Highlights
-
•
System-level approaches will provide a more complete picture of the immune response.
-
•
Omic studies will allow identification of signatures of immune protection.
-
•
Network-based methods are powerful tools for multi-omic data integration.
-
•
Discovery of host–virus interactions will improve vaccine development.
-
•
An emphasis on producing clinically actionable findings is necessary.
Abstract
Numerous challenges have been identified in vaccine development, including variable efficacy as a function of population demographics and a lack of characterization and mechanistic understanding of immune correlates of protection able to guide delivery and dosing. There is tremendous opportunity in recent technological and computational advances to elucidate systems level understanding of pathogen–host interactions and correlates of immunity. A systems biology approach to vaccinology provides a new paradigm for rational vaccine design in a ‘precision medicine’ context.
Current Opinion in Immunology 2013, 25:551–555
This review comes from a themed issue on Systems biology and bioinformatics
Edited by Anna Karolina Palucka and Bali Pulendran
For a complete overview see the Issue and the Editorial
Available online 23rd October 2013
0952-7915/$ – see front matter, Published by Elsevier Ltd.
Introduction
The historical emphasis on an empirical model for vaccine development has been largely ineffective for some of the most rapidly evolving pathogens, such as HIV and tuberculosis, suggesting the need for a new direction in vaccine strategies [1]. New approaches to vaccine development must be able to encompass host genetic and demographic variability, pathogen variability, as well as the interactions between host and pathogen including the diverse immune cell subsets that can be involved.
The power of a systems perspective
The immune response to vaccination depends on interactions between a multitude of factors, including genetic, epigenetic, physiologic and environmental factors, such as coinfections and the microbiome. This view, first proposed by Poland and colleagues [2, 3], known as the immune response network theory, illustrates the complexity of the immune response and provides the rationale for systems level approaches to vaccine development.
For example, one of the most important and difficult areas of vaccine research is the discovery of biomarkers (e.g. omic signatures) capable of predicting an individual's response to vaccination. The identification of these immune correlates of protection may allow for the development of more individualized vaccination strategies. Systems level data analyses, such as the integration of multiple high-throughput omics data sets in combination with network-based methods, hold particular promise for this line of research [4, 5].
Recently, systems level approaches have been successful in identifying genomic signatures predictive of the response to both yellow fever and influenza vaccines [6, 7, 8]. In these studies, advanced machine learning approaches were used to identify gene expression signatures predictive of the immune response to vaccination, including the CD8+T cell and antibody response.
The findings from these studies are significant in that they provide strong evidence of the ability to identify biomarkers of vaccine protection soon after vaccine administration. Biomarkers that are predictive of immune response, if found to be reliable across different patient populations, could prove invaluable for the design of clinical trials for new vaccines [9].
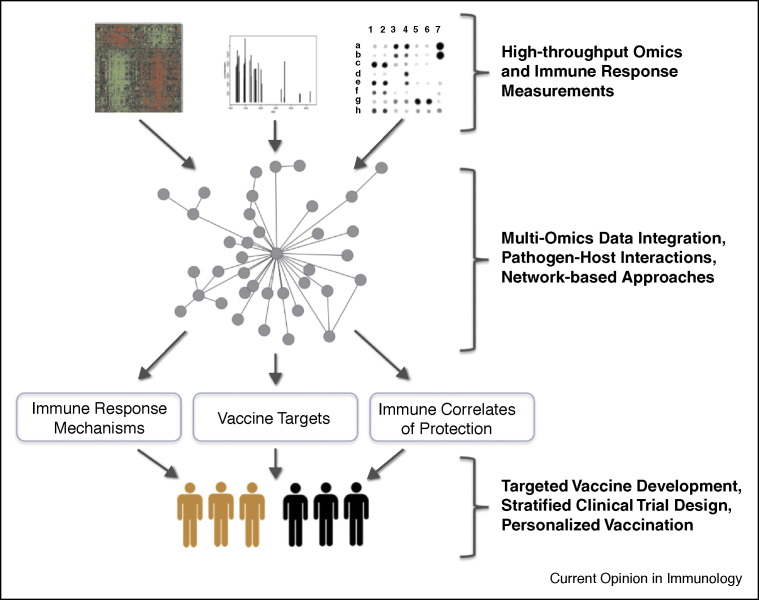
An overview of the systems biology workflow for vaccine development, from multi-omic measurement to discovery of immune correlates of protection and improved clinical trial design, is shown in Figure 1 .
Figure 1.
System-level approach to vaccine development from bench to bedside. The integration of multi-omic measurements (proteomic, transcriptomic, etc.) along with information about host–pathogen interactions will allow for a system-level view of the host reponse to infection (or vaccination). Analysis with network-based approaches (module identification, differential network analysis, etc.) will enable discoveries about the host immune response, including insights into the mechanism of action of vaccines, biomarkers of immune protection, and potentially new vaccine candidates (targets), leading to improved vaccine development and delivery.
Data integration: finding a path forward
The ability to integrate information from a diversity of data sources, such as genome-wide DNA variation along with transcript and protein abundance measures, is what makes systems biology methods so powerful. However, data integration remains a major challenge in the field. Immunology and vaccine research present additional complexities given the need to model both host and pathogen systems. And the need to track the immune response over time greatly increases the amount of data produced.
Nakaya and colleagues provide a comprehensive overview of the methods of systems vaccinology, including the benefits gained from integrating multiple sources of omics data, using research on the yellow fever vaccine as a proof of concept [10].
Expression microarray experiments, which measure genome-wide transcript abundances, have been the main focus of many systems biology studies of vaccines so far [11, 12, 13, 14, 15, 16]. These studies have provided new insights relevant to two major goals in vaccinology: the elucidation of a vaccine's mechanism of action, and the identification of a molecular signature able to predict a patient's response to vaccination (i.e. whether or not the vaccine will confer protection). For instance, Obermoser et al. recently used blood transcriptome measurements to investigate the differences in immune response after vaccination with influenza and pneumococcal vaccines. They observed significant differences in the gene expression profiles elicited by the two vaccines, with the influenza vaccine producing a strong interferon signature and the pneumococcal vaccine producing an increase in inflammation-related transcripts [17]. The authors suggest that ‘comparing global immune response elicited by different vaccines will be critical to our understanding of the immune mechanisms underpinning successful vaccination.’
Methods that can model the interactions between multiple genes are crucial for providing a truly system-level view of the transcriptome and its response to vaccination (or infection). Regev, Hacohen, and colleagues have used a system-level perturbation strategy to reconstruct regulatory networks involved in the immune response. In dendritic cells they measured gene expression profiles after stimulation with pathogen components to identify candidate regulators of immune response. They then perturbed each candidate regulator using shRNA knockdown, again stimulated the cells with pathogen components, and observed the resulting changes in a gene expression signature of immune response. The direct responses to regulator perturbation allowed construction of regulatory networks [18, 19].
Network-based approaches [20] are ideally suited for large data integration problems and have become a powerful tool in systems biology research. Network methods particularly relevant for infectious disease research include differential network analysis and cross-species interaction networks [21, 22], which can be used to model system-level changes during the progression of infection, as well as host–pathogen protein interactions (discussed further below). Bisson et al. recently reported on a mass spectrometry-based method for measuring the changes in protein–protein interactions in response to a stimulus [23]. If applied on a system level, this type of method could vastly improve our understanding of immune network remodeling in response to infection or vaccination.
Multiple systems: modeling host and pathogen interactions
The response to viral infection depends on the many interactions between viral and host proteins [24]. Modeling these interactions will be essential for developing predictive models of pathogen virulence and host response.
The human viral infectome is an effort to model all interactions that occur during human viral infection [25]. This infectome was constructed by integrating 416 viral proteins into a human protein–protein interaction network through manually curated virus–host protein interactions. Navratil and colleagues found that viral proteins interact with approximately 5% of human proteins, and that a significant number of these targeted proteins interact with multiple viruses or virus families. These observations suggest common molecular mechanisms of infection across viruses.
A particularly interesting finding that has come from research on the human virome (the collection of all viruses that infect humans) is the impact of coinfections and the bacterial microbiome on the host immune response [26]. Gaining a better understanding of the human virome [27], and the ways in which viruses interact with the immune system will provide valuable information relevant for vaccine development, including the identification of new vaccine targets and insights into the variability of immune responses.
The National Institute of Allergy and Infectious Disease (NIAID) created the Systems Biology for Infectious Diseases Research program specifically to investigate the interactions between viruses and the host immune system [28]. Transcriptomic and proteomic analyses done at two of the program's centers, the Systems Virology Center and the Center for Systems Influenza, have identified transcriptome changes in response to infection with influenza and SARS-CoV, as well as novel interactions between the H5N1 influenza polymerase and a number of host proteins [29, 30, 31].
Efforts to identify the complete set of mechanisms by which viruses interact with host immune systems will provide numerous benefits for vaccine development. Not only will this virus–host interaction data provide potential targets for current vaccine development, but it may also speed the development of vaccines or treatments for newly emergent viral infections. Moreover, the identification of sequence variation among interacting proteins, which may play a role in modifying the response to infection or the protective effect of a vaccine, may allow for more personalized vaccination regimens (altered dosing schedules, the use of adjuvants, etc.).
Moving toward clinically actionable results
Given the great need for new interventions to fight infectious diseases, particularly those like malaria and tuberculosis where drug resistance is a major issue, drug repurposing/repositioning is becoming an important area of research. In cancer research, drug repositioning is becoming an effective path for improving treatment, particularly as researchers gain a better understanding of specific oncogenic mutations [32]. The repurposing of Imatinib and Crizotinib for additional cancer types (but with the same mutations) is example of the benefits gained from precision medicine, particularly the improved understanding of the genomic factors influencing disease development [33, 34].
So far, in infectious disease research most work in the area of drug repositioning has been done for antimicrobial treatments [35, 36, 37]. However, with increasing knowledge about host–virus interactions and the evolutionary relationships between viruses, along with improved development of targeted vaccine adjuvants [38, 39], could repurposing techniques play a role in vaccine development as well? This is an intriguing question that remains to be answered, but which could provide promising opportunities for new vaccine candidates.
While development of new interventions is crucial, techniques for improving delivery of treatments available now should also be a priority. With the advent of precision medicine, there is an emphasis on the discovery of clinically actionable information to ensure the well-timed delivery of the correct drug at the accurate dose specific for a given patient [40]. Precision medicine embraces the notion that molecular information improves the precision with which patients are stratified and treated [41] and has clear implications for vaccine development and delivery. Recent studies have begun to focus on examining the genetic variation related to vaccine-specific immune responses. For example, Ovsyannikova et al. have reported polymorphisms in CD46 and SLAM, both cellular receptors for the measles virus, are significantly associated with the immune response to measles vaccine [42, 43]. Studies like these will help shape omics-guided stratification and individualized delivery/dosing.
Conclusion
The advent of systems-level omics characterization, as well as the computational and bioinformatics methods to analyze, integrate and model this data offers an unprecedented opportunity for vaccine discovery, development, and delivery. Characterizing how genetic variation can shape innate and adaptive immune responses will guide omics-driven population stratification for vaccine delivery. If this framework is embraced, it could lead to a substantial decrease in vaccine failure and adverse events, providing a significant benefit to global health.
Acknowledgements
Funding: NIH/NIAID (5U54AI081680 and 1U19AI100625); NIH/NCI (5P30CA069533); NIH/NCATS (5UL1RR024140) and NIH/NLM (2T15LM007088).
References
- 1.Rueckert C., Guzmán C.A. Vaccines: from empirical development to rational design. PLoS Pathog. 2012;8:e1003001. doi: 10.1371/journal.ppat.1003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poland G.A., Kennedy R.B., McKinney B.A., Ovsyannikova I.G., Lambert N.D., Jacobson R.M., Oberg A.L. Vaccinomics, adversomics, and the immune response network theory: individualized vaccinology in the 21st century. Semin Immunol. 2013;25:89–103. doi: 10.1016/j.smim.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poland G.A., Ovsyannikova I.G., Kennedy R.B., Haralambieva I.H., Jacobson R.M. Vaccinomics and a new paradigm for the development of preventive vaccines against viral infections. OMICS. 2011;9:625–636. doi: 10.1089/omi.2011.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li S., Nakaya H.I., Kazmin D.A., Oh J.Z., Pulendran B. Systems biological approaches to measure and understand vaccine immunity in humans. Semin Immunol. 2013 doi: 10.1016/j.smim.2013.05.003. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C.C., Zhu B., Fan X., Gicquel B., Zhang Y. Systems approach to tuberculosis vaccine development. Respirology. 2013;3:412–420. doi: 10.1111/resp.12052. [DOI] [PubMed] [Google Scholar]
- 6.Querec T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D., Pirani A., Gernert K., Deng J., Marzolf B. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;1:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakaya H.I., Wrammert J., Lee E.K., Racioppi L., Marie-Kunze S., Haining W.N., Means A.R., Kasturi S.P., Khan N., Li G.M. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaucher D., Therrien R., Kettaf N., Angermann B.R., Boucher G., Filali-Mouhim A., Moser J.M., Mehta R.S., Drake D.R., 3rd., Castro E. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corey L., Nabel G.J., Dieffenbach C., Gilbert P., Haynes B.F., Johnston M., Kublin J., Lane H.C., Pantaleo G., Picker L.J., Fauci A.S. HIV-1 vaccines and adaptive trial designs. Sci Transl Med. 2011;3:ps13. doi: 10.1126/scitranslmed.3001863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakaya H.I., Li S., Pulendran B. Systems vaccinology: learning to compute the behavior of vaccine induced immunity. Wiley Interdiscip Rev Syst Biol Med. 2012;2:193–205. doi: 10.1002/wsbm.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy R.B., Oberg A.L., Ovsyannikova I.G., Haralambieva I.H., Grill D., Poland G.A. Transcriptomic profiles of high and low antibody responders to smallpox vaccine. Genes Immun. 2013;5:277–285. doi: 10.1038/gene.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu H., Nau M., Ehrenberg P., Chenine A.L., Macedo C., Zhou Y., Daye Z.J., Wei Z., Vahey M., Michael N.L. Distinct gene-expression profiles associated with the susceptibility of pathogen-specific CD4 T cells to HIV-1 infection. Blood. 2013;121:1136–1144. doi: 10.1182/blood-2012-07-446278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richert L., Hue S., Hocini H., Raimbault M., Lacabaratz C., Surenaud M., Wiedemann A., Tisserand P., Durier C., Salmon D., ANRS Vaccine Network/Vaccine Research Institute Cytokine and gene transcription profiles of immune responses elicited by HIV lipopeptide vaccine in HIV-negative volunteers. AIDS. 2013;27:1421–1431. doi: 10.1097/QAD.0b013e32835f5b60. [DOI] [PubMed] [Google Scholar]
- 14.Sessions O.M., Tan Y., Goh K.C., Liu Y., Tan P., Rozen S., Ooi E.E. Host cell transcriptome profile during wild-type and attenuated dengue virus infection. PLoS Negl Trop Dis. 2013;7:e2107. doi: 10.1371/journal.pntd.0002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popper S.J., Gordon A., Liu M., Balmaseda A., Harris E., Relman D.A. Temporal dynamics of the transcriptional response to dengue virus infection in Nicaraguan children. PLoS Negl Trop Dis. 2012;6:e1966. doi: 10.1371/journal.pntd.0001966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bucasas K.L., Franco L.M., Shaw C.A., Bray M.S., Wells J.M., Niño D., Arden N., Quarles J.M., Couch R.B., Belmont J.W. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J Infect Dis. 2011;203:921–929. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obermoser G., Presnell S., Domico K., Xu H., Wang Y., Anguiano E., Thompson-Snipes L., Ranganathan R., Zeitner B., Bjork A. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amit I., Garber M., Chevrier N., Leite A.P., Donner Y., Eisenhaure T., Guttman M., Grenier J.K., Li W., Zuk O., Schubert L.A., Birditt B., Shay T., Goren A., Zhang X., Smith Z., Deering R., McDonald R.C., Cabili M., Bernstein B.E., Rinn J.L., Meissner A., Root D.E., Hacohen N., Regev A. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chevrier N., Mertins P., Artyomov M.N., Shalek A.K., Iannacone M., Ciaccio M.F., Gat-Viks I., Tonti E., DeGrace M.M., Clauser K.R., Garber M., Eisenhaure T.M., Yosef N., Robinson J., Sutton A., Andersen M.S., Root D.E., von Andrian U., Jones R.B., Park H., Carr S.A., Regev A., Amit I., Hacohen N. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell. 2011;147:853–867. doi: 10.1016/j.cell.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barabási A.L., Gulbahce N., Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ideker T., Krogan N.J. Differential network biology. Mol Syst Biol. 2012;8:565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzosa E.A., Garamszegi S., Xia Y. Toward a three-dimensional view of protein networks between species. Front Microbiol. 2012;3:428. doi: 10.3389/fmicb.2012.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bisson N., James D.A., Ivosev G., Tate S.A., Bonner R., Taylor L., Pawson T. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat Biotechnol. 2011;29:653–658. doi: 10.1038/nbt.1905. [DOI] [PubMed] [Google Scholar]
- 24.Shapira S.D., Gat-Viks I., Shum B.O., Dricot A., de Grace M.M., Wu L., Gupta P.B., Hao T., Silver S.J., Root D.E. A physical and regulatory map of host–influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navratil V., de Chassey B., Combe C.R., Lotteau V. When the human viral infectome and diseasome networks collide: towards a systems biology platform for the aetiology of human diseases. BMC Syst Biol. 2011 doi: 10.1186/1752-0509-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wylie K.M., Weinstock G.M., Storch G.A. Virome genomics: a tool for defining the human virome. Curr Opin Microbiol. 2013;16:479–484. doi: 10.1016/j.mib.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wylie K.M., Weinstock G.M., Storch G.A. Emerging view of the human virome. Transl Res. 2012;160:283–290. doi: 10.1016/j.trsl.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontana J.M., Alexander E., Salvatore M. Translational research in infectious disease: current paradigms and challenges ahead. Transl Res. 2012;159:430–453. doi: 10.1016/j.trsl.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C., Bankhead A., 3rd., Eisfeld A.J., Hatta Y., Jeng S., Chang J.H., Aicher L.D., Proll S., Ellis A.L., Law G.L., Waters K.M., Neumann G., Katze M.G., McWeeney S., Kawaoka Y. Host regulatory network response to infection with highly pathogenic H5N1 avian influenza virus. J Virol. 2011;85:10955–10967. doi: 10.1128/JVI.05792-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradel-Tretheway B.G., Kelley Z., Chakraborty-Sett S., Takimoto T., Kim B., Dewhurst S. The human H5N1 influenza A virus polymerase complex is active in vitro over a broad range of temperatures, in contrast to the WSN complex, and this property can be attributed to the PB2 subunit. J Gen Virol. 2008;89:2923–2932. doi: 10.1099/vir.0.2008/006254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng X., Gralinski L., Armour C.D., Ferris M.T., Thomas M.J., Proll S., Bradel-Tretheway B.G., Korth M.J., Castle J.C., Biery M.C., Bouzek H.K., Haynor D.R., Frieman M.B., Heise M., Raymond C.K., Baric R.S., Katze M.G. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1 doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y.Y., Jones S.J. Drug repositioning for personalized medicine. Genome Med. 2012;4:27. doi: 10.1186/gm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw A.T., Yasothan U., Kirkpatrick P. Crizotinib. Nat Rev Drug Discov. 2011;10:897–898. doi: 10.1038/nrd3600. [DOI] [PubMed] [Google Scholar]
- 34.Druker B.J. Imatinib as a paradigm of targeted therapies. Adv Cancer Res. 2004;91:1–30. doi: 10.1016/S0065-230X(04)91001-9. [DOI] [PubMed] [Google Scholar]
- 35.Shahinas D., Liang M., Datti A., Pillai D.R. A repurposing strategy identifies novel synergistic inhibitors of Plasmodium falciparum heat shock protein 90. J Med Chem. 2010;53:3552–3557. doi: 10.1021/jm901796s. [DOI] [PubMed] [Google Scholar]
- 36.Eastman R.T., Pattaradilokrat S., Raj D.K., Dixit S., Deng B., Miura K., Yuan J., Tanaka T.Q., Johnson R.L., Jiang H. A class of tricyclic compounds blocking malaria parasite oocyst development and transmission. Antimicrob Agents Chemother. 2013;57:425–435. doi: 10.1128/AAC.00920-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong E.B., Cohen K.A., Bishai W.R. Rising to the challenge: new therapies for tuberculosis. Trends Microbiol. 2013;21:493–501. doi: 10.1016/j.tim.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pashine A., Valiante N.M., Ulmer J.B. Targeting the innate immune response with improved vaccine adjuvants. Nat Med. 2005;11(4 Suppl):S63–S68. doi: 10.1038/nm1210. [DOI] [PubMed] [Google Scholar]
- 39.Levitz S.M., Golenbock D.T. Beyond empiricism: informing vaccine development through innate immunity research. Cell. 2012;148:1284–1292. doi: 10.1016/j.cell.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desmond-Hellmann S. Toward precision medicine: a new social contract? Sci Transl Med. 2012;4:ed3. doi: 10.1126/scitranslmed.3003473. [DOI] [PubMed] [Google Scholar]
- 41.Katsnelson A. Momentum grows to make ‘personalized’ medicine more ‘precise’. Nat Med. 2013;19:249. doi: 10.1038/nm0313-249. [DOI] [PubMed] [Google Scholar]
- 42.Katsnelson A. Vaccines shoot for more precise population targets. Nat Med. 2012;18:478. doi: 10.1038/nm0412-478. [DOI] [PubMed] [Google Scholar]
- 43.Ovsyannikova I.G., Haralambieva I.H., Vierkant R.A., O’Byrne M.M., Jacobson R.M., Poland G.A. The association of CD46, SLAM and CD209 cellular receptor gene SNPs with variations in measles vaccine-induced immune responses: a replication study and examination of novel polymorphisms. Hum Hered. 2011;72:206–223. doi: 10.1159/000331585. [DOI] [PMC free article] [PubMed] [Google Scholar]