.6 g, 14.6 mmol) in 50 mL of acetic anhydride was stirred at 120 °C for 1 h. Solvent was removed under reduced pressure, and the residue was purified by column chromatography (CH2Cl2/MeOH, 100:0 to 90:10) to afford 7.37 g (93%) of 22. 1H NMR (500 MHz, CDCl3): δ 9.17 (s, 1H), 8.32 (d, J = 5.5 Hz, 1H), 7.76 (br s, 1H), 7.22 (d, J = 8.3 Hz, 4H), 6.85 (d, J = 8.3 Hz, 4H), 4.73 (s, 4H), 3.84 (s, 6H), 3.79 (s, 6H), 2.16 (s, 3H). MS (m/z): [M + H]+ 563.2.
N-(2-((2-Amino-4,6-dimethoxypyrimidin-5-yl)thio)pyrimidin-4-yl)acetamide (23)
A solution of 22 (7.0 g, 12.9 mmol) in 30 mL of TFA was heated at 60 °C for 12 h. Excess TFA and solvent were removed under reduced pressure, and the residue was purified by column chromatography (CH2Cl2/MeOH–NH3 (7 N), 20:1) to afford 4.16 g (85%) of 23. 1H NMR (500 MHz, CDCl3/MeOH-d4): δ 10.23 (br s, 1H), 8.21 (s, 1H), 7.75 (s, 1H), 5.98 (br s, 2H), 3.88 (s, 6H), 2.14 (s, 3H). 13C NMR (125 MHz, CDCl3/MeOH-d4): δ 170.8, 170.2, 170.0, 161.8, 157.5, 157.4, 105.1, 80.2, 53.7, 23.8. HRMS (m/z): [M + H]+ calcd for C12H15N6O3S, 323.0926; found, 323.0913. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 5.56 min.
N-(2-((2-Fluoro-4,6-dimethoxypyrimidin-5-yl)thio)pyrimidin-4-yl)acetamide (24)
23 (1.72 g, 5.32 mmol) was added to a plastic tube fitted with a stir bar and cooled to 0 °C followed by addition of a solution of HF/pyridine (3.6 mL, 144 mmol). NaNO2 (0.545 g, 7.9 mmol) was added in portions over a period of 20 min with stirring. The resulting solution was vigorously stirred for an additional 50 min at 0 °C and 2 h at rt. CaCO3 (14.4 g, 144 mmol) was added to destroy excess HF. The mixture was extracted with CH2Cl2 and purified by column chromatography (CH2Cl2/MeOH, 20:1) to afford 0.99 g (57%) of 24. 1H NMR (500 MHz, CDCl3): δ 8.34 (d, J = 5.6 Hz, 1H), 7.82 (d, J = 5.6 Hz, 1H), 4.00 (s, 6H), 2.21 (s, 3H). 13C NMR (125 MHz, CDCl3): δ 173.2, 169.2, 162.8, 161.1, 159.0, 157.1, 105.9, 90.7, 55.8, 24.8. HRMS (m/z): [M + H]+ calcd for C12H13FN5O3S, 326.0723; found, 326.0732. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 7.75 min.
N-(2-((4,6-Dimethoxy-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)acetamide (25a)
To a solution of 24 (213 mg, 0.655 mmol) in DMF (10 mL) was added 1-methylpiperazine (0.291 mL, 262 mg, 2.62 mmol), and the resulting solution was heated at 90 °C for 1 h. Solvent was removed under reduced pressure, and the residue was purified by chromatography (CH2Cl2/MeOH–NH3 (7 N), 50:1 to 30:1) to afford 0.253 g (95%) of 25a. 1H NMR (500 MHz, CDCl3): δ 8.34 (d, J = 5.6 Hz, 1H), 8.09 (br s, 1H), 7.75 (br s, 1H), 3.90 (s, 6H), 3.88 (m, 4H), 2.48 (m, 4H), 2.38 (s, 3H), 2.18 (s, 3H). HRMS (m/z): [M + H]+ calcd for C17H24N7O3S, 406.1661; found, 406.1661. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 5.75 min.
2-((4,6-Dimethoxy-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-amine (26a)
25a (253 mg, 0.624 mmol) in MeOH (2 mL) and 1 N NaOH(aq) (10 mL) was stirred at 60 °C for 80 min. Solvent was removed under reduced pressure, and the residue was purified by chromatography (CH2Cl2/MeOH–NH3 (7 N), 50:1 to 25:1) to afford 221 mg (97%) of 26a. 1H NMR (500 MHz, CDCl3): δ 7.95 (d, J = 5.8 Hz, 1H), 6.04 (d, J = 5.8 Hz, 1H), 5.01 (s, 2H), 3.89 (br s, 10H), 2.48 (m, 4H), 2.36 (s, 3H). HRMS (m/z): [M + H]+ calcd for C15H22N7O2S, 364.1556; found, 364.1542. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 4.90 min.
N-(2-((4,6-Dimethoxy-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (27a)
To a solution of 26a (50 mg, 0.138 mmol) and Et3N (69 mg, 0.688 mmol) in CH2Cl2 (2 mL) was added acryloyl chloride (62 mg, 0.688 mmol) under a water bath. The resulting mixture was stirred at rt for 24 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to afford 42 mg (72%) of 27a. 1H NMR (500 MHz, CDCl3): δ 8.37 (d, J = 5.6 Hz, 1H), 8.07 (s, 1H), 7.85 (d, J = 5.6 Hz, 1H), 6.47 (d, J = 16.9 Hz, 1H), 6.23 (dd, J = 16.9, 10.3 Hz, 1H), 5.86 (d, J = 10.3 Hz, 1H), 3.89 (m, 10H), 2.49 (m, 4H), 2.37 (s, 3H). 13C NMR (125 MHz, CDCl3): δ 171.5, 171.1, 164.0, 160.2, 159.0, 157.1, 130.3, 129.9, 105.8, 79.5, 54.9, 54.3, 46.2, 43.6. HRMS (m/z): [M + H]+ calcd for C18H24N7O3S, 418.1661; found, 418.1666.
N-(2-((4,6-Dimethoxy-2-morpholinopyrimidin-5-yl)thio)pyrimidin-4-yl)acetamide (25b)
To a solution of 24 (100 mg, 0.307 mmol) in DMF (4 mL) was added morpholine (107 mg, 1.23 mmol), and the resulting solution was heated at 90 °C for 1 h. Solvent and excess reagent were removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to yield 102 mg (85%) of 25b. 1H NMR (500 MHz, CDCl3): δ 8.35 (d, J = 5.6 Hz, 1H), 8.10 (s, 1H), 7.75 (br s, 1H), 3.89 (s, 6H), 3.84 (m, 4H), 3.77 (m, 4H), 2.18 (s, 3H). 13C NMR (125 MHz, CDCl3): δ 168.6, 161.0, 159.2, 158.7, 130.6, 129.3, 113.8, 82.3, 71.5, 55.2, 47.9, 29.5. HRMS (m/z): [M + H]+ calcd for C16H21N6O4S, 393.1345; found, 393.1332. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 7.57 min.
2-((4,6-Dimethoxy-2-morpholinopyrimidin-5-yl)thio)pyrimidin-4-amine (26b)
A mixture of 25b (100 mg, 0.255 mmol) and 1 N NaOH(aq) (4 mL) in 8 mL of methanol was stirred at 60 °C for 1 h. Solvents were removed under reduced pressure, and the residue was purified by preparatory TLC to afford 84 mg (94%) of 26b. 1H NMR (500 MHz, CDCl3): δ 7.95 (d, J = 5.7 Hz, 1H), 6.05 (d, J = 5.7 Hz, 1H), 4.96 (s, 2H), 3.90 (s, 6H), 3.82 (m, 4H), 3.76 (m, 4H). 13C NMR (125 MHz, CDCl3): δ 171.3, 171.1, 162.5, 160.2, 156.2, 101.1, 80.5, 66.8, 54.2, 44.2. HRMS (m/z): [M + H]+ calcd for C14H19N6O3S, 351.1239; found, 351.1248. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 6.30 min.
N-(2-((4,6-Dimethoxy-2-morpholinopyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (27b)
To a solution of 26b (50 mg, 0.143 mmol) and Et3N (72 mg, 0.713 mmol) in CH2Cl2 (2 mL) was added acryloyl chloride (65 mg, 0.713 mmol) under a water bath. The resulting mixture was stirred at rt for 24 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to afford 43 mg (74%) of 27b. 1H NMR (500 MHz, CDCl3): δ 8.37 (d, J = 5.6 Hz, 1H), 8.03 (s, 1H), 7.85 (d, J = 5.6 Hz, 1H), 6.47 (d, J = 16.9 Hz, 1H), 6.22 (dd, J = 16.9, 10.3 Hz, 1H), 5.86 (d, J = 10.3 Hz, 1H), 3.89 (s, 6H), 3.85 (m, 4H), 3.77 (m, 4H). HRMS (m/z): [M + H]+ calcd for C17H21N6O4S, 405.1345; found, 405.1338. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 8.07 min.
N-(2-((4,6-Dimethoxy-2-(piperidin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)acetamide (25c)
To a solution of 24 (100 mg, 0.307 mmol) in DMF (4 mL) was added piperidine (105 mg, 1.23 mmol), and the resulting solution was heated at 90 °C for 1 h. Solvent and excess reagent were removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to yield 104 mg (87%) of 25c. 1H NMR (500 MHz, CDCl3): δ 8.35 (d, J = 5.6 Hz, 1H), 8.00 (br s, 1H), 7.73 (br s, 1H), 3.88 (s, 6H), 3.80 (m, 4H), 2.18 (s, 3H), 1.66 (m, 2H), 1.59 (m, 4H). 13C NMR (125 MHz, CDCl3): δ 171.6, 171.0, 160.1, 158.9, 156.9, 105.3, 82.5, 54.1, 44.8, 29.7, 25.8, 24.8. HRMS (m/z): [M + H]+ calcd for C17H23N6O3S, 391.1552; found, 391.1549. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 9.18 min.
2-((4,6-Dimethoxy-2-(piperidin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-amine (26c)
A mixture of 25c (100 mg, 0.256 mmol) and 1 N NaOH(aq) (4 mL) in 8 mL of methanol was stirred at 60 °C for 1 h. Solvents were removed under reduced pressure, and the residue was purified by preparatory TLC to afford 83 mg (93%) of 26c. 1H NMR (500 MHz, CDCl3): δ 7.95 (d, J = 5.6 Hz, 1H), 6.03 (d, J = 5.6 Hz, 1H), 4.95 (s, 2H), 3.88 (s, 6H), 3.79 (m, 4H), 1.66 (m, 2H), 1.60 (m, 4H). 13C NMR (125 MHz, CDCl3): δ 171.5, 171.0, 162.5, 160.0, 156.2, 101.0, 54.1, 44.8, 25.7, 24.8. HRMS (m/z): [M + H]+ calcd for C15H21N6O2S, 349.1447; found, 349.1434. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 7.60 min.
N-(2-((4,6-Dimethoxy-2-(piperidin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (27c)
To a solution of 26c (50 mg, 0.144 mmol) and Et3N (72 mg, 0.718 mmol) in CH2Cl2 (2 mL) was added acryloyl chloride (65 mg, 0.718 mmol) under a water bath. The resulting mixture was stirred at rt for 24 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to afford 45 mg (78%) of 27c. 1H NMR (500 MHz, CDCl3): δ 8.37 (d, J = 5.6 Hz, 1H), 7.95 (s, 1H), 7.83 (d, J = 5.6 Hz, 1H), 6.48 (dd, J = 16.9, 0.8 Hz, 1H), 6.23 (dd, J = 16.9, 10.4 Hz, 1H), 5.85 (dd, J = 10.3, 0.8 Hz, 1H), 3.88 (s, 6H), 3.81 (m, 4H), 1.69 (m, 2H), 1.62 (m, 4H). 13C NMR (125 MHz, CDCl3): δ 171.7, 171.0, 164.0, 160.1, 158.9, 157.0, 130.3, 105.6, 54.1, 44.8, 25.8, 24.8. HRMS (m/z): [M + H]+ calcd for C18H23N6O3S, 403.1552; found, 403.1541. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 9.23 min.
N-(2-((4,6-Dimethoxy-2-(pyrrolidin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)acetamide (25d)
To a solution of 24 (100 mg, 0.307 mmol) in DMF (4 mL) was added pyrrolidine (87 mg, 1.23 mmol), and the resulting solution was heated at 90 °C for 1 h. Solvent and excess reagent were removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to yield 95 mg (82%) of 25d. 1H NMR (500 MHz, CDCl3): δ 8.34 (d, J = 5.6 Hz, 1H), 8.10 (s, 1H), 7.72 (br s, 1H), 3.89 (s, 6H), 3.59 (m, 4H), 2.17 (s, 3H), 1.96 (m, 4H). 13C NMR (125 MHz, CDCl3): δ 171.7, 170.8, 159.0, 158.9, 156.9, 105.3, 78.3, 54.0, 46.6, 25.4, 24.7. HRMS (m/z): [M + H]+ calcd for C16H21N6O3S, 377.1396; found, 377.1383. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 8.85 min.
2-((4,6-Dimethoxy-2-(pyrrolidin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-amine (26d)
A mixture of 25d (90 mg, 0.239 mmol) and 1 N NaOH(aq) (4 mL) in 8 mL of methanol was stirred at 60 °C for 1 h. Solvents were removed under reduced pressure, and the residue was purified by preparatory TLC to afford 77 mg (96%) of 26d. 1H NMR (500 MHz, CDCl3): δ 7.96 (d, J = 5.7 Hz, 1H), 6.03 (d, J = 5.7 Hz, 1H), 4.88 (s, 2H), 3.90 (s, 6H), 3.58 (t, J = 6.0 Hz, 4H), 1.96 (t, J = 6.0 Hz, 4H). 13C NMR (125 MHz, CDCl3): δ 171.7, 170.9, 162.4, 159.0, 156.3, 100.9, 79.0, 54.0, 46.5, 25.4. HRMS (m/z): [M + H]+ calcd for C14H19N6O2S, 335.1290; found, 335.1291. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 7.32 min.
N-(2-((4,6-Dimethoxy-2-(pyrrolidin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (27d)
To a solution of 26d (50 mg, 0.150 mmol) and Et3N (76 mg, 0.748 mmol) in CH2Cl2 (2 mL) was added acryloyl chloride (68 mg, 0.748 mmol) under a water bath. The resulting mixture was stirred at rt for 24 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to afford 43 mg (73%) of 27d. 1H NMR (500 MHz, CDCl3): δ 8.37 (d, J = 5.6 Hz, 1H), 7.94 (s, 1H), 7.83 (d, J = 5.6 Hz, 1H), 6.47 (dd, J = 16.9, 0.8 Hz, 1H), 6.22 (dd, J = 16.9, 10.4 Hz, 1H), 5.85 (dd, J = 10.4, 0.8 Hz, 1H), 3.91 (s, 6H), 3.61 (m, 4H), 1.98 (m, 4H). 13C NMR (125 MHz, CDCl3): δ 171.6, 171.0, 164.7, 159.2, 158.8, 130.4, 129.7, 105.9, 54.2, 46.7, 25.5. HRMS (m/z): [M + H]+ calcd for C17H21N6O3S, 389.1396; found, 389.1398. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 10 min), tR = 9.15 min.
4-(5-((4-Acrylamidopyrimidin-2-yl)thio)-4,6-dimethoxypyrimidin-2-yl)-1-methylpiperazine 1-Oxide (28)
To a mixture of 27a (20 mg, 0.0479 mmol) and NaHCO3 (40 mg, 0.479 mmol) in CH2Cl2 (2 mL) cooled to −78 °C was added m-CPBA (9.1 mg, 0.0527 mmol). The reaction mixture was stirred for 4 h at −78 °C. Then Na2S2O3 was added in one portion. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 20:1) to afford 17 mg (83%) of 28. 1H NMR (500 MHz, CDCl3): δ 9.47 (br s, 1H), 8.33 (d, J = 5.7 Hz, 1H), 7.89 (d, J = 5.7 Hz, 1H), 6.43–6.49 (m, 2H), 5.81 (dd, J = 7.7, 3.8 Hz, 1H), 4.67–4.75 (m, 2H), 3.85–3.95 (m, 8H), 3.41–3.51 (m, 7H). 13C NMR (125 MHz, CDCl3): δ 171.2, 170.8, 164.8, 159.6, 158.8, 157.7, 130.6, 129.7, 106.2, 81.1, 65.3, 60.2, 54.5, 38.6. HRMS (m/z): [M + H]+ calcd for C18H24N7O4S, 434.1610; found, 434.1592. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (40–95% ACN in 10 min), tR = 6.35 min.
2-((2,4,6-Trimethoxypyrimidin-5-yl)thio)pyrimidin-4-amine (29)
To a solution of 24 (100 mg, 0.307 mmol) in MeOH (5 mL) was added NaOCH3 (36 mg, 0.675 mmol), and the resulting solution was heated at 90 °C for 1 h. Then 1 N NaOH(aq) (2 mL) was added and heating continued at 60 °C for 1 h. Solvent and excess reagent were removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 10:1) to yield 68 mg (75%) of 29. 1H NMR (500 MHz, DMSO-d6): δ 7.77 (d, J = 5.8 Hz, 1H), 6.86 (br s, 2H), 6.11 (d, J = 5.8 Hz, 1H), 3.95 (s, 3H), 3.87 (s, 6H). MS (m/z): [M + H]+ 295.9.
N-(2-((2,4,6-Trimethoxypyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (30)
To a solution of 29 (50 mg, 0.169 mmol) and Et3N (26 mg, 0.254 mmol) in CH2Cl2 (2 mL) was added acryloyl chloride (23 mg, 0.254 mmol) under a water bath. The resulting mixture was stirred at rt for 2 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 20:1) to afford 40 mg (68%) of 30. 1H NMR (500 MHz, CDCl3): δ 8.29 (d, J = 5.6 Hz, 1H), 7.81 (d, J = 5.6 Hz, 1H), 7.78 (s, 1H), 6.40 (d, J = 16.9 Hz, 1H), 6.15 (dd, J = 16.9, 10.3 Hz, 1H), 5.80 (d, J = 10.3 Hz, 1H), 3.97 (s, 3H), 3.95 (s, 6H). HRMS (m/z): [M + H]+ calcd for C14H16N5O4S, 350.0923; found, 350.0936.
N-(6-Amino-2-((2-amino-4,6-dimethoxypyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (31)
To a solution of 11a (50 mg, 0.093 mmol) and Et3N (14 mg, 0.140 mmol) in CH2Cl2 (2 mL) was added acryloyl chloride (12.7 mg, 0.140 mmol) under a water bath. The resulting mixture was stirred at rt for 2 h. Solvent was removed under reduced pressure, the residue was dissolved in 1 mL of TFA/CHCl3 (1:1), and the resulting solution was heated at 62 °C for 12 h. TFA and solvent were removed under reduced pressure, and the residue was purified by preparatory TLC (CH2Cl2/MeOH–NH3 (7 N), 10:1) to afford 18 mg (55%) of 31. 1H NMR (500 MHz, CDCl3/MeOH-d4): δ 7.04 (s, 1H), 6.32–6.41 (m, 2H), 5.80 (dd, J = 10.3, 0.8 Hz, 1H), 3.88 (s, 6H). HRMS (m/z): [M + H]+ calcd for C13H16N7O3S, 350.1035; found, 350.1025. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (5–95% ACN in 12 min), tR = 5.92 min.
4,6-Dimethoxy-2-(4-methylpiperazin-1-yl)pyrimidine (33a)
A 5 g (0.0286 mol) portion of 2-chloro-4,6-dimethoxypyrimidine (32) and 7.93 mL (7.16 g, 0.0715 mol) of N-methylpiperazine in 22 mL of DMF were heated at 90 °C for 2.5 h. The reaction mixture was concentrated under reduced pressure, and the residue was taken up into 200 mL of CH2Cl2. This was washed with brine (3 × 50 mL), dried over MgSO4, filtered, and concentrated to give 6.51 g (95%) of 33a. 1H NMR (500 MHz, CDCl3): δ 5.37 (s, 1H), 3.85 (s, 6H), 3.82 (m, 4H), 2.44 (m, 4H), 2.33 (s, 3H). 13C NMR (125 MHz, CDCl3): δ 172.0, 160.8, 77.8, 55.0, 53.4, 46.3, 43.7. MS (m/z): [M + H]+ 239.2.
5-Iodo-4,6-dimethoxy-2-(4-methylpiperazin-1-yl)pyrimidine (34a)
To 1.59 g (6.67 mmol) of 33a in 40 mL of acetonitrile were added 1.80 g (8.01 mmol) of NIS and 0.771 mL (1.14 g, 10.0 mmol) of TFA, and the resulting solution was stirred at rt for 1.5 h. The reaction mixture was concentrated to dryness, and the residue was taken up into 100 mL of CH2Cl2, washed with 10% sodium thiosulfate (50 mL) and 5% NaHCO3 (3 × 50 mL), dried over MgSO4, filtered, and concentrated to give a residue which was purified by column chromatography (CH2Cl2/MeOH–NH3 (7 N), 1:0 to 20:1) to yield 2.06 g (86%) of 34a. 1H NMR (500 MHz, CDCl3): δ 3.93 (s, 6H), 3.82 (m, 4H), 2.47 (m, 4H), 2.36 (s, 3H). MS (m/z): [M + H]+ 365.1.
3-((4,6-Dimethoxy-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)aniline (35a)
A mixture of 34a (0.200 g, 0.549 mmol), 3-aminothiophenol (70 μL, 0.082 g, 0.659 mmol), neocuproine (0.023 g, 0.11 mmol), CuI (0.038 g, 0.011 mmol), and K2CO3 (0.152 g, 1.10 mmol) in DMF (7 mL) was stirred at 130 °C for 16 h. Solvent was removed under reduced pressure, and the residue was purified by column chromatography (CH2Cl2/MeOH–NH3 (7 N), 200:1 to 40:1) to afford 0.133 g (67%) of 35a. 1H NMR (500 MHz, CDCl3): δ 6.97 (t, J = 8.0 Hz, 1H), 6.46–6.51 (m, 1H), 6.36–6.40 (m, 2H), 3.88–4.03 (m, 10H), 3.58 (br s, 2H), 2.52 (m, 4H), 2.38 (s, 3H). 13C NMR (125 MHz, CDCl3): δ 171.8, 160.1, 146.9, 139.5, 129.6, 116.2, 112.2, 112.1, 81.1, 55.0, 54.6, 46.3, 43.8. HRMS (m/z): [M + H]+ calcd for C17H24N5O2S, 362.1651; found, 362.1649.
N-(3-((4,6-Dimethoxy-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)phenyl)acrylamide (36a)
To a solution of 35a (50 mg, 0.138 mmol) and Et3N (21 mg, 0.207 mmol) in CH2Cl2 (2 mL) was added acryloyl chloride (19 mg, 0.207 mmol) under a water bath. The resulting mixture was stirred at rt for 2 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 20:1) to afford 44 mg (77%) of 36a. 1H NMR (500 MHz, CDCl3): δ 7.49 (m, 1H), 7.36 (m, 1H), 7.14 (m, 2H), 6.84 (d, J = 7.5 Hz, 1H), 6.36 (m, 1H), 6.22 (m, 1H), 5.73 (m, 1H), 4.02 (m, 4H), 3.90 (s, 6H), 2.70 (m, 4H), 2.50 (s, 3H). HRMS (m/z): [M + H]+ calcd for C20H26N5O3S, 416.1756; found, 416.1742.
N-(3-((4,6-Dimethoxy-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)phenyl)methacrylamide (36b)
To a solution of 35a (50 mg, 0.138 mmol) and Et3N (140 mg, 1.38 mmol) in CH2Cl2 (2 mL) was added methacryloyl chloride (144 mg, 1.38 mmol) dropwise under a water bath. The resulting mixture was stirred at rt for 16 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CH2Cl2/MeOH, 20:1) to afford 38 mg (65%) of 36b. 1H NMR (500 MHz, CDCl3): δ 7.40–7.47 (m, 2H), 7.12–7.18 (m, 2H), 6.79–6.83 (m, 1H), 5.74 (d, J = 1.2 Hz, 1H), 5.43 (d, J = 1.2 Hz, 1H), 3.99 (m, 4H), 3.91 (s, 6H), 2.65 (m, 4H), 2.47 (s, 3H), 2.03 (s, 3H). 13C NMR (125 MHz, CDCl3): δ 171.8, 166.7, 160.1, 141.2, 139.5, 138.4, 129.4, 121.9, 119.9, 117.1, 116.9, 81.3, 54.72, 54.67, 45.9, 43.3, 19.0. HRMS (m/z): [M + H]+ calcd for C21H28N5O3S, 430.1913; found, 430.1902. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (40–95% ACN in 10 min), tR = 3.02 min.
4-(4,6-Dimethoxypyrimidin-2-yl)morpholine (33b)
A 5 g (0.0286 mol) portion of 32 and 6.25 mL (6.23 g, 0.0715 mol) of morpholine in 22 mL of DMF were heated at 90 °C for 2.5 h. The reaction mixture was concentrated under reduced pressure, and the residue was taken up into 200 mL of CH2Cl2. This was washed with brine (3 × 50 mL), dried over MgSO4, filtered, and concentrated to give 6.00 g (93%) of 33b. 1H NMR (500 MHz, CDCl3): δ 5.40 (s, 1H), 3.86 (s, 6H), 3.77 (m, 4H), 3.75 (m, 4H). 13C NMR (125 MHz, CDCl3): δ 172.0, 160.9, 78.2, 66.8, 53.5, 44.2. MS (m/z): [M + H]+ 226.2.
4-(5-Iodo-4,6-dimethoxypyrimidin-2-yl)morpholine (34b)
To 1.5 g (6.66 mmol) of 33b in 40 mL of acetonitrile was added 1.80 g (8.00 mmol) of NIS, and the resulting solution was stirred at rt for 2 h. The reaction mixture was concentrated to dryness, and the residue was taken up into 100 mL of CH2Cl2, washed with 10% sodium thiosulfate (50 mL) and 5% NaHCO3 (3 × 50 mL), dried over MgSO4, filtered, and concentrated to give a residue which was purified by column chromatography (hexane/EtOAc, 80:20) to yield 1.99 g (85%) of 34b. 1H NMR (500 MHz, CDCl3): δ 3.92 (s, 6H), 3.71–3.79 (m, 8H). MS (m/z): 352.1 [M + H]+.
3-((4,6-Dimethoxy-2-morpholinopyrimidin-5-yl)thio)aniline (35b)
A mixture of 34b (1.33 g, 3.8 mmol), 3-aminothiophenol (0.48 mL, 0.571 g, 4.56 mmol), neocuproine (0.172 g, 0.76 mmol), CuI (0.144 g, 0.76 mmol), and K2CO3 (1.57 g, 11.3 mmol) in 30 mL of DMF was stirred at 120 °C for 16 h. Solvent was removed under reduced pressure, and the residue was purified by column chromatography (hexane/EtOAc, 1:1) to afford 0.92 g (70%) of 35b. 1H NMR (500 MHz, CDCl3): δ 6.97 (m, 1H), 6.49 (d, J = 8.0 Hz, 1H), 6.37–6.40 (m, 2H), 3.90 (s, 6H), 3.83–3.86 (m, 4H), 3.77–3.80 (m, 4H). HRMS (m/z): [M + H]+ calcd for C16H21N4O3S, 349.1334; found, 349.1341. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (40–95% ACN in 10 min), tR = 3.16 min.
N-(3-((4,6-Dimethoxy-2-morpholinopyrimidin-5-yl)thio)phenyl)acrylamide (37)
To a solution of 35b (50 mg, 0.144 mmol) and Et3N (22 mg, 0.215 mmol) in CH2Cl2 (2 mL) was added acryloyl chloride (19 mg, 0.215 mmol) under a water bath. The resulting mixture was stirred at rt for 2 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CHCl3/MeOH–NH3 (7 N), 20:1) to afford 36 mg (63%) of 37. 1H NMR (500 MHz, CDCl3): δ 7.46–7.50 (m, 2H), 7.12–7.16 (m, 2H), 6.81 (d, J = 7.4 Hz, 1H), 6.38 (d, J = 16.9 Hz, 1H), 6.20 (dd, J = 16.9, 10.3 Hz, 1H), 5.71 (d, J = 10.3 Hz, 1H), 3.90 (s, 6H), 3.82–3.86 (m, 4H), 3.75–3.79 (m, 4H). 13C NMR (125 MHz, CDCl3): δ 171.5, 163.5, 160.1, 139.3, 138.2, 135.3, 131.2, 129.2, 127.7, 121.7, 116.7, 80.9, 66.7, 54.4, 44.2. HRMS (m/z): [M + Na]+ calcd for C19H22N4O4SNa, 425.1259; found, 425.1260. HPLC: (a) H2O + 0.1% TFA, (b) ACN + 0.1% TFA (40–95% ACN in 10 min), tR = 7.15 min.
4,6-Dimethyl-2-(4-methylpiperazin-1-yl)pyrimidine (39a)
A 1.43 g (10 mmol) portion of 2-chloro-4,6-dimethylpyrimidine (38) and 2.50 g (25 mmol) of N-methylpiperazine in 20 mL of DMF were heated at 90 °C for 2.5 h. The reaction mixture was concentrated under reduced pressure, and the residue was taken up into 100 mL of CH2Cl2. This was washed with brine (3 × 25 mL), dried over MgSO4, filtered, and concentrated to give 1.92 g (93%) of 39a. MS (m/z): [M + H]+ 207.0.
5-Iodo-4,6-dimethyl-2-(4-methylpiperazin-1-yl)pyrimidine (40a)
To 1.61 g (7.8 mmol) of 39a in 40 mL of acetonitrile were added 3.7 g (16.4 mmol) of NIS and 2.4 mL (3.56 g, 31.2 mmol) of TFA, and the resulting solution was stirred at rt for 17 h. The reaction mixture was concentrated to dryness, and the residue was taken up into 150 mL of CH2Cl2, washed with 10% sodium thiosulfate (50 mL) and 5% NaHCO3 (3 × 50 mL), dried over MgSO4, filtered, and concentrated to give a residue which was purified by column chromatography (CH2Cl2/MeOH, 1:0 to 10:1) to yield 2.32 g (90%) of 40a. 1H NMR (500 MHz, CDCl3): δ 3.82 (m, 4H), 2.52 (s, 6H), 2.43 (m, 4H), 2.33 (s, 3H). MS (m/z): [M + H]+ 332.9.
2-((4,6-Dimethyl-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-amine (41a)
A mixture of 40a (350 mg, 1.05 mmol), 4-amino-2-mercaptopyrimidine (147 mg, 1.15 mmol), neocuproine (47 mg, 0.21 mmol), CuI (40 mg, 0.21 mmol), and K2CO3 (1.46 g, 10.5 mmol) in 20 mL of DMF was stirred at 110 °C for 16 h. Solvent was removed under reduced pressure, and the residue was purified by column chromatography (CH2Cl2/MeOH, 100:0 to 90:10) to afford 261 mg (75%) of 41a. MS (m/z): [M + H]+ 332.4.
N-(2-((4,6-Dimethyl-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (42a)
To a solution of 41a (50 mg, 0.151 mmol) and Et3N (76 mg, 0.755 mmol) in CH2Cl2 (2 mL) was added acryloyl chloride (68 mg, 0.755 mmol) dropwise under a water bath. The resulting mixture was stirred at rt for 16 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CH2Cl2/MeOH–NH3 (7 N), 10:1) to afford 44 mg (75%) of 42a. 1H NMR (500 MHz, CDCl3): δ 8.40 (d, J = 5.6 Hz, 1H), 7.90 (d, J = 5.6 Hz, 1H), 7.79 (br s, 1H), 6.49 (d, J = 16.9 Hz, 1H), 6.22 (dd, J = 16.9, 10.4 Hz, 1H), 5.89 (d, J = 10.4 Hz, 1H), 3.92 (m, 4H), 2.48 (m, 4H), 2.45 (s, 6H), 2.36 (s, 3H). HRMS (m/z): [M + H]+ calcd for C18H24N7OS, 386.1763; found, 386.1762.
(E)-N-(2-((4,6-Dimethyl-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)but-2-enamide (42b)
To a solution of 41a (50 mg, 0.151 mmol) and Et3N (152 mg, 1.51 mmol) in CH2Cl2 (2 mL) was added crotonyl chloride (158 mg, 1.51 mmol) dropwise under a water bath. The resulting mixture was stirred at rt for 16 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CH2Cl2/MeOH, 20:1) to afford 41 mg (67%) of 42b. 1H NMR (500 MHz, CDCl3): δ 8.37 (d, J = 5.7 Hz, 1H), 7.88 (d, J = 5.7 Hz, 1H), 7.67 (s, 1H), 7.01–7.10 (m, 1H), 5.92 (dd, J = 15.2, 1.7 Hz, 1H), 3.96 (m, 4H), 2.52 (m, 4H), 2.45 (s, 6H), 2.43 (s, 3H), 1.94 (dd, J = 7.0, 1.7 Hz, 3H). HRMS (m/z): [M + H]+ calcd for C19H26N7OS, 400.1920; found, 400.1920.
N-(2-((4,6-Dimethyl-2-(4-methylpiperazin-1-yl)pyrimidin-5-yl)thio)pyrimidin-4-yl)methacrylamide (42c)
To a solution of 41a (50 mg, 0.151 mmol) and Et3N (152 mg, 1.51 mmol) in CH2Cl2 (2 mL) was added methacryloyl chloride (158 mg, 1.51 mmol) dropwise under a water bath. The resulting mixture was stirred at rt for 16 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CH2Cl2/MeOH, 20:1) to afford 60 mg (75%) of 42c. 1H NMR (500 MHz, CDCl3): δ 8.37 (d, J = 5.7 Hz, 1H), 8.08 (br s, 1H), 7.87 (d, J = 5.7 Hz, 1H), 5.85 (s, 1H), 5.60 (s, 1H), 3.95 (m, 4H), 2.52 (m, 4H), 2.45 (s, 6H), 2.38 (s, 3H), 2.04 (s, 3H). HRMS (m/z): [M + H]+ calcd for C19H26N7OS, 400.1920; found, 400.1909.
4-(4,6-Dimethylpyrimidin-2-yl)morpholine (39b)
A 1.43 g (10 mmol) portion of 38 and 2.18 g (25 mmol) of morpholine in 20 mL of DMF were heated at 90 °C for 2.5 h. The reaction mixture was concentrated under reduced pressure, and the residue was taken up into 100 mL of CH2Cl2. This was washed with brine (3 × 25 mL), dried over MgSO4, filtered, and concentrated to give 1.84 g (95%) of 39b. 1H NMR (500 MHz, CDCl3): δ 6.30 (s, 1H), 3.79 (m, 4H), 3.75 (m, 4H), 2.29 (s, 6H). 13C NMR (125 MHz, CDCl3): δ 167.1, 161.9, 109.4, 67.0, 44.3, 24.0. MS (m/z): [M + Na]+ 216.1.
4-(5-Iodo-4,6-dimethylpyrimidin-2-yl)morpholine (40b)
To 1.50 g (7.8 mmol) of 39b in 40 mL of acetonitrile were added 2.1 g (9.4 mmol) of NIS and 0.898 mL (1.33 g, 11.7 mmol) of TFA, and the resulting solution was stirred at rt for 2 h. The reaction mixture was concentrated to dryness, and the residue was taken up into 100 mL of CH2Cl2, washed with 10% sodium thiosulfate (50 mL) and 5% NaHCO3 (3 × 50 mL), dried over MgSO4, filtered, and concentrated to give a residue which was purified by column chromatography (CH2Cl2/MeOH, 1:0 to 20:1) to yield 2.09 g (84%) of 40b. 1H NMR (500 MHz, CDCl3): δ 3.77 (m, 4H), 3.73 (m, 4H), 2.53 (s, 6H). MS (m/z): [M + Na]+ 341.9.
2-((4,6-Dimethyl-2-morpholinopyrimidin-5-yl)thio)pyrimidin-4-amine (41b)
A mixture of 40b (1.0 g, 3.13 mmol), 4-amino-2-mercaptopyrimidine (0.477 g, 3.76 mmol), neocuproine (0.140 g, 0.626 mmol), CuI (0.122 g, 0.626 mmol), and K2CO3 (0.864 g, 6.26 mmol) in 60 mL of DMF was stirred at 110 °C for 16 h. Solvent was removed under reduced pressure, and the residue was purified by column chromatography (CH2Cl2/MeOH, 100:0 to 90:10) to afford 0.690 g (69%) of 41b. MS (m/z): [M + H]+ 319.1.
N-(2-((4,6-Dimethyl-2-morpholinopyrimidin-5-yl)thio)pyrimidin-4-yl)acrylamide (43)
To a solution of 41b (50 mg, 0.157 mmol) and Et3N (79 mg, 0.786 mmol) in CH2Cl2 (2 mL) was added acryloyl chloride (71 mg, 0.786 mmol) dropwise under a water bath. The resulting mixture was stirred at rt for 16 h. Solvent was removed under reduced pressure, and the residue was purified by preparatory TLC (CH2Cl2/MeOH–NH3 (7 N), 10:1) to afford 58 mg (68%) of 43. 1H NMR (500 MHz, CDCl3/MeOH-d4): δ 8.39 (d, J = 5.7 Hz, 1H), 6.49 (d, J = 16.9 Hz, 1H), 6.27 (dd, J = 16.9, 10.3 Hz, 1H), 5.86 (d, J = 10.3 Hz, 1H), 3.89 (m, 4H), 3.78 (m, 4H), 2.46 (s, 6H). 13C NMR (125 MHz, CDCl3): δ 172.2, 170.4, 164.6, 160.4, 159.0, 157.8, 130.1, 130.0, 109.2, 106.2, 66.8, 44.1, 23.7. MS (m/z): [M + H]+ 373.0.
Biological Testing
Cell Lines
SKBr3 cells were a gift from Dr. Neal Rosen (Memorial Sloan-Kettering Cancer Center, MSKCC) and Kasumi-1 and MOLM-13 from Dr. S. Nimer (MSKCC). Cells were cultured routinely in DME/F12 (SKBr3) or in RPMI (Kasumi-1, MOML-13) supplemented with 10% fetal bovine serum, 1% l-glutamine, 1% penicillin, and streptomycin.
Western Blotting
Cells were grown to 60–70% confluence and treated with inhibitor or DMSO vehicle for the indicated times. Protein lysates were prepared in 50 mM Tris, pH 7.4, 150 mM NaCl, and 1% NP-40 lysis buffer. Protein concentrations were measured using the BCA kit (Pierce) according to the manufacturer’s instructions. Protein lysates (10–50 μg) were resolved by SDS–PAGE, transferred onto a nitrocellulose membrane, and incubated with the indicated primary antibodies: anti-HER2 from rabbit (1:250, 28-0004, Zymed), anti-Hsp70 from mouse (1:500, SPA-810, Stressgen), anti-Raf-1 from rabbit (1:500, sc-133, Santa Cruz), anti-PARP (p85 fragment) from rabbit (1:500, G7341, Promega), anti-HOP from mouse (1:500, SRA-1500, Enzo) and anti-β-actin from mouse (1:2500, A1978, Sigma-Aldrich). Membranes were then incubated with a corresponding peroxidase-conjugated secondary antibody (1:3000 dilution).
Hsp90 Binding Assay
For the competition studies, fluorescence polarization (FP) assays were performed as previously reported.49 Briefly, FP measurements were performed on an Analyst GT instrument (Molecular Devices, Sunnyvale, CA). Measurements were taken in black 96-well microtiter plates (Corning no. 3650) where both the excitation and the emission occurred from the top of the wells. A stock of 10 μM GM-cy3B was prepared in DMSO and diluted with Felts buffer (20 mM HEPES (K), pH 7.3, 50 mM KCl, 2 mM DTT, 5 mM MgCl2, 20 mM Na2MoO4, and 0.01% NP40 with 0.1 mg/mL BGG). To each 96-well plate were added 6 nM fluorescent GM (GM-cy3B), 3 μg of SKBr3 lysate (total protein), and tested inhibitor (initial stock in DMSO) in a final volume of 100 μL of HFB buffer. Drugs were added in triplicate wells. For each assay, background wells (buffer only), tracer controls (free, fluorescent GM only), and bound GM controls (fluorescent GM in the presence of SKBr3 lysate) were included on each assay plate. GM was used as a positive control. The assay plate was incubated on a shaker at 4 °C for 24 h, and the FP values (mP) were measured. The fraction of tracer bound to Hsp90 was correlated to the FP value and plotted against the values of competitor concentrations. The inhibitor concentration at which 50% of bound GM was displaced was obtained by fitting the data. All experimental data were analyzed using SOFTmax Pro 4.3.1 and plotted using Prism 4.0 (Graphpad Software Inc., San Diego, CA).
Chemical Precipitation in Cell Extracts
Protein lysates were prepared using 20 mM Tris, pH 7.4, 25 mM NaCl, 0.1% NP-40 lysis buffer. Streptavidin agarose beads (50 μL) (Thermo Scientific) were washed three times with lysis buffer, 44b and 45b were added at the indicated concentrations, and the complexes were incubated at 4 °C for 1 h. Upon a three-time wash with buffer, the beads were added to the indicated total cellular protein in the binding buffer. Samples were incubated at 4 °C overnight, washed five times with the lysis buffer, and applied to SDS–PAGE (see Figure 4a).
Chemical Precipitation in a Live Cell
Cancer cells were treated with 44b (25 μM) or d-biotin for 6 h and then lysed in a buffer containing 20 mM Tris, pH 7.4, 25 mM NaCl, and 0.1% NP-40. Aliquots (500 μg total protein) were incubated with streptavidin beads for 1 h at 4 °C. Purified protein complexes were washed with 20 mM Tris, pH 7.4, 1 M NaCl, 0.1% NP-40 (high salt) buffer and applied to SDS–PAGE. The gel was stained with the SilverQuest staining kit (Invitrogen) (see Figure 4b).
Immunoprecipitation
Cancer cells were collected and lysed in a buffer containing 20 mM Tris, pH 7.4, 25 mM NaCl, and 0.1% NP-40. The anti-Hsp70 antibody (BB70; 5 μL) was added to 500 μg of extract together with protein G agarose beads (30 μL) (Upstate), and the mixture was incubated at 4 °C overnight. Samples were washed with lysis buffer and applied to SDS–PAGE. The gel was stained with the SilverQuest staining kit (Invitrogen) (see Figure 4b).
Competition Assay
Cancer cells were pretreated for 2 h with increasing concentrations of 17a and then lysed in a buffer containing 20 mM Tris, pH 7.4, 25 mM NaCl, and 0.1% NP-40. Meanwhile, to prepare the 44b–streptavidin bead conjugates, 44b (50 μM) was added to high-capacity streptavidin agarose beads (Thermo Scientific) (30 μL per experimental sample), incubated for 1 h at 4 °C, and then washed with lysis buffer. Protein extracts (500 μg of total protein) were then incubated overnight with the 44b–streptavidin bead conjugate. The complexes were washed in high-salt buffer, applied to SDS–PAGE, and stained with the SilverQuest staining kit (see Figure 4c).
Hsp70–HOP Complex Analysis
SKBr3 cells were treated with the indicated concentrations of the inhibitor for 24 h. Samples were collected and lysed in 20 mM Tris, pH 7.4, 25 mM NaCl, 0.1% NP-40 buffer with protease inhibitors added. Aliquots of 500 μg of total protein adjusted to 100 μL with the lysis buffer were prepared. Samples were incubated with 5 μL of BB70 antibody (Stressmarq) or normal IgG (as a negative control) and 20 μL of protein G agarose beads (Upstate) at 4 °C overnight. Samples were washed five times with the lysis buffer and applied to SDS–PAGE followed by a standard Western blotting procedure to detect levels of HOP protein in the Hsp70 complexes upon treatment.
Growth Inhibition Assay
We evaluated the antiproliferative effects of inhibitors using the dye Alamar Blue. This reagent offers a rapid objective measure of cell viability in cell culture, and it uses the indicator dye resazurin to measure the metabolic capacity of cells, an indicator of cell viability. Briefly, cells were plated on Costar 96-well plates. For attached cells (such as SKBr3), 8000 cells/well were used. For suspension cells (such as Kasumi-1), 20000 cells/well were plated. Cells were allowed to incubate for 24 h at 37 °C before drug treatment. Drugs were added in triplicate at the indicated concentrations, and the plate was incubated for 72 h. Alamar Blue (50 μM) was added and the plate read 6 h later using the Analyst GT (fluorescence intensity mode, excitation 530 nm, emission 580 nm, with a 560 nm dichroic mirror). Results were analyzed using the Softmax Pro software. The percentage cell growth inhibition was calculated by comparing fluorescence readings obtained from treated versus control cells, accounting for the initial cell population (time zero). IC50 was calculated as the drug concentration that inhibits cell growth by 50%.
Caspase-3,7 Activation.50
MOLM-13 cells (30000 cells/well) were plated in black 96-well plates (Corning no. 3603) in 40 μL of RPMI medium and left in an incubator (37 °C, 5% CO2) for up to 24 h. Cells were treated for 16 h with compounds or DMSO (control) at the desired concentrations in 50 μL of medium. Drugs were added in triplicate wells. Following exposure of cells to Hsp70 inhibitors, 50 μL of buffer containing 10 mM HEPES (pH 7.5), 2 mM EDTA, 0.1% CHAPS, and the caspase substrate Z-DEVD-R110 at 25 μM was added to each well. Plates were incubated until the signal stabilized, and then the fluorescence signal of each well was measured in an Analyst GT microplate reader. The percentage increase in apoptotic cells was calculated by comparison of the fluorescence reading obtained from treated versus control cells.
Kinase Screen
For most assays, kinase-tagged T7 phage strains were grown in parallel in 24-well blocks in an Escherichia coli host derived from the BL21 strain. E. coli were grown to log phase and infected with T7 phage from a frozen stock (multiplicity of infection 0.4) and incubated with shaking at 32 °C until lysis (90–150 min). The lysates were centrifuged (6000g) and filtered (0.2 μm) to remove cell debris. The remaining kinases were produced in HEK-293 cells and subsequently tagged with DNA for qPCR detection. Streptavidin-coated magnetic beads were treated with biotinylated small-molecule ligands for 30 min at room temperature to generate affinity resins for kinase assays. The liganded beads were blocked with excess biotin and washed with blocking buffer (SeaBlock (Pierce), 1% BSA, 0.05% Tween 20, 1 mM DTT) to remove unbound ligand and to reduce nonspecific phage binding. Binding reactions were assembled by combining kinases, liganded affinity beads, and test compounds in 1× binding buffer (20% SeaBlock, 0.17× PBS, 0.05% Tween 20, 6 mM DTT). Test compounds were prepared as 40× stocks in 100% DMSO and directly diluted into the assay. All reactions were performed in polypropylene 384-well plates in a final volume of 0.04 mL. The assay plates were incubated at room temperature with shaking for 1 h, and the affinity beads were washed with wash buffer (1× PBS, 0.05% Tween 20). The beads were then resuspended in elution buffer (1× PBS, 0.05% Tween 20, 0.5 μM nonbiotinylated affinity ligand) and incubated at room temperature with shaking for 30 min. The kinase concentration in the eluates was measured by qPCR. KINOMEscan’s selectivity score (S) is a quantitative measure of compound selectivity. It is calculated by dividing the number of kinases that bind to the compound by the total number of distinct kinases tested, excluding mutant variants. TREEspot is a proprietary data visualization software tool developed by KINOMEscan. Kinases found to bind are marked with red circles, where larger circles indicate higher affinity binding. The kinase dendrogram was adapted and is reproduced with permission from Science and Cell Signaling Technology, Inc.
Computational Studies
All computations were carried out on an HP workstation xw8200 with the Ubuntu 8.10 operating system using Maestro v8.5 (Schrodinger LLC, New York). Grids were prepared using the Receptor Grid Generation tool in Glidev4.0. Docking calculations were run in the standard precision (SP) mode of Glide v4.0. The maxkeep variable, which sets the maximum number of poses generated during the initial phase of the docking calculation, was set to 5000, and the keep best variable, which sets the number of poses per ligand that enter the energy minimization, was set to 1000. The energy minimization protocol includes a dielectric constant of 4.0 and 1000 steps of conjugate gradient. Upon completion of each docking calculation, at most 100 poses per ligand were allowed to generate. The best docked conformation was chosen considering the orientation and Glidescore (G-score).22
Acknowledgments
We are supported in part by MSKCC’s Technology Transfer Fund (G.C., A.R., Y.K.), Department of Defense Grant W81XWH-10-1-0490 (T.T.), Susan G. Komen for the Cure (T.T., G.C.), the SPORE Pilot Award and Research & Therapeutics Program in Prostate Cancer (G.C.), NIH Grant R01 CA119001 (G.C.), Clinical and Translational Science Center (CTSC) Grant UL1 RR024996 (A.G.), the Breast Cancer Research Fund (G.C.), the Leukemia and Lymphoma Society (G.C.), the Hirshberg Foundation for Pancreatic Cancer Research (G.C.), and NIH Grants 1U01 AG032969-01A1 (G.C.), 1R01 CA172546-01A1 (G.C.), and 1R01 CA155226-01 (G.C.). We thank Dr. George Sukenick and Dr. Hui Liu of the NMR Analytical Core Facility at MSKCC for expert mass spectral analysis.
Glossary
Abbreviations Used
- Hsp70
heat shock protein 70
- Hsp90
heat shock protein 90
- HOP
heat shock organizing protein
- Hsc70
heat shock cognate 70
- Hsp40
heat shock protein 40
- BAG
Bcl-2-associated anthogene
- Hsp110
heat shock protein 110
- IP
Immunoprecipitation
- NBD
nucleotide-binding domain
- PARP
poly(ADP-ribose) polymerase
- HER2
human epidermal growth factor receptor 2
- P-gp/MDR-1
P-glycoprotein/multidrug resistance protein 1
Supporting Information Available
Description of the synthesis of compounds 44a,b and 45a,b. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
The PDB ID codes of crystal structures used as a starting point for building the homology model are 1S3X, 2KHO, and 2P32.
Author Present Address
⊥ Y.K.: BioZone Pharmaceuticals Inc., 710 Fox Run Dr., Plainsboro, NJ 08536.
Author Present Address
# A.G.: Albany Medical Center, 43 New Scotland Ave., Albany, NY 12208.
Author Present Address
∇ C.C.C.: Albert Einstein College of Medicine, 1300 Morris Park Ave., Bronx, NY 10461.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Daugaard M.; Rohde M.; Jäättelä M. The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007, 581, 3702–3710. [DOI] [PubMed] [Google Scholar]
- Brodsky J. F.; Chiosis G. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr. Top. Med. Chem. 2006, 6, 1215–1225. [DOI] [PubMed] [Google Scholar]
- Lanneau D.; Brunet M.; Frisan E.; Solary E.; Fontenay M.; Garrido C. Heat shock proteins: essential proteins for apoptosis regulation. J. Cell Mol. Med. 2008, 12, 743–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers M. V.; Clarke P. A.; Workman P. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell 2008, 14, 250–262. [DOI] [PubMed] [Google Scholar]
- Mayer M. P.; Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. C.; Agashe V. R.; Siegers K.; Hartl F. U. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004, 5, 781–791. [DOI] [PubMed] [Google Scholar]
- Evans C. G.; Chang L.; Gestwicki J. E. Heat shock protein 70 (hsp70) as an emerging drug target. J. Med. Chem. 2010, 53, 4585–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patury S.; Miyata Y.; Gestwicki J. E. Pharmacological targeting of the Hsp70 chaperone. Curr. Top. Med. Chem. 2009, 9, 1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu J. I.-J.; Pimkina J.; Frank A.; Murphy M. E.; George D. L. A small molecule inhibitor of inducible heat shock protein 70. Mol. Cell 2009, 36, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu J. I.-J.; Pimkina J.; Pandey P.; Murphy M. E.; George D. L. HSP70 inhibition by the small-molecule 2-phenylethynesulfonamide impairs protein clearance pathways in tumor cells. Mol. Cancer Res. 2011, 9, 936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M. J.; Scott S. S.; Scott C. M.; Behrman S.; Walter P.; Wipf P.; Coplan J. D.; Chrico W.; Joseph D.; Brodsky J. L.; Batuman O. Antimyeloma effects of the heat shock protein 70 molecular chaperone inhibitor MAL3-101. J. Oncol. 2011, 2011, ID 232037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.; Miyata Y.; Ung P. M.; Bertelsen E. B.; McQuade T. J.; Carlson H. A.; Zuiderweg E. R.; Gestwicki J. E. Chemical screens against a reconstituted multiprotein complex: myricetin blocks DnaJ regulation of DnaK through an allosteric mechanism. Chem Biol. 2011, 18, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousaki A.; Miyata Y.; Jinwal U. K.; Dickey C. A.; Gestwicki J. E.; Zuiderweg E. R. Allosteric drugs: the interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J. Mol. Biol. 2011, 411, 614–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H. J.; Gee H. Y.; Baek K. H.; Ko S. K.; Park J. M.; Lee H.; Kim N. D.; Lee M. G.; Shin I. A small molecule that binds to an ATPase domain of Hsc70 promotes membrane trafficking of mutant cystic fibrosis transmembrane conductance regulator. J. Am. Chem. Soc. 2011, 133, 20267–20276. [DOI] [PubMed] [Google Scholar]
- Williams D. R.; Ko S. K.; Park S.; Lee M. R.; Shin I. An apoptosis-inducing small molecule that binds to heat shock protein 70. Angew. Chem., Int. Ed. 2008, 47, 7466–7469. [DOI] [PubMed] [Google Scholar]
- Kawakami M.; Koya K.; Ukai T.; Tatsuta N.; Ikegawa A.; Ogawa K.; Shishido T.; Chen L. B. Structure–activity of novel rhodacyanine dyes as antitumor agents. J. Med. Chem. 1998, 41, 130–142. [DOI] [PubMed] [Google Scholar]
- Kawakami M.; Koya K.; Ukai T.; Tatsuta N.; Ikegawa A.; Ogawa K.; Shishido T.; Chen L. B. Synthesis and evaluation of novel rhodacyanine dyes that exhibit antitumor activity. J. Med. Chem. 1997, 40, 3151–3160. [DOI] [PubMed] [Google Scholar]
- Williamson D. S.; Borgognoni J.; Clay A.; Daniels Z.; Dokurno P.; Drysdale M. J.; Foloppe N.; Francis G. L.; Graham C. J.; Howes R.; Macias A. T.; Murray J. B.; Parsons R.; Shaw T.; Surgenor A. E.; Terry L.; Wang Y.; Wood M.; Massey A. J. Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design. J. Med. Chem. 2009, 52, 1510–1513. [DOI] [PubMed] [Google Scholar]
- Jhaveri K.; Taldone T.; Modi S.; Chiosis G. Advances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancers. Biochim. Biophys. Acta 2012, 1823, 742–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A. J. ATPases as drug targets: insights from heat shock proteins 70 and 90. J. Med. Chem. 2010, 53, 7280–7286. [DOI] [PubMed] [Google Scholar]
- Massey A. J.; Williamson D. S.; Browne H.; Murray J. B.; Dokurno P.; Shaw T.; Macias A. T.; Daniels Z.; Geoffroy S.; Dopson M.; Lavan P.; Matassova N.; Francis G. L.; Graham C. J.; Parsons R.; Wang Y.; Padfield A.; Comer M.; Drysdale M. J.; Wood M. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother. Pharmacol. 2010, 66, 535–545. [DOI] [PubMed] [Google Scholar]
- Rodina A.; Patel P. D.; Kang Y.; Patel Y.; Baaklini I.; Wong M. J. H.; Taldone T.; Yan P.; Yang C.; Maharaj R.; Gozman A.; Patel M. R.; Patel H. J.; Chirico W.; Erdjument-Bromage H.; Talele T. T.; Young J. C.; Chiosis G. Identification of an allosteric pocket on human Hsp70 reveals a mode of inhibition of this therapeutically important protein. Chem. Biol. 2013, 20, 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taldone T.; Kang Y.; Patel H. J.; Patel M. R.; Patel P. D.; Rodina A.; Patel Y.; Gozman A.; Maharaj R.; Clement C. C.; Lu A.; Young J. C.; Chiosis G.. Heat shock protein 70 inhibitors. 2. 2,5′-thiodipyrimidines, 5-(phenylthio)pyrimidines, 2-(pyridin-3-ylthio)pyrimidines, and 3-(phenylthio)pyridines as reversible binders to an allosteric site on heat shock protein 70. J. Med. Chem. 2014, DOI: 10.1021/jm401552y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram M.; Osipiuk J.; Freeman B.; Morimoto R.; Joachimiak A. Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure 1997, 5, 403–414. [DOI] [PubMed] [Google Scholar]
- Stevens S. Y.; Cai S.; Pellecchia M.; Zuiderweg E. R. The solution structure of the bacterial HSP70 chaperone protein domain DnaK (393–507) in complex with the peptide NRLLLTG. Protein Sci. 2003, 12, 2588–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall L. J.; Walkinshaw M. D. Crystal structure of the C-terminal three-helix bundle subdomain of C. elegans Hsp70. Biochem. Biophys. Res. Commun. 2007, 357, 105–110. [DOI] [PubMed] [Google Scholar]
- Wisniewska M.; Karlberg T.; Lehtio L.; Johansson I.; Kotenyova T.; Moche M.; Schuler H. Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B′, and HSPA5/BiP/GRP78. PLoS One 2010, 5, e8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A.; Kurochkin A. V.; Yip G. N.; Zhang Y.; Bertelsen E. B.; Zuiderweg E. R. Allostery in Hsp70 chaperones is transduced by subdomain rotations. J. Mol. Biol. 2009, 388, 475–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H. J.; Jiang J.; Lafer E. M.; Sousa R. ATP-induced conformational changes in Hsp70: molecular dynamics and experimental validation of an in silico predicted conformation. Biochemistry 2009, 48, 11470–11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Levy E. J.; Chirico W. J. N-Ethylmaleimide inactivates a nucleotide-free Hsp70 molecular chaperone. J. Biol. Chem. 1996, 271, 29937–29944. [DOI] [PubMed] [Google Scholar]
- Kamal A.; Thao L.; Sensintaffar J.; Zhang L.; Boehm M. F.; Fritz L. C.; Burrows F. J. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature 2003, 425, 407–410. [DOI] [PubMed] [Google Scholar]
- Moulick K.; Ahn J. H.; Zong H.; Rodina A.; Cerchietti L.; Gomes-DaGama E. M.; Caldas-Lopes E.; Beebe K.; Perna F.; Hatzi K.; Vu L. P.; Zhao X.; Zatorska D.; Taldone T.; Smith-Jones P.; Alpaugh M.; Gross S. S.; Pillarsetty N.; Ku T.; Lewis J. S.; Larson S. M.; Levine R.; Erdjument-Bromage H.; Guzman M. L.; Nimer S. D.; Melnick A.; Neckers L.; Chiosis G. Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nat. Chem. Biol. 2011, 7, 818–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe K.; Mollapour M.; Scroggins B.; Prodromou C.; Xu W.; Tokita M.; Taldone T.; Pullen L.; Zierer B. K.; Lee M. J.; Trepel J.; Buchner J.; Bolon D.; Chiosis G.; Neckers L. Posttranslational modification and conformational state of heat shock protein 90 differentially affect binding of chemically diverse small molecule inhibitors. Oncotarget 2013, 4, 1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman P.; Burrows F.; Neckers L.; Rosen N. Drugging the cancer chaperone HSP90: combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann. N. Y. Acad. Sci. 2007, 1113, 202–216. [DOI] [PubMed] [Google Scholar]
- Caldas-Lopes E.; Cerchietti L.; Ahn J. H.; Clement C. C.; Robles A. I.; Rodina A.; Moulick K.; Taldone T.; Gozman A.; Guo Y.; Wu N.; Stanchina E.; White J.; Gross S. S.; Mab Y.; Varticovski L.; Melnick A.; Chiosis G. Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc. Natl. Acad. Sci. U. S. A. 2008, 106, 8368–8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H.; Zatorska D.; Kim J.; Aguirre J.; Llauger L.; She Y.; Wu N.; Immormino R. M.; Gewirth D. T.; Chiosis G. Identification of potent water soluble purine-scaffold inhibitors of the heat shock protein 90. J. Med. Chem. 2006, 49, 381–390. [DOI] [PubMed] [Google Scholar]
- Whitesell L.; Mimnaugh E. G.; De Costa B.; Myers C. E.; Neckers L. M. Inhibition of heat shock protein Hsp90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. U. S. A. 1994, 91, 8324–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilenchik M.; Solit D.; Basso A.; Huezo H.; Lucas B.; He H.; Rosen N.; Spampinato C.; Modrich P.; Chiosis G. Targeting wide-range oncogenic transformation via PU24FCl, a specific inhibitor of tumor Hsp90. Chem. Biol. 2004, 11, 787–797. [DOI] [PubMed] [Google Scholar]
- Moulick K.; Clement C. C.; Aguirre J.; Kim J.; Kang Y.; Felts S.; Chiosis G. Synthesis of a red-shifted fluorescence polarization probe for Hsp90. Bioorg. Med. Chem. Lett. 2006, 16, 4515–4518. [DOI] [PubMed] [Google Scholar]
- Fabian M. A.; Biggs W. H. 3rd; Treiber D. K.; Atteridge C. E.; Azimioara M. D.; Benedetti M. G.; Carter T. A.; Ciceri P.; Edeen P. T.; Floyd M.; Ford J. M.; Galvin M.; Gerlach J. L.; Grotzfeld R. M.; Herrgard S.; Insko D. E.; Insko M. A.; Lai A. G.; Lélias J. M.; Mehta S. A.; Milanov Z. V.; Velasco A. M.; Wodicka L. M.; Patel H. K.; Zarrinkar P. P.; Lockhart D. J. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005, 23, 329–336. [DOI] [PubMed] [Google Scholar]
- Karaman M. W.; Herrgard S.; Treiber D. K.; Gallant P.; Atteridge C. E.; Campbell B. T.; Chan K. W.; Ciceri P.; Davis M. I.; Edeen P. T.; Faraoni R.; Floyd M.; Hunt J. P.; Lockhart D. J.; Milanov Z. V.; Morrison M. J.; Pallares G.; Patel H. K.; Pritchard S.; Wodicka L. M.; Zarrinkar P. P. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008, 26, 127–132. [DOI] [PubMed] [Google Scholar]
- Hurwitz J. L.; Scullin P.; Campbell L. Afatinib treatment in advanced non-small cell lung cancer. Lung Cancer: Targets Ther. 2011, 2, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein H. J.; Sun Y.; Dirix L. Y.; Jiang Z.; Paridaens R.; Tan A. R.; Awada A.; Ranade A.; Jiao S.; Schwartz G.; Abbas R.; Powell C.; Turnbull K.; Vermette J.; Zacharchuk C.; Badwe R. Neratinib, an irreversible ErbB Receptor tyrosine kinase inhibitor, in patients with advanced ErbB2-positive breast cancer. J. Clin. Oncol. 2010, 28, 1301–1307. [DOI] [PubMed] [Google Scholar]
- Sequist L. V.; Besse B.; Lynch T. J.; Miller V. A.; Wong K. K.; Gitlitz B.; Eaton K.; Zacharchuk C.; Freyman A.; Powell C.; Ananthakrishnan R.; Quinn S.; Soria J.-C. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non-small-cell lung cancer. J. Clin. Oncol. 2010, 28, 3076–3083. [DOI] [PubMed] [Google Scholar]
- Ou S.-H. I. Second-generation irreversible epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs): a better mousetrap? A review of the clinical evidence. Crit. Rev. Oncol. Hematol. 2012, 83, 407–421. [DOI] [PubMed] [Google Scholar]
- Kwak E. The role of irreversible HER family inhibition in the treatment of patients with non-small cell lung cancer. Oncologist 2011, 16, 1498–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner C.; Vejda S.; Gelbmann D.; Bayer E.; Gotzmann J.; Schulte-Hermann R.; Mikulits W. Concomitant determination of absolute values of cellular protein amounts, synthesis rates, and turnover rates by quantitative proteome profiling. Mol. Cell Proteomics 2002, 1, 528–537. [DOI] [PubMed] [Google Scholar]
- Singh J.; Petter R. C.; Baillie T. A.; Whitty A. The resurgence of covalent drugs. Nat. Rev. Drug Discovery 2011, 10, 307–317. [DOI] [PubMed] [Google Scholar]
- Du Y.; Moulick K.; Rodina A.; Aguirre J.; Felts S.; Dingledine R.; Fu H.; Chiosis G. High-throughput screening fluorescence polarization assay for tumor-specific Hsp90. J. Biomol. Screening 2007, 12, 915–924. [DOI] [PubMed] [Google Scholar]
- Rodina A.; Vilenchik M.; Moulick K.; Aguirre J.; Kim J.; Chiang A.; Litz J.; Clement C. C.; Kang Y.; She Y.; Wu N.; Felts S.; Wipf P.; Massague J.; Jiang X.; Brodsky J. L.; Krystal G. W.; Chiosis G. Selective compounds define Hsp90 as a major inhibitor of apoptosis in small-cell lung cancer. Nat. Chem. Biol. 2007, 3, 498–507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.