Abstract
Catalytic hairpin assembly (CHA) has previously proven useful as a transduction and amplification method for nucleic acid detection. However, the two hairpin substrates in a CHA circuit can potentially react non-specifically even in the absence of a singles-stranded catalyst, and this non-specific background degrades signal-to-noise. The introduction of mismatched base-pairs that impede uncatalyzed strand exchange reactions greatly decreased background signal while only partially damping signal in the presence of catalyst. Various types and lengths of mismatches were assayed by fluorimetry and in many instances our MismatCHA designs yielded 100-fold signal-to-background ratios compared to a similar ratio of 4 with the perfectly matched substrates. These observations may prove to be of general utility for the design of non-enzymatic nucleic acid circuits.
Keywords: Catalyzed hairpin assembly, Mismatch, Signal: background ratio
Nucleic acid circuits based on toehold-mediated strand exchange reactions have yielded interesting approaches to computation, nanotechnology, and diagnostics.[1] An example of a common amplification reaction known as the catalytic hairpin assembly (CHA) is shown in Figure 1. Originally developed by Pierce and Yin,[1c] this circuit has been subsequently adapted to a variety of applications, including acting as a monitor of isothermal amplification reactions, both end-point[2] and real-time.[3]
Figure 1.
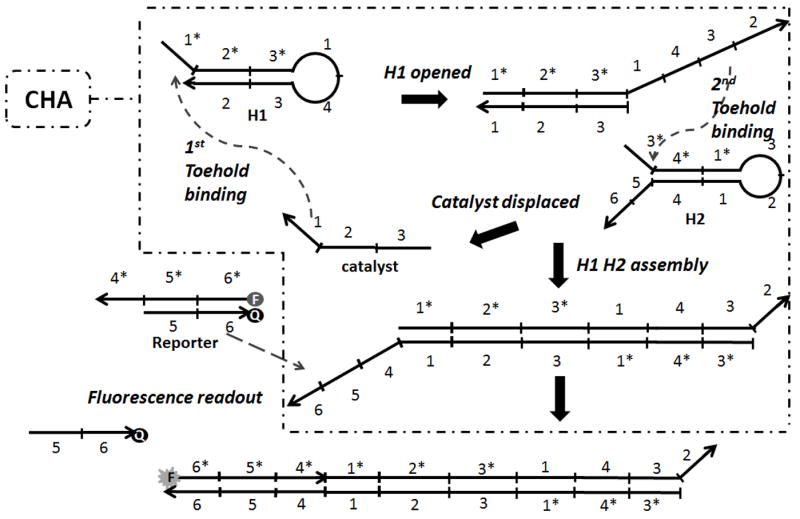
Catalytic hairpin assembly reaction with fluorescence read-out. Briefly, one short linear oligonucleotide, ‘catalyst’, will react with H1 via toehold binding and then initiate a branch migration reaction. The partially-opened H1 can interact with a toehold on H2 and similarly initiate a branch migration reaction. At the conclusion of the second branch migration the catalyst will be completely displaced from H1 and will be available for additional reaction cycles. Numbers in the figure stand for different sequence domains; each domain includes 8 bases.
Unfortunately, CHA circuits have also been shown to execute non-specifically, even in the absence of particular inputs.[4] This background leakage is characterized by an initial burst of signal followed by a steady-state, non-catalyzed rate of circuit execution. In our previous work, the rate constant of the steady-state leakage of a typical CHA circuit was about 200 M-1s-1 while the corresponding catalytic rate with 5 nM catalyst was 4000 M-1s-1.[4a] While the 20-fold enhancement observed in the catalytic rate allowed for robust signal detection, any accompanying background leakage can potentially make quantitation of lower concentrations of inputs more difficult. For example, we have found that while CHA circuits can be designed for a variety of sequence targets and applications, the signal-to-noise ratio for these circuits (that is, the catalyzed reaction relative to the uncatalyzed reaction) seldom exceeds 100-fold.
The background leakage can be attributed to a number of factors, including the purity of DNA samples[5] and the mis-folding of nucleic acids into alternative conformers. Underlying many of these mechanisms, though, is the uncatalyzed binding of an otherwise occluded toehold to its hybridization partner, the subsequent initiation of strand exchange, and the continued propagation of the hairpin assembly reaction. For example, when the kinetically trapped hairpin substrates in CHA ‘breathe’ they inadvertently reveal binding sites that can then initiate CHA even in the absence of a catalyst strand.
In order to reduce the prevalence of uncatalyzed strand exchange, we hypothesized that it might be possible to ‘block’ either the revealed, inadvertent binding reaction and / or its continuation as a strand exchange reaction. In turn, the simplest way to introduce a block was to introduce mismatched nucleotides into the regions thought to breathe and / or adjacent positions that might be involved in strand exchange. Since the ends of helices are more likely to ‘breathe’ than internal base-pairs[6] we chose to introduce mismatches into these portions of the hairpin substrates. A CHA circuit (Circuit A) similar to those we and others have previously used[4a] was designed, except that domains 1 and 1* were shortened from 10 nucleotides to 8, a length that we found could still act as an efficient toehold. In addition, mismatches were introduced at the 3’ end of domain 2 in H2 (CircA-H2D2M2, where CircA refers to the overall circuit, H2 refers to the hairpin substrate, D2 refers to the domain, and M2 refers to the type of mutation, i.e. single, double, etc.; see also Table 1) to reduce its ability to hybridize to the complementary domain 2* in H1. In order to probe the potential contribution of different mismatches to background suppression, two consecutive mismatches were introduced at each of four sites (Figure 2; Table 1).
Table 1.
Wild-type sequence and mismatched sequences*.
| Name | Sequence |
|---|---|
| CircA-H1 | AGAGGCAT CAATGGGA ATGGGATC ATGCCTCT AACCTAGC GATCCCAT TCCCATTG |
| CircA-H2 | ATGGGATC GCTAGGTT AGAGGCAT GATCCCAT TCCCATTG ATGCCTCT AACCTAGC CCTTGTCA TAGAGCAC |
| CircA-H2D2M2 | ATGGGATC GCTAGGTT AGAGGCAT GATCCCAT TCCCATac ATGCCTCT AACCTAGC CCTTGTCA TAGAGCAC |
| CircA-H2D3M2 | ATGGGATC GCTAGGTT AGAGGCAT atTCCCAT TCCCATTG ATGCCTCT AACCTAGC CCTTGTCA TAGAGCAC |
| CircA-H1D4M2 | AGAGGCAT CAATGGGA ATGGGATC ATGCCTCT AACCTAcg GATCCCAT TCCCATTG |
| CircA-H1D1M2 | AGAGGCAT CAATGGGA ATGGGATC taGCCTCT AACCTAGC GATCCCAT TCCCATTG |
Mismatches in lowercase
Figure 2.
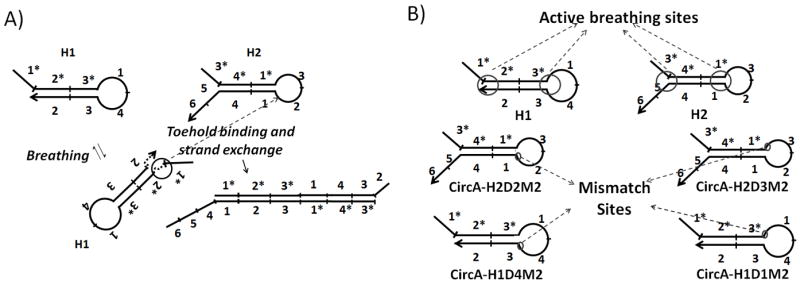
Possible pathways for leakage and positions of potential active breathing sites relative to the introduced mismatches: A) When the left-end of the stem of H1 ‘breathes’, the 3’-end of domain 2 will be transiently exposed, revealing a partial toehold that is complementary to domain 2 of H2. This transient toehold exposure would permit H1 and H2 to react in the absence of catalyst; B) The four mismatch positions correspond to the revealed interactions that could initiate strand displacement reactions between H1 and H2. For example, mismatches at the 3’ end of domain 2 of H2 would disrupt binding and / or strand exchange with domain 2* of H1; similarly, mismatches at the 5’ end of domain 1 of H1 would disrupt binding and / or strand exchange with domain 1* of H2.
The resultant ‘MismatCHA’ circuits were assayed by monitoring the release of a fluorescent oligonucleotide from a quencher (‘Reporter’ in Figure 1, see sequence in Table S6). For example, CircA-H2D2M2 was paired with H1 and the development of a fluorescent signal was followed as a function of time (Figure 3). When compared with the perfectly paired wild-type Circuit A (CircA-H1 paired with CircA-H2), the introduction of a double mismatch into domain 2 (CircA-H2D2M2, Table 1) (Figure 3A) led to a significant diminution of background signal development in the absence of catalyst. In contrast, when mismatches were introduced into domains 3 and 1 (CircA-H2D3M2, CircA-H1D1M2, Table 1) there was little effect on the performance of the circuit (Figure 3B & D), and the double mismatch in domain 4 (CircA-H1D4M2, Table 1) severely compromised the catalytic reaction rate (Figure 3C). We also studied a single mismatch on domain 4 and found that both the catalytic rate and the background leakage were one-fifth that of the wild-type constructs(Figure S1, Table S1).
Figure 3.
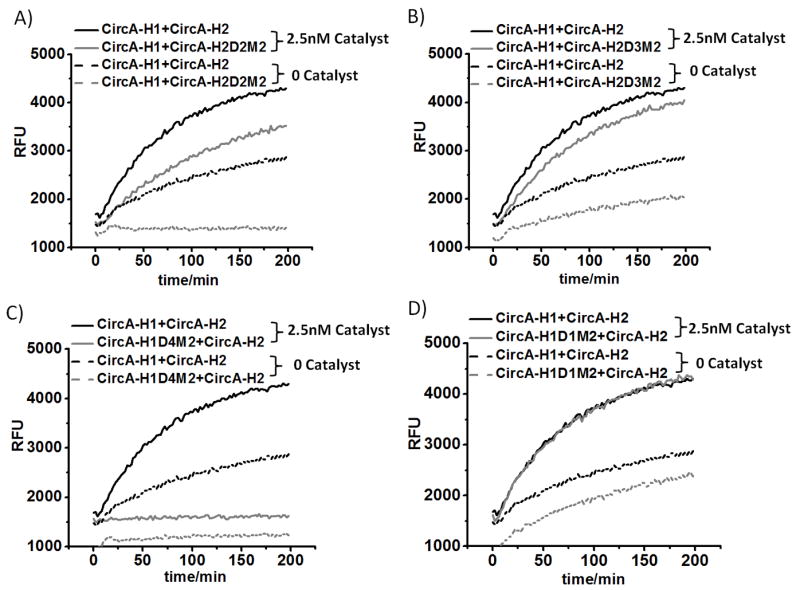
Signal generation with four different mismatches. A) Wild-type circuit and CircA-H2D2M2 circuit; B) Wild-type circuit and CircA-H2D3M2 circuit; C) Wild-type circuit and CircA-H1D4M2 circuit; D) Wild-type circuit and CircA-H1D1M2 circuit. The wild-type data are the same for each comparison; they are simply broken out for ease of viewing.
While the rate of the reaction with CircA-H2D2M2 was slightly compromised (Figure 4), it nonetheless had a roughly 23-fold improved signal-to-noise ratio relative to the wild-type reaction. Although these initial results represent only a single set of designs, they can be rationalized by making two assumptions: first, that breathing is more significant at the termini of helices than at positions adjacent to loops. This would mean that there was more opportunity for the suppression of background for constructs CircA-H2D2M2 and CircA-H1D4M2 than for CircA-H2D3M2 and CircA-H1D1M2 since CircA-H2D2M2 and CircA-H1D4M2 should decrease background due to breathing at the terminus of the H1 helix while CircA-H2D3M2 and CircA-H1D1M2 should reduce background interactions due to breathing adjacent to the loop in H1. Second, in the case of CircA-H1D4M2 the double mismatch may effectively prevent not only the uncatalyzed reaction due to breathing, but also initiation of the second strand exchange reaction that occurs following the opening of H1 (Figure 1). In contrast, the double mismatch in domain 2 would not interfere with the initiation but only the propagation of the second strand exchange reaction.
Figure 4.
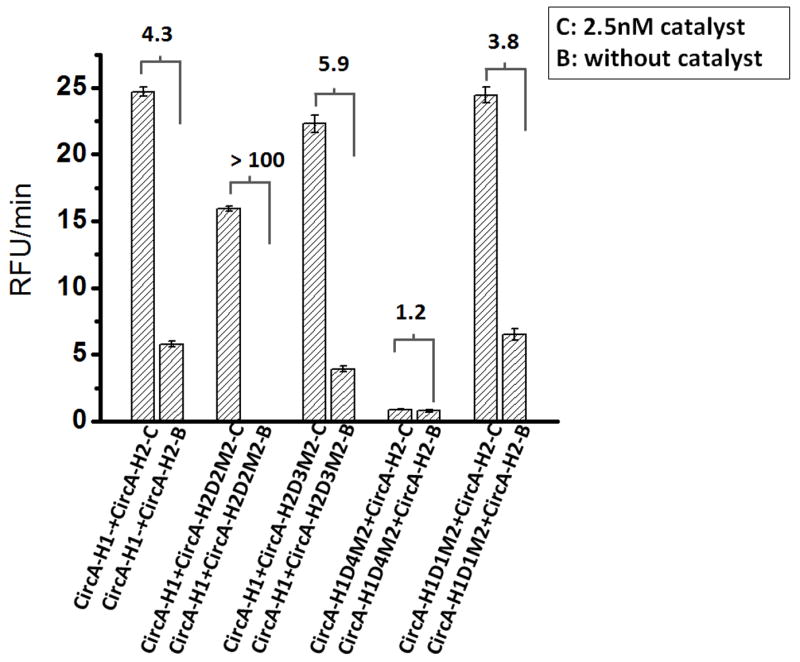
Signal-to-background ratios for four different mismatches. C denotes ‘with 2.5 nM catalyst’; B denotes ‘background’ or ‘without catalyst’. Signal:noise ratios were calculated from the linear portion of each fluorescence curve (such as those seen in Figure 3); each set of CHA reactions has been repeated at least three times. The numbers at the tops of columns represent the ratio of the catalyzed rate to background leakage.
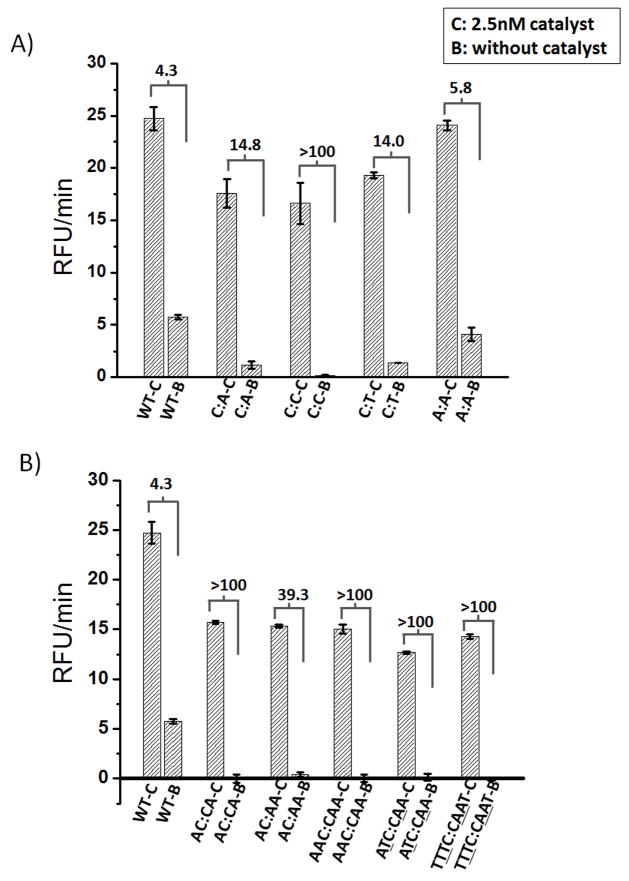
Building on the observation that mismatches in H2 domain 2 (adjacent to the loop) can lead to better signal-to-noise characteristics; we generated a series of Circuit A variants that changed the number, position, and identity of the introduced mismatches (Figure 5, Table S2). In Figure 5A and Figure S2A, we tested circuits with a variety of single mismatches between H1 and H2. The mismatches (C:A, C:C, and C:T) were positioned at either the 3’-end of domain 2 or at the penultimate residue adjacent to the 3’-end of domain 2 (A:A). All mismatches improved the signal-to-background ratio, and the C:C mismatch gave a signal-to-background ratio of over 100. The performance of the C:C mismatch may be due to the fact that it is one of the strongest mismatches[7], or may be due to the increase in the length of the H2 stem, due to a fortuitous pairing with an opposing guanosine in the loop. In Figure 5B and Figure S2B, we compared various multiple mismatches between H1 and H2. Four different double mismatches were assayed, either adjacent to one another (AC:CA, AC:AA) or separated by potentially paired residues (ATC:CAA, TTTC:CAAT; paired residues underlined). The double mismatches had generally higher signal-to-background ratios than the single mismatches, and three of the four were over 100. Three mismatches (AAC:CAA) also yielded a high signal-to-background ratio. Both the double and triple mismatches decreased the catalytic rate of the CHA circuit while generally improving the signal-to-background ratio.
Figure 5.
Signal generation in Circuit A with nine different domain 2 mismatches. Calculated signal-to-background ratios for A) single mismatches in CircA-H2 (naming according to mismatches; see also Table S2); and B) double mismatches and triple mismatches. Each CHA reaction was carried out with 50 nM of H2 (either wild-type or mismatched), 50 nM CircA-H1, and 50nM CircA-reporter.
To ensure that the circuits with mismatches were executing similarly to more well-known CHA circuits without mismatches, we also examined the strand exchange reactions by native gel electrophoresis of the reaction between CircA-H2 and CircA-H2D2M2 (Figure S3). Products were observed at the same sizes, irrespective of the presence or absence of mismatches. Consistent with the fluorescence assays, there is more background product from the perfectly paired CircA-H2 relative to the double mismatched CircA-H2D2M2 (roughly twice as much at end-point).
In order to generally demonstrate that mismatches on domain 2 can improve signal:noise ratios we used NUPACK[8] to generate a second circuit (Circuit B) with the same domain organization as in Figure 1, but with completely different sequences for the hairpin substrates. We again assayed circuit variants that contained mismatches at four positions (Table S3). Once more we found that only CircB-H2D2M2 (Figures S4 & S5) improved the signal-to-background ratio to over 100. In order to again show the generality of these results, we also introduced different mismatches into Circuit B. While the wild-type Circuit B had a signal:background ratio of 4.5 (Figure S6C), a single mismatched H2 (CircB-H2D2M1, A:A, Figure S6A & C, Table S5) increased the signal:background ratio to 12.6 and a double mismatch (CircB-H2D2M2, CA:AC, Figure S6B & C, Table S5) increased the signal:background ratio to over 100.
Overall, MismatCHA designs substantially decreased the uncatalyzed background in CHA amplification reactions. For a typical CHA circuit, introducing mismatches at the 3’ end of domain 2 generally gave higher signal-to-background ratios, with multiple mismatches almost always yielding much larger signal-to-background ratios while only modestly decreasing rates. Since MismatCHA circuits can increase the signal-to-background ratio from single digits to over 100 they should prove useful for the sequence-specific signal transduction with amplicons arising from isothermal amplification reactions.[9] In essence, CHA can now be used with isothermal amplification reactions in the same way a TaqMan probe is used for PCR. The rules developed herein should now greatly extend the utility of CHA probes for many different amplicon sequences and for different types of isothermal amplification reactions.
In addition, these results continue to highlight the utility of noncanonical pairings in DNA nanotechnology. We have previously shown that defects in DNA nanostructures can potentially be detected using DNA circuits sensitive to mismatches[4a] (although in this instance the presence of a mismatch in the structure led to a lower rate of reaction, rather than to the higher signal-to-background ratio observed here with MismatCHA). Mismatches have also been shown to be specifically incorporated into DNA nanostructures, with regular DNA crystals being formed in part from non-Watson-Crick pairings[10] and even into larger structures, such as oligonucleotides hybridized to the surfaces of gold nanoparticles.[11] As the rules for the use of mismatches are further elucidated, the wealth of rationally designed DNA circuits and structures will continue to expand.
Supplementary Material
Footnotes
This work was supported by the Bill and Melinda Gates Foundation (OPP1028808); the Defense Advanced Research Projects Agency (HR0011-12-2-0001, 5-35830); and the National Institute of Health TR01 (5 R01 AI092839).
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.a) Zhang DY, Winfree E. J Am Chem Soc. 2009;131:17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]; b) Ma C, Wang Z, Li Z, Cao L, Wang Q. Anal Biochem. 2012;429:99–102. doi: 10.1016/j.ab.2012.07.009. [DOI] [PubMed] [Google Scholar]; c) Yin P, Choi HMT, Calvert CR, Pierce NA. Nature. 2008;451:318–U314. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]; d) Zhang DY, Turberfield AJ, Yurke B, Winfree E. Science. 2007;318:1121–1125. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]; e) Zhang H, Li F, Dever B, Li X, Li XC. Chem Rev. 2013;113:2812–2841. doi: 10.1021/cr300340p. [DOI] [PubMed] [Google Scholar]; f) Liu J, Cao Z, Lu Y. Chem Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li B, Chen X, Ellington AD. Anal Chem. 2012;84:8371–8377. doi: 10.1021/ac301944v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang Y, Li B, Milligan JN, Bhadra S, Ellington AD. J Am Chem Soc. 2013;135:7430–7433. doi: 10.1021/ja4023978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Li B, Jiang Y, Chen X, Ellington AD. J Am Chem Soc. 2012;134:13918–13921. doi: 10.1021/ja300984b. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Huang J, Su X, Li Z. Anal Chem. 2012;84:5939–5943. doi: 10.1021/ac3004727. [DOI] [PubMed] [Google Scholar]; c) Ren J, Wang J, Han L, Wang E, Wang J. Chem Commun. 2011;47:10563–10565. doi: 10.1039/c1cc13973h. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Briggs N, McLain JR, Ellington AD. Proc Natl Acad Sci U S A. 2013;110:5386–5391. doi: 10.1073/pnas.1222807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) SantaLucia J. Proc Natl Acad Sci U S A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) SantaLucia J, Hicks D. Annu Rev Cell Dev Biol. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 7.a) Kwok S, Chang SY, Sninsky JJ, Wang A. Genome Res. 1994;3:S39–S47. doi: 10.1101/gr.3.4.s39. [DOI] [PubMed] [Google Scholar]; b) Kwok S, Kellogg DE, McKinney N, Spasic D, Goda L, Levenson C, Sninsky JJ. Nucleic Acids Res. 1990;18:999–105. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. J Comput Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]; b) Dirks RM, Bois JS, Schaeffer JM, Winfree E, Pierce NA. Siam Rev. 2007;49:65–88. [Google Scholar]; c) Dirks RM, Pierce NA. J Comput Chem. 2003;24:1664–1677. doi: 10.1002/jcc.10296. [DOI] [PubMed] [Google Scholar]; d) Dirks RM, Pierce NA. J Comput Chem. 2004;25:1295–1304. doi: 10.1002/jcc.20057. [DOI] [PubMed] [Google Scholar]
- 9.a) Compton J. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]; b) Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP. Nucleic Acids Res. 1992;20:1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Nucleic Acids Res. 2000;28 doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paukstelis PJ, Nowakowski J, Birktoft JJ, Seeman NC. Chem Biol. 2004;11:1119–1126. doi: 10.1016/j.chembiol.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 11.Hill HD, Hurst SJ, Mirkin CA. Nano Lett. 2009;9:317–321. doi: 10.1021/nl8030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.