Summary
Production of type I interferons (IFN-I) is a crucial innate immune mechanism against viral infections. IFN-I induction is subject to negative regulation by both viral and cellular factors, but the underlying mechanism remains unclear. We report that the noncanonical NF-κB pathway was stimulated along with innate-immune cell differentiation and viral infections and had a vital role in negatively regulating IFN-I induction. Genetic deficiencies in major components of the noncanonical NF-κB pathway caused IFN-I hyper-induction and rendered cells and mice substantially more resistant to viral infection. Noncanonical NF-κB suppressed signal-induced histone modifications at the Ifnb promoter, an action that involved attenuated recruitment of the transcription factor RelA and a histone demethylase, JMJD2A. These findings reveal an unexpected function of the noncanonical NF-κB pathway and highlight an important mechanism regulating antiviral innate immunity.
Keywords: NF-κB, noncanonical NF-κB, NIK, antiviral innate immunity, type I interferon
INTRODUCTION
Innate immunity serves as the first line of host defense against invading microorganisms. In response to an infection, germ-line-encoded pattern recognition receptors (PRRs), such as toll-like receptors (TLRs) and RIG-I–like receptors, detect pathogen-associated molecular patterns and rapidly induce the production of proinflammatory cytokines and type I interferons (IFN-Is) (Dunn et al., 2006; Kawai and Akira, 2006). The IFN-Is, including IFN-α and IFN-β, are produced in various cell types, such as macrophages, dendritic cells (DCs), and fibroblasts, and function as critical effector molecules in an innate immune response against viral infections (Stetson and Medzhitov, 2006). Under normal conditions, the induction of IFN-Is is under tight control, since their deregulated production may promote the development of immunological disorders under certain circumstances (Trinchieri, 2010). Moreover, viruses have mechanisms to negatively regulate IFN-I induction, thereby weakening the innate immune responses (Katze et al., 2002). However, the molecular mechanisms that negatively control IFN-I induction remain incompletely understood.
Induction of IFN-Is involves a signaling pathway leading to the activation of two homologous kinases, TBK1 and IKKε, and the subsequent activation of the transcription factors IFN-regulatory factor (IRF) 3 and IRF7 (Fitzgerald et al., 2003; Sharma et al., 2003). In addition, the IFN gene induction also requires NF-κB, which cooperates with IRF3 and IRF7 in the assembly of an enhanceosome in the IFN-β promoter and the recruitment of coactivators and chromatin-remodeling factors (Bartlett et al., 2012; Falvo et al., 2000; Panne et al., 2007; Wang et al., 2010).
NF-κB represents a family of transcription factors, including RelA, RelB, c-Rel, NF-κB1 p50, and NF-κB2 p52, which can be activated by canonical and noncanonical pathways (Hayden and Ghosh, 2008; Sun, 2012; Vallabhapurapu and Karin, 2009). The canonical NF-κB members, which typically include p50-RelA and p50-c-Rel heterodimers, are activated by various immune receptors and known to participate in the induction of IFN-Is and prolinflammatory cytokines (Hayden and Ghosh, 2008; Wang et al., 2010). In contrast, the noncanonical NF-κB, the p52-RelB dimer, is thought to respond to selective receptor signals that mediate adaptive immune functions, such as lymphoid organ development and B-cell survival (Sun, 2012). A central step in noncanonical NF-κB signaling is stabilization of the protein kinase NIK (Liao et al., 2004), which together with its downstream kinase IKKα, induce phosphorylation-dependent processing of p100, an NF-κB precursor protein that also functions as a cytoplasmic inhibitor of NF-κB (Senftleben et al., 2001; Sun, 2012; Xiao et al., 2001). Under normal conditions, the signaling function of NIK is suppressed through its constant degradation by a TRAF3-dependent mechanism that involves recruitment of NIK to the E3 ubiquitin ligase c-IAP1 or c-IAP2 (Liao et al., 2004; Vallabhapurapu et al., 2008; Zarnegar et al., 2008). Signal-induced noncanonical NF-κB activation involves ubiquitin-dependent degradation of TRAF3 and accumulation of NIK (Sun, 2012).
Although the noncanonical NF-κB pathway plays a crucial role in the regulation of lymphoid organ development and adaptive immunity, it is currently unclear whether this pathway also has a role in the regulation of innate immunity. In the present study, we discovered an unexpected role for the noncanonical NF-κB pathway in the negative regulation of IFN-I induction. Genetic deficiency in the signaling components of this pathway caused IFN-I hyper-induction, whereas the deregulated activation of noncanonical NF-κB contributed to the attenuated IFN-I induction. The noncanonical NF-κB regulated histone modifications at the Ifnb promoter, which involved attenuated recruitment of the canonical NF-κB RelA and a histone demethylase, JMJD2A, to the Ifnb promoter. These findings reveal an unexpected mechanism that controls type I IFN induction and establish a crucial regulatory role of the noncanonical NF-κB pathway in antiviral innate immunity.
RESULTS
NIK is a negative regulator of antiviral innate immunity
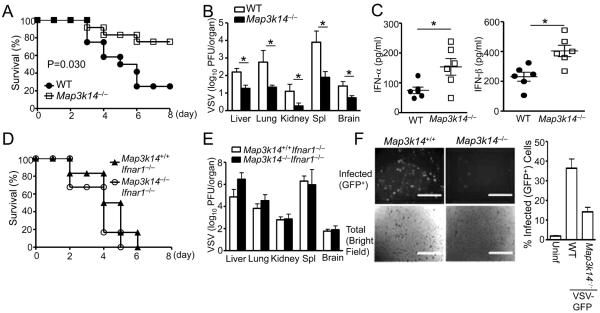
Since NIK is a central component of the noncanonical NF-κB pathway, we examined the role of NIK in the regulation of innate immunity against viral infections. We crossed the Map3k14−/− mice (deficient in the NIK-encoding gene Map3k14) onto Rag1−/−background to eliminate their adaptive cellular components, lymphocytes, and challenged them with a well-characterized viral pathogen, the vascular stomatitis virus (VSV). Interestingly, the Map3k14−/− mice were substantially more resistant to VSV infection than the Map3k14+/+ (wild-type, or WT) mice, as shown by increased survival rate (Figure 1A) and reduced viral load in different organs (Figure 1B). Moreover, the Map3k14−/− mice produced significantly more IFN-α and IFN-β (Figure 1C). The elevated IFN-I production contributed to the VSV-resistance phenotype of the Map3k14−/− mice, since the Map3k14−/− and WT mice displayed similar sensitivity to VSV infection when crossed to the Infar1−/−background (Figures 1D and 1E). In the Infar1−/− background, NIK deficiency promoted the production of IFN-β, but not IFN-α (Figure S1A). Since IFN-β-stimulated IFNAR1 signaling is known to mediate IFN-α induction (Asano et al., 1990; Marie et al., 1998), these results indicate that the effect of NIK deficiency on IFN-α induction may involve the autocrine action of IFN-β. To further confirm that NIK negatively regulates antiviral innate immunity, we performed in vitro experiments using control and Map3k14−/− mouse embryonic fibroblasts (MEFs). The Map3k14−/− MEFs were substantially more resistant to VSV infection in vitro than the WT MEFs (Figure 1F). These results suggest a negative role for NIK in regulating antiviral innate immunity.
Figure 1. NIK deficiency potentiates antiviral immunity.
(A–C) Age-matched (6–8 weeks old) WT and Map3k14−/− mice, bred to the Rag1−/− background, were infected i.v. with VSV (2×107 PFU per mouse). (A) Survival curves (n=12). (B) Tissue VSV titers on day 3 of infection. (C) Serum concentrations of IFN-α and IFN-β at 12 h of infection. Data are presented as mean ± S.D. of six animals.
(D–E) WT and Map3k14−/− mice, bred to the Rag1−/−-Ifnar1−/− background, were infected i.v. with VSV (2×107 PFU per mouse). (D) Survival curves (n=6). (E) Tissue VSV titers on day 3 of infection. Data are presented as mean ± S.D. of six animals.
(F) WT or Map3k14−/− primary MEFs were infected with GFP-expressing VSV (VSV-GFP) at a MOI of 0.1 for 24 hr. Data are presented as a representative picture, showing the infected (GFP+) and total (bright field) cells (left). The sacle bar indicated as length of 1000μm. The summary graph of flow cytometric quantification of the infected cells (right). Data are representative of 3 independent experiments, and statistical analyses represent variations in technical replicates. *P<0.05. See also Figure S1.
NIK negatively regulates IFN-I induction by viruses and TLR ligands
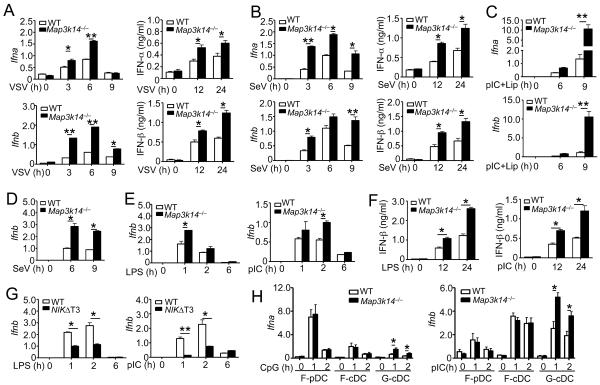
To examine whether NIK directly regulates IFN-I induction, we infected the WT and NIK-deficient MEFs in vitro using two different RNA viruses, VSV and Sendai virus (SeV). Compared to the WT MEFs, the NIK-deficient MEFs were hyperresponsive to both viruses in the induction of IFN-α and IFN-β at both the mRNA and protein levels (Figures 2A and 2B). NIK deficiency also promoted IFN-I induction by liposome-delivered poly(I:C), a synthetic double-stranded RNA known to stimulate the RIG-I signaling pathway when delivered into the cytoplasm by transfection (Figure 2C).
Figure 2. NIK negatively regulates IFN-I induction.
(A and B) WT or Map3k14−/− primary MEFs were infected with VSV (A) or SeV (B). The relative mRNA amount and protein concentration were determined by QPCR and ELISA, respectively. Data are presented as mean ± S.D.
(C) QPCR analysis of relative Ifna and Ifnb mRNA amounts in WT and Map3k14−/− MEF cells stimulated with Lipofectamine-transfected poly(I:C) (pIC) for the indicated time periods. (D–F) BMDMs derived from WT or Map3k14−/− mice were infected with SeV (D) or stimulated with LPS and poly(I:C) (E and F). Relative mRNA amounts (D and E) and protein concentration (F) of IFNβ were determined by QPCR and ELISA, respectively.
(G) QPCR analysis of Ifnb mRNA expression in WT and NIKΔT3 BMDMs stimulated with LPS or poly(I:C).
(H) QPCR analysis of Ifna and Ifnb mRNA expression in FLT3L-differentiated pDC (F-pDC) and cDC (F-cDC) cells and GM-CSF-differentiated cDC (G-cDC) cells stimulated as indicated. All QPCR data are presented as fold relative to the amount of an internal control, Actb. Data are representative of three-four independent experiments, and statistical analyses represent variations in technical replicates. *P<0.05 and **P<0.01. See also Figure S1.
Macrophages serve as an important cellular component in innate immunity and respond to various microbial components, such as viral RNA and bacterial lipopolysaccharides (LPS). We thus analyzed the effect of NIK deficiency on IFN-I induction in bone marrow derived macrophages (BMDMs). NIK deficiency did not influence BMDM differentiation (Figure S1B) but significantly promoted induction of Ifnb gene expression by SeV (Figure 2D) and VSV (Figure S1C). NIK deficiency also elevated IFN-I induction by ligands for TLR4 (LPS), TLR3 [poly(I:C)], TLR7 (R848), and TLR9 (CpG) (Figures 2E and 2F; Figure S1D). Furthermore, although IKKα has a positive role in IRF7-mediated IFN-α induction in pDCs (Hoshino et al., 2006), the loss of the IKKα-encoding gene Chuk in macrophages promoted IFN-β induction by LPS (Figure S1E). As a complementary approach, we employed a transgenic mouse expressing a stable form of NIK lacking its TRAF3-binding motif (NIKΔT3) (Sasaki et al., 2008). The induction of Ifnb gene expression by LPS and poly(I:C) as well as VSV was profoundly suppressed in the NIKΔT3-expressing BMDMs (Figure 2G and Figure S1F).
We next analyzed the role of NIK in IFN-I induction in DCs, including FLT3 ligand (FLT3L)-induced plasmacytoid DCs (pDCs) and conventional DCs (cDCs) as well as granulocyte macrophage-colony stimulating factor (GM-CSF)-induced cDCs. Interestingly, NIK deficiency did not promote IFN-α induction in pDCs or FLT3L-induced cDCs, but it resulted in hyper-induction of IFN-α and IFN-β in GM-CSF-differentiated cDCs (Figure 2H). Collectively, these results suggest that NIK functions as a potent negative regulator of IFN-I induction by both RNA viruses and TLRs, although this function was not seen in the FLT3L-induced DCs.
NIK regulates IFN-I induction via activation of noncanonical NF-κB
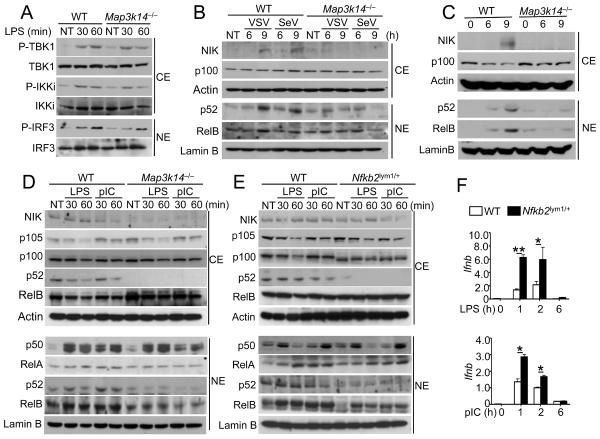
To understand how NIK negatively regulates IFN-I induction, we examined the effect of NIK deficiency on the activation of signaling factors involved in this innate immune response. The loss of NIK did not affect LPS-induced activation of TBK1 and IKKε, as assessed by phospho-specific immunoblotting (IB) assays (Figure 3A). The phosphorylation of IRF3 was also comparable between the WT and Map3k14−/− cells (Figure 3A). Similar results were obtained with poly(I:C)-stimulated cells (Figure S2A). NIK deficiency also did not influence the activation of MAP kinases (Figure S2B).
Figure 3. NIK-dependent noncanonical NF-κB signaling negatively regulates IFN-I induction.
(A) Immunoblot analysis of the indicated phosphorylated (P-) and total proteins in the cytoplasmic (CE) and nuclear (NE) extracts of WT and Map3k14−/− BMDMs stimulated with LPS.(B and C) Immunoblot analysis of the indicated proteins in the cytoplasmic (CE) and nuclear (NE) extracts of WT and Map3k14−/− MEFs infected with VSV or SeV (B) or stimulated with Lipofectamine-transfected poly(I:C) (C).
(D and E) Immunoblot analysis of the indicated proteins in the cytoplasmic (CE) and nuclear (NE) extracts of Map3k14−/− (D), Nfkb2lym1/+ (E) or their WT control BMDMs stimulated with LPS and poly(I:C) as indicated.
(F) QPCR analysis of Ifnb mRNA amounts (fold relative to the internal control Actb mRNA) of WT and Nfkb2lym1/+BMDMs stimulated with LPS and poly(I:C). *P<0.05 and **P<0.01. Data in all panels are representative of three independent experiments, and statistical analyses represent variations in technical replicates. *P<0.05 and **P<0.01. See also Figures S2 and S3.
NIK is a central mediator of the noncanonical NF-κB. Although the role of noncanonical NF-κB in innate immunity is unclear, a recent study indicates the induction of this pathway by inducers of RIG-I (Liu et al., 2008). We found that infection of MEFs with VSV or SeV led to the induction of NIK and nuclear expression of p52 and RelB (Figure 3B). Furthermore, NIK deficiency abrogated the production and nuclear translocation of p52 and partially inhibited the nuclear translocation of RelB. The virus-induced NIK upregulation was readily detected at 9h (Figure 3B). An immunoblot using excessive cell lysates also revealed the induction of NIK as early as 3 h after viral infection as well as a low basal amount of NIK in the WT MEFs (Figure S2C). Similarly, liposome-transfected poly(I:C) stimulated the NIK protein abundance and noncanonical NF-κB nuclear expression in WT MEFs, but not in Map3k14−/− MEFs (Figure 3C). These molecular events were associated with degradation of the NIK inhibitor TRAF3 (Figure S2D). Thus, like the other known noncanonical NF-κB stimuli, such as BAFF, lymphotoxin, and CD40 ligand, RNA viruses stimulate the NIK pathway through induction of TRAF3 degradation. The relative slow kinetics of noncanonical NF-κB induction by viruses and liposome-delivered poly(I:C) (Figures 3B and 3C) was correlated with that of IFN-I induction (Figures 2A–2C). Furthermore, NIK deficiency in MEFs did not promote virus-induced phosphorylation of TBK1 or IRF3 (Figure S2E).
Since TLR ligands could induce rapid IFN-I expression in macrophages, we were curious about their ability to stimulate noncanonical NF-κB signaling under such conditions. Neither LPS nor poly(I:C) induced the accumulation of NIK or generation of p52 (Figure 3D). Interestingly, however, macrophages had basal amounts of NIK expression and p52 production, and the production of p52 was blocked in the Map3k14−/− macrophages (Figure 3D). Furthermore, p52 was detected in both the cytoplasm and the nucleus of the WT cells, and the nuclear translocation of p52 was further induced upon LPS and poly(I:C) stimulation (Figure 3D). Although the expression of RelB was comparable in the Map3k14−/− macrophages, its nuclear translocation was reduced in these mutant cells (Figure 3D). In contrast, the induction of canonical NF-κB members, p50 and RelA, was not influenced by NIK deficiency (Figure 3D). These results suggest that p100 processing may be stimulated during macrophage differentiation, thereby contributing to the negative regulation of TLR-stimulated IFN-I expression.
To further examine the role of p100 processing in IFN-I gene induction, we employed a mouse strain carrying an Nfkb2 gene mutation (called lym1), which causes the production of a nonprocessible form of p100 due to the loss of its C-terminal phosphorylation site (Tucker et al., 2007). Both homozygous Nfkb2lym1/lym1 and heterozygous (Nfkb2lym1/+) mice have severe defect in p52 generation and immune functions (Tucker et al., 2007). Since the efficiency of breeding Nfkb2lym1/lym1mice was low, we used the Nfkb2lym1/+heterozygous BMDMs in the experiments. Like NIK deficiency, the Nfkb2 lym1 mutation did not appreciably influence the differentiation of macrophages (data not shown). We found that p52 was produced in the WT macrophages but barely in the Nfkb2lym1/+macrophages (Figure 3E). The nuclear translocation of RelB was also attenuated in the Nfkb2lym1/+cells (Figure 3E). The activation of canonical NF-κB members, p50 and RelA, was either not affected or only weakly reduced in the Map3k14−/− or Nfkb2lym1/+ macrophages (Figures 3D and 3E). Moreover, the defect in noncanonical NF-κB activation in the Nfkb2lym1/+macrophages was associated with a striking increase in the induction of Ifnb gene expression (Figure 3F). Consistent with the macrophage results, the Nfkb2lym1/lym1MEFs were hyper-responsive to stimulation by liposome-transfected poly(I:C) in IFN-I gene induction (Figure S3). We also found that preincubation of WT MEFs, but not Nfkb2lym1/lym1MEFs, with a non-canonical NF-kB inducer (anti-LTβR) significantly suppressed IFN-I gene induction (Figure S3). These results further confirm a crucial role for the noncanonical NF-κB signaling pathway in the negative control of IFN-I gene induction.
Hematopoietic cytokines stimulate noncanonical NF-κB along with innate immune cell differentiation
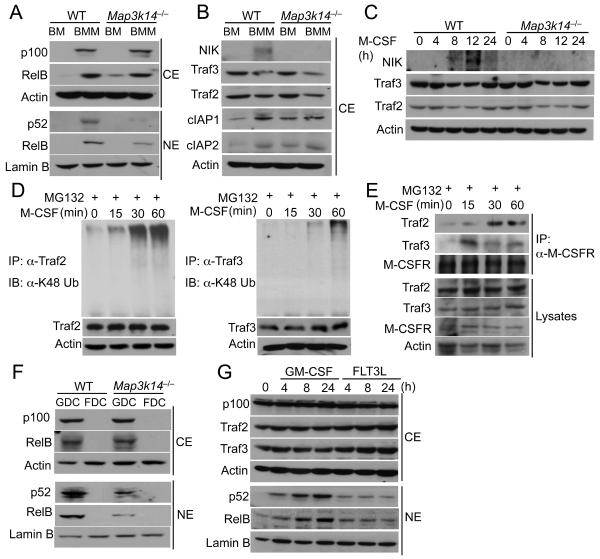
The finding that NIK controlled the production of p52 in macrophages suggested an intriguing possibility that NIK might regulate noncanonical NF-κB signaling during macrophage differentiation. Since macrophage colony stimulating factor (M-CSF) receptor (M-CSFR) is the primary receptor that mediates macrophage differentiation from bone marrow cells (Inaba et al., 1993), we examined the possible role of M-CSFR in the induction of noncanonical NF-κB signaling. Cultivation of bone marrow in M-CSF-containing medium led to the potent induction of p100 and RelB protein amount (Figure 4A). M-CSF also induced the nuclear translocation of both p52 and RelB. Furthermore, the induction of p52 and, to a lesser extent RelB, was dependent on NIK (Figure 4A). This result suggested a role for M-CSF in the induction of noncanonical NF-κB signaling along with the macrophage differentiation. Indeed, the M-CSF-induced BMDM differentiation was also associated with accumulation of NIK and reduction in the NIK negative regulators TRAF2 and TRAF3 (Figure 4B). The E3 ubiquitin ligases cIAP1 and cIAP2, which are involved in the degradation of TRAF2 and TRAF3 (Vallabhapurapu et al., 2008), were also induced (Figure 4B).
Figure 4. M-CSF induces noncanonical NF-κB signaling.
(A and B) Immunoblot analysis of the indicated proteins in the cytoplasmic (CE) and nuclear (NE) extracts of bone marrow (BM) cells or M-CSF-induced BMDMs (BMM) from WT and Map3k14−/− mice.
(C) Immunoblot analysis of the indicated proteins in whole-cell lysates of WT or Map3k14−/− bone marrow cells stimulated with M-CSF for the indicated time points.
(D) Bone marrow cells were stimulated with M-CSF in the presence of the proteasome inhibitor MG132. TRAF2 and TRAF3 were immunoprecipitated from denatured cell lysates, and ubiquitinated TRAF2 and TRAF3 was detected by immunoblot using an antibody detecting K48-linked polyubiquitin chains.
(E) WT bone marrow cells were stimulated with M-CSF in the presence of the proteasome inhibitor MG132. Whole-cell lysates were subjected to M-CSFR IP followed by detecting MCSFR-associated TRAF2 and TRAF3 by IB (top two panels). Cell lysates were also directly subjected to direct IB (lower four panels).
(F) Immunoblot analysis of the indicated proteins in the cytoplasmic (CE) and nuclear (NE) extracts of FLT3L or GM-CSF generated dendritc cells from WT and Map3k14−/− mice. (G) D2SC/1 dendritic cell line constituted with FLT3 (D2SC/1-FLT3) was stimulated with GM-CSF or FLT3L as indicated and subjected to immunoblot analyses. Data in all panels are representative of two-three independent experiments. See also Figure S4.
To further examine the capability of M-CSFR to induce noncanonical NF-κB activation, we stimulated bone marrow cells with M-CSF in a time course and analyzed the degradation of TRAF2 and TRAF3 and accumulation of NIK, crucial events of noncanonical NF-κB signaling (Sun, 2012). M-CSF stimulated gradual loss of both TRAF2 and TRAF3 and accumulation of NIK (Figure 4C). These signaling events peaked around 12 h and reversed by 24 h following M-CSF stimulation (Figure 4C). However, more persistent signaling could be achieved upon replacement of medium with newly added M-CSF (Figures S4A and S4B). Parallel experiments revealed that M-CSF stimulated conjugation of K48-linked polyubiquitination chains to TRAF2 and TRAF3 (Figure 4D). CD40 ligand-induced degradation of TRAF2 and TRAF3 is thought to involve their recruitment to the signaling receptor CD40, along with the E3 ubiquitin ligases c-IAP1 and c-IAP2 (Vallabhapurapu et al., 2008). Similarly, M-CSFR also recruited TRAF2 and TRAF3 (Figure 4E) as well as c-IAPs (Figure S4C) upon M-CSF stimulation. In addition, the MCSF-stimulated degradation of TRAF2 and TRAF3 was blocked by a small molecule c-IAP inhibitor, Smac (Figure S4D). These results suggest that M-CSFR mediates noncanonical NF-κB signaling via induction of ubiquitin-dependent degradation of TRAF2 and TRAF3 and accumulation of NIK.
We next examined the noncanonical NF-κB activation along with DC differentiation driven by GM-CSF and FLT3L. Similar to M-CSF, GM-CSF stimulated NIK-dependent activation of p52 and RelB during DC differentiation (Figure 4F). In sharp contrast, FLT3L-mediated DC differentiation did not stimulate noncanonical NF-κB activation (Figure 4F). To further confirm this finding, we examined the noncanonical NF-κB activation by GM-CSF and FLT3L in a DC cell line stably expressing murine FLT3 (D2SC-mFLT3). As seen in the primary DCs, GM-CSF, but not FLT3L, stimulated the nuclear translocation of p52 and RelB (Figure 4G). GM-CSF also induced partial loss of TRAF3 (Figure 4G), although it was less profound than that stimulated by M-CSF (Figure 4C). These findings suggest that GM-CSF, but not FLT3L, stimulates the noncanonical NF-κB signaling, thus explaining the different function of NIK in DCs differentiated by GM-CSF and FLT3L (Figure 2H).
Noncanonical NF-κB suppresses IFN-I gene induction
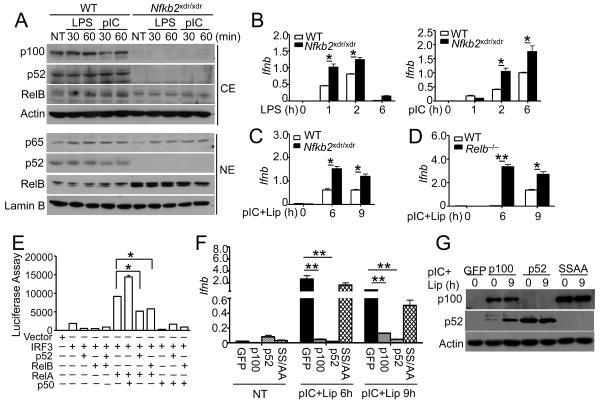
The data described above strongly suggest the involvement of noncanonical NF-κB members in the negative regulation of IFN-I gene induction. We further examined this possibility using BMDMs derived from a mutant mouse (Nfkb2xdr) deficient in NF-κB2 expression due to a splicing-disruptive Nfkb2 gene mutation (Miosge et al., 2002). Consistent with the previous study (Miosge et al., 2002), BMDMs from homozygous Nfkb2xdr/xdr mice lack both p100 and p52 (Figure 5A). Unlike the Map3k14−/− and Nfkb2lym1/+BMDMs, the Nfkb2xdr/xdrBMDMs did not have attenuated, but rather had elevated, nuclear translocation of RelB, which was apparently due to the loss of the RelB inhibitor p100 (Figure 5A). The Nfkb2xdr/xdrmacrophages also had competent nuclear translocation of the canonical NF-κB RelA (Figure 5A). Thus, these mutant cells allowed us to specifically address the role of p52 in the regulation of Ifnb gene induction. Despite their high amounts of nuclear RelB, the Nfkb2xdr/xdrmacrophages were hyperresponsive to LPS and poly(I:C) in the induction of Ifnb gene expression (Figure 5B). Similar results were obtained by stimulating Nfkb2xdr/xdr MEFs using the RIG-I stimulator, liposome-delivered poly(I:C) (Figure 5C). Parallel experiments revealed that loss of RelB also caused hyper-induction of Ifnb gene expression (Figure 5D). These findings suggest that negative regulation of IFN-I gene induction requires both of the noncanonical NF-κB members, p52 and RelB.
Figure 5. Noncanonical NF-κB members suppress IFN-I induction.
(A) Immunoblot analysis of the indicated proteins in the cytoplasmic (CE) and nuclear (NE) extracts of WT or Nfkb2xdr/xdr BMDMs stimulated with LPS and poly(I:C).
(B) QPCR analysis of relative amounts of Ifnb mRNAs (fold of the internal control Actb mRNA) in the WT and Nfkb2xdr/xdr BMDMs stimulated with LPS and poly(I:C) (pIC).
(C and D) QPCR analysis of relative amounts of Ifnb mRNA in xdr/xdr and WT control MEFs (C) or Relb−/− and WT control MEFs (D) stimulated with Lipofectamine-transfected poly(I:C).
(E) HEK293 cells were transfected with an Ifnb-luciferase reporter plasmid in the presence (+) or absence (−) of the indicated empty vector or expression plasmids. Luciferase assays were performed as fold based on empty vector group 36 h after transfection.
(F and G) Nfkb2xdr/xdr MEFs were reconstituted with GFP, p100, p52 and p100SSAA. The cells were either not treated (NT) or stimulated with Lipofectamine-transfected poly(I:C) and subjected QPCR (F) or IB (G) analysis. Data in all panels are representative of 2–3 independent experiments, and statistical analyses represent variations in technical replicates. *P<0.05 and **P<0.01.
To further examine the role of noncanonical NF-κB in the regulation of Ifnb gene expression, we examined the effect of different NF-κB members on the Ifnb promoter activity using luciferase reporter gene assays in transiently transfected HEK293 cells. As expected, the canonical NF-κB member RelA potently synergized with the IRF3 in the induction of Ifnb promoter (Figure 5E). Expression of p50 further promoted the induction of Ifnb promoter. In contrast, expression of p52 or RelB led to the inhibition of RelA-IRF3-stimulated Ifnb promoter activation (Figure 5E). We also examined whether overexpressed p52 or p100 inhibited the induction of endogenous Ifnb gene expression. Interestingly, reconstitution of the Nfkb2xdr/xdrMEFs with either p52 or p100 led to efficient inhibition of Ifnb induction (Figure 5F). Furthermore, expression of a processing-defective p100 mutant, p100SSAA (Xiao et al., 2001), did not appreciably inhibit the Ifnb gene induction (Figure 5F). Parallel immunoblot analyses revealed comparable amounts of expression of p100, p52 and p100SSAA (Figure 5G). These data further emphasize a negative role for the noncanonical NF-κB members in the regulation of Ifnb gene induction.
Noncanonical NF-κB suppresses the binding of RelA to the Ifnb promoter
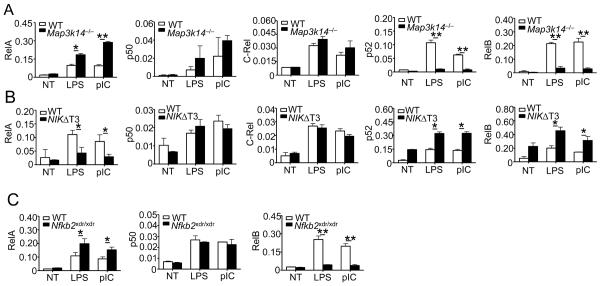
Signal-induced Ifnb gene expression requires recruitment of a number of transcription factors, including IRF3 and canonical NF-κB, to the positive regulatory domains (PRD) of the Ifnb promoter (Bartlett et al., 2012; Falvo et al., 2000; Panne et al., 2007; Wang et al., 2010). To elucidate the mechanism by which noncanonical NF-κB suppresses Ifnb gene induction, we examined the effect of noncanonical NF-κB deficiency on the recruitment of IRF3 and canonical NF-κB members to the Ifnb promoter. Chromatin immunoprecipitation (ChIP) assays revealed that NIK deficiency or the Nfkb2 lym1 mutation did not affect the recruitment of IRF3 to the Ifnb promoter following LPS or poly(I:C) stimulation (Figure S5A). In contrast, these genetic alterations in the noncanonical NF-κB pathway potently enhanced the recruitment of the canonical NF-κB RelA to the Ifnb promoter (Figure 6A and Figures S5B and S5C). Similar results were obtained with MEFs stimulated with liposome-transfected poly(I:C) (Figure S5D). On the other hand, the NIK deficiency and Nfkb2 lym1 mutation had little or no effect on the binding of the other canonical NF-κB subunits, p50 and c-Rel (Figure 6A and Figure S5C). We found that the noncanonical NF-κB members, p52 and RelB, were also recruited to the Ifnb promoter in response to LPS and poly(I:C) stimulation, which was greatly attenuated in the NIK-deficient and Nfkb2lym1/+cells (Figure 6A and Figure S5C). In line with these findings, the BMDMs derived from the NIKΔT3 transgenic mice, which overexpressed a stabilized form of NIK, NIKΔT3, had increased recruitment of p52 and RelB and decreased recruitment of RelA to the Ifnb promoter (Figure 6B and Figure S5B). These findings have important implications, since RelA has been implicated as an important transactivator of IFN-I (Perez de Diego et al., 2010; Wang et al., 2010). Indeed, the RelA deficiency in MEFs reduced the induction of Ifna and Ifnb genes (Figure S5E).
Figure 6. RelA physically interacts with JMJD2A and binds to Ifnb promoter in a manner that is suppressed by noncanonical NF-κB.
(A–C) BMDMs prepared from Map3k14−/− (A), NIKΔT3 transgenic (B), or Nfkb2xdr/xdr (C) mice and their WT littermate controls were stimulated for 1 h with LPS or poly(I:C). ChIP assays were performed and quantified by QPCR to detect the binding of NF-κB family members to the Ifnb promoter. Data are presented as percentage of the total input DNA.
Data are representative of two-three independent experiments, and statistical analyses represent variations in technical replicates. *P<0.05 and **P<0.01. See also Figure S5.
The results described above indicated that the noncanonical NF-κB members might inhibit the binding of RelA to the Ifnb promoter. We further examined this possibility by performing EMSA to test whether p52 and RelB inhibits the binding of RelA to the Ifnb NF-κB-binding site. When co-expressed in HEK293 cells, p52 and RelB dose-responsively suppressed the binding of RelA to the Ifnb κB probe (Figure S5F). Moreover, this experiment also revealed that the p52-RelB dimer binds to the Ifnb κB probe more strongly than RelA (Figure S5F). As a complementary approach, we examined whether loss of p52 promoted the binding of RelA to the Ifnb promoter by ChIP assays. Indeed, the p52 deficiency in the Nfkb2xdr/xdmacrophages significantly promoted the binding of RelA to the Ifnb promoter (Figure 6C). Interestingly, the p52 deficiency also attenuated the binding of RelB to the Ifnb promoter (Figure 6C), a finding that was consistent with the requirement of partner proteins for the DNA-binding function of RelB (Fusco et al., 2008). These results indicate that the noncanonical NF-κB members may compete with RelA for binding to the κB site at the Ifnb promoter.
Noncanonical NF-κB appears to regulate histone modifications in the Ifnb promoter
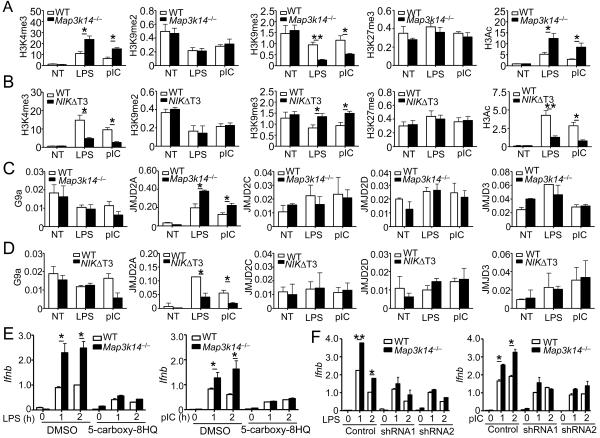
Histone modifications represent a critical epigenetic mechanism that regulates gene transcription (Berger, 2002). Some histone modifications, including dimethylation of H3K9 (H3K9me2), trimethylation of H3K9 (H3K9me3), and trimethylation of H3K27 (H3K27me3), are associated with gene repression, whereas other modifications, such as H3K4me3 and H3 acetylation (H3Ac), are associated with active chromatin structures (Cedar and Bergman, 2009). To obtain more insight into the mechanism by which noncanonical NF-κB suppresses IFN-I induction, we performed ChIP assays to examine histone modifications in the Ifnb promoter. Stimulation of WT macrophages with LPS and poly(I:C) led to the induction of H3K4me3 and H3Ac, coupled with reduction in H3K9me2 and H3K9me3, at the Ifnb promoter (Figure 7A and Figure S6A). The abundance of H3K27me3 was also moderately increased. Similar amounts of H3K9me2 and H3K27me3 were detected in the WT and NIK-deficient macrophages (Figure 7A). Interestingly, NIK deficiency profoundly promoted the loss of H3K9me3 following stimulation with LPS or poly(I:C) (Figure 7A and Figure S6A). In the contrary, the NIK-deficient macrophages had a significant increase in the activating type of histone modifications H3Ac and H3K4me3. Similar results were obtained with the LPS- and poly(I:C)-stimulated Nfkb2lym1/+macrophages (Figure S4B) as well as Map3k14−/− MEFs stimulated by liposome-transfected poly(I:C) (Figure S6C).
Figure 7. NIK regulates histone modifications and JMJD2A binding at the Ifnb promoter.
(A–D) BMDMs derived from Map3k14−/−, NIKΔT3 transgenic, or their WT control mice were stimulated with LPS or poly(I:C) for 1h. ChIP assays were performed to detect histone modifications (A and B) and binding of the indicated factors (C and D) at the Ifnb promoter. The Y axis is percentage (%) based on total H3 for A–B, and percentage (%) based on total input DNA for C–D. Data are representative of three independent experiments, and statistical analyses represent variations in technical replicates.
(E) WT and Map3k14−/− BMDMs were pretreated for 12 h with 20 mM of a JMJD2 inhibitor (5-carboxy-8HQ) and then stimulated with LPS or poly(I:C) as indicated. The relative amount of Ifnb mRNAs were quantified by QPCR and presented as fold relative to the internal Actb mRNA control.
(F) WT and Map3k14−/− BMDMs were infected with two different JMJD2A shRNAs or a non-silencing control shRNA and then stimulated with LPS and poly(I:C) as indicated. The relative amount of Ifnb mRNAs were quantified by QPCR and presented as fold relative to the internal Actb mRNA control. Data in all panels are representative of two-three independent experiments, and statistical analyses represent variations in technical replicates. *P<0.05 and **P<0.01. See also Figures S6 and S7
We next employed the NIKΔT3 transgenic mice to examine the effect of NIK overexpression on the histone modifications in the Ifnb promoter. The NIK overexpression had little or no effect on the amount of H3K9me2 and H3K27me3 but significantly attenuated the loss of H3K9me3 and induction of H3Ac in cells treated with LPS and poly(I:C) (Figure 7B and Figure S6D). The NIK overexpression also attenuated poly(I:C)-mediated induction of H3K4me3 (Figure 7B and Figure S6D). These results indicate that NIK and the noncanonical NF-κB pathway possibly regulate histone modifications, particularly H3K9me3 and H3Ac, at the Ifnb promoter.
JMJD2A engages the Ifnb promoter in a NIK-regulated manner
Histone methylations are conversely regulated by lysine methyltransferases and lysine demethylases (Black et al., 2012). In particular, the lysine methyltransferase G9a and the Jumonji domain 2 (JMJD2) family of lysine demethylases play an important role in regulating the methylation status of H3K9 (Gray et al., 2005; Tachibana et al., 2001; Whetstine et al., 2006). Our finding that the noncanonical NF-κB pathway inhibited the TLR-stimulated erase of H3K9me3 at the Ifnb promoter indicated a role for this pathway in regulating the recruitment of a lysine methyltransferase or a lysine demethylase. We found that the WT and Map3k14−/− macrophages had a similar amount of G9a binding to the Ifnb promoter (Figure 7C). Interestingly, e NIK deficiency significantly promoted the recruitment of JMJD2A, although not the other JMJD2 members, to the Ifnb promoter (Figure 7C and Figure S6E). Similar results were obtained with Map3k14−/− MEFs (Figure S6C). Consistently, NIK overexpression in the NIKΔT3 transgenic macrophages inhibited the TLR-stimulated binding of JMJD2A to the Ifnb promoter (Figures 7D and S6E).
In searching for potential mechanism by which NIK regulates the recruitment of JMJD2A to the Ifnb promoter, we found that RelA and JMJD2A physically interacted under both transfection (Figure S7A) and endogenous conditions (Figure S7B), the latter of which occurred in response to stimulation by LPS or poly(I:C). Under the same conditions, several other NF-κB members did not bind JMJD2A (Figure S7C). Furthermore, RelA deficiency attenuated the engagement of JMJD2A to the Ifnb promoter and inhibited the erase of H3K9me3 in cells stimulated with lipofectamine-delivered poly(I:C) (Figure S7D). These results suggest a possibility that RelA may recruit JMJD2A to the Ifnb promoter, although this idea is still quite preliminary and requires additional explorations. Nevertheless, JMJD2A appeared to have an important role in mediating IFN-I induction, since JMJD2 inhibition by a pharmacological inhibitor, 5-carboxy-8HQ (Figure 7E), or JMJD2A silencing by shRNA (Figure S7E and Figure 7F) inhibited the induction of Ifnb gene expression in both the WT and Map3k14−/− macrophages. The JMJD2A silencing also prevented the LPS- and poly(I:C)-stimulated downregulation of H3K9me3 at the Ifnb promoter (Figure S7F). Similar results were obtained with JMJD2A-knockdown MEFs (Figures S7G and S7H). Furthermore, reconstitution of the JMJD2Asilenced cells with an RNAi-resistant form of JMJD2A, but not a catalytically inactive JMJE2A mutant (H188A), restores e IFN-I induction (Figures S7G and S7H). These results indicate that noncanonical NF-κB may regulate the recruitment of JMJD2A to the Ifnb promoter and modulate histone modifications, which contribute to the negative regulation of IFN-I induction.
DISCUSSION
The noncanonical NF-κB pathway has so far been linked to the regulation of specific adaptive immune functions, such as lymphoid organogenesis and B-cell survival. However, emerging evidence suggests that this pathway is also activated along with viral infection, although the functional significance in antiviral immunity has remained unclear (Manches et al., 2012; Ruckle et al., 2012). The data presented in this study demonstrated an unexpected role for the noncanonical NF-κB pathway in the regulation of antiviral innate immunity. We obtained genetic evidence that this signaling pathway controls the magnitude of IFN-I induction by RNA viruses and TLR ligands.
We found that both VSV and SeV could stimulate the activation of noncanonical NF-κB pathway. While MEFs had a very low basal amount of NIK, the infection with VSV or SeV led to profound upregulation of NIK. These RNA viruses induced the degradation of the NIK inhibitor, TRAF3, coupled with accumulation of NIK and induction of p100 processing. The virus-induced NIK upregulation and noncanonical NF-κB activation likely contribute to the suppression of IFN-I induction, as suggested by results obtained with different MEFs lacking NIK or p52. Infection of a lung epithelial cell line with the respiratory syncytial virus also induces NIK expression and p52 production (Choudhary et al., 2005). Although precisely how viruses stimulate the NIK pathway remains unclear, the RNA helicase RIG-I appears to be involved (Liu et al., 2008). Consistently, we found that lipofectamine-delivered poly(I:C), a known stimulator of RIG-I, induced NIK stabilization and p100 processing. Furthermore, both viral infections and poly(I:C) transfection induced TRAF3 degradation, although the underlying mechanism is incompletely understood. The c-IAP1 and c-IAP2 E3 ubiquitin ligases are known to mediate TRAF3 ubiquitination and degradation stimulated by the typical noncanonical NF-κB inducers LTβR and CD40 (Hacker et al., 2011). The c-IAPs have also been shown to mediate RIG-I- and virus-induced TRAF3 ubiquitination (Mao et al., 2010), although another E3 ligase, Triad3A, has been suggested to mediate the degradation of TRAF3 in virus-infected cells (Nakhaei et al., 2009). It remains to be examined whether these E3 ubiquitin ligases mediate virus-induced NIK induction and p100 processing.
Compared to MEFs, macrophages had a much higher basal amount of NIK, which explains why the NIK deficiency in macrophages promotes the rapid induction of IFN-I by TLR ligands. We discovered a unexpected pathway of noncanonical NF-κB signaling triggered by M-CSF, a cytokine that mediates the differentiation and growth of macrophages. M-CSF stimulated the recruitment of TRAF2 and TRAF3, as well as the E3 ubiquitin ligases c-IAP1 and c-IAP2, to its receptor, M-CSFR, which was associated with K48 ubiquitination and degradation of these TRAF molecules and concomitant accumulation of NIK. Thus, as implicated for CD40 (Vallabhapurapu et al., 2008), M-CSFR appeared to mediate the activation of noncanonical NF-κB signaling via destruction of TRAF2 and TRAF3 in the receptor complex. Like M-CSF, GM-CSF stimulated the activation of noncanonical NF-κB, which appeared to also involve degradation of TRAF3. Consistently, NIK and the nocanonical NF-κB pathway also inhibit IFNI-induction in GM-CSF-differentiated DCs. On the other hand, loss of NIK did not enhance IFNI-induction in FLT3L-differentiated DCs. This was correlated with the inability of FLT3L to induce noncanonical NF-κB activation.
TBK1 and IKKε are known as innate immune regulators that mediate induction of IFNs in response to TLR stimulation and viral infection(Fitzgerald et al., 2003; Sharma et al., 2003). Our data revealed that the noncanonical NF-κB deficiency did not promote the activation of TBK1 and IKKε or their downstream transcription factor IRF3. Instead, our data suggest a crosstalk between the canonical and noncanonical NF-κB pathways, in which the noncanonical NF-κB appeared to repress the binding of the canonical NF-κB RelA to the Ifnb promoter. This mechanism of noncanonical NF-κB function was demonstrated by both EMSA and ChIP assays as well as by luciferase reporter assays.
Our data also suggest a role for the noncanonical NF-κB pathway in the regulation of histone modifications at the Ifnb gene promoter, although this line of studies is somewhat preliminary and requires additional experiments for understanding the underlying mechanism. We found that genetic deficiencies in NIK or the noncanonical NF-κB p52 promoted the induction of the transcriptionally active histone mark H3Ac and the erase of the repressive mark H3K9me3. Since the acetylation and methylation at the lysines of histones are mutually exclusive (Wang et al., 2008), it is possible that the upregulation of H3Ac in Map3k14−/− and Nfkb2llym1/+ macrophages was a result of downregulation of methylation. The H3K9me3 downregulation in the mutant macrophages was associated with enhanced recruitment of a lysine demethylase, JMJD2A. To date, the function of JMJD2A has been mainly studied in cancer cell lines and linked to cancer cell growth, whereas its role in the regulation of immunity has remained unclear. Our data indicate a role for JMJD2A in the regulation of IFN-I in macrophages. One intriguing possibility is that RelA binds to JMJD2A and recruits this demethylase to the Ifnb promoter. Our data indeed suggest the physical interaction between RelA and JMJD2A, although additional studies are required to examine whether this interaction contributes to the histone modification at the Ifnb promoter.
In summary, we have identified the noncanonical NF-κB pathway as an important mechanism that controls IFN-I induction and antiviral immunity. Our findings provide functional insight into the induction of this signaling pathway along with viral infection. We have also provided genetic evidence that the noncanonical NF-κB pathway is stimulated along with innate immune cell differentiation, providing an example for how a differentiation-associated signaling event programs cells for the regulation of innate immunity. Our studies have also identified a potential function for the noncanonical NF-κB in regulating histone modifications in the Ifnb promoter. These findings not only provide important insight into the mechanism regulating IFN-I induction but also assigned a crucial function of the noncanonical NF-κB pathway.
EXPERIMENTAL PROCEDURES
Mice
Nfκb2Lym1 mutant mice were provided by Dr. Robyn Starr and The Water and Eliza Hall Institute of Medical Research. The nfkb2xdr/xdr mice were provided by The Australian National University and are available from the Australian Phenome Bank (ID #93). NIKΔT3-floxed mice (Jackson Lab) were crossed with lysozyme 2 Cre transgenic mice (Lyz2-Cre, Jackson Lab) to produce age-matched NIKΔT3-lyz2+/+ (termed WT) and NIKΔT3fl/fllyz2Cre/+ (termed NIKΔT3) mice for experiments. The Map3k14−/− mice were provided by Amgen Inc and the Relb−/− mice were from Bristol-Myers Squibb Pharmaceutical Research Institute. In some experiments, Map3k14−/− mice were further crossed to the Rag1−/− or Rag1−/−-Ifnar1−/− background. The mice were maintained in specific pathogen-free facility of The University of Texas MD Anderson Cancer Center, and all animal experiments were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee.
Viral Infection
For gene induction and signaling analyses, MEFs were seeded into 12-well plates (3.75 × 105 cells per well) and infected with VSV-AV1 mutant or SeV strains in serum-free medium for 1 h. The cells were washed once and cultured in growth medium for the indicated time periods and then collected for immunoblot or QPCR assays, and the supernatant were used for ELISA. To determine the infection efficiency, MEFs were infected with VSV-GFP for 12 h, and the infected (GFP+) cells were visualized under a fluorescence microscope and quantified by flow cytometry. For mouse infection, age-matched (6–8 old) mutant and WT control mice were housed in microisolater cages in a biosafety level 2 facility and infected i.v. with VSV (2×107 PFU pre mouse in 250 μl). The infected mice were monitored for lethality for up to 8 days or, where indicated, bled for collecting blood or perfused using sterile phosphate-buffered saline (PBS) for collecting tissues. The sera were used for measuring IFNs by ELISA, and the tissues were homogenized for determining the VSV titers in the supernatants using BHK cells.
Flow cytometry and cell sorting
Flow cytometry and cell sorting were performed using FACSAria (BD Bioscience). The fluorescence-labeled antibodies that were used included PE-conjugated anti-F4/80 (eBioscience, BM8) and anti-B220 (BD Bioscience, RA3-6B2) and APC-conjugated anti-CD11b (eBioscience, M1/70).
Cells and stimulation
Bone marrows were prepared from the femurs of adult mice and cultured in a MCSF-conditional medium for BMDM differentiation as previously described(Waterfield et al., 2003). To prepare FL3L-induced DCs (called F-DCs), we cultured the bone marrow cells in RPMI medium supplemented with FLT3L (50 ng/ml) for 7 days and then purified the F-cDC and F-pDC by flow cytometric cell sorting based on specific surface markers, CD11c+CD11b+B220− for F-cDCs and CD11c+CD11b−B220+ for F-pDCs. GM-CSF-induced DCs were prepared using the same procedure, except that the cells were cultured in the presence of GM-CSF (5 ng/ml). To prepare primary MEFs, we bred heterozygous mice for obtaining the KO and WT embryos from the same pregnant female mice(Chang et al., 2011). Immortalized RelA-KO MEFs were provided by Drs. Lin-Feng Chen and Paul Chiao. All MEFs, except the RelA-KO MEFs, were used as primary cell culture. The MEFs, BMDMs, and DCs were starved overnight in medium supplemented with 0.5% FCS before being stimulated with LPS (1 μg/ml for immunoblot experiments and 100 ng/ml for cytokine induction experiments), poly(I:C) (20 μg/ml), Lipofectamine-transfected poly(I:C) (20μg/ml), CpG-A (25 mM), or R848 (1 μg/ml) for the indicated times. Total and subcellular extracts were prepared for immunoblot assays, and total RNA was prepared for QPCR assays.
shRNA-mediated gene silencing
The pGIPZ lentiral vectors encoding shRNAs for NIK, JMJD2A, or a nonsilencing control shRNA were obtained from Open Biosystems, and the lentiral vector pLKO.1 encoding NIK shRNAs or the control luciferase shRNA were from Sigma. To produce lentiviral particles, the lentiviral vectors were transfected into HEK293T cells (using calcium method) along with the packaging vectors psPAX2 and pMD2 (Chang et al., 2011). MEFs or the dendritic cell line D2SC/1 were infected with the lentiviruses and selected with puromycin (for pLKO.1 and pGIPZ vectors) or by flow cytometric cell sorting based on GFP (for pGIPZ; pGIPZ carries both the puromycin-resistance gene and the GFP gene).
Real-time quantitative RT-PCR (QPCR)
Total RNA was isolated using TRI reagent (Molecular Research Center, Inc.) and subjected to cDNA synthesis using RNase H-reverse transcriptase (Invitrogen) and oligo (dT) primers. QPCR was performed in triplicates, using iCycler Sequence Detection System (Bio-Rad) and iQTM SYBR Green Supermix (Bio-Rad). The expression of individual genes was calculated by a standard curve method and normalized to the expression of Actb. The gene-specific PCR primers (all for mouse genes) are shown in Table S1.
Chromatin immunoprecipitation (ChIP) assays
BMDMs (6×107) were stimulated for 1 h with LPS or poly(I:C), or MEFs were stimulated with poly(I:C) plus lipofectamine for 9 h. And then these cells were fixed with 1% formaldehyde and sonicated as previously described(Nelson et al., 2006). Lysates (from 2×107 cells in 3 ml) were subjected to immunoprecipitation with the indicated antibodies, and the precipitated DNA was then purified by Qiaquick columns (Qiagen) and quantified by QPCR using a pair of primers that amplify the PRDIII-II region of the Ifnb promoter (Table S1). The precipitated DNA is presented as percentage of the total input DNA (Y axis). For histone modification analyses, the DNA bound by modified histone 3 is presented as percentage of total histone 3-bound DNA. The repeated results of ChIP assays are presented in supplementary figures.
Statistical analysis
Prism software was used for two-tailed unpaired t-tests. P-values < 0.05 and 0.01 are considered significant and very significant, respectively.
Supplementary Material
Highlights
Noncanonical NF-κB is stimulated by viral infection and macrophage differentiation
Noncanonical NF-κB negatively regulates IFN-I induction and antiviral immunity
Noncanonical NF-κB inhibits the recruitment of RelA to Ifnb promoter
Noncanonical NF-κB regulates histone modifications at the Ifnb promoter
ACKNOWLEDGEMENTS
We would like to thank Amgen Inc, Bristol-Myers Squibb Pharmaceutical Research Institute, R Starr and Water Eliza Hall Institute of Medical Research for mutant mice; L Schmitz (Justus-Liebig-University), HJ Kung (University of California, Davis), J Pagano (University of North Carolina), and D Thanos (Academy of Athens) for plasmid vectors; LF Chen (University of Illinois) and Paul Chiao (MD Anderson Cancer Center) for Rela−/− MEFs; and G Barber (University of Miami) and J Bell (University of Ottawa) for VSV viruses. We also thank the personnel from the NIH/NCI-supported resources (Flow Cytometry, DNA Analysis, and Genetically Engineered Mouse core facilities) under award number P30CA016672 at The MD Anderson Cancer Center. This study was supported by grants from National Institutes of Health (AI057555, AI064639, AI104519, and GM84459).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS J.J. designed and performed the research, prepared the Figures, and wrote the manuscript; H.H., H.L., J.Y., Y.X., G.C.B., Q.Z., and X.C. contributed experiments; F.A.M. and S.S.W. contributed reagents, and S.S.W. also supervised H.L.; and S.-C.S. supervised the work and wrote the manuscript.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
SUPPLEMENTAL INFORMATION Supplemental information includes Extended Experimental Procedures, 7 figures, and 1 table.
REFERENCES
- Asano M, Hayashi M, Yoshida E, Kawade Y, Iwakura Y. Induction of interferon-alpha by interferon-beta, but not of interferon-beta by interferon-alpha, in the mouse. Virology. 1990;176:30–38. doi: 10.1016/0042-6822(90)90227-i. [DOI] [PubMed] [Google Scholar]
- Bartlett NW, Slater L, Glanville N, Haas JJ, Caramori G, Casolari P, Clarke DL, Message SD, Aniscenko J, Kebadze T, et al. Defining critical roles for NF-kappaB p65 and type I interferon in innate immunity to rhinovirus. EMBO Mol. Med. 2012;4:1244–1260. doi: 10.1002/emmm.201201650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chang M, Jin W, Chang JH, Xiao Y, Brittain GC, Yu J, Zhou X, Wang YH, Cheng X, Li P, et al. The ubiquitin ligase Peli1 negatively regulates T cell activation and prevents autoimmunity. Nat. Immunol. 2011;12:1002–1009. doi: 10.1038/ni.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary S, Boldogh S, Garofalo R, Jamaluddin M, Brasier AR. Respiratory syncytial virus influences NF-kappaB-dependent gene expression through a novel pathway involving MAP3K14/NIK expression and nuclear complex formation with NF-kappaB2. J. Virol. 2005;79:8948–8959. doi: 10.1128/JVI.79.14.8948-8959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- Falvo JV, Parekh BS, Lin CH, Fraenkel E, Maniatis T. Assembly of a functional beta interferon enhanceosome is dependent on ATF-2-c-jun heterodimer orientation. Mol. Cell. Biol. 2000;20:4814–4825. doi: 10.1128/mcb.20.13.4814-4825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Fusco AJ, Savinova OV, Talwar R, Kearns JD, Hoffmann A, Ghosh G. Stabilization of RelB requires multidomain interactions with p100/p52. J. Biol. Chem. 2008;283:12324–12332. doi: 10.1074/jbc.M707898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SG, Iglesias AH, Lizcano F, Villanueva R, Camelo S, Jingu H, Teh BT, Koibuchi N, Chin WW, Kokkotou E, Dangond F. Functional characterization of JMJD2A, a histone deacetylase- and retinoblastoma-binding protein. J. Biol. Chem. 2005;280:28507–28518. doi: 10.1074/jbc.M413687200. [DOI] [PubMed] [Google Scholar]
- Hacker H, Tseng PH, Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 2011;11:457–468. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Hoshino K, Sugiyama T, Matsumoto M, Tanaka T, Saito M, Hemmi H, Ohara O, Akira S, Kaisho T. IkappaB kinase-alpha is critical for interferon-alpha production induced by Toll-like receptors 7 and 9. Nature. 2006;440:949–953. doi: 10.1038/nature04641. [DOI] [PubMed] [Google Scholar]
- Inaba K, Inaba M, Deguchi M, Hagi K, Yasumizu R, Ikehara S, Muramatsu S, Steinman RM. Granulocytes, macrophages, and dendritic cells arise from a common major histocompatibility complex class II-negative progenitor in mouse bone marrow. Proc. Natl. Acad. Sci. U S A. 1993;90:3038–3042. doi: 10.1073/pnas.90.7.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze MG, He Y, Gale M., Jr. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Liao G, Zhang M, Harhaj EW, Sun SC. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- Liu P, Li K, Garofalo RP, Brasier AR. Respiratory syncytial virus induces RelA release from cytoplasmic 100-kDa NF-kappa B2 complexes via a novel retinoic acid-inducible gene-I{middle dot}NF- kappa B-inducing kinase signaling pathway. J. Biol. Chem. 2008;283:23169–23178. doi: 10.1074/jbc.M802729200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manches O, Fernandez MV, Plumas J, Chaperot L, Bhardwaj N. Activation of the noncanonical NF-kappaB pathway by HIV controls a dendritic cell immunoregulatory phenotype. Proc. Natl. Acad. Sci. U S A. 2012;109:14122–14127. doi: 10.1073/pnas.1204032109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao AP, Li S, Zhong B, Li Y, Yan J, Li Q, Teng C, Shu HB. Virus-triggered ubiquitination of TRAF3/6 by cIAP1/2 is essential for induction of interferon-beta (IFN-beta) and cellular antiviral response. J. Biol. Chem. 2010;285:9470–9476. doi: 10.1074/jbc.M109.071043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miosge LA, Blasioli J, Blery M, Goodnow CC. Analysis of an ethylnitrosourea-generated mouse mutation defines a cell intrinsic role of nuclear factor kappaB2 in regulating circulating B cell numbers. J. Exp. Med. 2002;196:1113–1119. doi: 10.1084/jem.20020959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin. Immunol. 2009;21:215–222. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez de Diego R, Sancho-Shimizu V, Lorenzo L, Puel A, Plancoulaine S, Picard C, Herman M, Cardon A, Durandy A, Bustamante J, et al. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 2010;33:400–411. doi: 10.1016/j.immuni.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle A, Haasbach E, Julkunen I, Planz O, Ehrhardt C, Ludwig S. The NS1 protein of influenza A virus blocks RIG-I-mediated activation of the noncanonical NF-kappaB pathway and p52/RelB-dependent gene expression in lung epithelial cells. J. Virol. 2012;86:10211–10217. doi: 10.1128/JVI.00323-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Calado DP, Derudder E, Zhang B, Shimizu Y, Mackay F, Nishikawa S, Rajewsky K, Schmidt-Supprian M. NIK overexpression amplifies, whereas ablation of its TRAF3-binding domain replaces BAFF:BAFF-R-mediated survival signals in B cells. Proc. Natl. Acad. Sci. USA. 2008;105:10883–10888. doi: 10.1073/pnas.0805186105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senftleben U, Cao Y, Xiao G, Kraehn G, Greten F, Chen Y, Hu Y, Fong A, Sun S-C, Karin M. Activation of IKKa of a second, evolutionary conserved, NF-kB signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Sun SC. The noncanonical NF-kappaB pathway. Immunol. Rev. 2012;246:125–140. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. Type I interferon: friend or foe? J. Exp. Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker E, O'Donnell K, Fuchsberger M, Hilton AA, Metcalf D, Greig K, Sims NA, Quinn JM, Alexander WS, Hilton DJ, et al. A novel mutation in the Nfkb2 gene generates an NF-kappa B2 “super repressor”. J. Immunol. 2007;179:7514–7522. doi: 10.4049/jimmunol.179.11.7514. [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, Vignali DA, Bergsagel PL, Karin M. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat. Immunol. 2008;9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Basagoudanavar SH, Wang X, Hopewell E, Albrecht R, Garcia-Sastre A, Balachandran S, Beg AA. NF-kappa B RelA subunit is crucial for early IFN-beta expression and resistance to RNA virus replication. J Immunol. 2010;185:1720–1729. doi: 10.4049/jimmunol.1000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfield M, Zhang M, Norman LP, Sun SC. NF-kappaB1/p105 Regulates Lipopolysaccharide-Stimulated MAP Kinase Signaling by Governing the Stability and Function of the Tpl2 Kinase. Mol. Cell. 2003;11:685–694. doi: 10.1016/s1097-2765(03)00070-4. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Xiao G, Harhaj EW, Sun SC. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol. Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, Shiba T, Yang X, Yeh WC, Mak TW, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 2008;9:1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.