Abstract
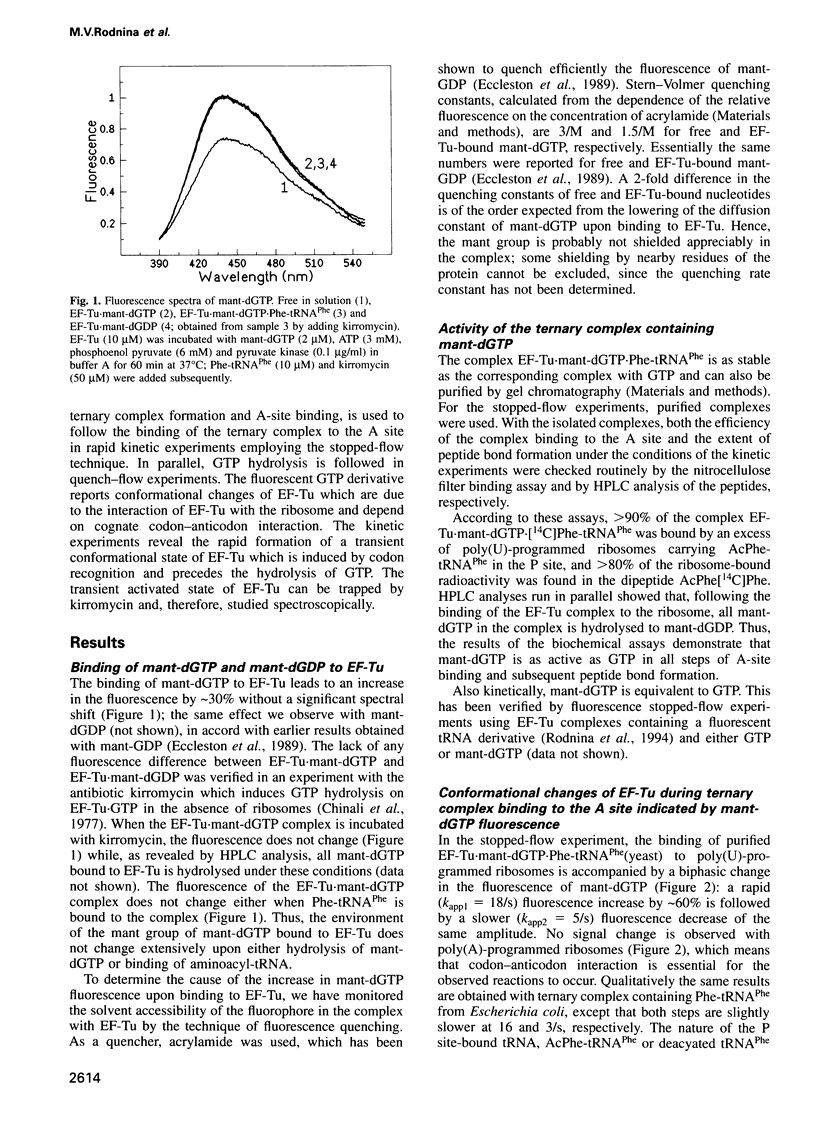
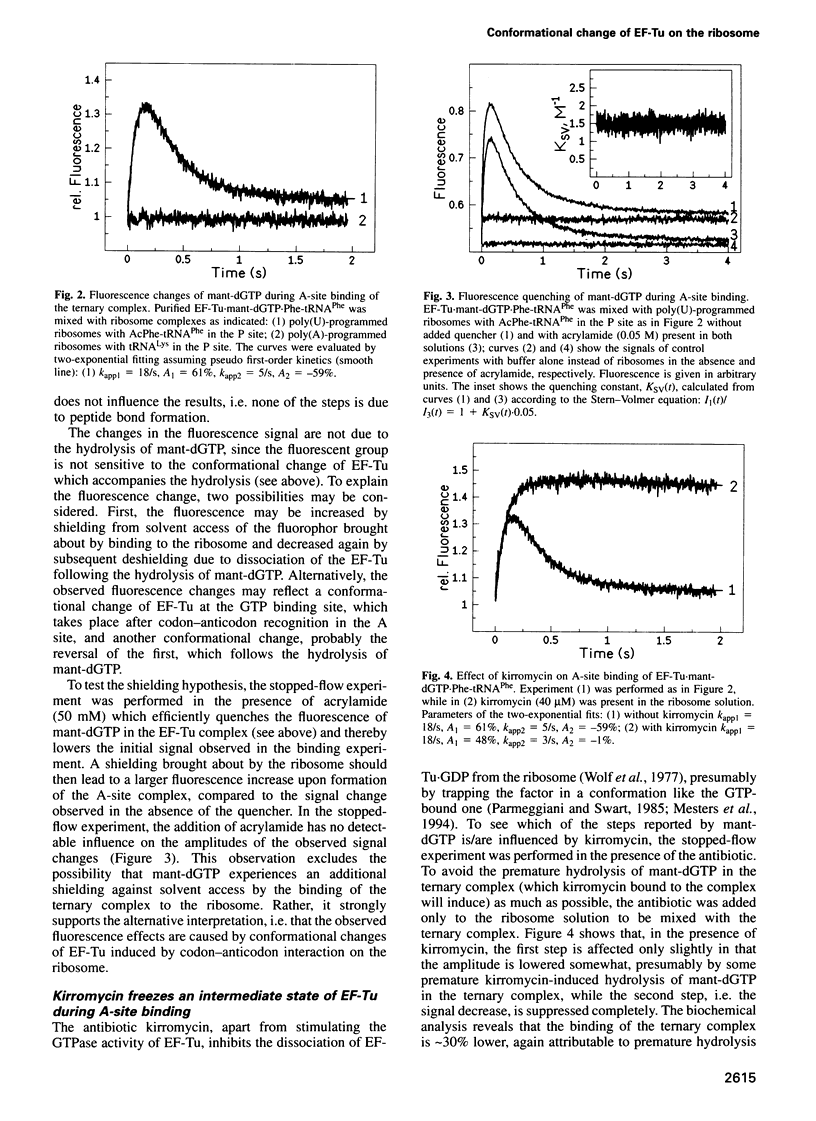
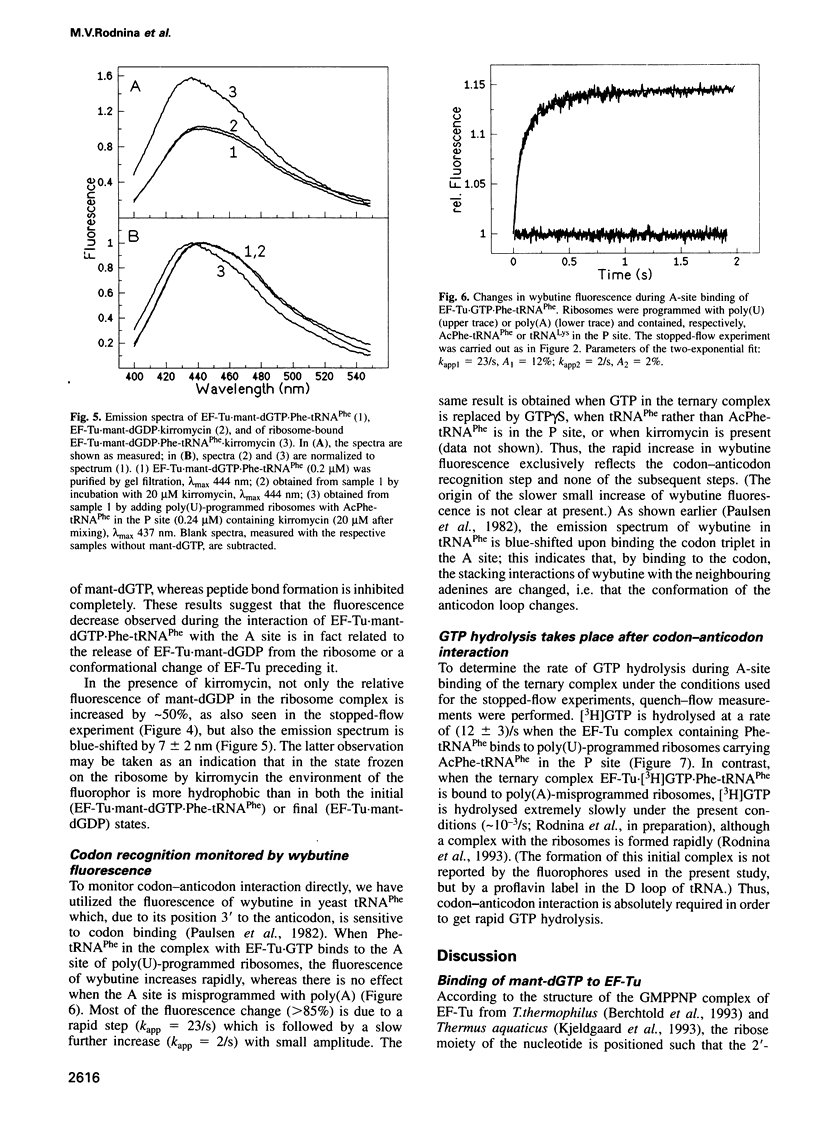
The mechanisms by which elongation factor Tu (EF-Tu) promotes the binding of aminoacyl-tRNA to the A site of the ribosome and, in particular, how GTP hydrolysis by EF-Tu is triggered on the ribosome, are not understood. We report steady-state and time-resolved fluorescence measurements, performed in the Escherichia coli system, in which the interaction of the complex EF-Tu.GTP.Phe-tRNAPhe with the ribosomal A site is monitored by the fluorescence changes of either mant-dGTP [3'-O-(N-methylanthraniloyl)-2-deoxyguanosine triphosphate], replacing GTP in the complex, or of wybutine in the anticodon loop of the tRNA. Additionally, GTP hydrolysis is measured by the quench-flow technique. We find that codon-anticodon interaction induces a rapid rearrangement within the G domain of EF-Tu around the bound nucleotide, which is followed by GTP hydrolysis at an approximately 1.5-fold lower rate. In the presence of kirromycin, the activated conformation of EF-Tu appears to be frozen. The steps following GTP hydrolysis--the switch of EF-Tu to the GDP-bound conformation, the release of aminoacyl-tRNA from EF-Tu to the A site, and the dissociation of EF-Tu-GDP from the ribosome--which are altogether suppressed by kirromycin, are not distinguished kinetically. The results suggest that codon recognition by the ternary complex on the ribosome initiates a series of structural rearrangements resulting in a conformational change of EF-Tu, possibly involving the effector region, which, in turn, triggers GTP hydrolysis.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdulkarim F., Liljas L., Hughes D. Mutations to kirromycin resistance occur in the interface of domains I and III of EF-Tu.GTP. FEBS Lett. 1994 Sep 26;352(2):118–122. doi: 10.1016/0014-5793(94)00937-6. [DOI] [PubMed] [Google Scholar]
- Berchtold H., Reshetnikova L., Reiser C. O., Schirmer N. K., Sprinzl M., Hilgenfeld R. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993 Sep 9;365(6442):126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- Bilgin N., Claesens F., Pahverk H., Ehrenberg M. Kinetic properties of Escherichia coli ribosomes with altered forms of S12. J Mol Biol. 1992 Apr 20;224(4):1011–1027. doi: 10.1016/0022-2836(92)90466-w. [DOI] [PubMed] [Google Scholar]
- Chinali G., Wolf H., Parmeggiani A. Effect of kirromycin on elongation factor Tu. Location of the catalytic center for ribosome-elongation-factor-Tu GTPase activity on the elongation factor. Eur J Biochem. 1977 May 2;75(1):55–65. doi: 10.1111/j.1432-1033.1977.tb11503.x. [DOI] [PubMed] [Google Scholar]
- Clark B. F., Kjeldgaard M., la Cour T. F., Thirup S., Nyborg J. Structural determination of the functional sites of E. coli elongation factor Tu. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):203–208. doi: 10.1016/0167-4781(90)90167-z. [DOI] [PubMed] [Google Scholar]
- Hausner T. P., Atmadja J., Nierhaus K. H. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie. 1987 Sep;69(9):911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- Kaziro Y. The role of guanosine 5'-triphosphate in polypeptide chain elongation. Biochim Biophys Acta. 1978 Sep 21;505(1):95–127. doi: 10.1016/0304-4173(78)90009-5. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M., Nissen P., Thirup S., Nyborg J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993 Sep 15;1(1):35–50. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M., Nyborg J. Refined structure of elongation factor EF-Tu from Escherichia coli. J Mol Biol. 1992 Feb 5;223(3):721–742. doi: 10.1016/0022-2836(92)90986-t. [DOI] [PubMed] [Google Scholar]
- Mesters J. R., Zeef L. A., Hilgenfeld R., de Graaf J. M., Kraal B., Bosch L. The structural and functional basis for the kirromycin resistance of mutant EF-Tu species in Escherichia coli. EMBO J. 1994 Oct 17;13(20):4877–4885. doi: 10.1002/j.1460-2075.1994.tb06815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Robertson J. M., Noller H. F. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988 Jul 28;334(6180):362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- Moras D., Dock A. C., Dumas P., Westhof E., Romby P., Ebel J. P., Giegé R. The structure of yeast tRNA(Asp). A model for tRNA interacting with messenger RNA. J Biomol Struct Dyn. 1985 Dec;3(3):479–493. doi: 10.1080/07391102.1985.10508436. [DOI] [PubMed] [Google Scholar]
- Pai E. F., Krengel U., Petsko G. A., Goody R. S., Kabsch W., Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990 Aug;9(8):2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmeggiani A., Swart G. W. Mechanism of action of kirromycin-like antibiotics. Annu Rev Microbiol. 1985;39:557–577. doi: 10.1146/annurev.mi.39.100185.003013. [DOI] [PubMed] [Google Scholar]
- Paulsen H., Robertson J. M., Wintermeyer W. Effect of ribosome binding and translocation on the anticodon of tRNAPhe as studied by wybutine fluorescence. Nucleic Acids Res. 1982 Apr 24;10(8):2651–2663. doi: 10.1093/nar/10.8.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen H., Robertson J. M., Wintermeyer W. Topological arrangement of two transfer RNAs on the ribosome. Fluorescence energy transfer measurements between A and P site-bound tRNAphe. J Mol Biol. 1983 Jun 25;167(2):411–426. doi: 10.1016/s0022-2836(83)80342-8. [DOI] [PubMed] [Google Scholar]
- Peter M. E., Schirmer N. K., Reiser C. O., Sprinzl M. Mapping the effector region in Thermus thermophilus elongation factor Tu. Biochemistry. 1990 Mar 20;29(11):2876–2884. doi: 10.1021/bi00463a033. [DOI] [PubMed] [Google Scholar]
- Powers T., Noller H. F. Evidence for functional interaction between elongation factor Tu and 16S ribosomal RNA. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1364–1368. doi: 10.1073/pnas.90.4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., Noller H. F. The 530 loop of 16S rRNA: a signal to EF-Tu? Trends Genet. 1994 Jan;10(1):27–31. doi: 10.1016/0168-9525(94)90016-7. [DOI] [PubMed] [Google Scholar]
- Rigler R., Wintermeyer W. Dynamics of tRNA. Annu Rev Biophys Bioeng. 1983;12:475–505. doi: 10.1146/annurev.bb.12.060183.002355. [DOI] [PubMed] [Google Scholar]
- Robertson J. M., Wintermeyer W. Effect of translocation on topology and conformation of anticodon and D loops of tRNAPhe. J Mol Biol. 1981 Sep 5;151(1):57–79. doi: 10.1016/0022-2836(81)90221-7. [DOI] [PubMed] [Google Scholar]
- Rodnina M. V., Fricke R., Wintermeyer W. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry. 1994 Oct 11;33(40):12267–12275. doi: 10.1021/bi00206a033. [DOI] [PubMed] [Google Scholar]
- Rodnina M. V., Wintermeyer W. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc Natl Acad Sci U S A. 1995 Mar 14;92(6):1945–1949. doi: 10.1073/pnas.92.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M. V., Wintermeyer W. Two tRNA-binding sites in addition to A and P sites on eukaryotic ribosomes. J Mol Biol. 1992 Nov 20;228(2):450–459. doi: 10.1016/0022-2836(92)90834-7. [DOI] [PubMed] [Google Scholar]
- Swart G. W., Parmeggiani A., Kraal B., Bosch L. Effects of the mutation glycine-222----aspartic acid on the functions of elongation factor Tu. Biochemistry. 1987 Apr 7;26(7):2047–2054. doi: 10.1021/bi00381a038. [DOI] [PubMed] [Google Scholar]
- Tapprich W. E., Dahlberg A. E. A single base mutation at position 2661 in E. coli 23S ribosomal RNA affects the binding of ternary complex to the ribosome. EMBO J. 1990 Aug;9(8):2649–2655. doi: 10.1002/j.1460-2075.1990.tb07447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. C., Dix D. B., Karim A. M. The reaction of ribosomes with elongation factor Tu.GTP complexes. Aminoacyl-tRNA-independent reactions in the elongation cycle determine the accuracy of protein synthesis. J Biol Chem. 1986 Apr 15;261(11):4868–4874. [PubMed] [Google Scholar]
- Thompson R. C. EFTu provides an internal kinetic standard for translational accuracy. Trends Biochem Sci. 1988 Mar;13(3):91–93. doi: 10.1016/0968-0004(88)90047-3. [DOI] [PubMed] [Google Scholar]
- Tubulekas I., Hughes D. A single amino acid substitution in elongation factor Tu disrupts interaction between the ternary complex and the ribosome. J Bacteriol. 1993 Jan;175(1):240–250. doi: 10.1128/jb.175.1.240-250.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf H., Chinali G., Parmeggiani A. Mechanism of the inhibition of protein synthesis by kirromycin. Role of elongation factor Tu and ribosomes. Eur J Biochem. 1977 May 2;75(1):67–75. doi: 10.1111/j.1432-1033.1977.tb11504.x. [DOI] [PubMed] [Google Scholar]
- la Cour T. F., Nyborg J., Thirup S., Clark B. F. Structural details of the binding of guanosine diphosphate to elongation factor Tu from E. coli as studied by X-ray crystallography. EMBO J. 1985 Sep;4(9):2385–2388. doi: 10.1002/j.1460-2075.1985.tb03943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



