Abstract
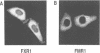
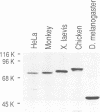
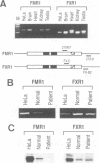
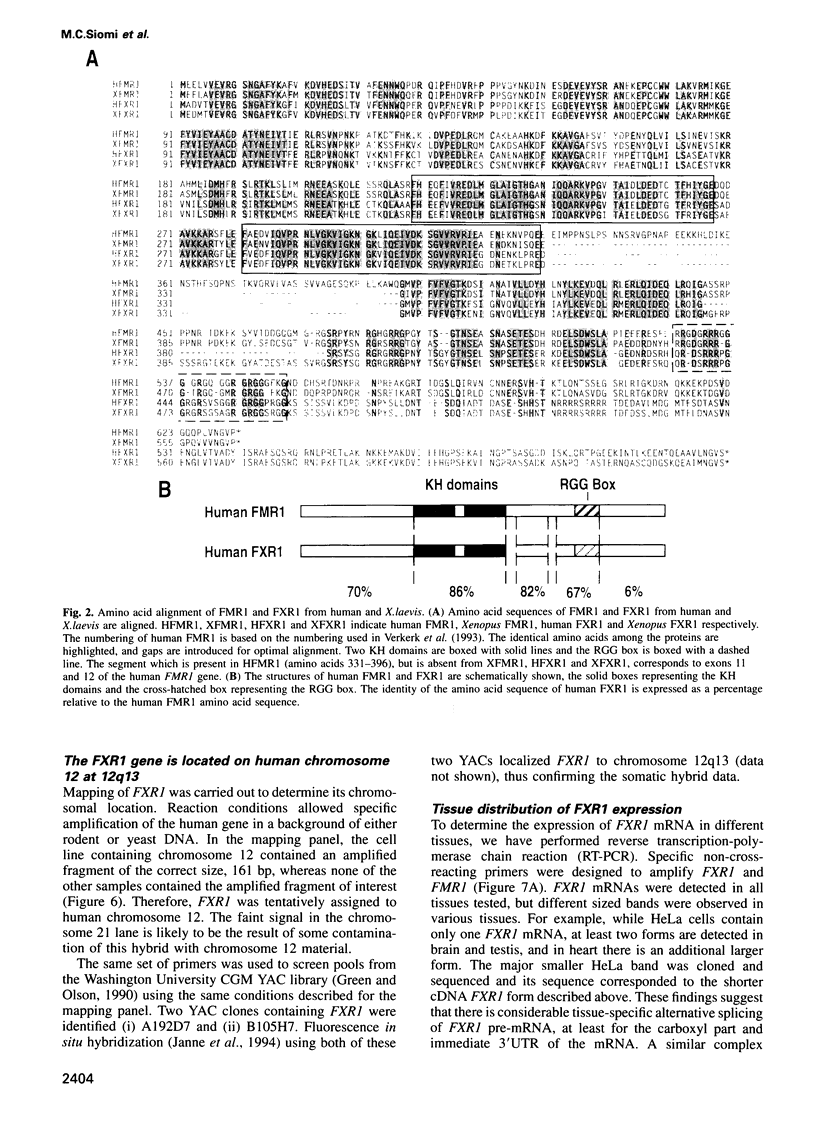
Fragile X mental retardation syndrome, the most common cause of hereditary mental retardation, is directly associated with the FMR1 gene at Xq27.3. FMR1 encodes an RNA binding protein and the syndrome results from lack of expression of FMR1 or expression of a mutant protein that is impaired in RNA binding. We found a novel gene, FXR1, that is highly homologous to FMR1 and located on chromosome 12 at 12q13. FXR1 encodes a protein which, like FMR1, contains two KH domains and is highly conserved in vertebrates. The 3' untranslated regions (3'UTRs) of the human and Xenopus laevis FXR1 mRNAs are strikingly conserved (approximately 90% identity), suggesting conservation of an important function. The KH domains of FXR1 and FMR1 are almost identical, and the two proteins have similar RNA binding properties in vitro. However, FXR1 and FMR1 have very different carboxy-termini. FXR1 and FMR1 are expressed in many tissues, and both proteins, which are cytoplasmic, can be expressed in the same cells. Interestingly, cells from a fragile X patient that do not have any detectable FMR1 express normal levels of FXR1. These findings demonstrate that FMR1 and FXR1 are members of a gene family and suggest a biological role for FXR1 that is related to that of FMR1.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abitbol M., Menini C., Delezoide A. L., Rhyner T., Vekemans M., Mallet J. Nucleus basalis magnocellularis and hippocampus are the major sites of FMR-1 expression in the human fetal brain. Nat Genet. 1993 Jun;4(2):147–153. doi: 10.1038/ng0693-147. [DOI] [PubMed] [Google Scholar]
- Ashley C. T., Jr, Wilkinson K. D., Reines D., Warren S. T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993 Oct 22;262(5133):563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Ashley C. T., Sutcliffe J. S., Kunst C. B., Leiner H. A., Eichler E. E., Nelson D. L., Warren S. T. Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG-repeat. Nat Genet. 1993 Jul;4(3):244–251. doi: 10.1038/ng0793-244. [DOI] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994 Jul 29;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Bächner D., Stéinbach P., Wöhrle D., Just W., Vogel W., Hameister H., Manca A., Poustka A. Enhanced Fmr-1 expression in testis. Nat Genet. 1993 Jun;4(2):115–116. doi: 10.1038/ng0693-115. [DOI] [PubMed] [Google Scholar]
- De Boulle K., Verkerk A. J., Reyniers E., Vits L., Hendrickx J., Van Roy B., Van den Bos F., de Graaff E., Oostra B. A., Willems P. J. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993 Jan;3(1):31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- Devys D., Lutz Y., Rouyer N., Bellocq J. P., Mandel J. L. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993 Aug;4(4):335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Eichler E. E., Richards S., Gibbs R. A., Nelson D. L. Fine structure of the human FMR1 gene. Hum Mol Genet. 1993 Aug;2(8):1147–1153. doi: 10.1093/hmg/2.8.1147. [DOI] [PubMed] [Google Scholar]
- Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell. 1994 Jul 15;78(1):23–33. [PubMed] [Google Scholar]
- Fu Y. H., Kuhl D. P., Pizzuti A., Pieretti M., Sutcliffe J. S., Richards S., Verkerk A. J., Holden J. J., Fenwick R. G., Jr, Warren S. T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991 Dec 20;67(6):1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Gahrton G., Robert K. H., Friberg K., Juliusson G., Biberfeld P., Zech L. Cytogenetic mapping of the duplicated segment of chromosome 12 in lymphoproliferative disorders. Nature. 1982 Jun 10;297(5866):513–514. doi: 10.1038/297513a0. [DOI] [PubMed] [Google Scholar]
- Gibson T. J., Rice P. M., Thompson J. D., Heringa J. KH domains within the FMR1 sequence suggest that fragile X syndrome stems from a defect in RNA metabolism. Trends Biochem Sci. 1993 Sep;18(9):331–333. doi: 10.1016/0968-0004(93)90068-x. [DOI] [PubMed] [Google Scholar]
- Gibson T. J., Thompson J. D., Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 1993 Jun 21;324(3):361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- Green E. D., Olson M. V. Systematic screening of yeast artificial-chromosome libraries by use of the polymerase chain reaction. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1213–1217. doi: 10.1073/pnas.87.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds H. L., Ashley C. T., Sutcliffe J. S., Nelson D. L., Warren S. T., Housman D. E., Schalling M. Tissue specific expression of FMR-1 provides evidence for a functional role in fragile X syndrome. Nat Genet. 1993 Jan;3(1):36–43. doi: 10.1038/ng0193-36. [DOI] [PubMed] [Google Scholar]
- Jackson R. J. Cytoplasmic regulation of mRNA function: the importance of the 3' untranslated region. Cell. 1993 Jul 16;74(1):9–14. doi: 10.1016/0092-8674(93)90290-7. [DOI] [PubMed] [Google Scholar]
- Jänne P. A., Dutra A. S., Dracopoli N. C., Charnas L. R., Puck J. M., Nussbaum R. L. Localization of the 75-kDa inositol polyphosphate-5-phosphatase (INPP5B) to human chromosome band 1p34. Cytogenet Cell Genet. 1994;66(3):164–166. doi: 10.1159/000133691. [DOI] [PubMed] [Google Scholar]
- Kiledjian M., Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992 Jul;11(7):2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens F., Johansson B., Mandahl N., Heim S., Bennet K., Rydholm A., Willén H., Mitelman F. Clonal chromosome abnormalities in two liposarcomas. Cancer Genet Cytogenet. 1987 Sep;28(1):137–144. doi: 10.1016/0165-4608(87)90363-3. [DOI] [PubMed] [Google Scholar]
- Murty V. V., Houldsworth J., Baldwin S., Reuter V., Hunziker W., Besmer P., Bosl G., Chaganti R. S. Allelic deletions in the long arm of chromosome 12 identify sites of candidate tumor suppressor genes in male germ cell tumors. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):11006–11010. doi: 10.1073/pnas.89.22.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlé I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boué J., Bertheas M. F., Mandel J. L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991 May 24;252(5009):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Pieretti M., Zhang F. P., Fu Y. H., Warren S. T., Oostra B. A., Caskey C. T., Nelson D. L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991 Aug 23;66(4):817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Rastinejad F., Blau H. M. Genetic complementation reveals a novel regulatory role for 3' untranslated regions in growth and differentiation. Cell. 1993 Mar 26;72(6):903–917. doi: 10.1016/0092-8674(93)90579-f. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Sachs A. B. Messenger RNA degradation in eukaryotes. Cell. 1993 Aug 13;74(3):413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer R. H. RNA zipcodes for cytoplasmic addresses. Curr Biol. 1993 Oct 1;3(10):719–721. doi: 10.1016/0960-9822(93)90079-4. [DOI] [PubMed] [Google Scholar]
- Siomi H., Choi M., Siomi M. C., Nussbaum R. L., Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994 Apr 8;77(1):33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Siomi H., Matunis M. J., Michael W. M., Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993 Mar 11;21(5):1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Siomi M. C., Nussbaum R. L., Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993 Jul 30;74(2):291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Sonenberg N. mRNA translation: influence of the 5' and 3' untranslated regions. Curr Opin Genet Dev. 1994 Apr;4(2):310–315. doi: 10.1016/s0959-437x(05)80059-0. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Swanson M. S., Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988 May;8(5):2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turc-Carel C., Dal Cin P., Boghosian L., Leong S. P., Sandberg A. A. Breakpoints in benign lipoma may be at 12q13 or 12q14. Cancer Genet Cytogenet. 1988 Nov;36(1):131–135. doi: 10.1016/0165-4608(88)90083-0. [DOI] [PubMed] [Google Scholar]
- Verheij C., Bakker C. E., de Graaff E., Keulemans J., Willemsen R., Verkerk A. J., Galjaard H., Reuser A. J., Hoogeveen A. T., Oostra B. A. Characterization and localization of the FMR-1 gene product associated with fragile X syndrome. Nature. 1993 Jun 24;363(6431):722–724. doi: 10.1038/363722a0. [DOI] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Verkerk A. J., de Graaff E., De Boulle K., Eichler E. E., Konecki D. S., Reyniers E., Manca A., Poustka A., Willems P. J., Nelson D. L. Alternative splicing in the fragile X gene FMR1. Hum Mol Genet. 1993 Apr;2(4):399–404. doi: 10.1093/hmg/2.4.399. [DOI] [PubMed] [Google Scholar]
- Wilson A. C., Carlson S. S., White T. J. Biochemical evolution. Annu Rev Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]
- Yu S., Pritchard M., Kremer E., Lynch M., Nancarrow J., Baker E., Holman K., Mulley J. C., Warren S. T., Schlessinger D. Fragile X genotype characterized by an unstable region of DNA. Science. 1991 May 24;252(5009):1179–1181. doi: 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]