Abstract
Pulmonary arterial hypertension (PAH) is a rare and devastating disease, resulting from progressive obliteration of small caliber pulmonary arteries by proliferating vascular cells, and leading to cardiac failure, with an untreated mean survival of less than three years 1,2. PAH can complicate other pathological conditions, or can occur in the context of genetic mutations causing heritable PAH, or can be considered as idiopathic (iPAH), which represents approximately 40% of all PAH 3,4. Low penetrance dominant BMPR2 mutations are found in ~70% of familial PAH (fPAH), and in ~15% of iPAH which are thereafter considered as heritable PAH 5,6. We conducted a Genome-Wide Association Study (GWAS) based on two independent case-control studies for iPAH and fPAH (without BMPR2 mutations) totaling 625 patients and 1,525 healthy individuals, to identify novel genetic factors associated with iPAH and fPAH (i/fPAH) in the absence of BMPR2 mutations. A genome wide significant association was detected at the CBLN2 locus mapping to 18q22.3, the risk allele being associated with an odds ratio for i/fPAH of 1.97 [1.59 – 2.45] (P = 7.47 x 10−10). CBLN2 is expressed in the lung, particularly in pulmonary vascular endothelial cells, and its expression is increased in explanted lungs from PAH patients and in endothelial cells cultured from explanted PAH lungs.
In our quest to identify novel genes predisposing to idiopathic and familial pulmonary hypertension (iPAH and fPAH, respectively), we conducted a two-stage genome-wide association study (GWAS) of patients with iPAH and fPAH (i/fPAH) without detectable BMPR2 mutations. During the discovery stage of our analysis, 472,421 SNPs were typed and quality controlled using the Illumina 610-Quad DNA beadchip, and were tested for association with i/fPAH in a sample of 340 i/fPAH patients and 1,068 controls of French origin (see Methods and Supplementary Table 1). The application of the Eigenstrat program in order to test for SNP association with i/fPAH 7 did not reveal any evidence of population stratification, with a genomic inflation coefficient of 1.02 (Supplementary Figure 1). No SNP achieved the statistical threshold for genome-wide significance (P < 1 x 10−7) (Supplementary Figure 2) but the 384 most significant SNPs with corresponding association P-values ranging from 1.82 x 10−6 to 6.87 x 10−4, were nevertheless selected for further association validation in an independent replication sample population of 285 patients and 457 controls. Among the 384 SNPs genotyped by a dedicated Illumina GoldenGate assay (see Methods), 319 SNPs passed quality controls. Following statistical Bonferroni correction for the number of tested SNPs, two of the 319 SNPs showed significant association with i/fPAH (Supplementary Table 2), rs2217560 (P = 1.63 x 10−5) and rs9916909 (P = 3.50 x 10−5). These two SNPs are located 52 kb downstream of the CBLN2 gene at locus 18q22.3, and are in almost complete linkage disequilibrium (r2 = 0.99).
As indicated in Table 1, the association pattern of rs2217560 with i/fPAH was homogeneous in both discovery and replication cohorts. In the discovery GWAS, the rs2217560-G allele was more frequent in cases than in controls (0.123 vs 0.070) and was associated with an increased risk for i/fPAH of 1.87 [1.41 – 2.48] (P = 3.88 x 10−4 ). In the replication population, the rs2217560-G allele had a higher frequency in cases (0.132 vs 0.066) and the corresponding odds ratio (OR) was 2.16 [1.51 – 3.09] (P = 1.63 x 10−5). In the combined sample, the rs2217560-G allele was associated with an increased risk for i/fPAH of 1.97 [1.59 – 2.45] with overall statistical significance reaching P = 7.47 x 10−10 (Table 1). No evidence for any sex-specific association was observed (Supplementary Table 3). We further examined whether the observed association could be explained by untyped SNP(s) located in the vicinity of rs2217560. Using the August 2010 release of the 1000 Genomes dataset as a reference, we conducted imputation analysis in the GWAS discovery cohort (see Methods). No single imputed SNP at the 18q22.3 locus showed stronger evidence of association than rs2217560 (Supplementary Table 4; Supplementary Figure 3). Additional conditional regression analysis on the effect of rs2217560 did not detect any other independent association signal at the CBLN2 locus (Supplementary Table 4).
Table 1.
Association of CBLN2 rs2217560 with i/fPAH in two independent case-control studies
| Discovery Cohort | Replication Cohort | |||
|---|---|---|---|---|
| Controls | i/fPAH | Controls | i/fPAH | |
| rs2217560 | N = 1,068 | N = 340 | N = 456 | N = 284 |
| AA | 925 (87%) | 262 (77%) | 400 (88%) | 211 (74%) |
| AG | 136 (13%) | 72 (21%) | 52 (11%) | 71 (25%) |
| GG | 7 (<1%) | 6 (2%) | 4 (1%) | 2 (1%) |
| MAF (G) | 0.070 | 0.123 | 0.066 | 0.132 |
| P(1) | P = 1.56 x 10−5 | P = 1.63 x 10−5 | ||
| Allelic odds ratio | 1.866 [1.407 – 2.475] | 2.160 [1.511 – 3.089] | ||
| 1.973 [1.587 – 2.453] | ||||
| P= 7.47 10−10 | ||||
MAF: Minor Allele Frequency
P-value of the Cochran-Armitage trend test. The P-value of the association test in the discovery cohort after controlling for any underlying population stratification (EIGENSTRAT software) was P = 3.9 x 10−4.
The Cochran Q statistical test did not detect any heterogeneity between the odds ratios obtained from the two cohorts (P = 0.811). The Mantel-Haenszel estimate of the common allelic odds ratio was 1.977 [1.584 – 2.467], P = 1.649 x 10−9.
Due to the rarity of the disease, it was not possible to collect a larger sample of patients for GWAS analysis, which is the current practice for common complex diseases 8. As a consequence, the power of the single discovery sample was rather limited 9, for example, there was only ~40 % power to detect an allelic OR of 2.0 associated with a minor allele frequency (MAF) of 0.10, at the genome-wide significance level of 1.0 x 10−7. Accordingly, we utilized a two-stage GWAS strategy which provided ~90 % power to detect such genetic effects. However, the two-stage strategy did not have enough power to detect moderate genetic effects, with a maximum power of ~40 % to detect an OR of 1.5 or less, associated with a MAF of 0.20.
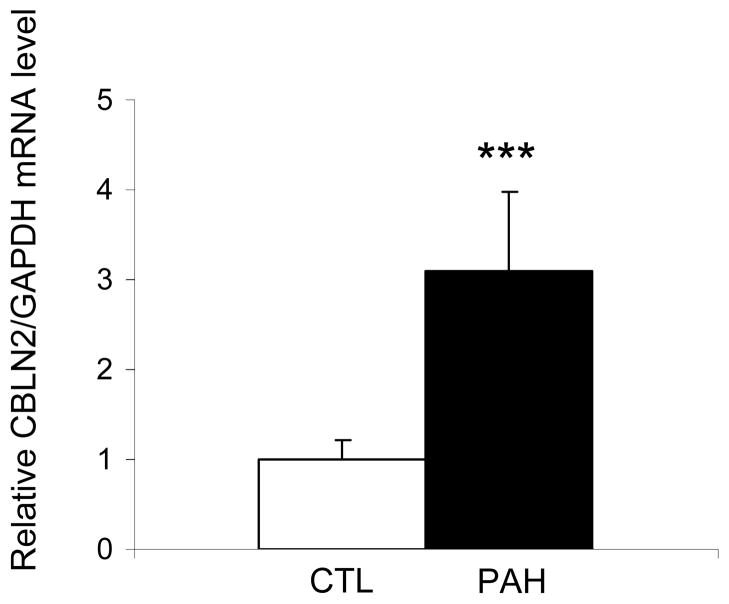
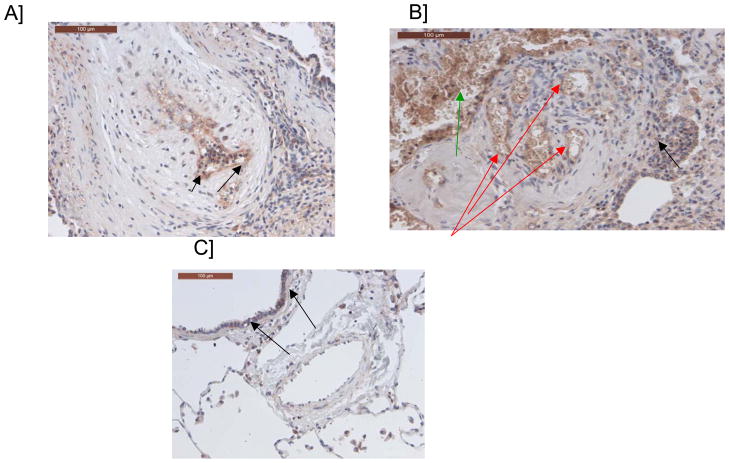
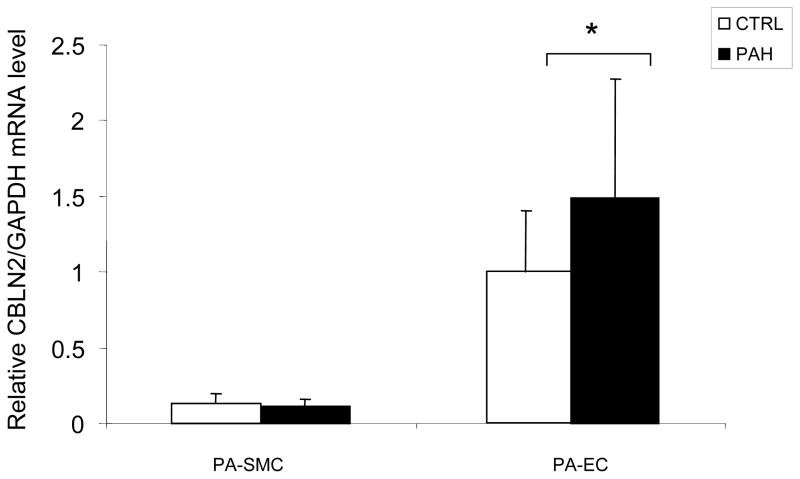
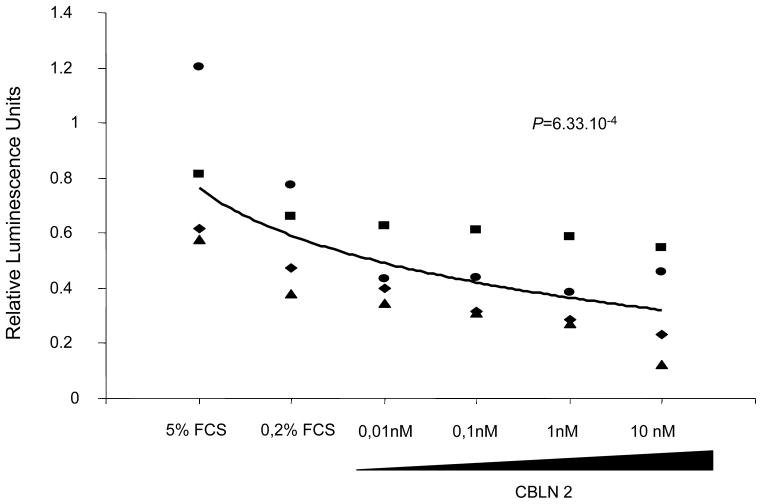
The rs2217560 SNP identified through our two-stage GWAS strategy lies 52 kb downstream from the CBLN2 gene. CBLN2 belongs to the cerebellin gene family, a group of secreted neuronal glycoproteins (Cbln1-4) and encodes the precursor of CBLN-2, an hexadecapeptide with 94 % and 44 % sequence homology to the cerebellin 1 and 3 peptides, respectively 10. CBLN2 has been previously reported to be mainly expressed in different regions of the brain 11, but because CBLN2 is in close proximity to the associated SNPs, we aimed to determine CBLN2 expression in the lung and other cell types. We found that CBLN2 mRNA is expressed in the whole lung and at lower levels in circulating cells, such as lymphocytes. CBLN2 mRNA levels were significantly higher in lungs explanted from PAH patients than from histologically normal lung tissue derived from subjects undergoing surgery for lung cancer (control lungs) (Figure 1). CBLN2 was detected in endothelial, epithelial and in circulating cells from patient and control lung samples via immunohistochemistry, but the levels of expression varied widely from sample to sample (Figure 2). In cultured pulmonary vascular cells, we found that CBLN2 mRNA was expressed in endothelial cells (EC) isolated from pulmonary artery (PA-EC), in contrast with pulmonary artery vascular smooth muscle cells (PA-SMC) where expression was detected at very low levels (Figure 3). The CBLN2 expression levels in primary cultured EC explanted from PAH lung samples collected during lung transplantation were higher than in EC from control lung samples (Figure 3). We hypothesized that CBLN2 synthesized by the EC might act on vascular SMC proliferation in a paracrine fashion. Therefore we treated serum-deprived PA-SMC in primary cultures with the CBLN2 peptide at concentrations ranging from 0.01 to 10 nM. A significant log-linear trend to proliferation inhibition was observed in cultured PA-SMC with increasing concentrations of CBLN2 (Figure 4). A generic toxic effect of CBLN2 peptide on PA-SMC was excluded by measuring cell viability on CBLN2 treated cells (Supplementary Figure 5).
Figure 1. CBLN2 mRNA levels are increased in lungs from PAH patients.
CBLN2 mRNA levels were measured by real time RT-PCR in lungs from control (CTL, white bars, n = 9) and PAH patients (PAH, black bars, n = 6) and normalized to GAPDH mRNA levels. *** P < 0.001 vs controls.
Figure 2. Immunostaining of pulmonary arteries with a cerebellin 2 antibody.
A/ Endothelial labeling of a pulmonary artery with intimal fibrosis (see arrows) B/ Endothelial labeling of a plexiform lesion (red arrows), labeling of intravascular cellular elements (erythrocytes, leukocytes) (green arrow), background-staining in some areas (black arrow); C/ Pulmonary artery from a control patient without labeling. Scattered bronchial cells are labeled (black arrows).
Figure 3. CBLN2 mRNA levels in pulmonary vascular cells from PAH patients.
CBLN2 mRNA levels were measured by real time RT-PCR in cultured pulmonary artery endothelial cells (PA-EC) and pulmonary artery smooth muscle cells (PA-SMC) from controls (CTL, white bars, n = 10 for PA-EC and n = 10 for PA-SMC) and PAH patients (PAH, black bars, n = 8 for PA-EC and n = 5 for PA-SMC) and normalized to GAPDH mRNA levels. * P < 0.05 vs controls.
Figure 4. Cell proliferation in pulmonary artery smooth muscle cells (PA-SMC) treated with CBLN2.
Cell proliferation was measured by BrdU incorporation in PA-SMC cultured in complete medium with 5 % fetal calf serum (FCS) or depleted medium (0.2 % FCS) and treated with CBLN2 peptide at concentrations ranging from 0 to 10 nM. A significant decrease in the proliferation of PA-SMC was observed when cells were treated with increasing doses of CBLN2 peptide. Each symbol represents the value for one experiment. The log-linear regression line is indicated (n = 4, log-linear regression test, P = 6.33 x 10−4).
Preliminary data (not shown) did not reveal any evidence for an association between the rs2217560 genotype and CBLN2 mRNA levels in monocytes, but these results do not preclude a tissue specific effect in lungs.
CBLN2 was previously shown to bind the S4 domain of β-neurexin (a pre-synaptic cell adhesion molecule encoded by NRXN1) as well as the post-synaptic δ2 glutamate receptor (GluD2), contributing to synapse formation by bridging the two molecules 12. NRXN1, which encodes both α- and β-neurexin, is expressed in the vascular wall. The β-neurexin isoform, transcribed from an internal promoter and containing a specific N-terminal peptide, is present in a subset of SMCs and also in chicken embryo arteries 13. Our results show that β-neurexin is expressed in the lung without a significant difference between control and PAH lung samples (Supplementary Figure 4). Anti-β-neurexin antibody was shown to decrease FGF-2 induced angiogenesis in the chorioallantoic membrane model, and decreased noradrenalin induced vessel tension on isolated chick embryo arteries 13. In recent years, several molecules have been implicated that participate both in neurogenesis and angiogenesis 14.
Altogether, these results strongly support a role for the CBLN2 locus as a novel contributor to the physiopathology of i/fPAH. The CBLN2 gene is a promising candidate because of its vascular expression and its effects in vascular cells. CBLN2, that we find to be produced by EC, was shown to bind an adhesion molecule present in SMC. Further studies are needed to document the putative function of CBLN2 in pulmonary vessels.
In conclusion, we report the first GWAS in BMPR2 mutation negative i/fPAH, which identified an allele associated with two-fold increased risk. The associated locus is completely novel with respect to i/fPAH pathogenesis. Fine-mapping and deep-sequencing of the entire locus is now required to identify and characterize the functional variant(s) responsible for the observed association. These results pave the way for improved pathophysiological understanding as well as new therapeutic approaches for pulmonary hypertension.
Methods
Study population
Patients and controls
The diagnosis of PAH was defined by hemodynamic measurement during right-heart catheterization for all patients included in the study (discovery stage and replication stage), including those identified by the French PAH Network between January 1, 2003 and April 1, 2010. For all patients, PAH was defined as a mean pulmonary arterial pressure equal to or exceeding 25 mm Hg associated with normal pulmonary capillary wedge pressure. Hemodynamic evaluation by right-heart catheterization was performed at baseline in all subjects according to previously described protocols 15,16.
PAH was considered to be idiopathic (iPAH) after clinical and biological investigation eliminated all known causes. Patients with iPAH were screened for BMPR2 mutations, and patients with a family history of PAH, were screened for BMPR2 and ACVRL1 mutations. Screening for point mutations and large rearrangements was performed as previously reported 5,17. Patients carrying a mutation in either of these genes were excluded. When patients had a family history of PAH without evidence of either BMPR2 or ACVRL1 mutation, a single index case from the family was included in the GWAS analysis.
Discovery stage
A total of 378 patients meeting these criteria were included. All clinical characteristics at PAH diagnosis and follow-up were stored in the Registry of the French PAH Network 3. This registry was set up in agreement with French bioethics laws (French Commission Nationale de l’Informatique et des Libertés), and informed patient consent 5. The control group was composed of a random sample of 1,140 subjects free of any chronic diseases from the 3C Study 18. The 3C Study is a population-based prospective cohort with a four-year follow up carried out in three French cities: Bordeaux (southwest France), Montpellier (southeast France) and Dijon (central eastern France). This study has served as a control population for several French GWAS projects 19–22.
Replication stage
Specimens (297 PAH patients and 479 healthy controls) from participants in the Vanderbilt Prospective Pulmonary Hypertension Research Cohort study, and from the Columbia University Pulmonary Hypertension Center were included. These subjects were recruited via the Vanderbilt and Columbia Pulmonary Hypertension Centers, the NIH Clinical Trials website (http://clinicaltrials.gov), and in collaboration with the Pulmonary Hypertension Association Conference Research Recruitment Room (2010 Conference, Anaheim, California). The Vanderbilt University Medical Center and Columbia University Institutional Review Boards approved all study protocols. All participants gave informed written consent to participate in genetic and clinical studies and underwent genetic counseling when appropriate, in accordance with the guidelines of the American College of Chest Physicians23.
As with the Discovery stage, PAH was defined as a mean pulmonary arterial pressure equal to or exceeding 25 mm Hg associated with normal pulmonary capillary wedge pressure. Hemodynamic evaluation by right-heart catheterization was performed at baseline in all subjects according to previously described protocols 15,16. PAH was considered to be idiopathic (iPAH) after clinical and biological investigation eliminated all known causes, or familial as appropriate. Screening for mutations in BMPR2 and ACVRL1 was the same as for the Discovery stage cohort.
Genotyping
Discovery stage
The sample of 378 i/fPAH patients and 1,140 healthy controls were typed with the Illumina Human 610-Quad Beadchip. Individuals with genotyping success rates lower than 95% were excluded from the analyses, as were individuals demonstrating close relatedness. The latter instance was assessed by pairwise identity clustering by state distance (IBS) and multi-dimensional scaling (MDS) using the PLINK software 24. The Eigenstrat program 7 was further used to detect individuals of non-European ancestry. A total of 104 subjects (32 patients and 72 healthy controls) were thus excluded from the analysis because of close relatedness and/or evidence of non-European ancestry.
SNPs showing either a significant deviation (P < 10−5) from the Hardy-Weinberg Equilibrium (HWE) in controls, or those SNPs with a minor allele frequency (MAF) < 1% in controls or < 5% in cases, or where genotyping call rates were < 99%, were filtered out. This resulted in a final analysis of 462,499 autosomal and 9,922 X-linked SNPs in a sample of 340 i/fPAH patients and 1,068 healthy individuals.
Replication stage
The 384 SNPs showing the strongest association with i/fPAH and assigned an Illumina ScoreDesign greater than 0.4 were selected for genotyping in an independent sample of 297 i/fPAH patients and 479 controls using the Illumina GoldenGate assay. A total of 34 individuals (12 patients and 22 controls) were discarded due to low genotype calling rates (< 80%). SNPs showing significant deviation (P < 10−5) from HWE in controls and call rates of < 99% were removed from the analysis, resulting in the statistical analysis of 319 SNPs in a sample of 285 patients and 457 controls.
Statistical analysis
At the discovery stage, the genome-wide association analysis of SNPs was conducted using the Eigenstrat program that corrects for any uncontrolled population stratification 7. The genomic control (GC) inflation factor was also computed according to the median test statistic 25. At the replication stage, the association of SNPs with i/fPAH was assessed by use of the Cochran-Armitage trend test 26. Homogeneity of associations across the two stages was tested using the Mantel-Haenszel method 27.
The imputation of 11,572,501 autosomal SNPs was conducted using the MACH (http://www.sph.umich.edu/csg/abecasis/mach/) and minimac softwares (http://genome.sph.umich.edu/wiki/Minimac) according to the August 2010 release of the 1000G CEU reference dataset. The association of each imputed SNP with i/fPAH was tested by logistic regression analysis in which allele dosage (from 0 to 2 copies of the minor allele) for imputed SNPs was assessed. Analyses were adjusted for the first four principal components and were performed using the mach2dat (v 1.08.18) software (http://www.sph.umich.edu/csg/abecasis/MACH/download/).
Expression of Cerebellin-2 in pulmonary tissue
Study of Cerebellin-2 expression and effect on proliferation in pulmonary vascular cells
Tissues and cells
Lung specimens were obtained at the time of lung transplantation from patients with idiopathic iPAH. Control lung specimens were obtained from patients without any evidence of pulmonary vascular disease who underwent lobectomy or pneumonectomy for localized lung cancer, with normal tissue collected at a distance from the tumors. Both control and PAH lung specimens were collected under the same conditions by the same surgical department. (Supplementary Table 5).
This study was approved by the local ethics committee (Comité de Protection des Personnes Ile-de-France, Le Kremlin-Bicêtre, France), and patients gave informed consent before the study.
For RNA extraction, lung samples were washed in PBS, and then immediately frozen in liquid nitrogen and stored at −80 °C.
Human pulmonary artery endothelial cells (PA-EC) and human pulmonary artery smooth muscle cells (PA-SMC) were isolated from iPAH patient and control samples (Supplementary Table 5), and cultured as previously described 28,29. Cells were used between passages 3–6.
Real-time RT-PCR assay
Real-time RT PCR assays were performed as previously described 30 using primers described in Supplementary Table 6. Data are expressed as mean fold change ± standard deviation (SD) from at least three independent experiments. Statistical analysis was performed using a non parametric Mann-Whitney U test for single comparisons and ANOVA for multiple comparisons (XLSTAT software, Addinsoft, Paris, France). P values less than 0.05 were considered to be significant.
Immunohistochemical studies
Lungs sections were stained and analyzed as described in Montani et al 31.
Proliferation assay
PA-SMC purchased from Lonza (Walkersville, USA) were seeded in SmGm2 Bullekit media (Lonza, Walkersville, USA) at a density of 5000 cells/well in 96-well plates. The following day, cells were starved in 0.2 % SmGm2 media for 24h before being stimulated with the CBLN2 peptide (SGSAKVAFSATRSTNH) (ProteoGenix, Oberhausbergen, France) at 0.01, 0.1, 1 and 10 nM. After 24 h of incubation with the CBLN2 peptide, proliferation was measured by a BrdU incorporation assay using a BrdU cell proliferation Elisa kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s instructions.. Statistical analysis was performed using a log-linear regression test.
Supplementary Material
Acknowledgments
The PHAAR project was financially supported by the Agence Nationale pour la Recherche (Project ANR-07-MRAR-021), and by PHRC AOM07-041 PHAM, INSERM and UPMC. The 3C Study is conducted under a partnership agreement between INSERM, the Victor Segalen –Bordeaux II University and Sanofi-Synthelabo. The Fondation pour la Recherche Médicale funded the preparation and first phase of the study. The 3C-Study is also supported by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, Mutuelle Générale de l’Education Nationale, the Institut de la Longévité, Agence Française de Sécurité Sanitaire des Produits de Santé, the Regional Governments of Aquitaine, Bourgogne and Languedoc-Roussillon and, the Fondation de France, the Ministry of Research-INSERM Programme “Cohorts and collection of biological material”. The Lille Genopole received an unconditional grant from Eisai. The financial supporters had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
MG was funded by a grant from the Agence Nationale pour la Recherche (Project PHAAR, ANR-07-MRAR-021) and the Program Hospitalier de recherche Clinique (PHRC2009 RENOVA-TV;). Statistical analyses utilized the C2BIG computing centre funded by the Fondation pour la Recherche Médicale and Région Ile de France. Collection and management of samples from Vanderbilt University was supported by NIH grant P01 HL072058, NIH grant K23 HL0987431, and GCRC RR000095. Collection of the samples from Columbia University was supported by NIH grant R01 HL060056.
References
- 1.Rabinovitch M. Molecular pathogenesis of pulmonary arterial hypertension. J Clin Invest. 2008;118:2372–9. doi: 10.1172/JCI33452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubin LJ. Primary pulmonary hypertension. N Engl J Med. 1997;336:111–7. doi: 10.1056/NEJM199701093360207. [DOI] [PubMed] [Google Scholar]
- 3.Humbert M, et al. Pulmonary arterial hypertension in France: results from a national registry. Am J Respir Crit Care Med. 2006;173:1023–30. doi: 10.1164/rccm.200510-1668OC. [DOI] [PubMed] [Google Scholar]
- 4.Simonneau G, et al. Updated clinical classification of pulmonary hypertension. J Am Coll Cardiol. 2009;54:S43–54. doi: 10.1016/j.jacc.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Sztrymf B, et al. Clinical outcomes of pulmonary arterial hypertension in carriers of BMPR2 mutation. Am J Respir Crit Care Med. 2008;177:1377–83. doi: 10.1164/rccm.200712-1807OC. [DOI] [PubMed] [Google Scholar]
- 6.Girerd B, et al. Absence of influence of gender and BMPR2 mutation type on clinical phenotypes of pulmonary arterial hypertension. Respir Res. 2010;11:73. doi: 10.1186/1465-9921-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 8.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 10.Yiangou Y, Burnet P, Nikou G, Chrysanthou BJ, Bloom SR. Purification and characterisation of cerebellins from human and porcine cerebellum. J Neurochem. 1989;53:886–9. doi: 10.1111/j.1471-4159.1989.tb11787.x. [DOI] [PubMed] [Google Scholar]
- 11.Miura E, Iijima T, Yuzaki M, Watanabe M. Distinct expression of Cbln family mRNAs in developing and adult mouse brains. Eur J Neurosci. 2006;24:750–60. doi: 10.1111/j.1460-9568.2006.04950.x. [DOI] [PubMed] [Google Scholar]
- 12.Matsuda K, Yuzaki M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur J Neurosci. 2011;33:1447–61. doi: 10.1111/j.1460-9568.2011.07638.x. [DOI] [PubMed] [Google Scholar]
- 13.Bottos A, et al. The synaptic proteins neurexins and neuroligins are widely expressed in the vascular system and contribute to its functions. Proc Natl Acad Sci U S A. 2009;106:20782–7. doi: 10.1073/pnas.0809510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichmann A, Le Noble F, Autiero M, Carmeliet P. Guidance of vascular and neural network formation. Curr Opin Neurobiol. 2005;15:108–15. doi: 10.1016/j.conb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Sitbon O, et al. Long-term response to calcium channel blockers in idiopathic pulmonary arterial hypertension. Circulation. 2005;111:3105–11. doi: 10.1161/CIRCULATIONAHA.104.488486. [DOI] [PubMed] [Google Scholar]
- 16.Loyd JE, Primm RK, Newman JH. Familial primary pulmonary hypertension: clinical patterns. Am Rev Respir Dis. 1984;129:194–7. doi: 10.1164/arrd.1984.129.1.194. [DOI] [PubMed] [Google Scholar]
- 17.Eyries M, et al. ACVRL1 germinal mosaic with two mutant alleles in hereditary hemorrhagic telangiectasia associated with pulmonary arterial hypertension. Clin Genet. 2011 doi: 10.1111/j.1399-0004.2011.01727.x. [DOI] [PubMed] [Google Scholar]
- 18.3C Study Group. Vascular factors and risk of dementia: design of the Three-City Study and baseline characteristics of the study population. Neuroepidemiology. 2003;22:316–25. doi: 10.1159/000072920. [DOI] [PubMed] [Google Scholar]
- 19.Allanore Y, et al. Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 2011;7:e1002091. doi: 10.1371/journal.pgen.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Germain M, et al. Genetics of venous thrombosis: insights from a new genome wide association study. PLoS One. 2011;6:e25581. doi: 10.1371/journal.pone.0025581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollingworth P, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–35. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saad M, et al. Genome-wide association study confirms BST1 and suggests a locus on 12q24 as the risk loci for Parkinson’s disease in the European population. Hum Mol Genet. 2011;20:615–27. doi: 10.1093/hmg/ddq497. [DOI] [PubMed] [Google Scholar]
- 23.McGoon M, et al. Screening, early detection, and diagnosis of pulmonary arterial hypertension: ACCP evidence-based clinical practice guidelines. Chest. 2004;126:14S–34S. doi: 10.1378/chest.126.1_suppl.14S. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 26.Sasieni PD. From genotypes to genes: doubling the sample size. Biometrics. 1997;53:1253–61. [PubMed] [Google Scholar]
- 27.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 28.Guignabert C, et al. Dichloroacetate treatment partially regresses established pulmonary hypertension in mice with SM22{alpha}-targeted overexpression of the serotonin transporter. Faseb J. 2009 doi: 10.1096/fj.09-131664. [DOI] [PubMed] [Google Scholar]
- 29.Tu L, et al. Autocrine fibroblast growth factor-2 signaling contributes to altered endothelial phenotype in pulmonary hypertension. Am J Respir Cell Mol Biol. 2011;45:311–22. doi: 10.1165/rcmb.2010-0317OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dupuis M, et al. Profiling of aortic smooth muscle cell gene expression in response to chronic inhibition of nitric oxide synthase in rats. Circulation. 2004;110:867–73. doi: 10.1161/01.CIR.0000138850.72900.FE. [DOI] [PubMed] [Google Scholar]
- 31.Montani D, et al. C-kit-positive cells accumulate in remodeled vessels of idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med. 2011;184:116–23. doi: 10.1164/rccm.201006-0905OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.