Abstract
Whether MHC restriction by the T cell receptor (TCR) is a product of evolutionary pressures leading to germline-encoded ‘rules of engagement’ remains avidly debated. Structural results derived from analysis of TCR–peptide–MHC complexes appear to support a model of physical specificity between TCR germline V regions and MHC. Yet, some recent evidence suggests that thymic selection, and co-receptors may have misled us into thinking the TCR is exclusively MHC-specific, when in fact, TCRs can robustly engage non-MHC ligands when given the chance. Here, I propose that seemingly contradictory data and hypotheses for, and against, germline bias are, in fact, compatible and can be reconciled into a unifying model.
Structural basis for TCR recognition of peptide and MHC
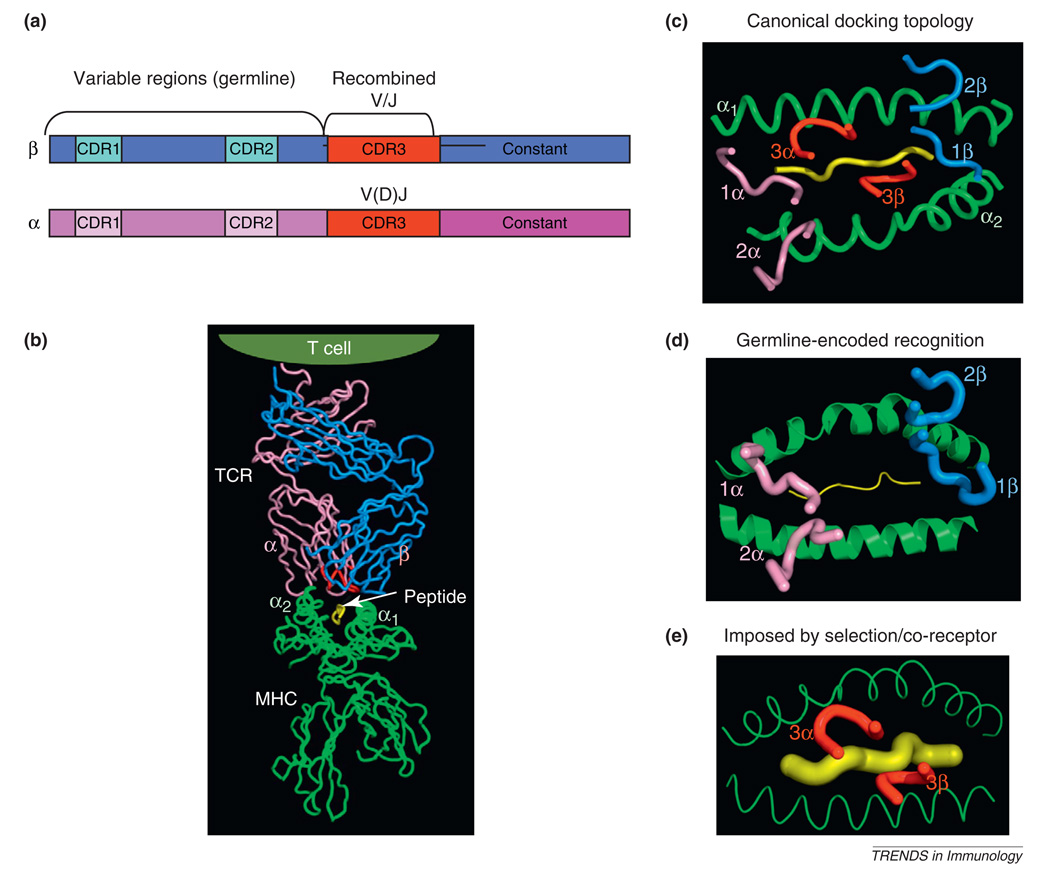
The conundrum that TCRs can apparently recognize only MHC, yet at the same time any MHC, is an intriguing concept for structural biologists [1–10]. There is no protein–protein interaction system in nature where two different structural families of proteins are specific for one another, yet at the same time, nearly entirely cross-reactive with one another. At first, models of TCR–MHC recognition were inferred from the structures of antigen receptor genes, which are segregated into germline-derived (V regions) and recombined segments (V/D/J) that yield germline-encoded CDR1 and CDR2 (germline) and CDR3 (recombined) loops in the protein products, respectively (Figure 1a, b) [11–13]. Subsequently, a wealth of structural data has shown us that the TCR recognizes peptide MHC such that the variable portions of the TCR (CDR3s) and MHC (the antigenic peptide) are apposed within the TCR–pMHC interface, whereas the conserved parts (the germline-encoded CDR1s and CDR2s, and the tops of the MHC helices) are also matched across the interface (Figure 1b, c) [7,14–17]. The segregation of CDR3 contact with peptide and CDRs 1 and 2 contact with MHC are not absolute as most structures usually contain some degree of CDR3 interaction with the MHC helices, and CDR1 and 2 interactions with peptide [18]. It is a fair generalization, however, that the structural segregation of the TCR and MHC genes (Figure 1a), which contain regions of high sequence diversity and regions of high conservation [12], are complementarily manifested in the binding interface of the TCR–pMHC complex (Figure 1c). Variations on this theme exist [19,20], but this is how ‘conventional’ TCRs usually engage peptide–MHC molecules.
Figure 1.
Structural models of T cell receptor (TCR) recognition of peptide–MHC. (a) Schematic of TCR α and β chain genes showing segregation of the germline and recombined regions. (b) Structure of an αβ TCR bound to a class I MHC presenting a peptide antigen [56]. The CDR3 loops in the TCR binding site are colored red. (c) Classical ‘footprint’ view of the CDR loops of the TCR overlaying the peptide and MHC. This view shows the roughly diagonal binding orientation with the germline-derived CDRs 1 and 2 primarily engaging the MHC helices, and the TCR CDR3s engaging the peptide. The angular orientation of these CDR loops to the long axis of the MHC groove is referred to as the docking angle or docking topology. (d) The germline hypothesis is indicated by the primacy of the CDR1 and CDR2 germline derived TCR loops forming the principal interactions with the MHC independently of CDR3. (e) In contrast to (d), the alternative model proposes that the CDR3–peptide interaction, or the influence of co-receptor during thymic selection, underlies TCR–MHC specificity irrespective of germline influences.
Here, I discuss disparate viewpoints on the subject of how the TCR seems biased for recognizing MHC molecules presenting peptides [1–10], in contrast to antibodies, which are not restricted to recognizing certain classes of ligands. My views on this are decidedly reductionist and structurally based, deriving from both evolutionary arguments and my understanding of chemical principles governing protein–protein interactions. For a counterpoint to the opinion that TCR–MHC recognition is in large driven by germline-encoded interactions, see [21].
Germline-encoded MHC recognition
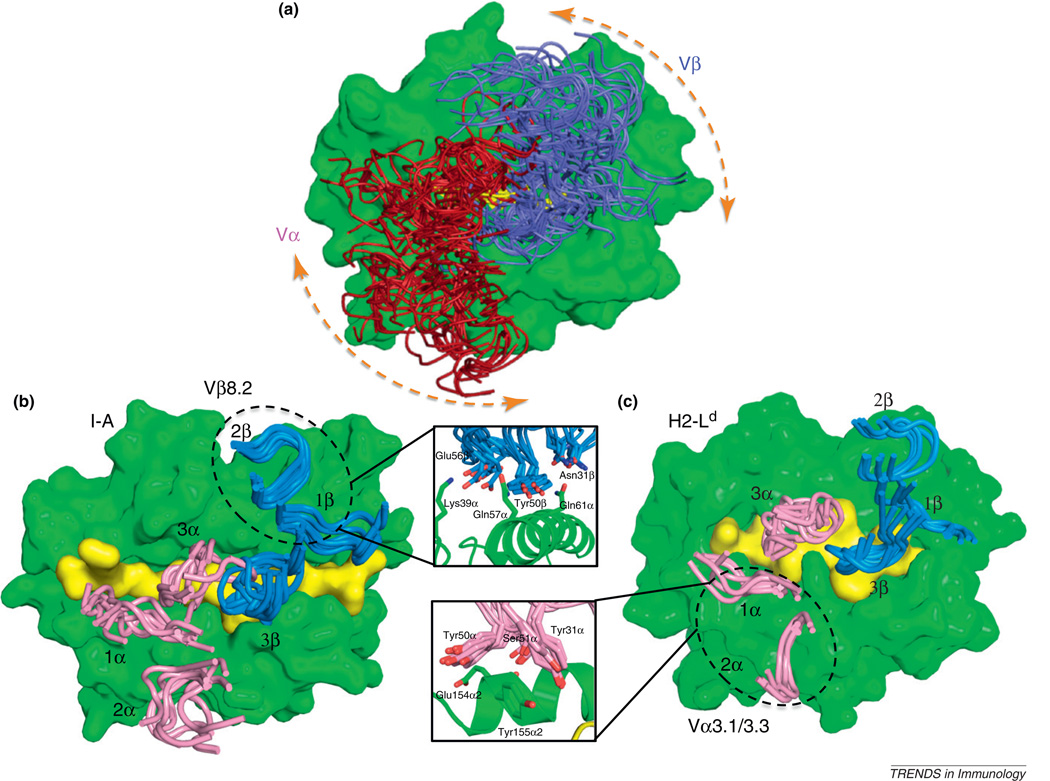
The conundrum at issue here is whether the TCR–MHC recognition mode is determined by co-evolved germline TCR–MHC amino acid contacts [6,8], mediated by CDRs 1 and 2 (Figure 1d). Or, alternatively, whether the TCR repertoire is actually not innately specific for MHC, but instead is subject to other externally imposed influences that lead to the appearance of a recognition bias that may not really exist (Figure 1e) [5,22]. One possibility is that TCR–MHC engagement is simply a product of CDR3–peptide interactions that are selected during thymic education [23], and that the germline components are ‘along for the ride’ (Figure 1e). Another possibility is that the TCR and MHC are compelled to interact as a result of co-receptor engagement, such as with CD8 or CD4; a view advocated in several intriguing publications from the Singer group [22,24], which are further elaborated upon in this issue of Trends in Immunology [21]. What has confused a clear structural resolution of this issue is that, although this basic structural paradigm (Figure 1b, c) holds for the vast majority of the TCR–pMHC structures determined so far, there is a great deal of variation in the binding geometry, or ‘docking angle’ in the ~50 different complexes seen so far (nearly 120 degrees) (Figure 2a). Clearly conserved interactions across the complexes that would give us a clue as to the basis of TCR–MHC bias have not been evident from this type of analysis. Yet, even with this degree of variability in binding solutions, one constant is that the Vα and Vβ regions of the TCR are always oriented over the MHC with a defined polarity: Vα or the α2 helix and Vβ over α1 or β1 helices (Figures 1c and 2a). Thus, there is at the same time convergence and divergence. How can we explain TCR–MHC germline specificity in the absence of conserved interactions?
Figure 2.
Interaction codons mediating germline specificity of the T cell receptor (TCR) for MHC. (a) This panel shows the wide range of TCR docking angles on MHC seen from an overlap for ~40 different class I TCR–MHC complex crystal structures. Conserved interactions that might denote germline contacts have not been evident from this type of analysis. (b) Focused structural studies of Vβ8.2 containing TCRs recognizing similar type of MHC (class II I-A allotypes) reveal a highly convergent mode of germline contact between the TCR CDR1β and CDR2β with the MHC, which we refer to as an interaction codon. The inset shows the amino acids involved in this contact, and the degree of superposition, and also wobble, in the manner by which approximately 10 Vβ8.2 TCRs engage with I-A MHC. (c) Another germline codon, involving a TCR Vα3 and class I MHC H-2Ld also indicates a close structural superposition.
My own view that TCR–MHC recognition is driven by germline-encoded interactions has been influenced by previous functional data [25–28], and by experiments in my laboratory where structural analysis has been combined with protein engineering. I have been guided by a simple principle that is agnostic about the biological origins of the system: Res ipsa loquitor (‘the thing speaks for itself’). In my opinion, basic principles of structural energetics tell us that the germline contacts from CDRs 1 and 2 with the MHC helices, as resolved in TCR–pMHC complex crystal structures (e.g., Figure 1c), have been evolutionarily refined, and engrafted in the germline. I have difficulty accepting that these germline contacts, which comprise almost 75% of most TCR–pMHC binding interfaces (Figure 1d), are externally imposed, or are simply opportunistic bystanders that secondarily form as a result of a very small portion of the interface (CDR3–peptide) (Figure 1e) engaging in the key interactions.
About 10 years ago, I concluded that the different TCR–pMHC complexes solved contained different combinations of TCR V regions, peptides and MHC, and so could not be compared directly to elucidate regions of conservation (e.g., Figure 2a). Therefore, we set out in my laboratory to characterize structurally TCR containing a single commonly occurring V region, Vβ8.2, complexed with defined class I and II peptide–MHC ligands to determine if we could penetrate this question in the old fashioned way, by changing one variable at a time. In the class II system, my group and that of John Kappler and Pippa Marrack independently determined that Vβ8.2 in different TCR from different systems, when paired with distinct Vα chains and possessing different CDR3 sequences recognizing different peptides, converged on a similar binding mode with the tops of the helices of class II MHC I-A allotypes (I-Au, I-Ab) (Figure 2b) [26,29]. This motif was nearly identical to the Vβ8.2 interaction seen earlier from the Reinherz group in the first TCR complex structure with a class II pMHC ligand (I-AK) [30]. A striking feature of this contact was that a tyrosine residue at the tip of CDR2β, together with several other amino acids at the tips of CDR1β and CDR2β, played central roles in contacting the I-A helix (Figure 2b, inset). My group euphemistically referred to this conserved motif as a codon of MHC restriction [31], to highlight the mutual specificity between TCR germline and MHC in this set of amino acid contacts. There are now a total of nearly 10 resolved complexes (some unpublished) of Vβ8.2 TCRs complexed with I-A-peptide complexes [26,30–32], and all show similar binding chemistry in the Vβ8.2–MHC points of contact (Figure 2b). A recent study has claimed to show that pairing the Vβ8.2 with a different Vα results in the Vβ8.2–MHC interactions changing substantially [32]. However, a close inspection of this structure shows the Vβ8.2–I-A contacts still to be similar to those that had been determined previously (Figure 2b).
Importantly, the germline-derived amino-acid contacts in the various Vβ8.2-containing TCR complexes are not atomically identical (Figure 2b, inset). There is flexibility in the bonding patterns to accommodate differences in peptide and Vα partners, but the similarities are more pronounced than the differences. The flexibility of this codon to recognize different MHC ligands is enhanced by the presence of the tyrosine at the tip of CDR2β, which is an amino acid with the ability to engage in a wide range of different types of bonding interactions. In fact, the TCR–MHC interface does seem to tolerate structural ‘wobble’ in the pairwise contacts on the order of several angstroms and degrees in translational and rotational shifts, respectively, while maintaining an overall docking convergence [33]. Thus, the nature of the germline interactions is such that they can tolerate some conformational ‘slippage’ in accommodation with the changing peptide, Vα and CDR3 environment [34]. Nevertheless, the convergence of this structural motif, or codon, provides strong evidence that there is, indeed, chemical specificity between germline TCR domains and the MHC helices mediated by mutual complementary shape and amino acid contacts. I say mutual because specificity is a product of mutual compensatory bonding interactions; in this case, on the germline CDR loops and the MHC helices. Therefore, the evolution has not been one-sided, and the TCR and MHC have co-evolved a good fit for one another. Several additional codons have been observed when structures have been analyzed where a given Vα or Vβ engages a common MHC [26,35]. For example, there appears to be a highly conserved codon for Vα3 recognition of H-2Ld (Figure 2c, and inset) [35]. This latter codon is interesting given that germline amino acids in Vα3 have been shown to play a role in CD8 versus CD4 lineage choice; presumably through distinct interactions with class I versus class II MHC [9].
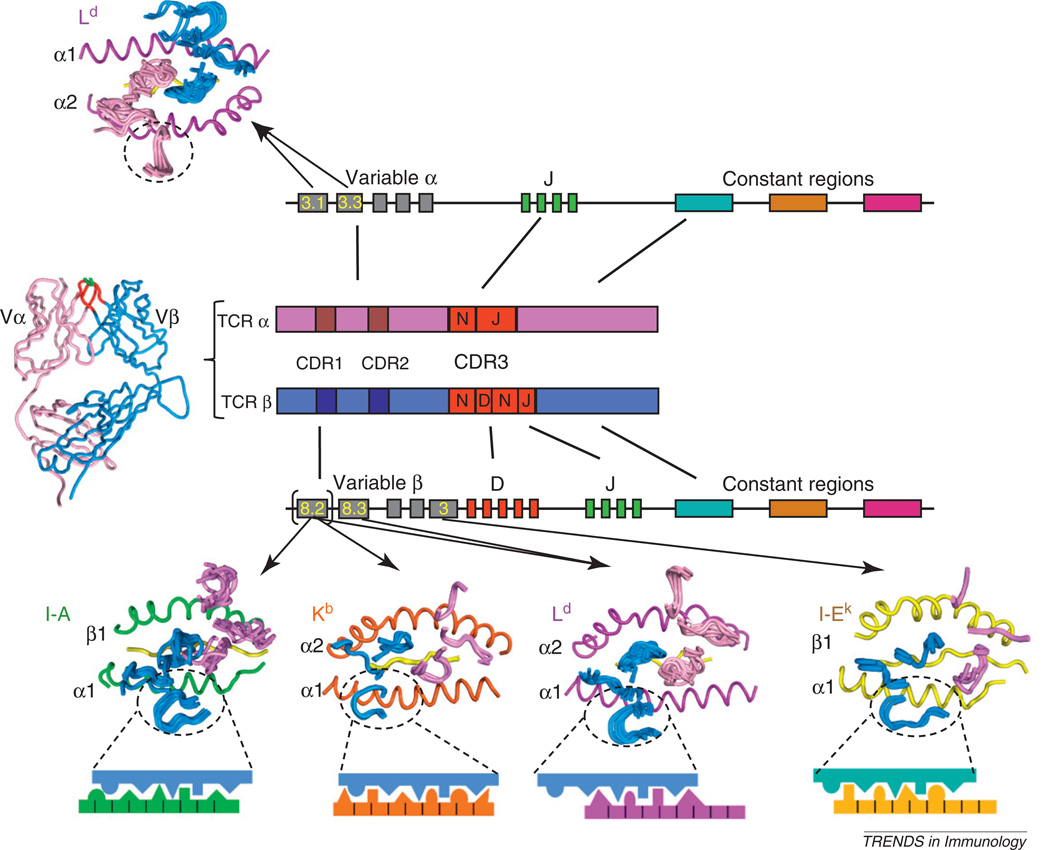
Although the codon hypothesis [6] derived from the Vβ8.2–I-A series of studies was satisfactory to explain how a certain V region might engage one type of MHC, it did not explain the fact that TCR variable regions are cross-reactive and appear to have the capacity to recognize many different types of MHC molecules. That is, a given TCR using, for example a Vβ8.2 germline component, can engage essentially any type of MHC, class I or class II. So far, clear segregation of certain V regions for recognition of certain types of MHC has not been observed. Conversely, the TCR repertoire of germline chains found in a given immune response to a specific peptide–MHC is generally rather broad, encompassing many different V-region germline types, although certain dominant V regions in TCRs are expressed in response to certain antigens. So, how do we square the codon specificity model with this apparent lack of MHC allotype specificity? To address this, we have determined crystal structures of additional Vβ8.2-containing TCRs complexed with other types of MHC, namely self and foreign class I pMHC [35–37]. In a nutshell, what we found was that the Vβ8.2 engaged in entirely distinct binding chemistries to these MHC compared to that seen from the class II complexes. Nevertheless, as we noted in the class II analysis, there is a clear convergence on common binding motifs between Vβ8.2 and each one of these other type of MHC [35–37], but each of these alternative MHCs engaged the Vβ8.2 using its own distinct binding chemistry. This convergence occurred in multiple different complexes despite the fact that the TCRs crystallized had different CDR3 sequences and/or the MHC presented different peptides. From these data, I propose that each TCR V region has evolved a distinct mode (or chemistry) of interaction with each type of MHC [6]. That is, there does not exist a shared structural signature, or epitope, present on all MHCs that TCR germline V regions seek out. Rather, each V region appears to have co-evolved unique interaction chemistries with each MHC. This explains how a given V region can cross-react with many different types of MHC, yet only react with MHC molecules, and not other non-MHC proteins. This type of cross-reactivity is not promiscuity, but rather polyspecificity, as has been discussed previously [38]. An important caveat to this model is that there is probably not a single codon for each germline TCR–MHC pair. There probably exist alternative modes, or ‘menus’ of codons from which a V region can choose to engage in germline contacts with the MHC. The selection of codon is probably influenced by the Vα pairing partner and CDR3–peptide contacts. Models of TCR scanning the MHC helices [27] suggest that a single TCR can probably ‘click’ into several different germline codons to engage a given peptide–MHC.
Functional interrogation of the germline hypothesis
It nevertheless remains possible that thymic selection, not germline bias, of certain CDR3–peptide interactions could be responsible for the TCR–MHC binding we see in ‘educated’ or thymically selected TCRs that have been exposed to MHC during development. Perhaps the prethymic αβ TCR repertoire is actually not MHC specific, but that thymic selection somehow excludes non-MHC-binding TCR, and allows only those TCRs that bind MHC (by virtue of CDR3–peptide complementarity) to exit into the periphery. Given that the TCRs we studied in my laboratory were of the ‘garden-variety’, thymically selected type, perhaps we were simply ‘looking for the keys under the lamp post.’ What was needed was a way to inspect unselected TCRs. Two groups have addressed this issue with a series of very elegant experiments [26,39,40]. They have isolated TCRs from single-chain peptide–MHC transgenic mice, which essentially had ‘leaky’ negative selection that allowed TCRs that would normally have been deleted to survive and enter the periphery; essentially slipping through due to a lack of appropriate negatively selecting peptide ligands. These prethymic Vβ8.2-containing TCRs were then isolated and analyzed for their recognition properties and structure [26,39]. Strikingly, these TCRs not only recognized MHC molecules, but crystal structures of complexes with I-A MHC showed that they also bound in a nearly indistinguishable manner from thymically selected TCRs that my group has resolved. Additional studies have shown that the central Tyr50 on the TCR Vβ8.2 is crucial for thymic selection of the TCR repertoire [1], and that the same motif is found in TCRβ chains from bony fish, and is necessary for generic recognition of mouse MHC [41]. These results, derived from both structural and functional approaches, clearly indicate that MHC specificity, and the Vβ8.2/I-A structural codon, are resident in the TCR repertoire before thymic selection.
In my own group, we carried out an analogous experiment that did not involve mice, but instead used combinatorial biology and protein engineering. The concept of the experiment was to ‘wag the dog’ of selected TCRs, where we refer to the tail as the recombined CDR3–peptide interactions, and the dog equates to the CDR1/2–MHC germline interactions. We asked if we remodeled the CDR3 sequences that were interacting with the peptide (i.e., if we wagged the tail), would the dog move? Conversely, would remodeling the peptide sequence engaging the CDR3s (again wagging the tail) disrupt the germline contacts (move the dog). Collectively, we have determined many structures of TCRs with entirely different CDR3s that have been randomized and selected for pMHC binding by yeast display [36,37,42]. Recently, we have also developed a yeast peptide–MHC library method to discover alternative peptide sequences so that we can wag the tail using the peptide instead of CDR3 [34]. The collective results of these studies have shown that, in all cases but one (discussed below), the TCR persists in canonical germline docking modes (with slight slippage), regardless of variations in the CDR3 and peptide interactions. Here again, this provides strong evidence that the germline contacts are exerting a dominant structural pressure to persist in a certain docking geometry, and are not assuming a passive role in deference to CDR3. These results are especially compelling given that these engineering experiments took place using recombinant molecules in a cell-free environment, in the absence of any co-receptors that might, in principle, have exerted some influence on the TCR binding mode.
Even with the collective weight of the structural results, there remain several unresolved problems with this model that so far defy explanation. There exist several unconventional complexes in which the TCR binding mode is difficult to explain by germline specificity. For example, a human autoreactive TCR binds far down to one end of the MHC groove, in such a way that only minimal contacts are made with the CDR1 and CDR2 loops, and almost all of the interface contacts are mediated by CDR3 [19]. Another curious example is a human class I TCR–MHC complex in which the peptide is radically bulged out of the groove [20,43]. The TCR binds to the protruding peptide almost entirely through CDR3 contact, much in the manner of an antibody–peptide interaction, whereas the germline loops engage the MHC minimally through a triad of pairwise contacts. These systems are unconventional compared to the vast majority of complexes and they cannot be rationalized easily based on the codon model, unless the minimal degree of germline contact seen in these structures is enough to generate specificity [18]. They do share one important feature with all other TCR–pMHC complexes determined, however. The TCR–MHC docking polarity (i.e., the locations of the Vα and Vβ chains with respect to the α1/β1 and α2 helices) remains consistent with the current paradigm, so these complexes have not ‘officially’ broken a very important tenet of germline specificity, but they do push the envelope of the germline concept. Confusion also arises when trying to distinguish wobble from an alternative codon [18,33,32]. How much wobble is allowed before it is no longer a germline motif? How do we delineate between codon slippage, from transition into a completely distinct binding chemistry that may, or may not be germline encoded? For those opposed to germline bias, these tolerances might be small, whereas for those supporting the primacy of germline bias, the tolerances might be more liberally interpreted.
CD4 and CD8 co-receptors and MHC restriction
Recently, a new and intriguing challenge to the germline specificity model has arisen from the Singer group [22,24]. They have generated ‘quad-deficient’ mice lacking CD8, CD4, MHC-I and MHC-II, and have found that T cells from these mice can be activated by non-MHC ligands [22]. One of these ligands was recently isolated and identified as CD155, which is a murine homolog of the human poliovirus receptor [24]. Rigorous biochemical and cellular experiments clearly show that TCRs from these quad-deficient mice bind to recombinant CD155 with high affinity (KD ~200 nM), that the interaction requires αβ chain pairing and CD155 glycosylation, and both TCR CDR3 and CDR1/2 germline residues are important for the interaction. They have concluded that the αβ TCR is binding to the non-MHC ligand in a manner reminiscent of an antibody–antigen interaction. The conceptual interpretation of this elegant and convincing work is that MHC bias is externally imposed on the TCR during thymic selection; principally by the fact that the CD8 and CD4 co-receptors bind to MHC, and essentially limit the TCR from seeing other potential ligands. The co-receptors also funnel the TCR to only signal-inducing ligands (i.e., MHC) by virtue of their association with Lck44. By sequestering intracellular Lck, the co-receptors essentially co-opt the signaling kinase from the intracellular pool, which excludes non-co-receptor associated TCRs from signaling when they might engage a non-MHC ligand. In a cell with no co-receptors, where the Lck pool is free floating and available to all TCRs, Singer et al. have proposed that the reactivity of TCR with non-MHC ligands is revealed.
This is a fascinating result. Non-MHC ligand recognition by αβ TCRs was also shown as early as 1992 [44]. However, I think it remains perfectly compatible with notions of a physical germline bias consistent with the structural and functional evidence I have discussed. Germline bias does not denote a negative bias towards non-MHC ligands, but rather a positive bias for MHC. The TCR is an antibody-like molecule with highly diverse CDR3 loops. Certainly from a structure and chemistry standpoint, a repertoire of αβ TCRs with diverse CDR3 loops can, in principle, recognize other ligands if exposed to enough of them. For example, phage libraries exist for antibodies and TCRs where CDR3 loops are randomized, and the libraries are selected for binding to any desired ligand. There is no question that these libraries are capable of selecting antibody or TCR variants with reasonable, or even, high affinity for virtually any ligand. Antibodies have also been shown to possess dual specificity [45]. I would imagine this is also true in the context of the extracellular milieu of a TCR. When an environment is devoid of MHC and co-receptors, TCRs will no doubt find other ligands to engage; probably including secreted proteins. If the other ligand is multivalent, such as would be the case for a membrane protein such as CD155 that is presented on another cell surface, then one could easily imagine the TCR being activated after CD155 binding, although the evidence from the Singer studies falls short of demonstrating that CD155 is a specific signaling ligand for T cells that is important in T cell biology. Clearly, elimination of co-receptors and MHC places the TCR in an ideal environment for engaging other ligands where cross-reactivity will be unmasked. However, it is important to distinguish spurious cross-reactivity of a binding site that is ‘tuned’ for ligand recognition, from a bona fide intended and specific interaction. The question remains whether non-MHC ligands are physiologically relevant or whether the unmasking of non-MHC reactivity is a relic of an early, ancient form of a TCR that possesses germline antibody-like ligand recognition characteristics, which are no longer functionally important. There may turn out to be cases in which non-MHC ligands could be physiologically important in some way, although this remains to be proven. However, the salient issue for this essay is that I do not think that the identification of non-MHC ligands that activate αβ TCRs negates, or even weakens, the notion that the TCR germline is evolved to recognize MHC. The idea of non-MHC engagement by the TCR is entirely compatible with germline specificity for MHC and I still am inclined to believe that the vast majority of TCRs rely on MHC for their ‘day jobs’.
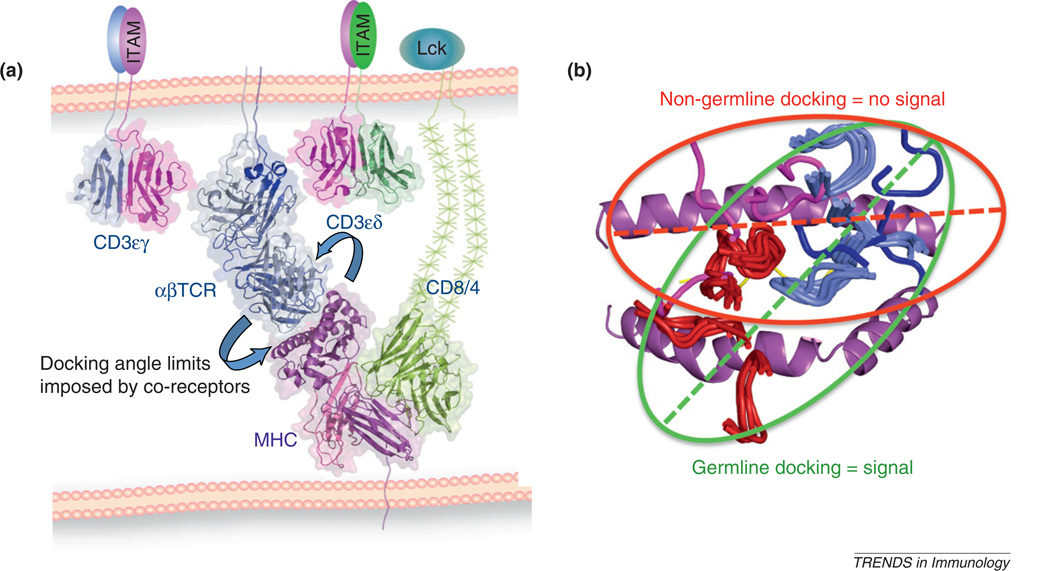
A different role for co-receptors in influencing TCR recognition of MHC has also been suggested from a structural study from the Collins group [46]. A handful of CD8-dependent and independent TCR–MHC docking angles were analyzed and it was concluded that there were differences between these two groups; possibly because of CD8 exerting a steric influence on the docking mode for CD8-dependent TCRs, and physically participating in positioning the TCR on the MHC. The difficulty with this model (Figure 3) is that there is scant evidence for direct contact between TCRs and CD8 or CD4, and in fact the architecture of MHC–co-receptor complexes makes this scenario unlikely within a unitary TCR–pMHC–CD8 or CD4 complex [47–49]. In fact, a recent landmark structure determination of the full ternary complex of TCR–pMHC and CD4 unequivocally shows the relative orientations of these molecules in a signaling complex and it appears virtually impossible for the TCR to have simultaneous contact with the co-receptor [49]. However, this ternary complex does, indeed, make a strong argument that steric influences can, and probably do, influence how the TCR docks to pMHC to generate a signaling response by virtue of the relative proximities of the CD4-associated Lck to the CD3 immunoreceptor tyrosine-based activation motif (ITAM) motifs. This relative distance is influenced by both co-receptor and TCR–pMHC engagement geometry (see Figure 5 of Yin et al. [49]). Second, a great deal more structural information has accumulated since 2003 showing that there are wide variations in TCR–MHC docking angles (Figure 2a) that exceed the small differences seen in the limited set of structures analyzed in Buslepp et al. [46], calling into question the significance of the observed differences with co-receptor dependence. However, I think it is likely that co-receptors do influence the docking topology of the TCR–MHC complex indirectly within the context of a clustered multimeric signaling complex, and recent results underscore this likelihood [49].
Figure 3.
Model for germline specificity of the T cell receptor (TCR) V region repertoire for MHC. TCR–MHC binding footprints, represented by the TCR CDR loops only, are shown for several different TCR germline types that appear illustrative of distinct specificities of each type of V region for each type of MHC. Note that, for the example of Vβ8.2 recognizing three different types of MHC, the chemistry of the interactions differ (cartoon blowups) yet within each MHC the interactions are similar.
A unifying hypothesis
I conclude by proposing a unifying hypothesis that reconciles apparently conflicting models (Figure 4a). I believe that the fully assembled oligomeric TCR–CD3–CD4–CD8–MHC signaling complex imposes steric requirements for concurrent association of all of the components – either through direct subunit contact or through indirect geometric requirements (Figure 4a) [50,51]. The TCR associates with both CD3 and MHC and MHC is also engaged with its co-receptor. This ternary complex then dimerizes [52,53], at least for signaling to occur, in what appears to be a geometrically precise manner [54]. The classical Janeway anti-TCR antibody experiments have shown that the oligomeric geometry is not completely permissive [55], although it is probably rather forgiving as studies with defined pMHC oligomers seem to indicate [53]. The multi-point attachments within the complex, taken together with some liberal limits on the architecture of the association modes between the oligomers, probably restricts the range of TCR–MHC docking angles compatible with multi-subunit and multivalent assembly (Figure 4a). In this sense, the TCR–MHC docking angle is probably limited by extrinsic factors imposed by its structural surroundings, as illustrated in the ternary complex structure [49]. Extending this idea, it also seems probable that the specific germline-derived chemistry between TCR and MHC probably evolved from TCR–MHC docking topologies that are compatible with concurrent co-receptor association in the signaling complex. My group recently has reported a result that appears to support this conclusion (Figure 4b) [35]. We have discovered a peptide, in association with MHC, that the TCR recognizes in a highly divergent docking mode from the canonical germline codon used by the TCR to recognize the natural endogenous antigen. In contrast to agonist peptides that all induce signaling, and also assuming a common docking mode, we have found that this particular peptide fails to induce signaling (Figure 4b). It is tempting to speculate from this admittedly singular result that germline specificity serves a functional role to ensure that a binding event takes place in a manner that is conducive to signaling, and this reflects the evolutionary imprint of co-receptor influence. This interpretation brings us full circle to the question of why germline bias might be important for TCR function beyond simply promoting recognition of many different MHCs.
Figure 4.
Constraints on germline-encoded T cell receptor (TCR)–MHC engagement imposed by co-receptors. (a) Model of a signaling complex of TCR–CD3–peptide–MHC and CD8 intended to indicate the geometric constraints imposed by simultaneous association of all of the subunits that might limit the TCR–pMHC docking angle. (b) Docking footprints of signaling-productive, presumably germline-encoded TCR docking footprints that are highly convergent shown compared to a non-signaling docking footprint that has potentially exceeded the geometric tolerances highlighted in panel A.
Concluding remarks
In summary, I think that the many lines of evidence for, and against, the concept of TCR germline specificity for MHC can be reconciled. Certainly, exceptions to these rules exist, and will continue to emerge. However for most conventional TCR–pMHC interactions these rules of engagement probably represent a sound basis for understanding the physical basis of MHC restriction. No doubt the Singer group will interrogate the functional relevance of their fascinating case of a non-MHC ligand activating a TCR. The results of such efforts could further modify our view of this complex and, one might even say, existential issue.
Acknowledgments
I gratefully acknowledge many helpful discussions with Jay Adams, Michael Birnbaum, David Kranz, Pippa Marrack, Mark Davis and Paul Allen. I am funded by NIH (AI48540) and HHMI.
References
- 1.Scott-Browne JP, et al. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huseby E, et al. TCR-MHC/peptide interactions: kissing-cousins or a shotgun wedding? Eur. J. Immunol. 2004;34:1243–1250. doi: 10.1002/eji.200425000. [DOI] [PubMed] [Google Scholar]
- 3.Felix NJ, Allen PM. Specificity of T-cell alloreactivity. Nat. Rev. Immunol. 2007;7:942–953. doi: 10.1038/nri2200. [DOI] [PubMed] [Google Scholar]
- 4.Germain RN. Immunology. Making a molecular match. Nature. 1990;344:19–22. doi: 10.1038/344019a0. [DOI] [PubMed] [Google Scholar]
- 5.Collins EJ, Riddle DS. TCR-MHC docking orientation: natural selection, or thymic selection? Immunol. Res. 2008;41:267–294. doi: 10.1007/s12026-008-8040-2. [DOI] [PubMed] [Google Scholar]
- 6.Garcia KC, et al. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat. Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Housset D, Malissen B. What do TCR–pMHC crystal structures teach us about MHC restriction and alloreactivity? Trends Immunol. 2003;24:429–437. doi: 10.1016/s1471-4906(03)00180-7. [DOI] [PubMed] [Google Scholar]
- 8.Marrack P, et al. Evolutionarily conserved amino acids that control TCR–MHC interaction. Annu. Rev. Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sim BC, et al. Control of MHC restriction by TCR Valpha CDR1 and CDR2. Science. 1996;273:963–966. doi: 10.1126/science.273.5277.963. [DOI] [PubMed] [Google Scholar]
- 10.Jerne NK. The somatic generation of immune recognition. Eur. J. Immunol. 1971;1:1–9. doi: 10.1002/eji.1830010102. [DOI] [PubMed] [Google Scholar]
- 11.Bjorkman PJ, Davis MM. Model for the interaction of T-cell receptors with peptide/MHC complexes. Cold Spring Harb. Symp. Quant. Biol. 1989;54:365–373. doi: 10.1101/sqb.1989.054.01.045. [DOI] [PubMed] [Google Scholar]
- 12.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 13.Jorgensen JL, et al. Mapping T-cell receptor-peptide contacts by variant peptide immunization of single-chain transgenics. Nature. 1992;355:224–230. doi: 10.1038/355224a0. [DOI] [PubMed] [Google Scholar]
- 14.Garcia KC, Adams EJ. How the T cell receptor sees antigen—a structural view. Cell. 2005;122:333–336. doi: 10.1016/j.cell.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 15.Rudolph MG, et al. How TCRs bind MHCs, peptides, and coreceptors. Ann. Rev. Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 16.Yin Y, Mariuzza RA. The multiple mechanisms of T cell receptor cross-reactivity. Immunity. 2009;31:849–851. doi: 10.1016/j.immuni.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Turner SJ, et al. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 2006;6:883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 18.Burrows SR, et al. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10608–10613. doi: 10.1073/pnas.1004926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn M, et al. Unconventional topology of self peptide–major histocompatibility complex binding by a human autoimmune T cell receptor. Nat. Immunol. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tynan FE, et al. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat. Immunol. 2005;6:1114–1122. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- 21.Van Laethem F, et al. MHC restriction is imposed on a diverse T cell receptor repertoire by CD4 and CD8 co-receptors during thymic selection. Trends Immunol. 2012;33:437–441. doi: 10.1016/j.it.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Laethem F, et al. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity. 2007;27:735–750. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Bevan MJ. In thymic selection, peptide diversity gives and takes away. Immunity. 1997;7:175–178. doi: 10.1016/s1074-7613(00)80520-8. [DOI] [PubMed] [Google Scholar]
- 24.Tikhonova AN, et al. αβ T cell receptors that do not undergo major histocompatibility complex-specific thymic selection possess antibody-like recognition specificities. Immunity. 2012;36:79–91. doi: 10.1016/j.immuni.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zerrahn J, et al. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 26.Dai S, et al. Crossreactive T cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu LC, et al. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 2002;418:552–556. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- 28.Merkenschlager M, et al. How many thymocytes audition for selection? J. Exp. Med. 1997;186:1149–1158. doi: 10.1084/jem.186.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng J, et al. Convergence on a distinctive assembly mechanism by unrelated families of activating immune receptors. Immunity. 2005;22:427–438. doi: 10.1016/j.immuni.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinherz EL, et al. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- 31.Feng D, et al. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat. Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 32.Stadinski BD, et al. A role for differential variable gene pairing in creating T cell receptors specific for unique major histocompatibility ligands. Immunity. 2011;35:694–704. doi: 10.1016/j.immuni.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borbulevych OY, et al. TCRs used in cancer gene therapy cross-react with MART-1/Melan-A tumor antigens via distinct mechanisms. J. Immunol. 2011;187:2453–2463. doi: 10.4049/jimmunol.1101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borbulevych OY, et al. Conformational melding permits a conserved binding geometry in TCR recognition of foreign and self molecular mimics. J. Immunol. 2011;186:2950–2958. doi: 10.4049/jimmunol.1003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams JJ, et al. T cell receptor signaling is limited by docking geometry to peptide–major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colf LA, et al. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 37.Jones LL, et al. Distinct CDR3 conformations in TCRs determine the level of cross-reactivity for diverse antigens, but not the docking orientation. J. Immunol. 2008;181:6255–6264. doi: 10.4049/jimmunol.181.9.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wucherpfennig KW, et al. Polyspecificity of T cell and B cell receptor recognition. Semin. Immunol. 2007;19:216–224. doi: 10.1016/j.smim.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huseby ES, et al. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 40.Ignatowicz L, et al. The repertoire of T cells shaped by a single MHC/peptide ligand. Cell. 1996;84:521–529. doi: 10.1016/s0092-8674(00)81028-4. [DOI] [PubMed] [Google Scholar]
- 41.Scott-Browne JP, et al. Evolutionarily conserved features contribute to alphabeta T cell receptor specificity. Immunity. 2011;35:526–535. doi: 10.1016/j.immuni.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holler PD, et al. TCRs with high affinity for foreign pMHC show self-reactivity. Nat. Immunol. 2003;4:55–62. doi: 10.1038/ni863. [DOI] [PubMed] [Google Scholar]
- 43.Tynan FE, et al. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat. Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- 44.Ganju RK, et al. Similarity between fluorescein-specific T-cell receptor and antibody in chemical details of antigen recognition. Proc. Natl. Acad. Sci. U.S.A. 1992;89:11552–11556. doi: 10.1073/pnas.89.23.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bostrom J, et al. Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site. Science. 2009;323:1610–1614. doi: 10.1126/science.1165480. [DOI] [PubMed] [Google Scholar]
- 46.Buslepp J, et al. A correlation between TCR Valpha docking on MHC and CD8 dependence: implications for T cell selection. Immunity. 2003;19:595–606. doi: 10.1016/s1074-7613(03)00269-3. [DOI] [PubMed] [Google Scholar]
- 47.Wang JH, et al. Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10799–10804. doi: 10.1073/pnas.191124098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao GF, et al. Assembly and crystallization of the complex between the human T cell coreceptor CD8alpha homodimer and HLA-A2. Protein Sci. 1998;7:1245–1249. doi: 10.1002/pro.5560070520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin Y, et al. Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5405–5410. doi: 10.1073/pnas.1118801109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuhns MS, Davis MM. Disruption of extracellular interactions impairs T cell receptor–CD3 complex stability and signaling. Immunity. 2007;26:357–369. doi: 10.1016/j.immuni.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 51.Kuhns MS, et al. Deconstructing the form and function of the TCR/CD3 complex. Immunity. 2006;24:133–139. doi: 10.1016/j.immuni.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 52.Boniface JJ, et al. Initiation of signal transduction through the T cell receptor requires the multivalent engagement of peptide/MHC ligands [corrected] [Erratum in Immunity 1998;9891] Immunity. 1998;9:459–466. doi: 10.1016/s1074-7613(00)80629-9. [DOI] [PubMed] [Google Scholar]
- 53.Cochran JR, et al. Receptor proximity, not intermolecular orientation, is critical for triggering T-cell activation. J. Biol. Chem. 2001;276:28068–28074. doi: 10.1074/jbc.M103280200. [DOI] [PubMed] [Google Scholar]
- 54.Kuhns MS, et al. Evidence for a functional sidedness to the alphabetaTCR. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5094–5099. doi: 10.1073/pnas.1000925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon ST, et al. Both high and low avidity antibodies to the T cell receptor can have agonist or antagonist activity. Immunity. 1994;1:563–569. doi: 10.1016/1074-7613(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 56.Garcia KC, et al. An αβ T cell receptor structure at 2.5 Å and its orientation in the TCR–MHC complex. Science. 1996;274:209–219. [PubMed] [Google Scholar]