Abstract
Optimization of the lactam ω-aminoalkyl substituents in a series of 7-azaindenoisoquinolines resulted in new anticancer agents with improved Top1 inhibitory potencies and cancer cell cytotoxicities. The new compounds 14–17 and 19 exhibited mean graph midpoint cytotoxicity (GI50) values of 21–71 nM in the NCI panel of 60 human cancer cell cultures. Ternary 7-azaindenoisoquinoline–DNA–Top1 cleavage complexes that persist for up to 6 h were detected in HCT116 colon cancer cells. Ternary complexes containing 7-azaindenoisoquinolines were significantly more stable than those in which camptothecin was incorporated. DNA content distribution histograms showed S-phase block 3 h after drug removal. Drug-induced DNA damage in HCT116 cells was revealed by induction of the histone γ-H2AX marker. The 7-azaindenoisoquinolines were able to partially overcome resistance in several drug-resistant cell lines, and they were not substrates for the ABCB1 drug efflux transporter. Molecular modeling studies indicate that the 7-azaindenoisoquinolines intercalate at the DNA cleavage site in DNA–Top1 covalent complexes with the lactam side chain projecting into the major groove. Overall, the results indicate that the 7-azaindenoisoquinolines are promising anticancer agents that merit further development.
Introduction
A variety of polycyclic aromatic molecules, including camptothecin [1 (Figure 1)]1 and the indenoisoquinoline NSC 314622 (2),2 are known for their ability to inhibit mammalian topoisomerase I (Top1) by stabilizing its covalent complex with DNA, the Top1–DNA cleavage complex (Top1cc), preventing further DNA religation and thus leading to the accumulation of DNA breaks.3−6
Figure 1.
Representative Top1 inhibitors.
Despite the usefulness of synthetic analogues of 1 in the clinical treatment of solid tumors, 1 and its derivatives suffer from a number of limitations mainly associated with hydrolytic instability of the lactone and rapid reversibility of 1–Top1–DNA ternary complexes. In comparison with camptothecins, indenoisoquinolines provide chemical stability, and cleavage complexes induced by 2 show greater persistence.2 Additionally, the DNA cleavage site specificity of indenoisoquinolines differs from that of camptothecin, suggesting that different genes and thus tumors could be targeted with indenoisoquinolines.7 Analogues of 2 possessing an ω-aminoalkyl substituent at position 6 (e.g., 3–5) were found to be potent Top1 inhibitors and cytotoxic agents.8,9 LMP400 (4) and LMP776 (5) were ultimately promoted to clinical study at the National Cancer Institute (NCI).10 Other indenoisoquinoline modifications have been almost exclusively confined to electron-donating alkoxy groups on the D-ring, and only a very limited number of electron-withdrawing substituents at positions 3 and 9 have been studied.11 Recently, a comprehensive structure–activity relationship (SAR) study of azaindenoisoquinolines containing a pyridine D-ring (e.g., 6–11) was reported.12,13 The rationale for incorporating nitrogen into the aromatic indenoisoquinoline system was driven by the hypothesis that it would stabilize drug–Top1–DNA ternary complexes through enhanced charge transfer interactions involving the donation of electron density to the drug from the flanking DNA base pairs.12,13 After nitrogen had been introduced into positions 7–10 within the D-ring of the indenoisoquinolines, the 7-azaindenoisoquinoline analogues were found to express the most potent Top1 inhibitory activity in the series while demonstrating an improvement in water solubility. Addition of a methoxy group at position 9 and a nitro group at position 3 further boosted the potencies of the azaindenoisoquinolines, resulting in compounds such as 10 and 11 with a level of Top1 inhibition close to or greater than that of 1 and nanomolar cytotoxicity. These modifications of 7-azaindenoisoquinolines were consistent with the SAR previously developed for indenoisoquinolines.11
Comparing the analogues 10 (Top1, +++; MGM of 104 nM) and 11 (Top1, ++++; MGM of 85 nM) to similarly substituted analogues 6 (Top1, +++; MGM of 1840 nM) and 7 (Top1, ++; MGM of 479 nM) shows the improvement in Top1 inhibitory activity and cytotoxicity that occurs with the replacement of the two methoxy groups on the isoquinolinone moiety with a 3-nitro group. Introduction of the methoxy group at position 9 led to the improvement in cytotoxicity of otherwise potent Top1 inhibitors 8 [Top1, +++(+); MGM of 2480 nM] and 9 (Top1, +++; MGM of >10000 nM), affording 10 and 11. Therefore, compounds 10 and 11 bearing both 3-nitro and 9-methoxy groups were chosen as lead compounds for further exploration.
This investigation was initiated to explore the possibility of improving the activity of 10 and 11 by modifying the lactam ω-aminopropyl group. Considering that the SAR of the azaindenoisoquinolines appears to be consistent with and complementary to the SAR of indenoisoquinolines, the potential candidate amines for replacement of the dimethylamine and morpholine moieties of 10 and 11, respectively, were drawn from previous studies of analogues of 2.2,9,14 The substituents on the lactam side chain of the indenoisoquinolines project out of the DNA major groove where they may bind to amino acid residues of the enzyme.4 In addition, in this study, the mechanism of action of the azaindenoisoquinolines was investigated in cellular systems, and in far greater detail than previously reported.12,13
Chemistry
The proposed series of analogues has been obtained as outlined in Schemes 1 and 2. The starting 7-azaindenoisoquinoline 12(13) was treated with sodium hydride in anhydrous DMF, followed by addition of 1,3-dibromopropane to provide 3-bromopropyl analogue 13 (Scheme 1). The azaindenoisoquinoline intermediate 12 is very hygroscopic and requires thorough drying under vacuum prior to its reaction with sodium hydride and 1,3-dibromopropane. If wet 12 is introduced into the reaction mixture, formation of 13 is suppressed or the resulting 13 is contaminated with dehydrobrominated byproducts. Compound 13 served as a common intermediate and was converted to target compounds 14–17 by displacement reactions with an excess of the corresponding amine (Scheme 1). Aminopropyl analogue 19 was obtained from 13 in two steps (Scheme 2). Reaction of 13 with sodium azide in DMSO afforded 18, which was further transformed into 19 by the Staudinger reaction. The desired compounds 14–17 and 19 were isolated and characterized as their hydrochloride salts.
Scheme 1.
Scheme 2.
Biological Results and Discussion
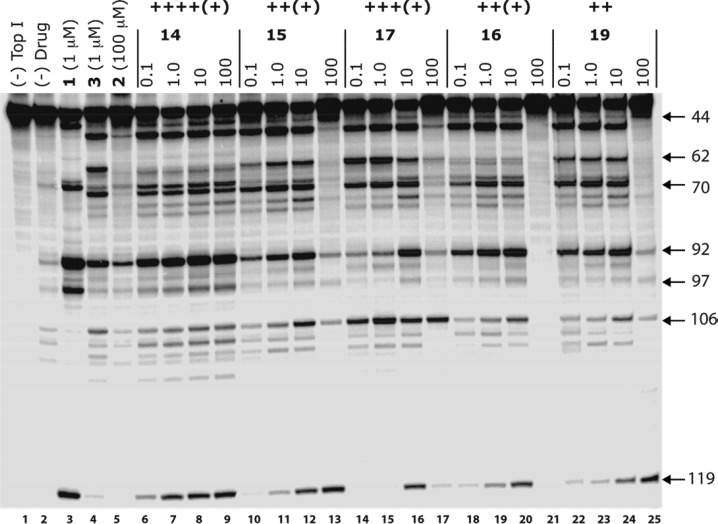
To assess the Top1 inhibitory potency, the target compounds 14–17 and 19 were incubated at different concentrations with a 32P 3′-end-labeled 117 bp DNA fragment and human recombinant Top1.15,16 The DNA fragments were separated on a denaturing gel (Figure 2), and the Top1 inhibitory activities of the drugs were assigned on the basis of visual inspection of the number and intensities of the DNA cleavage bands. They were expressed in semiquantitative fashion relative to the Top1 inhibitory activities of compounds 1 and 2: 0, no detectable activity; +, weak activity; ++, activity similar to that of 2; +++, activity greater than that of 2; ++++, equipotent to 1 (Figure 2 and Table 1). There is an attenuation in cleavage band density at high drug concentrations. This results from the indenoisoquinolines being Top1 poisons at low concentrations because of the intercalation of the drug at the cleavage site in the ternary cleavage complex, which blocks the DNA religation reaction. At high drug concentrations, the indenoisoquinolines intercalate into DNA in the absence of the enzyme, making the DNA a poorer substrate for the enzyme-catalyzed cleavage reaction.17 This causes some indenoisoquinolines to act as Top1 suppressors at high drug concentrations. The concentration at which this occurs differs depending on the structure of the particular indenoisoquinoline and seems to be related to the steric bulk of the substituent on the lactam, with larger substituents suppressing intercalation into free DNA.12
Figure 2.
Lane 1: DNA alone. Lane 2: Top1 alone. Lane 3: Top1 and 1 (1 μM). Lane 4: Top1 and 3 (1 μM). Lane 5: Top1 and 2 (100 μM). Lanes 6–9: Top1 and 14 at 0.1, 1, 10, and 100 μM, respectively. Lanes 10–13: Top1 and 15 at 0.1, 1, 10, and 100 μM, respectively. Lanes 14–17: Top1 and 17 at 0.1, 1, 10, and 100 μM, respectively. Lanes 18–21: Top1 and 16 at 0.1, 1, 10, and 100 μM, respectively. Lanes 22–25: Top1 and 19 at 0.1, 1, 10, and 100 μM, respectively. The numbers and arrows on the right show the cleavage site positions.
Table 1. Top1 Inhibitory and Antiproliferative Activities of 7-Azaindenoisoquinoline.
| cytotoxicity
[GI50 (μM)]c |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| compd | Top1 cleavagea | MGMb | lung, HOP-62 | colon, HCT-116 | CNS, SF-539 | melanoma, UACC-62 | ovarian, OVCAR-3 | renal, SN12C | prostate, DU-145 | breast, MCF7 |
| 1 | ++++ | 0.040 ± 0.0187 | 0.010 | 0.030 | 0.010 | 0.010 | 0.22 | 0.020 | 0.010 | 0.013 |
| 2 | ++ | 20.0 ± 14 | 1.3 | 35 | 41 | 4.2 | 73 | 68 | 37 | 1.58 |
| 6 | +++ | 1.84 ± 0.11 | 0.92 | 1.5 | 1.1 | 3.9 | 2.9 | 3.6 | 0.88 | 0.13 |
| 7 | ++ | 0.479 ± 0.011 | 0.24 | 0.33 | 0.27 | 0.22 | 0.31 | 0.34 | 0.34 | 0.10 |
| 8 | +++(+) | 2.48 ± 0.62 | 1.94 | 0.486 | 3.14 | 3.40 | 3.12 | 1.56 | 1.55 | 0.474 |
| 9 | +++ | NDd | NDd | NDd | NDd | NDd | NDd | NDd | NDd | NDd |
| 10 | +++ | 0.104 ± 0.0025 | 0.054 | 0.074 | 0.078 | 0.052 | 0.14 | 0.057 | 0.051 | 0.024 |
| 11 | ++++ | 0.085 ± 0.0059 | 0.051 | 0.050 | 0.035 | 0.040 | 0.11 | 0.043 | 0.040 | 0.020 |
| 14 | ++++(+) | 0.0208 ± 0.00045 | <0.01 | <0.01 | <0.01 | <0.01 | 0.025 | <0.01 | <0.01 | <0.01 |
| 15 | ++(+) | 0.0240 ± 0.0016 | <0.01 | <0.01 | 0.014 | <0.01 | 0.035 | <0.01 | <0.01 | <0.01 |
| 16 | ++(+) | 0.0708 ± 0.032 | 0.028 | 0.049 | 0.034 | 0.029 | 0.18 | 0.039 | 0.036 | 0.013 |
| 17 | +++(+) | 0.030 ± 0.0031 | <0.01 | 0.010 | 0.010 | <0.01 | 0.058 | <0.01 | <0.01 | <0.01 |
| 19 | ++ | 0.0712 ± 0.0082 | 0.019 | 0.036 | 0.073 | 0.024 | 0.098 | 0.040 | 0.035 | <0.01 |
The relative Top1 inhibitory potencies of the compounds are presented as follows: +, weak activity; ++, activity similar to that of compound 2; +++, activity greater than that of compound 2 but lower than that of camptothecin (1); ++++, activity similar to that of 1 μM 1.
Mean graph midpoint (MGM) for growth inhibition of all human cancer cell lines successfully tested.
The cytotoxicity GI50 values listed are the concentrations corresponding to 50% growth inhibition and are the result of single determinations.
GI50 value not determined because the low activities revealed in the initial single-concentration testing at 10 μM did not warrant the multiple-concentration testing required for the determination of GI50 values.
The antiproliferative activities of 14–17 and 19 were determined in the NCI60 cancer cell lines of the NCI-DTP screen (Table 1).18,19 Cancer cells were incubated with the test compounds at concentrations ranging from 100 μM to 10 nM. After the treated cells had been stained with sulforhodamine B dye, the percentage growth was plotted as a function of the common logarithm of the tested compound concentration. The 50% growth inhibition (GI50) values were determined by interpolation between the points located above and below the 50% growth. The average of GI50 across the entire panel of 60 cell lines for each compound was recorded as the MGM value. GI50 values above and below the tested range (10–4 to 10–8 M) are taken as the maximal (10–4 M) and minimal (10–8 M) drug concentrations, respectively, used in the screening test (Table 1). Respective data for lead compound 1,2 compound 2, and previously reported 7-azaindenoisoquinolines 6–11(12,13) are included in Table 1 for the sake of comparison.
The introduction of imidazole moiety into 14 [Top1, ++++(+); MGM of 21 nM] instead of the aliphatic amines in 10 and 11 significantly increased the level of Top1 inhibition and cytotoxicity, making 14 more potent than camptothecin (1, Top1, ++++; MGM of 40 nM). Compound 15, an ethanolamine analogue, also demonstrated high cytotoxicity with an MGM value of 24 nM. Although 15 still remained an active Top1 inhibitor, it was ranked only at ++(+). Comparison of 14 and 15 demonstrates the lack of a simple correlation between the Top1 inhibitory potency and cytotoxicity of some analogues. This discrepancy could potentially be explained by off-target activities of 15. One possible off-target effect could be intercalation into free DNA. Indeed, our previous studies demonstrated that 7-azaindenoisoquinoline analogues containing smaller amines such as dimethylamine (e.g., 6, 8, and 10), amine, and ethanolamine show an improved ability to intercalate into DNA in the absence of Top1.12 In the case of morpholine (e.g., 7, 9, and 11) or imidazole analogues, the binding to DNA in the absence of Top1 is attenuated or lacking altogether. Also, an earlier study of indenoisoquinoline 3 revealed the ability of 3 to induce cell death at high concentrations in otherwise Top1-deficient cells, also indicating its possible off-target effects.20N-Methylethanolamine analogue 16 retained the Top1cc activity of 15, whereas the cytotoxicity decreased by more than 2-fold.
A further increase in the bulk of the lactam aminoalkyl group in 17 [Top1, +++(+); MGM of 30 nM], a 4-hydroxypiperidine derivative, resulted in a Top1 inhibitory activity similar to that of 11. The similarity of 11 and 17 in their ability to inhibit Top1 could be explained by the similarity of the size and overall geometry of the amino group. The cytotoxicity of 17, on the other hand, improved approximately 3-fold relative to that of 11, resembling that of alcohol 15.
The aminopropyl analogue 19 (Top1, ++; MGM of 71 nM) as an inhibitor of Top1 was less potent than most of the other compounds in the series, although it retained strong cytotoxicity. The decrease in the Top1 inhibitory potency in the case of 19, bearing the smallest amine in this series of azaindenoisoquinolines, could be attributed to the strongest ability to intercalate into free DNA and thus suppress the ability of Top1 to form DNA breaks.
Compounds 11 and 14 have cytotoxicity profiles that are very similar to that of topotecan [20 (Figure 3)] in the NCI60 screen, each with Pearson correlation coefficients of 0.87 relative to 20, suggesting Top1 as their target (see Figure 1 of the Supporting Information, with data and plots from CellMiner21,22). Both drugs have submicromolar GI50 values with Top1 inhibitory activities comparable to that of topotecan (20). Moreover, they remain highly effective in OV:NCI_ADR_RES cells, indicating they are not substrates for the P-glycoprotein efflux pump (ABCB1).21
Figure 3.
Structural formulas of topotecan and doxorubicin.
The induction and stability of Top1cc’s produced by 11 and 14 were evaluated by DNA immunoblotting and DNA–protein cross-link (DPC) assays in human colon cancer carcinoma HCT116 cells (Figure 4).23 Figure 4A shows that 11 and 14 are more potent at trapping Top1cc’s than either 1 or 20 at the same concentration. Also, compounds 11 and 14 trap Top1cc’s at submicromolar concentrations. Reversal experiments after the drug had been removed from the culture medium were conducted to determine the stabilities of the Top1cc’s trapped by 11 and 14. Figure 4B shows the persistence of the cleavage complexes for up to 6 h following compound removal with complete reversal at 24 h. This cleavage complex stability is greater than that of 1 or 20, which is labile and reverses within 1 h.24
Figure 4.

(A) Top1cc’s induced after treatment for 1 h with 11 (0.1, 1, and 10 μM, bottom row of the left panel) or 14 (0.1, 1, and 10 μM, bottom row of the right panel), 1 μM 1, and 1 μM 20. ND means no drug. (B) Persistent Top1cc’s produced by treatment for 1 h with 11 and 14 and assayed at the indicated time points after incubation in drug-free medium. (C) Representative alkaline elution experiments showing persistent Top1cc’s measured as DNA protein cross-links (DPCs) in cells treated with 11 or 14. DPCs persisting 1 h after drug removal are shown. Rapidly reversible DPCs induced by CPT were used as a control. Untreated cells receiving only 30 Gy of irradiation to induce strand breaks were used as a negative control. The fraction of DNA remaining on the filter is plotted vs time (hours).
Alkaline elution, another method for measuring the formation and stability of Top1cc’s, was used to quantify Top1cc’s as DPCs (Figure 4C).23,24 Consistent with the immunocomplex of the enzyme (ICE) with DNAzol results (Figure 4A), 11 and 14 induced more DPCs than 1. In addition, the DPCs induced by 11 and 14 were significantly more persistent following drug removal than those induced by 1 (Figure 4C). Together, these results indicate the greater efficiency of 11 and 14 in trapping Top1cc’s compared to 1.
Drug-induced changes in the cell cycle were revealed in DNA content histograms using FACScan flow cytometry. Initially, no major change in the cell cycle occurs after drug exposure for 1 h and following removal of 11 and 14 for up to 1 h post-drug removal (Figure 5). However, at 3 h post-drug removal, S-phase block occurs and cells are arrested in S-phase. At 6 h post-drug removal, the cells appear to move toward G2, and at 24 h, the cells are arrested in G2. By 48 h post-drug removal, the 11- and 14-treated cells are arrested in G2 and there appears to be little apoptosis. The 48 h reversal time point includes an inset to show cells collected from the supernatant. These results show that azaindenoisoquinolines 11 and 14 are potent inhibitors of DNA replication, as expected from Top1 inhibition.23,25
Figure 5.
Cell cycle analysis of HCT116 cells treated with 1 μM 1, 11, or 14 for 1 h (column 1) followed by incubation in drug-free medium for the indicated times (columns 2–6). Fixed cells were stained with propidium iodide (PI) and analyzed for DNA content distribution histograms by flow cytometry. The 48 h reversal time point is included in the inset to show cells collected from the supernatant.
γ-H2AX-positive cells indicate the induction of DNA damage by Top1cc’s in response to 1 or azaindenoisoquinolines 11 and 14.25 As shown in Figure 6, both 11 and 14 induce γ-H2AX at levels similar to that of camptothecin (1). Flow cytometry demonstrates that DNA damage is observed throughout the S-phase, consistent with Top1cc’s inducing replication fork damage.26
Figure 6.

DNA damage induced by nitro-azaindenoisoquinolines. Drug-induced DNA damage was measured by induction of histone γ-H2AX following treatment for 1 h with 1 μM 1, 11, or 14. γ-H2AX-positive cells induced by 1, 11, or 14 were analyzed by flow cytometry. Numbers above each profile represent the percent cells that score for γ-H2AX. The DNA content determined by propidium iodide (PI) is on the bottom axis.
The development of drug resistance often occurs during treatment and presents a major obstacle to curing potentially sensitive cancers. Table 2 shows the testing of a panel of camptothecin-resistant and drug efflux transporter cells. The drug efflux ABC transporters ABCG2 (mitoxantrone resistance-associated/breast cancer resistance protein) and ABCB1 (MDR-1) confer a high degree of resistance to various anticancer drugs.27 Topotecan and irinotecan are substrates for both. Both 11 and 14 do not appear to be substrates for the ABCB1 pump, which is consistent with the activity of both drugs in the OV:NCI_ADR_RES cells (see Figure 1 of the Supporting Information). This is in contrast to the reference substrate doxorubicin (21). Both 11 and 14 also appear to be less of a substrate for the ABCG2 mitoxantrone-resistant transporter (Table 2) than the reference substrate 20.
Table 2. Cytotoxicities of 11 and 14 in Human Drug-Resistant Cancer Cell Lines As Measured by the MTT Assaya.
| GI50 (nM) |
||||
|---|---|---|---|---|
| compd | parental cell line | resistant subline | resistance ratio | mechanism of resistance |
| DU145 | RC0.1 | mutant Top128,29 | ||
| 1 | 18.20 ± 0.0128 | >1000 ± 0.1305 | >54.95b | |
| 11 | 141.25 ± 0.0501 | 1047.13 ± 0.0022 | 7.41b | |
| 14 | 45.71 ± 0.0622 | >1000 ± 0.2478 | >21.88b | |
| HCT116 | HCT116-siTop1 | siRNA Top130 | ||
| 1 | 17.38 ± 0.1195 | 35.48 ± 0.0307 | 2.04b | |
| 11 | 125.89 ± 0.0277 | 112.20 ± 0.0162 | 0.89 | |
| 14 | 21.88 ± 0.0083 | 70.79 ± 0.0256 | 3.24b | |
| KB3.1 cervical cancer | KBV.1/Vinbl | ABCB1 | ||
| 21 | 676.08 ± 0.2486 | >1,000 ± 0.1333 | >1.48 | |
| 11 | 562.34 ± 0.1112 | 416.8 ± 0.0834 | 0.74 | |
| 14 | 416.87 ± 0.0331 | 426.58 ± 0.1143 | 1.02 | |
| H460 NSCLC | H460/Mito | ABCG2 | ||
| 20 | 44.67 ± 0.0165 | >1,000 ± 0.0413 | >22.39b | |
| 11 | 33.88 ± 0.0020 | 177.83 ± 0.0214 | 5.25b | |
| 14 | 21.88 ± 0.0093 | 416.87 ± 0.0325 | 19.05b | |
The GI50 (concentration of drug required for 50% cell growth inhibition, based on at least two independent determinations) and relative resistances of 11 and 14 and the appropriate positive control (1, 20, or 21) in eight different cell lines. Relative resistances were calculated by dividing the GI50 of the mutant cell line by the GI50 of the parental cell line.
Significantly different.
Table 2 also shows that 11 is partially capable of overcoming resistance in two mutant camptothecin-resistant cell lines. In prostate cancer RC0.1 cells, Top1 is catalytically active but is highly resistant to inhibition by 1 and its derivatives because of an R364H mutation.29 Both 11 and 14 display cross-resistance in the RC0.1 cell line (Table 2). Analogue 11 appears to overcome this resistance more than 14 or reference substrate 1. HCT116 colon cancer cells with short hairpin RNA vectors expressing siRNA for Top1 are ∼2-fold resistant to 1, ∼3-fold resistant to 14, and not resistant to 11 [resistance ratio of 0.89 (Table 2)].
The ab initio calculations that were previously reported along with the SAR study of azaindenoisoquinolines13 revealed that the introduction of a 9-methoxy group produced an increase in the number of dispersion interactions between the azaindenoisoquinolines and the flanking base pairs at the binding site (Figure 7). This could potentially be attributed to the electron-donating nature of the methoxy group and its ability to participate in the resonance with the polyaromatic system of the azaindenoisoquinolines. The hypothetical binding mode of 14 suggested that the 3-nitro group would be facing the cut strand of the DNA and form a hydrogen bond with Asn722 of Top1 (Figure 7), while the 11-keto group acts as a hydrogen bond acceptor from the Arg364 side chain in the minor groove.13 This model is consistent with the ability of the R364H mutation to confer resistance [RC0.1 subline (Table 2)]. It also helps to explain the increase in potency conferred by the 3-nitro group through hydrogen bonding with Asn722 and facilitated charge transfer interactions that are expected to be conferred by the electronegativity of the nitro group. According to this binding mode, the lactam aminopropyl substituent is projected into the spacious major groove, allowing the accommodation of different amines inside the binding pocket.
Figure 7.

Hypothetical mode of binding of 14 (green) to Top1cc (gray). Hydrogen bonds are presented as distances between corresponding heavy atoms. A DNA base pair was removed from the top to clarify the view. This figure was generated by molecular modeling starting from the X-ray crystal structure of an indenoisoquinoline–Top1–DNA complex obtained from the Protein Data Bank (entry 1SC7).
In conclusion, the ω-aminoalkyl substituent has been optimized within a series of 7-azaindenoisoquinoline Top1 inhibitors bearing a 3-nitro group and a 9-methoxy group. The potent Top1 inhibitor and cytotoxic agent 14 was identified [Top1, ++++(+); MGM of 21 nM]. The synthesis of the target amines diverged at a late stage from a common 3-bromopropyl intermediate 13, thus enhancing the overall efficiency for synthesis of the series. This synthetic strategy will allow further exploration of the lactam ω-alkylamine side chain. The newly developed azaindenoisoquinolines 11 and 14 were found to possess a cytotoxicity profile similar to that of topotecan (20), a clinically useful Top1 inhibitor and anticancer agent. This investigation demonstrates that the azaindenoisoquinolines in question are capable of trapping Top1cc’s in stable ternary complexes, thus slowing and stopping DNA processing, which in turn leads to cell cycle arrest and double-strand DNA damage as shown by accumulation of histone γ-H2AX. Additionally, the ternary complexes formed by 11 and 14 were found to be more stable in a cellular system than those formed by camptothecin (1) and topotecan (20). It is also shown that azaindenoisoquinolines could be useful in the treatment of cancers that are otherwise resistant to camptothecins.
Experimental Section
General
Melting points were determined with a Mel-Temp apparatus using capillary tubes and are uncorrected. The proton nuclear magnetic resonance spectra (1H NMR) were recorded using an ARX300 300 MHz Bruker NMR spectrometer. IR spectra were recorded with a Perkin-Elmer 1600 series FTIR spectrometer. Purities of all tested compounds were ≥95%, as established by combustion and/or estimated by HPLC analysis. Combustion microanalyses were performed at Midwest Microlab, LLC, and the reported values are within 0.4% of the calculated values. HPLC analyses were performed on a Waters 1525 binary HPLC pump/Waters 2487 dual λ absorbance detector system. For purities estimated by HPLC, the major peak accounted for ≥95% of the combined total peak area when monitored by a UV detector at 254 nm. Analytical thin-layer chromatography was conducted on Baker-flex silica gel IB2-F plates, and compounds were visualized with UV light at 254 nm. Silica gel flash chromatography was performed using 230–400 mesh silica gel.
7-Aza-5,6-dihydro-6-(3-bromopropyl)-9-methoxy-3-nitro-5,11-dioxo-11H-indeno[1,2-c]isoquinoline (13)
Sodium hydride (60% in mineral oil, 20 mg, 0.5 mmol) was added to a suspension of 7-aza-5,6-dihydro-9-methoxy-3-nitro-5,11-dioxo-11H-indeno[1,2-c]isoquinoline (12, 80 mg, 0.25 mmol) in dry DMF (2 mL) at 0 °C. After the reaction mixture had been warmed to room temperature and stirred for 1.5 h, a dark-red solution was formed. The solution was cooled to 0 °C, and 1,3-dibromopropane (200 mg, 1 mmol) was added. The solution was stirred for 45 min and the reaction quenched with water (10 mL). The products were extracted with ethyl acetate (3 × 5 mL). The combined extracts were washed with water (3 × 5 mL) and brine (5 mL), dried with sodium sulfate, and evaporated to dryness under reduced pressure. The residue was subjected to flash column chromatography (silica gel), eluting with chloroform, to yield the solid product (37 mg, 33%): mp 170–172 °C; 1H NMR (300 MHz, CDCl3) δ 9.20 (d, J = 2.4 Hz, 1 H), 8.75 (d, J = 8.9 Hz, 1 H), 8.49 (dd, J = 9.0, 2.3 Hz, 1 H), 8.24 (d, J = 2.6 Hz, 1 H), 7.45 (d, J = 2.7 Hz, 1 H), 5.17–5.06 (m, 2 H), 3.99 (s, 3 H), 3.56 (t, J = 6.8 Hz, 2 H), 2.52–2.37 (m, 2 H); positive ion ESIMS m/z (relative intensity) 444/446 (MH+, 100/97).
General Procedure for the Preparation of 14–17
7-Aza-5,6-dihydro-6-(3-bromopropyl)-9-methoxy-3-nitro-5,11-dioxo-11H-indeno[1,2-c]isoquinoline (13, 100 mg, 0.23 mmol, 1 equiv), the appropriate amine (82.4–136.4 mg, 1.35 mmol, 5.9 equiv), and potassium carbonate (124 mg, 0.9 mmol, 3.9 equiv) were diluted with 1,4-dioxane (20 mL). The resulting mixture was heated to reflux for 4 h. The solvent was evaporated under reduced pressure, and the residue was redissolved in chloroform (30 mL). The chloroform solution was washed with water (3 × 5 mL) and brine (5 mL), dried with sodium sulfate, and evaporated to dryness. The solid residue was subjected to flash column chromatography (silica gel), eluting with 5–10% methanol in chloroform. The obtained product was redissolved in chloroform (20 mL), and HCl in methanol (3 M, 2 mL) was added. The mixture was stirred for 2 h. The precipitate was collected by filtration and washed with ether (3 × 10 mL) to afford 14–17 as a dark-red solids.
7-Aza-5,6-dihydro-6-[3-(1H-imidazol-1-yl)propyl]-9-methoxy-3-nitro-5,11-dioxo-11H-indeno[1,2-c]isoquinoline Hydrochloride (14)
The general procedure provided the desired compound as a dark-red solid (51.1 mg, 47%): mp 253–254 °C dec; IR (KBr) 3446, 3083, 2683, 2576, 1703, 1687, 1612, 1572, 1555, 1505, 1485, 1438, 1336, 1302 cm–1; 1H NMR (300 MHz, DMSO-d6) δ 9.15 (s, 1 H), 8.88 (d, J = 2.0 Hz, 1 H), 8.70–8.52 (m, 2 H), 8.16 (d, J = 2.7 Hz, 1 H), 7.82 (s, 1 H), 7.72 (s, 1 H), 7.66 (d, J = 2.7 Hz, 1 H), 4.82 (t, J = 6.8 Hz, 2 H), 4.36 (t, J = 6.9 Hz, 2 H), 3.99 (s, 3 H), 2.34 (m, 2 H); positive ion ESIMS m/z (relative intensity) 432 (MH+, 100). Anal. Calcd for C22H17N5O5·HCl·0.8H2O: C, 54.79; H, 4.10; N, 14.52. Found: C, 54.89; H, 3.93; N, 14.32. HPLC purity: 100% (C-18 reverse phase, MeOH).
7-Aza-5,6-dihydro-6-{3-[(2-hydroxyethyl)amino]propyl}-9-methoxy-3-nitro-5,11-dioxo-11H-indeno[1,2-c]isoquinoline Hydrochloride (15)
The general procedure provided the desired compound as a dark-red solid (40.7 mg, 38%): mp 278 °C dec; IR (KBr) 3422, 3247, 3085, 2928, 2691, 1702, 1690, 1612, 1508, 1482, 1339, 1305 cm–1; 1H NMR (300 MHz, DMSO-d6) δ 8.89 (s, 1 H), 8.69–8.56 (m, 3 H, NH2 plus one aromatic H), 8.38 (d, J = 2.6 Hz, 1 H), 7.68 (d, J = 2.6 Hz, 1 H), 5.26 (t, J = 4.8 Hz, 1 H, OH), 4.89 (t, J = 6.1 Hz, 2 H), 3.98 (s, 3 H), 3.74–3.58 (m, 2 H), 3.06 (s, 2 H), 2.97 (s, 2 H), 2.18 (s, 2 H); positive ion ESIMS m/z (relative intensity) 425 (MH+, 100). Anal. Calcd for C21H20N4O6·HCl: C, 54.73; H, 4.59; N, 12.16. Found: C, 54.43; H, 4.99; N, 12.49.
7-Aza-5,6-dihydro-6-{3-[(2-hydroxyethyl)(methyl)amino]propyl}-9-methoxy-3-nitro-5,11-dioxo-11H-indeno[1,2-c]isoquinoline Hydrochloride (16)
The general procedure provided the desired compound as a dark-red solid (105.2 mg, 96%): mp 257–260 °C dec; IR (KBr) 3394, 2946, 2632, 1704, 1683, 1614, 1505, 1486, 1338, 1303 cm–1; 1H NMR (300 MHz, DMSO-d6) δ 9.81 (s, 1 H), 8.85 (s, 1 H), 8.74–8.50 (m, 2 H), 8.36 (d, J = 2.7 Hz, 1 H), 7.64 (d, J = 2.7 Hz, 1 H), 5.33 (s, 1 H), 4.86 (d, J = 6.6 Hz, 2 H), 3.98 (s, 3 H), 3.71 (s, 2 H), 3.19 (s, 2 H), 3.09 (s, 2 H), 2.76 (d, J = 3.3 Hz, 3 H), 2.29–2.12 (m, 2 H); positive ion ESIMS m/z (relative intensity) 439 (MH+, 100). Anal. Calcd for C22H22N4O6·HCl·H2O: C, 53.61; H, 5.11; N, 11.37. Found: C, 53.66; H, 4.91; N, 11.47.
7-Aza-5,6-dihydro-6-[3-(4-hydroxypiperidin-1-yl)propyl]-9-methoxy-3-nitro-5,11-dioxo-11H-indeno[1,2-c]isoquinoline Hydrochloride (17)
The general procedure provided the desired compound as a solid (78.8 mg, 68%): mp 280 °C dec; IR (KBr) 3336, 2940, 2579, 1690, 1612, 1505, 1487, 1337, 1308 cm–1; 1H NMR (300 MHz, DMSO-d6) δ 9.83 (s, 1 H), 8.87 (d, J = 2.0 Hz, 1 H), 8.74–8.52 (m, 2 H), 8.34 (d, J = 11.5 Hz, 1 H), 7.66 (s, 1 H), 5.10–4.94 (m, 1 H), 4.87 (s, 2 H), 3.98 (s, 3 H), 3.27–3.04 (m, 4 H), 3.04–2.75 (m, 2 H), 2.27–2.11 (m, 2 H), 1.99–1.80 (m, 2 H), 1.78–1.51 (m, 2 H); positive ion ESIMS m/z (relative intensity) 465 (MH+, 100). Anal. Calcd for C24H24N4O6·HCl·H2O: C, 55.55; H, 5.24; N, 10.80. Found: C, 55.64; H, 4.87; N, 10.61. HPLC purity: 100% (C-18 reverse phase, MeOH).
7-Aza-5,6-dihydro-6-(3-aminopropyl)-9-methoxy-3-nitro-5,11-dioxo-11H-indeno[1,2-c]isoquinoline Hydrochloride (19)
7-Aza-5,6-dihydro-6-(3-bromopropyl)-9-methoxy-3-nitro-5,11-dioxo-11H-indeno[1,2-c]isoquinoline (13, 115 mg, 0.30 mmol) and sodium azide (110 mg, 1.69 mmol) were diluted with DMSO (20 mL). The resulting mixture was stirred for 18 h at room temperature. Water (60 mL) was added to the mixture, and the product was extracted with chloroform (3 × 20 mL). The combined extracts were washed with water (3 × 10 mL) and brine (15 mL) and dried with sodium sulfate. The chloroform solution was evaporated to dryness to provide crude organic azide 18 as an amorphous glassy solid (96.8 mg): IR (KBr) 3436, 2096, 1674, 1612, 1504, 1482, 1336, 1288 cm–1; 1H NMR (300 MHz, CDCl3) δ 9.53 (d, J = 2.2 Hz, 1 H), 9.08 (d, J = 8.9 Hz, 1 H), 8.82 (dd, J = 8.9, 2.1 Hz, 1 H), 8.58 (dd, J = 7.2, 2.6 Hz, 1 H), 7.78 (d, J = 2.7 Hz, 1 H), 5.46–5.36 (m, 2 H), 4.32 (s, 3 H), 3.84 (t, J = 6.6 Hz, 2 H), 2.63–2.38 (m, 2 H); positive ion ESIMS m/z (relative intensity) 407 (MH+, 77). Without additional purification, organic azide 18 was redissolved in benzene (20 mL), and triethyl phosphite (124 mg, 0.75 mmol) was added to the solution. The mixture was heated to reflux for 14 h. A solution of HCl in methanol (3 M, 5 mL) was added to the mixture, and heating was continued for 3 h. The precipitate was collected by filtration and washed with ether (3 × 10 mL) to afford a dark-red solid (70 mg, 56%): mp 286 °C dec; IR (KBr) 3446, 2976, 1683, 1613, 1506, 1482, 1336, 1304 cm–1; 1H NMR (300 MHz, DMSO-d6) δ 8.81 (s, 1 H), 8.54 (s, 2 H), 8.35 (d, J = 2.5 Hz, 1 H), 7.95 (s, 3 H), 7.59 (d, J = 2.5 Hz, 1 H), 4.85 (s, 2 H), 3.96 (s, 3 H), 2.91 (s, 2 H), 2.09 (s, 2 H); positive ion ESIMS m/z (relative intensity) 381 (MH+, 100). Anal. Calcd for C19H16N4O5·HCl: C, 54.75; H, 4.11; N, 13.44. Found: C, 54.95; H, 4.25; N, 13.06.
Topoisomerase I-Mediated DNA Cleavage Reactions
Human recombinant Top1 was purified from baculovirus as previously described.15,16 DNA cleavage reaction mixtures were prepared as previously reported.16 Briefly, a 117 bp DNA oligonucleotide (Integrated DNA Technologies) encompassing the previously identified Top1 cleavage sites in the 161 bp fragment from pBluescript SK(−) phagemid DNA was employed.16 This 117 bp oligonucleotide contains a single 5′-cytosine overhang, which was 3′-end-labeled by fill-in reaction with [α-32P]dGTP in React 2 buffer [50 mM Tris-HCl (pH 8.0), 100 mM MgCl2, and 50 mM NaCl] with 0.5 unit of DNA polymerase I (Klenow fragment, New England BioLabs). Unincorporated [32P]dGTP was removed using mini Quick Spin DNA columns (Roche, Indianapolis, IN), and the eluate containing the 3′-end-labeled DNA substrate was collected. Approximately 2 nM radiolabeled DNA substrate was incubated with recombinant Top1 in 20 μL of reaction buffer [10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 15 μg/mL BSA] at 25 °C for 20 min in the presence of various concentrations of compounds. The reactions were terminated by adding SDS (final concentration of 0.5%) followed by the addition of 2 volumes of loading dye (80% formamide, 10 mM sodium hydroxide, 1 mM sodium EDTA, 0.1% xylene cyanol, and 0.1% bromophenol blue). Aliquots of each reaction mixture were subjected to 20% denaturing polyacrylamide gel electrophoresis. Gels were dried and visualized by using a phosphoImager and ImageQuant (Molecular Dynamics). For the sake of simplicity, cleavage sites were numbered as previously described for the 161 bp fragment.
Induction and Stability of 7-Azaindenoisoquinoline-Top1cc’s in HCT116 Cells
Top1–DNA covalent complexes were isolated using a modified ICE with DNAzol method.23,31 Briefly, 106 HCT116 cells were treated with each compound at 1 μM for 1 h or left untreated. For reversal experiments, after drug treatment, cells were incubated in drug-free medium for the indicated time points. The medium was removed. Cells were washed twice with a phosphate-buffered solution (PBS) and lysed with DNAzol (Invitrogen) reagent. Genomic DNA was prepared according to the manufacturer’s instructions. Samples were sonicated briefly to shear the DNA. Serial dilutions of each DNA fraction were made and blotted on Immobilon-P membranes (Millipore) using a slot-blot vacuum. Top1–DNA complexes were detected using the C21 Top1 monoclonal antibody (a kind gift from Y.-C. Cheng, Yale University, New Haven, CT) and standard Western blotting procedures. Alkaline elutions were performed as described previously.23,25
Cell Cycle Arrest
HCT116 cells were treated with 1 μM 1, 11, or 14 for 1 h followed by incubation in drug-free medium for the indicated times. Fixed cells were stained with propidium iodide and analyzed for DNA content distribution histograms using a FACScan flow cytometer (Becton Dickinson). Cell cycle distributions were calculated using ModFit LT (Verity Software House, Inc.).
DNA Damage and Induction of Histone γ-H2AX
HCT116 cells were treated with 1 μM 1, 11, or 14 for 1 h. After treatment, cells were harvested, washed twice with ice-cold PBS, and fixed in 4% paraformaldehyde for 10 min at room temperature. Cell pellets were then washed with 1 mL of ice-cold PBS and permeabilized with 1 mL of prechilled (−20 °C) 70% ethanol for 20 min at room temperature. Cells were again washed in PBS and further permeabilized with ice-cold 0.25% Triton X-100 in PBS for 5 min on ice, washed in PBS, and incubated with the anti-γ-H2AX antibody at a 250-fold dilution in a PBS/1% BSA mixture for 30 min at room temperature. Cells were washed with PBS and resuspended in 500 mL of PBS containing 50 mg/mL propidium iodide (PI) and 0.5 mg/mL RNase A. Analyses of FL2-A (PI) versus FL1-H (γ-H2AX) were conducted using a FACScan flow cytometer (Becton Dickinson). Cell cycle distributions were calculated using ModFit LT (Verity Software House, Inc.).
Growth Inhibition of Drug-Resistant Cell Lines
Human colon HCT116 cells were obtained from the NCI Developmental Therapeutics Program. The stably transfected HCT116 Top1 short interfering RNA (siRNA; HCT116-siTop1) cells were derived in our laboratory as described previously.30 HCT116 cells were maintained in RPMI 1640 (Invitrogen) containing 10% FBS (Gemini Bio-Products) with the addition of 100 μg/mL hygromycin. H460 human lung cancer stable transfectants expressing wild-type ABCG2 and KB human cervical carcinoma expressing MDR-1/P-glycoprotein were a kind gift from M. D. Hall and M. M. Gottesman (Laboratory of Cell Biology, Center for Cancer Research, National Cancer Institute) and maintained in RPMI 1640 supplemented with 10% FBS and either 20 ng/mL mitoxantrone and 1 μg/mL vinblastine, respectively. The DU145 cell line was obtained from the American Type Culture Collection. The RC0.1 cell subline was derived from DU145 cells as previously described28,29 and maintained in RPMI 1640 and 10% FBS. Cells were seeded in 96-well plates 24 h before drug treatment. The cytotoxicities of 1, 20, 21, 11, and 14 in HCT116, DU145, KB3.1, and H460 cells and their resistant subclones were assessed by the MTS (Promega) colorimetric assay. Compound exposures were continuous for 72 h for all assays. The percentage of growth was calculated relative to the control (vehicle-treated cells) after cells had been cultured for 3 days with the control taken to be 100.
Molecular Modeling
The structure of 14 was prepared with Sybyl version 8.1 using the MMFF94s force field and MMFF94 charges for geometry optimization.32 The X-ray crystal structure coordinates of the template complex were obtained from the Protein Data Bank (entry 1SC7). Hydrogens were added to all atoms, and their positions were optimized with the MMFF94s force field. The original ligand was removed from the template structure of the ternary complex,4 and 14 was docked into the resulting Top1cc using the docking genetic algorithm and GoldScore fitness function within GOLD version 3.2.33 The best solution, as a result of 100 docking runs, was merged with the Top1cc. The position of 14 within the ternary complex was refined by geometry optimization with 100 iterations employing steepest descent minimization followed by 200 iterations with a conjugate gradient using the MMFF94s force field and MMFF94 charges within Sybyl version 8.1.
Acknowledgments
This work was made possible by the National Institutes of Health through support of this work by Research Grant UO1 CA89566, by a Purdue Research Foundation Research Grant, and by the Center for Cancer Research, Intramural Program of the National Cancer Institute.
Glossary
Abbreviations Used
- DMF
N,N-dimethylformamide
- DMSO
dimethyl sulfoxide
- DPC
DNA–protein cross-link
- ICE
immunocomplex of enzyme
- MGM
mean graph midpoint
- NCI
National Cancer Institute
- PI
propidium iodide
- TFA
trifluoroacetic acid
- Top1
topoisomerase I
- Top1cc
topoisomerase I–DNA cleavage complex
- SAR
structure–activity relationship
Supporting Information Available
Mean graphs of the cytotoxicity GI50 data for compounds 11, 14, and 20 in the NCI panel of human cancer cell lines. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
E.K. and D.S. contributed equally to this work.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Wall M. E.; Wani M. C.; Cook C. E.; Palmer K. H.; McPhail A. T.; Sim G. A. Plant Antitumor Agents. I. The Isolation and Structure of Camptothecin, a Novel Alkaloidal Leukemia and Tumor Inhibitor from Camptotheca acuminata. J. Am. Chem. Soc. 1966, 88, 3888–3890. [Google Scholar]
- Kohlhagen G.; Paull K.; Cushman M.; Nagafuji P.; Pommier Y. Protein-Linked DNA Strand Breaks Induced by NSC 314622, a Novel Noncamptothecin Topoisomerase I Poison. Mol. Pharmacol. 1998, 54, 50–58. [DOI] [PubMed] [Google Scholar]
- Khadka D. B.; Cho W.-J. Topoisomerase Inhibitors as Anticancer Agents: A Patent Update. Expert Opin. Ther. Pat. 2013, 23, 1033–1056. [DOI] [PubMed] [Google Scholar]
- Staker B. L.; Feese M. D.; Cushman M.; Pommier Y.; Zembower D.; Stewart L.; Burgin A. B. Structures of Three Classes of Anticancer Agents Bound to the Human Topoisomerase I-DNA Covalent Complex. J. Med. Chem. 2005, 48, 2336–2345. [DOI] [PubMed] [Google Scholar]
- Pommier Y.; Marchand C. Interfacial Inhibitors: Targeting Macromolecular Complexes. Nat. Rev. Drug Discovery 2012, 11, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y.; Cushman M. The Indenoisoquinoline Noncamptothecin Topoisomerase I Inhibitors: Update and Perspectives. Mol. Cancer Ther. 2009, 8, 1008–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. Topoisomerase I Inhibitors: Camptothecins and Beyond. Nat. Rev. Cancer 2006, 6, 789–802. [DOI] [PubMed] [Google Scholar]
- Antony S.; Jayaraman M.; Laco G.; Kohlhagen G.; Kohn K. W.; Cushman M.; Pommier Y. Differential Induction of Topoisomerase I-DNA Cleavage Complexes by the Indenoisoquinoline MJ-III-65 (NSC 706744) and Camptothecin: Base Sequence Analysis and Activity against Camptothecin-Resistant Topoisomerase I. Cancer Res. 2003, 63, 7428–7435. [PubMed] [Google Scholar]
- Nagarajan M.; Morrell A.; Ioanoviciu A.; Antony S.; Kohlhagen G.; Agama K.; Hollingshead M.; Pommier Y.; Cushman M. Synthesis and Evaluation of Indenoisoquinoline Topoisomerase I Inhibitors Substituted with Nitrogen Heterocycles. J. Med. Chem. 2006, 49, 6283–6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Study: 10-C-0056, a Phase I Study of Indenoisoquinolines LMP400 and LMP776 in Adults with Relapsed Solid Tumors and Lymphomas (http://clinicalstudies.info.nih.gov/cgi/detail.cgi?A_2010-C-0056.html) (accessed September 13, 2013).
- Morrell A.; Placzek M.; Parmley S.; Grella B.; Antony S.; Pommier Y.; Cushman M. Optimization of the Indenone Ring of Indenoisoquinoline Topoisomerase I Inhibitors. J. Med. Chem. 2007, 50, 4388–4404. [DOI] [PubMed] [Google Scholar]
- Kiselev E.; DeGuire S.; Morrell A.; Agama K.; Dexheimer T. S.; Pommier Y.; Cushman M. 7-Azaindenoisoquinolines as Topoisomerase I Inhibitors and Potential Anticancer Agents. J. Med. Chem. 2011, 54, 6106–6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev E.; Agama K.; Pommier Y.; Cushman M. Azaindenoisoquinolines as Topoisomerase I Inhibitors and Potential Anticancer Agents: A Systematic Study of Structure-Activity Relationships. J. Med. Chem. 2012, 55, 1682–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan M.; Xiao X.; Antony S.; Kohlhagen G.; Pommier Y.; Cushman M. Design, Synthesis, and Biological Evaluation of Indenoisoquinoline Topoisomerase I Inhibitors Featuring Polyamine Side Chains on the Lactam Nitrogen. J. Med. Chem. 2003, 46, 5712–5724. [DOI] [PubMed] [Google Scholar]
- Pourquier P.; Ueng L.-M.; Fertala J.; Wang D.; Park H.-J.; Essigmann J. M.; Bjornsti M.-A.; Pommier Y. Induction of Reversible Complexes Between Eukaryotic DNA Topoisomerase I and DNA-containing Oxidative Base Damages. 7,8-Dihydro-8-Oxoguanine and 5-Hydroxycytosine. J. Biol. Chem. 1999, 274, 8516–8523. [DOI] [PubMed] [Google Scholar]
- Dexheimer T. S.; Pommier Y. DNA Cleavage Assay for the Identification of Topoisomerase I Inhibitors. Nat. Protoc. 2008, 3, 1736–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony S.; Agama K. K.; Miao Z. H.; Hollingshead M.; Holbeck S. L.; Wright M. H.; Varticovski L.; Nagarajan M.; Morrell A.; Cushman M.; Pommier Y. Bisindenoisoquinoline Bis-1,3-{(5,6-dihydro-5,11-diketo-11H-indeno[1,2-c] isoquinoline)-6-propylamino}propane bis(trifluoroacetate) (NSC 727357), a DNA Intercalator and Topoisomerase Inhibitor with Antitumor Activity. Mol. Pharmacol. 2006, 70, 1109–1120. [DOI] [PubMed] [Google Scholar]
- Boyd M. R.; Paull K. D. Some Practical Considerations and Applications of the National Cancer Institute In Vitro Anticancer Drug Discovery Screen. Drug Dev. Res. 1995, 34, 91–109. [Google Scholar]
- Skehan P.; Storeng R.; Scudiero D.; Monks A.; McMahon J.; Vistica D.; Warren J. T.; Bokesch H.; Kenney S.; Boyd M. R. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [DOI] [PubMed] [Google Scholar]
- Antony S.; Kohlhagen G.; Agama K.; Jayaraman M.; Cao S.; Durrani F. A.; Rustum Y. M.; Cushman M.; Pommier Y. Cellular Topoisomerase I Inhibition and Antiproliferative Activity by MJ-III-65 (NSC 706744), an Indenoisoquinoline Topoisomerase I Poison. Mol. Pharmacol. 2005, 67, 523–530. [DOI] [PubMed] [Google Scholar]
- Reinhold W. C.; Sunshine M.; Liu H.; Varma S.; Kohn K. W.; Morris J.; Doroshow J.; Pommier Y. CellMiner: A Web-Based Suite of Genomic and Pharrmacologic Tools to Explore Transcript and Drug Patterns in the NCI-60 Cell Line Set. Cancer Res. 2012, 72, 3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CellMiner-Analysis Tools (http://discover.nci.nih.gov/cellminer/) (accessed September 13, 2013).
- Sooryakumar D.; Dexheimer T. S.; Teicher B. A.; Pommier Y. Molecular and Cellular Pharmacology of the Novel Noncamptothecin Topoisomerase I Inhibitor Genz-644282. Mol. Cancer Ther. 2011, 10, 1490–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa A.; Fujimori A.; Fujimori Y.; Pommier Y. Comparison of Topoisomerase I Inhibition, DNA Damage, and Cytotoxocity of Camptothecin Derivatives Presently in Clinical Trials. J. Natl. Cancer Inst. 1994, 86, 836–842. [DOI] [PubMed] [Google Scholar]
- Antony S.; Agama K. K.; Miao Z. H.; Takagi K.; Wright M. H.; Robles A. I.; Varticovski L.; Nagarajan M.; Morrell A.; Cushman M.; Pommier Y. Novel Indenoisoquinolines NSC 725776 and NSC 724998 Produce Persistent Topolsomerase I Cleavage Complexes and Overcome Multidrug Resistance. Cancer Res. 2007, 67, 10397–10405. [DOI] [PubMed] [Google Scholar]
- Furuta T.; Takemura H.; Liao Z. Y.; Aune G. J.; Redon C.; Sedelnikova O. A.; Pilch D. R.; Rogakou E. P.; Celeste A.; Chen H. T.; Nussenzweig A.; Aladjem M. I.; Bonner W. M.; Pommier Y. Phosphorylation of Histone H2AX and Activation of Mre11, Rad50, and Nbs1 in Response to Replication-dependent DNA Double-strand Breaks Induced by Mammalian DNA Topoisomerase I Cleavage Complexes. J. Biol. Chem. 2003, 278, 20303–20312. [DOI] [PubMed] [Google Scholar]
- Gottesman M. M.; Fojo T.; Bates S. E. Multidrug Resistance in Cancer: Role of ATP-Dependent Transporters. Nat. Rev. Cancer 2002, 2, 48–58. [DOI] [PubMed] [Google Scholar]
- Chatterjee D.; Wyche J. H.; Pantazis P. Induction of Apoptosis in Malignant and Camptothecin-resistant Human Cells. Ann. N.Y. Acad. Sci. 1996, 803, 143–156. [DOI] [PubMed] [Google Scholar]
- Urasaki Y.; Laco G. S.; Pourquier P.; Takebayashi Y.; Kohlhagen G.; Gioffre C.; Zhang H. L.; Chatterjee D.; Pantazis P.; Pommier Y. Characterization of a Novel Topoisomerase I Mutation from a Camptothecin-resistant Human Prostate Cancer Cell Line. Cancer Res. 2001, 61, 1964–1969. [PubMed] [Google Scholar]
- Miao Z. H.; Player A.; Shankavaram U.; Wang Y. H.; Zimonjic D. B.; Liao Z. Y.; Liu H.; Shimura T.; Zhang H. L.; Meng L. H.; Zhang Y. W.; Kawasaki E. S.; Popescu N. C.; Aladjem M. I.; Goldstein D. J.; Weinstein J. N.; Pommier Y. Nonclassic Functions of Human Topoisomerase I: Genome-wide and Pharmacologic Analyses. Cancer Res. 2007, 67, 8752–8761. [DOI] [PubMed] [Google Scholar]
- Zhang Y. W.; Regairaz M.; Seiler J. A.; Agama K. K.; Doroshow J. H.; Pommier Y. Poly(ADP-ribose) Polymerase and XPF-ERCC1 Participate in Distinct Pathways for the Repair of Topoisomerase I-Induced DNA Damage in Mammalian Cells. Nucleic Acids Res. 2011, 39, 3607–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SYBYL, version 8.1.1; Tripos International: St. Louis. [Google Scholar]
- Verdonk M. L.; Cole J. C.; Hartshorn M. J.; Murray C. W.; Taylor R. D. Improved Protein-Ligand Docking Using GOLD. Proteins: Struct., Funct., Genet. 2003, 52, 609–623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.