Abstract
Menin is a tumor suppressor protein whose loss or inactivation causes multiple endocrine neoplasia type 1 (MEN1), a hereditary autosomal dominant tumor syndrome characterized by tumorigenesis in multiple endocrine organs1. Menin interacts with a multitude of proteins and involves in a variety of cellular processes2–6. Menin binds the Jun family transcription factor JunD and inhibits its transcriptional activity7,8. Several MEN1 missense mutations disrupted the menin-JunD interaction suggestive of a correlation between menin’s tumor suppressor function and its interaction with JunD and suppression of JunD activated transcription8,9. Menin also interacts with mixed lineage leukemia protein 1 MLL1, a histone H3 lysine 4 (H3K4) methyltransferase, and functions as an oncogenic cofactor to upregulate gene (including HOX genes) transcription and promote MLL1 fusion protein (MFP)-induced leukemogenesis10–12. A recent report on menin tethering MLL1 to chromatin binding factor LEDGF indicates menin as a molecular adaptor to coordinate the functions of multiple proteins13. Despite the importance of menin, it still remains poorly understood how menin could interact with many distinct partners and control multiple functions. Here we present the crystal structures of menin, free and in complexes with MLL1 or JunD, or an MLL1-LEDGF heterodimer. These structures show that menin contains a deep pocket that binds short peptides of MLL1 or JunD in the same manner, but oppositely regulates transcription. The menin-JunD interaction blocks JNK kinase-meidated JunD phosphorylation, a crucial event for JunD activation.Moreover, menin functions as a scaffold molecule to promote gene transcription by binding MLL1 through the peptide-pocket yet interacting with LEDGF at a distinct surface.
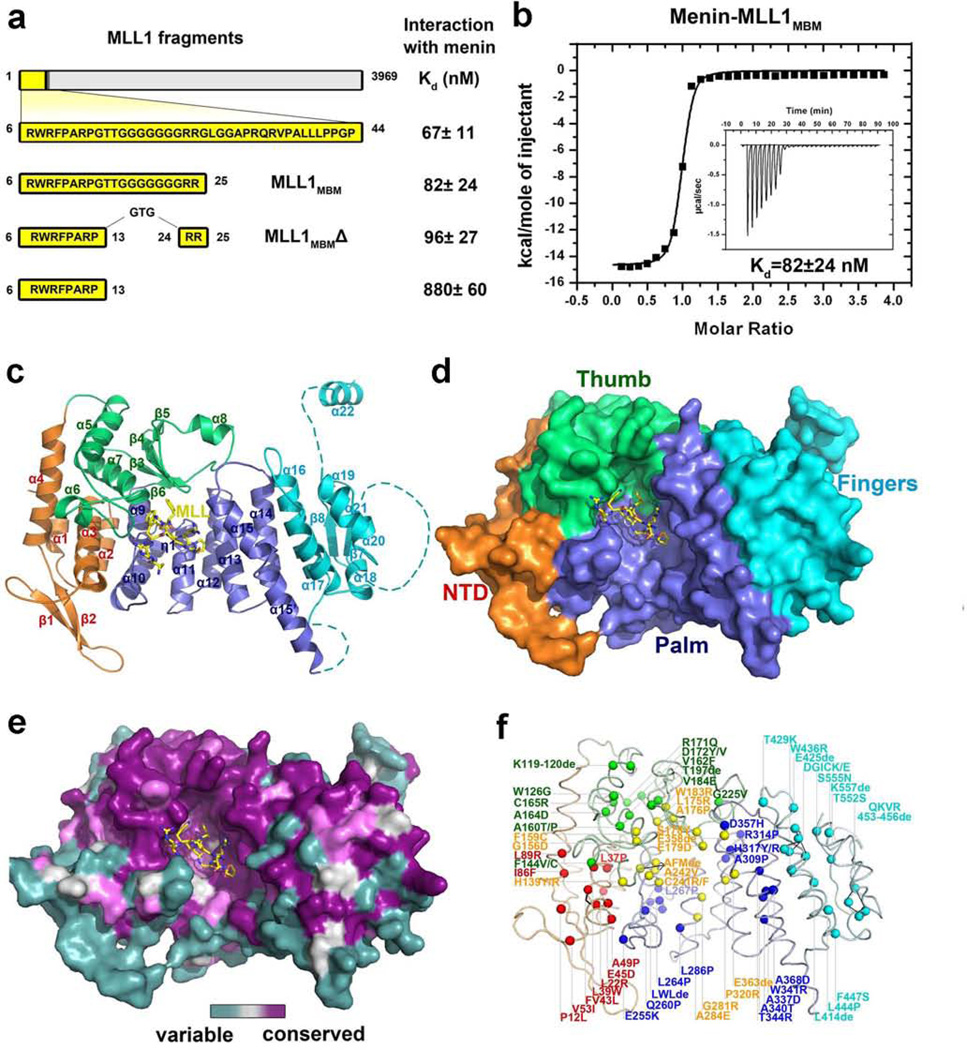
The N-terminal region of MLL1 was reported to interact with menin6,14,15. Isothermal titration calorimetry (ITC) measurements showed residues 6–25 of MLL1 (MLL1MBM: menin-binding motif) was necessary and sufficient for binding to menin with an affinity of 82 nM (Figs. 1a and 1b). Further mapping revealed that the internal non-conserved polyglycine does not contribute to the binding (Fig. 1a). MLL2, the closest relative of MLL1, contains an almost identical sequence as MLL1MBM at its N-terminus (Supplementary Fig. 2a); MLL216–35 (MLL2MBM) binds to menin equally well as MLL1MBM does, with an affinity of 120 nM (Supplementary Fig. 2b). To understand how MLL1 and MLL2 (collectively referred to as MLL) are recognized by menin, we determined the crystal structures of menin alone and in complex with MLL1MBM (Supplementary Fig. 3, Supplementary Table 1, and Supplementary text).
Figure 1. Crystal structure of the human menin-MLL1MBM complex.
a, Summary of ITC analysis of the interactions between menin and various MLL1 peptides (refer to Supplementary Fig. 1). b, ITC measurement of the interaction of menin with the MLL1MBM peptide. Insert is the ITC titration data. The binding curve was fit to a one-binding-site-per-menin model. c, Overall structure of the menin-MLL1MBM complex. The N-terminal domain (NTD) is colored in orange, the thumb domain in green, the palm domain in blue, the fingers domain in cyan, and the loop regions that are not included or visible in the crystal structure is shown as dashed lines. The secondary structure elements are labeled. The MLL1MBM peptide is shown in stick model and colored in yellow. d, Surface representation of menin indicates that menin adopts a curved left-hand-shaped conformation. The orientation of the menin-MLL1MBM complex is rotated by ~30° about a horizontal axis relative to the complex on the left panel in (c). e, Front view of the menin-MLL1MBM complex, colored according to amino acid conservation among all the identifiable menin homologues (purple, well conserved; cyan, highly variable). f, Positions of MEN1-related missense and in-frame-deletion mutations of menin are denoted by colored spheres. The color scheme is the same as in (c).
Menin adopts a rectangular-shaped conformation that resembles a curved left hand, with a deep pocket formed by the thumb and the palm (Figs. 1c and 1d). Menin consists of four associated domains, an N-terminal domain (NTD) characterized by a long β-hairpin, a transglutaminase-like domain that forms the thumb, a helical palm domain that contains three TPR motifs16, followed by a C-terminal fingers domain (Fig. 1c, Supplementary Fig. 4 and Supplementary text). Conserved residues of menin among different species are either buried in the hydrophobic core, or clustered together on a surface patch that covers both the thumb and palm domains (Fig. 1e). MEN1 disease-derived missense and in-frame deletion mutations are evenly distributed throughout the entire protein (Fig. 1f), indicating that all four domains are important for the in vivo function of menin (Fig. 1f and Supplementary Table 2).
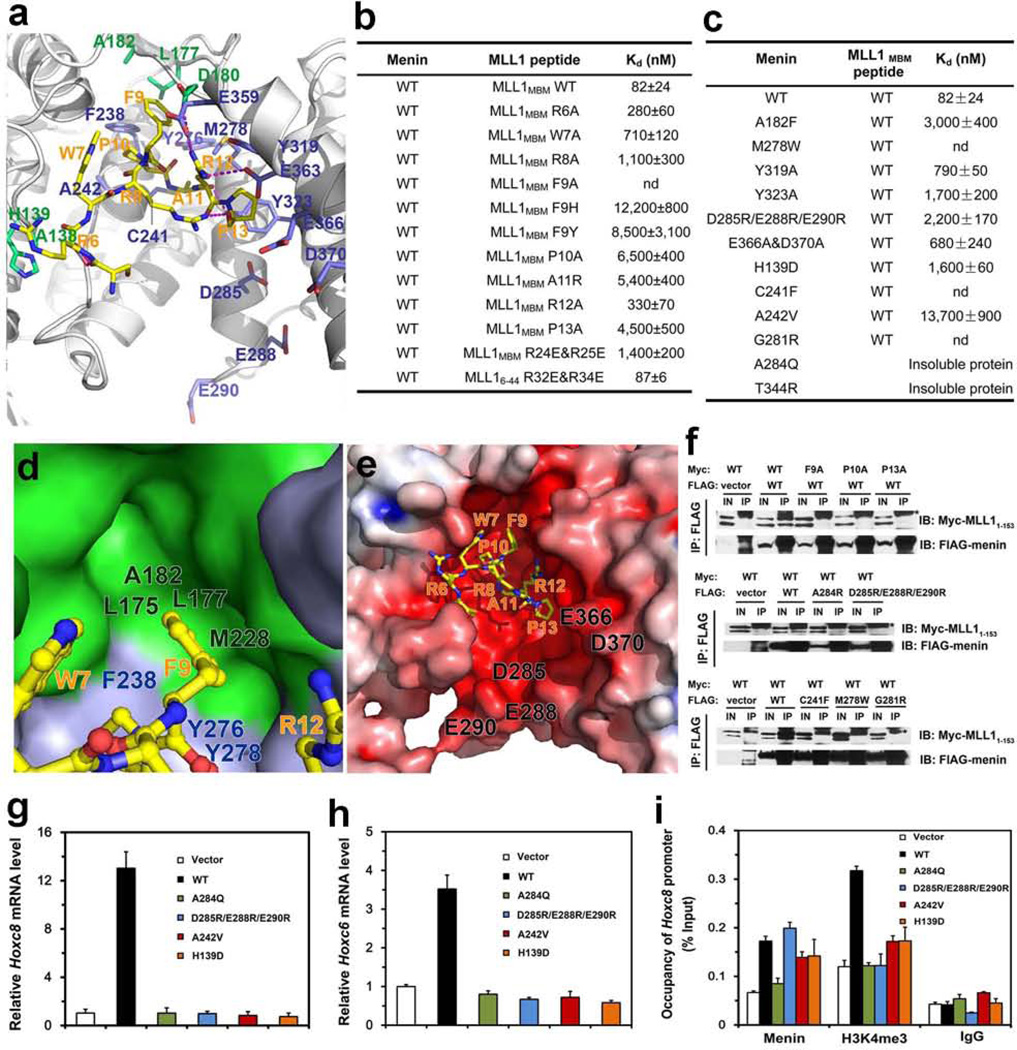
The MLL1MBM peptide is folded into a compact conformation and plugs into the deep pocket of menin (Fig. 2a). Mutagenesis data indicated that residues 6RWRFPARP13 in MLL1MBM contribute most of the interactions with menin (Fig. 2b). This motif adopts a cyclized conformation, stabilized by an intramolecular hydrogen bond between Arg8 and Pro13 (Fig. 2a). Pro13 is also sandwiched by menin Tyr319 and Tyr323 (Fig. 2a), and removal of this stacking contact greatly weakened the interaction (Figs. 2b and 2c). The cyclized configuration of MLL1MBM positions the phenol ring of Phe9 into a deep hydrophobic cavity formed by the thumb and palm (Fig. 2d). An M278W substitution altered the cavity shape and led to complete loss of binding (Fig. 2c). The MLL1MBM-binding pocket is highly acidic (Fig. 2e). The side-chain of Arg12MLL1 mediates three electrostatic contacts (Fig. 2a). The C-terminal two arginines (24RR25) in MLL1MBM are disordered, but they appear to be important for interaction, as substitution with two glutamates resulted in a 21-fold decrease in binding affinity (Fig. 2b). The relative position of the acidic residues of the MLL1MBM-binding pocket and MLL1 24RR 25 make it clear that electrostatic interactions occur. Consistent with this idea, mutation of these menin acidic residues also led to decreased binding (Fig. 2c).
Figure 2. Structural and mutational analyses of the menin-MLL1MBM interface.
a, detailed view of the menin-MLL1MBM interface. MLL1MBM (yellow) and the interacting residues in menin (green: residues in the thumb domain; blue: residues in the palm domain) are presented as stick models. The Menin-MLL1MBM intermolecular hydrogen bonds are shown as magenta dashed lines. b, In vitro ITC binding data of wild-type menin with mutant MLL1MBM (refer to Supplementary Fig. 5). c, In vitro ITC binding data of menin mutants with wild-type MLL1MBM (refer to Supplementary Fig. 6). d, The side chain of MLL1MBM Phe9, in ball-and-stick model (yellow), is nested in a deep hydrophobic pocket of menin formed by the thumb domain (green surface) and the palm domain (blue surface). e, Electrostatic surface potential of the MLL1MBM-binding pocket of menin (positive potential, blue; negative potential, red). f, Co-IP of the mutant menin-MLL11–153 interactions. Lanes marked “IN” represent 2.5% of input cell lysate used for the immunoprecipitation. Asterisks indicate the signals from IgG. Secondary structure prediction analysis showed that the N-terminal region of MLL1 (residues 1–153) is an intrinsically unstructured region (data not shown). Therefore, due to the difficulty of manipulation of full-length MLL1 (3969 residues), the menin-MLL11–153 interaction was used to represent the interaction between menin and full-length MLL1 in the Co-IP assay. g, Expression of Hoxc8 in menin-null MEFs complemented with control vector, wild-type or mutant menin was measured using quantitative real-time RT-PCR. h, Expression of Hoxc6 in menin-null MEFs complemented with control vector, wild type or mutant menin was measured using qRT-PCR. i, Menin binding and H3K4 trimethylation at the Hoxc8 promoter were detected by ChIP.
Next we examined the MLL1MBM-binding activity of several MEN1 disease-derived mutations (H139D, C241F, A242V, G281R, A284Q and T344R). Except for A284Q and T344R that yielded insoluble proteins, all the rest mutants impaired the menin-MLL1MBM interaction (Fig. 2c). To further examine the menin-MLL1 interaction in vivo, we studied the interactions of mutant proteins transiently expressed in human embryonic kidney 293T cells. Consistent with the ITC analysis, co-immunoprecipitation (Co-IP) data showed that mutations of the key residues at the interface completely abolished the menin-MLL1 interaction in cells (Fig. 2f).
Menin upregulates the expression of Hoxc6 and Hoxc8, presumably by recruiting the MLL complex and increasing H3K4 trimethylation at these gene loci10. To test the effect of the menin-MLL interaction on the expression levels of Hoxc6 and Hoxc8, wild-type (WT) and MLL-binding deficient mutants of menin (H139D, A242V, D285R/E288R/E290R, or A284Q) were individually used to complement menin-null mouse embryonic fibroblasts (MEFs). Western blots indicated a comparable expression of WT and mutant proteins in cells (Supplementary Fig. 7a). When Men1−/− cells were complemented with WT menin, expression of Hoxc8 and Hoxc6 was dramatically increased compared to vector-expressing cells (Figs. 2g and 2h). In contrast, overexpression of the menin mutants in Men1−/− cells failed to upregulate the mRNA levels of Hoxc6 or Hoxc8 (Figs. 2g and 2h), suggesting that the menin-MLL interaction is essential for Hoxc6 and Hoxc8 expression.
We next performed chromatin immunoprecipitation (ChIP) assay to determine the binding of mutant menin at the Hoxc8 promoter. Except for A284Q (a mutant that leads to insoluble proteins), all other mutants bound to the Hoxc8 promoter as effectively as WT menin (Fig. 2i). Notably, the Men1−/− cells complemented with WT menin exhibited a substantial increase in the level of the H3K4me3 mark at the Hoxc8 promoter compared with the vector-expressing cells (Fig. 2i). In contrast, the mutants failed to elevate the levels of H3K4me3 at the Hoxc8 promoter (Fig. 2i). Expression of WT or mutant menin did not significantly affect H3 distribution at the Hoxc8 promoter (Supplementary Fig. 7b). Thus, while the menin mutants were able to bind to the Hoxc8 promoter, their ability to recruit MLL and thus establish H3K4me3 at the Hoxc8 promoter was compromised, resulting in reduced Hoxc8 expression.
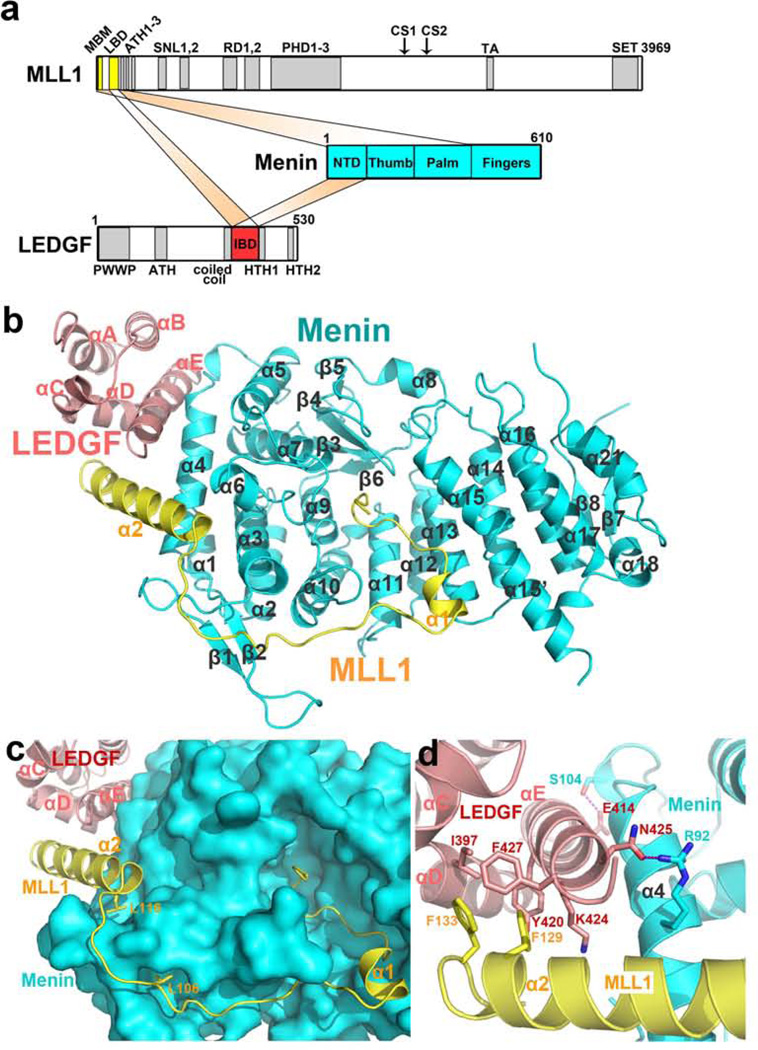
LEDGF, a chromatin-associated protein17, is required for MLL 1-dependent transcription and leukemic transformation through direct interactions with both MLL1 and menin13. ITC measurement showed that a complex composed of menin and an N-terminal fragment of MLL1 – MLL1MBM-LBM, residues 6–153 that include both MBM and the LEDGF-binding motif (LBM) – binds to the IBD domain of LEDGF(LEDGFIBD) with an affinity of 470 nM, whereas neither menin nor MLL1MBM-LBM individually interacts with LEDGFIBD suggestive of a cooperative binding among the three proteins (Fig. 3a and Supplementary Figs. 8a and 8b). We determined the menin-MLL1MBM-LBM-LEDGFIBD ternary complex structure at a resolution of 2.8 Å (Supplementary Fig. 8c and Supplementary Table 1).
Figure 3. Crystal structure of the menin-MLL1MBM-LBM-LEDGFIBD ternary complex.
a, Domain organization of menin, MLL1, and LEDGF. In MLL1, MBM and LBM are colored in yellow, and the rest domains and motifs in gray. Menin is colored in blue. In LEDGF, IBD is colored in red and the rest domains in gray. b, Ribbon diagram of the menin-MLL1MBM-LBM-LEDGFIBD complex. Menin is colored in cyan, MLL1 in yellow, and LEDGF in salmon. The secondary structure elements are labeled. c, Surface representation of menin and cartoon representation of MLL1MBM-LBM and LEDGFIBD indicate that the middle loop in MLL1MBM-LBM covers a large surface area of menin. Two leucine residues (Leu106 and Leu116) function as anchor points to define the path of the loop and properly fix one end of the α2 helix of MLL1MBM-LBM d, Detailed view of the intramolecular three-helix-bundle at the ternary interface. Key interacting residues are shown in stick models. The color scheme is the same as in (b).
In the complex, MLL1MBM-LBM has a non-globular, extended structure, which allows it to contact with a large surface area of menin (Fig. 3b). MLL1MBM-LBM binds to menin through two major sites. The N-terminal MLL1MBM coil folds into the high-affinity pocket in the same manner as in the menin-MLL1MBM structure (Supplementary Fig. 8d), whereas the C-terminal helix α2 loosely packs on the surface of meninNTD to form a V-shaped groove for LEDGFIBD binding (Fig. 3b). In contrast to the termini, the middle loop of MLL1MBM-LBM spans a large distance on menin without many specific interactions except for two leucine residues (Leu106 and Leu116), whose side-chains point to two shallow pockets on the menin surface, defining the path of the loop and fixing one end of the α2 helix (Fig. 3c).
Helix αE of LEDGFIBD is sandwiched between helices α2MLL1 and α4menin through both hydrophobic and electrostatic interactions (Fig. 3d). On one side, two hydrogen bonds anchor the LEDGFIBD αE helix onto α4 of meninNTD (Fig. 3d). On the other side, the aromatic rings of Phe129 and Phe133 of MLL1 insert into a hydrophobic pocket formed by three LEDGFIBD helices including αE (Fig. 3d). Consistent with the structure, an alanine substitution of MLL1 Phe129 completely abolished LEDGF binding and myeloid transformation13. Collectively, the ternary structure provides a strong and clear evidence that menin can function as an scaffold molecule to modulate gene expression by binding transcription regulators like MLL1 on one site yet interacting with other chromatin anchoring proteins such as LEDGF at a distinct surface. Although MLL1 and MLL2 share many functional domains and motifs including MBM (Supplementary Fig. 8e), MLL2 does not contain an LBM sequence and thus would not together with menin form a ternary complex with LEDGF. As the PWWP domain of LEDGF is required for MLL1-mediated leukemic transformation13, the inability of MLL2 to form a menin-MLL2-LEDGF complex provides an explanation of why to date only MLL1, but not MLL2 has been described as a proto-oncogene that can be activated by chromosomal translocations.
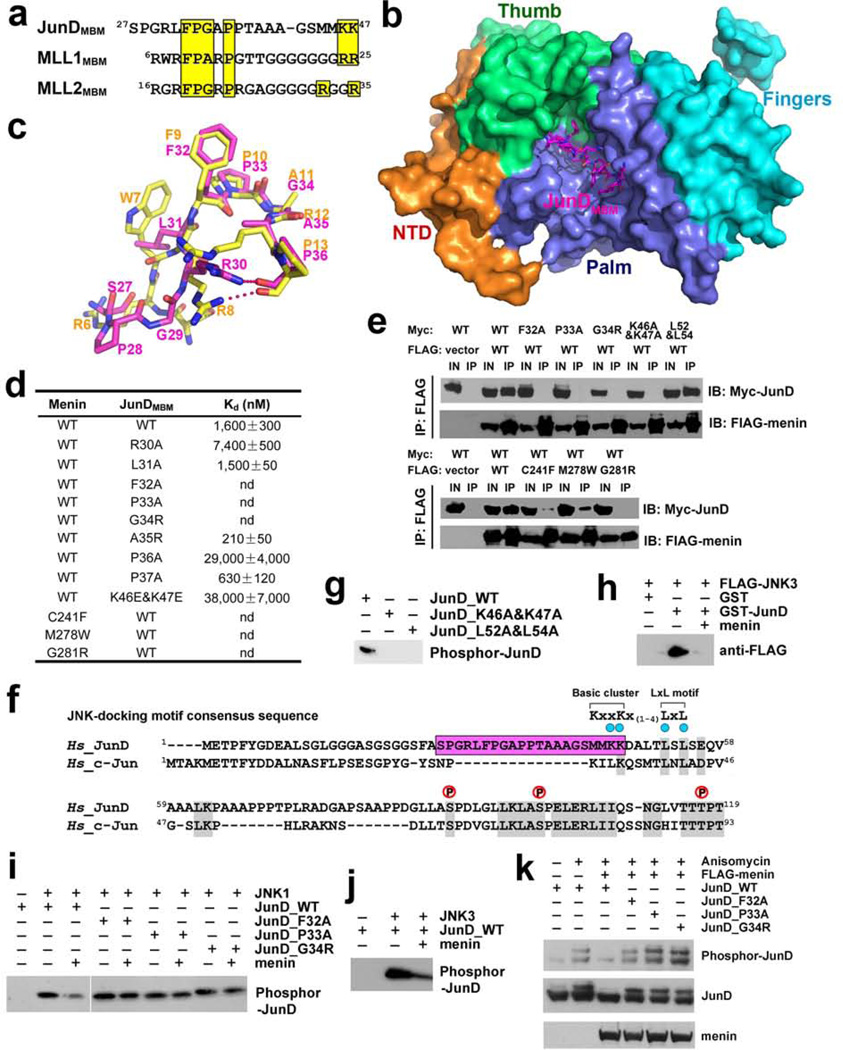
Menin also interacts with JunD by directly binding to its N-terminus7,8. We further defined JunD residues 27–47 as the menin-binding motif (JunDMBM) (Supplementary Fig. 9a). Sequence comparison of JunDMBM and MLL1MBM revealed a striking similarity (Fig. 4a), suggesting that JunDMBM might interact with menin through the same binding-pocket as MLL1MBM does. We determined the menin-JunDMBM complex structure that shows many similarities to menin-MLL1MBM (Figs. 4b and 4c and Supplementary Table 1). First, the F-P-A/G-R/A-P motifs in both MBM peptides are almost identical in overall conformation (Fig. 4c). Second, Phe32, Pro33, and Pro36 of JunD interact with menin in the same fashion as their counterparts in MLL1MBM (Supplementary Fig. 9f). Third, JunDMBM also adopts a cyclized conformation through a hydrogen bond between Arg30 and Pro36 (Fig. 4c). Notably, two C-terminal lysine residues (46KK47) in JunDMBM, equivalent to the disordered 24RR25 in MLL1MBM, are visible in the electron density map and point to an acidic surface on menin (Supplementary Fig. 9f). Mutation of these lysine residues and other key binding residues at the interface abolished or weakened the interaction both in vitro and in vivo (Figs. 4d and 4e).
Figure 4. Menin binds to JunD and blocks JNK-mediated JunD phosphrylation.
a, Sequence alignment of the MBM sequences of JunD, MLL1, and MLL2. Conserved residues are highlighted in yellow. b, Crystal structure of the menin-JunDMBM complex. Menin is in surface representation and colored as in Fig. 1c. The JunDMBM peptide is in stick model and colored in magenta. c, Overlay of JunDMBM (magenta) and MLL1MBM (yellow). d, In vitro ITC data of wild-type and mutant menin-JunDMBM interactions (refer to Supplementary Fig. 10). e, Co-IP of the mutant menin-JunD interactions. Lanes marked “IN” represent 2.5% of input cell lysate used for the immunoprecipitation. f, Sequence alignment of JunD and c-Jun. The MBM sequence of JunD is highlighted in magenta. Key residues in the JNK-docking motif are denoted by blue dots and three phosphorylation sites are labeled above the sequence. The consensus sequence of the JNK-docking motif is shown at the upper-right corner. g, Lys46, Lys47, Leu52, and L54 in the JNK-docking motif are required for JunD phosphorylation by JNK. h, GST-pull down assay of FLAG-JNK3 by GST-JunD in the presence or absence of full-length menin. i, The phosphorylation of wild-type or mutant JunD by JNK1 was examined with in vitro kinase assay. The phosphorylated JunD was detected with anti-JunD phosphor-Ser100 antibody. j, The phosphorylation of JunD by JNK3 was examined as described in (i). k, HEK-293T cells were transfected with wild type or mutant c-Myc-JunD plasmids, either alone or together with FLAG-menin plasmids. Immunoblots were performed with either anti-phosphor-JunD (upper panel), anti c-Myc (middle panel) or anti FLAG (lower panel) antibodies to examine the phosphorylation level of JunD in response to anisomycin stimulation.
It was reported that menin uncouples JunD phosphorylation from JNK activation, but the mechanism is poorly understood18. The consensus JNK-docking domain (DM) contains a cluster of basic amino acids preceding two leucine residues separated by a single amino acid (Fig. 4f)19. JunDMBM is partially overlapped with a putative JNK-docking motif of JunD (JunDDM) (Fig. 4f)20. In vitro kinase assay confirmed that both Lys46 and Lys47, and the leucine residues in JunDDM are indispensable for JNK mediated JunD phosphorylation (Fig. 4g). Thus, Lys46 and Lys47 are both required for menin binding and JunD phosphorylation by JNK. This led us to hypothesize that menin inhibits JunD phosphorylation through sequestering JunD from recognition by JNK. In the GST pull-down assay, GST-JunD can efficiently pull down JNK only when menin was not present, indicating that menin has a higher affinity to JunD (Fig. 4h). Furthermore, when menin was added in the in vitro kinase assay, phosphorylation of WT JunD was clearly inhibited (Figs. 4i and 4j). In contrast, phosphorylation of menin-binding deficient mutants of JunD was not affected by menin (Fig. 4i). Next, we examined whether menin could inhibit JunD phosphorylation in response to anisomycin activation of JNK in 293T cells18. While menin-binding deficient mutants were robustly phosphorylated in response to anisomycin, WT JunD phosphorylation was strongly suppressed by menin (Fig. 4k). Collectively, our findings reveal that menin binds JunD and blocks its JNK-induced phosphorylation. As phosphorylation of c-Jun, a close homologue of JunD, enhances its interaction with co-activators such as CBP/p300, transcription-activating histone acetyltransferases21,22, menin may repress JunD at the promoter by blocking its phosphorylation and association with co-activators, leading to suppression of gene transcription.
Here our structural and functional studies provide a mechanistic explanation of how menin could both positively and negatively regulate gene transcription. Our findings also provides concrete evidence that menin acts as a scaffold protein to assemble a menin-MLL1-LEDGF ternary complex to coordinating gene transcription and promoting MFP-induced leukemogenesis. This knowledge is important for understanding of how menin exerts its pleiotropic and sometime opposite functions, and also paves the way to developing novel therapy for the aggressive leukemia.
As menin interacts with multiple protein factors23, many of these partners (such as MLL and JunD) may interact with menin in a mutually exclusive manner. Hence, there should be multiple distinct menin complexes each with a different combination of components. This is consistent with our observation that in cells menin distributes quite heterogeneously and presumably exists in a large number of different complexes (Supplementary Fig. 11). Menin is an abundant protein with a relatively long half-life, and existence of menin-partner complexes could be dynamic, affected by cellular localities, context or cell types. In this regard, it is possible that the menin-MLL complex may play a major role in hematopoietic stem cells or leukemia cells as the menin-MLL targets, Hox genes, are highly expressed in these cells, especially in progenitors or stem cells11,24. On the other hand, the menin-JunD complex may be important for regulation of endocrine cells8. It is also possible that the different menin-partner complexes in the same type of the cells can function differently by interacting with loci of their target genes based on the landscape of the surrounding chromatin. We speculate that menin may function not only as a scaffold for its binding partners, but also as a coordinator, orchestrating complicated processes in a highly regulated manner.
Methods
Protein Expression and Purification
To facilitate crystallization, we genetically deleted an unstructured loop (resides 460–519) in menin, a short fragment (residues 40–45) in JunDMBM, and two loop regions (residues 16–22 and 36–102) in MLL1MBM-LBM. The resulting proteins, meninΔ and JunDMBMΔ retain wild-type binding affinities to MLL1MBM and menin, respectively (Supplementary Figs. 3d and 9d). The deletion mutant of MLL1MBM-LBM, MLL1MBM-LBMΔ, is still able to form a stable ternary complex with menin and LEDGFIBD (Supplementary Fig. 8c). For simplicity, MeninΔ, JunDMBMΔ, and MLL1MBM-LBMΔ are referred to as menin, JunDMBM, and MLL1MBM-LBM, respectively, unless stated otherwise.
Various human menin proteins and MLL and JunD peptides were expressed in E. coli BL21(DE3) using a modified pET28b vector with a SUMO protein fused at the N-terminus after the His6 tag. After induction for 16 hours with 0.1 mM IPTG at 25°C, the cells were harvested by centrifugation and the pellets were resuspended in lysis buffer (50 mM Tris-HCl pH 8.0, 50 mM NaH2PO4, 400 mM NaCl, 3 mM imidazole, 10% glycerol, 1 mM PMSF, 0.1 mg/ml lysozyme, 2 mM 2-mercaptoethanol, and home-made protease inhibitor cocktail). The cells were then lysed by sonication and the cell debris was removed by ultracentrifugation. The supernatant was mixed with Ni-NTA agarose beads (Qiagen) and rocked for 6 hours at 4˚C before elution with 250 mM imidazole. Then Ulp1 protease was added to remove the His6-SUMO tag. After Ulp1 digestion, the menin proteins, and the MLL and JunD peptides were further purified by gel-filtration chromatography on Hiload Superdex200 and Hiload Superdex75 columns (GE Healthcare) equilibrated with buffer A (25 mM Tris-HCl pH 8.0, 150 mM NaCl and 5 mM DTT) and buffer B (100 mM ammonium bicarbonate), respectively. The purified menin proteins were concentrated to 25 mg/ml and stored at −80˚C. The purified peptides were concentrated by Speed Vac system and then lyophilized. The lyophilization products were then resuspended in water at a concentration of 50 mg/ml and stored at −80°C.
For the menin-MLL1MBM-LBM-LEDGFIBD complex, LEDGFIBD were cloned into a modified pET28b vector with a SUMO protein fused at the N terminus after the His6 tag. MLL1MBM-LBM was cloned into a GST fusion protein expression vector, pGEX6p-1 (GE healthcare). The menin-MLL1MBM-LBM complex and LEDGFIBD itself were expressed in E. coli BL21 (DE3). The menin-MLL1MBM-LBM complex was purified by sequential affinity chromatography with Ni-NTA agarose beads and glutathione sepharose 4B beads (GE Healthcare). After removal of the His6-SUMO tag and GST tag respectively with Ulp1 and Protease 3C, the complex was further purified with gel-filtration chromatography on a Hiload Superdex 200. Meanwhile, LEDGFIBD was purified as an individual protein, and then was mixed with the purified menin-MLL1MBM-LBM complex with a molar ratio of 2:1. After one-hour incubation on ice, the protein mixtures were further purified with gel-filtration chromatography on a Hiload Superdex 200 column.
Crystallization, Data Collection and Structure Determination
Menin was crystallized by sitting-drop vapor diffusion at 4 °C. The precipitant solution contained 100 mM sodium cacodylate trihydrate (pH 6.5) and 1.4 M sodium acetate trihydrate. For the menin-MLL1MBM complex, purified menin was first mixed with the MLL1MBM peptide at a molar ratio of 1:2 and then the mixture was incubated on ice for 1 h to allow complex formation. Crystallization of the complex was achieved by sitting-drop vapor diffusion at 4 °C with the well solution containing 0.1 M Tris·HCl (pH 7.0), 0.2 M MgCl2 and 2.3 M NaCl. A similar procedure was also used for crystallization of the menin-JunDMBM complex. The menin-MLL1MBM-LBM-LEDGFIBD complex was crystallized by hanging-drop vapor diffusion at 4 °C with the well solution containing 0.05 M HEPES (pH 7.0), 1.6 M (NH4)2SO4 and 0.01 M MgCl2. The crystals were dehydrated with a solution containing 0.05 M HEPES (pH 7.0), 2.3 M (NH4)2SO4 and 0.01 M MgCl2.
All of the crystals were gradually transferred into a harvesting solution containing the respective precipitant solutions plus 5 M sodium formate, prior to being flash-frozen in liquid nitrogen for storage. Data were collected under cryogenic conditions (100 K). Se-Met-MAD data set of the menin-MLL1MBM complex at the Se peak and inflection wavelengths were collected at beamline 21ID-D at APS and processed using HKL200025. Seven selenium atoms were located and refined, and the MAD phases calculated using SHARP26. The initial MAD map of the menin-MLL1MBM complex was significantly improved by solvent flattening. A model was manually built into the modified experimental electron density using O27 and further refined in CNS28. Native datasets of menin, menin-MLL1MBM, menin-JunDMBM and menin-MLL1MBM-LBM-LEDGFIBD were collected at beamline 21ID-D at APS and processed using HKL2000. The structures were determined by molecular replacement using Phaser in the CCP4i suite29, and then further refined using simulated-annealing and positional refinement in CNS with manual rebuilding in O.
Isothermal Titration Calorimetry
The equilibrium dissociation constants of the menin-MLLMBM, menin-JunDMBM, and menin-MLL1MBM-LBM-LEDGFIBD interactions were determined by using a VP-ITC calorimeter (MicroCal). The enthalpies of binding between menin and the MLL1 or JunD peptides and between the menin-MLL1MBM-LBM complex and LEDGFIBD were measured at 20°C in 20 mM sodium phosphate (pH 7.0) and 100 mM NaCl. Two independent experiments were performed for every interaction described here. ITC data were subsequently analyzed and fit using Origin 7 software (OriginLab) with blank injections of peptides into buffer subtracted from the experimental titrations prior to data analysis.
Yeast Two-hybrid Assay
The yeast two-hybrid assays were performed using L40 strain harboring pBTM116 and PACT2 (Clontech) fusion plasmids. The colonies containing both plasmids were selected on –Leu –Trp plates. β-galactosidase activities were measured according to Clontech MATCHMAKER library protocol and the averages from three individual transformants were reported.
Coimmunoprecipitation (Co-IP)
Human 293T cells transfected with pcDNA3.1 vectors encoding c-myc-tagged MLL1 (residues 1–153) and FLAG-tagged menin were resuspended in 1 ml of lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1.0% Triton X-100, 1 mM EDTA and protease inhibitor cocktail). Immunoprecipitation of lysates was conducted using 20 µl anti-FLAG M2 affinity agarose (Sigma). After washing with lysis buffer, immunoprecipitated proteins were eluted with 2X loading buffer (50 mmol/l Tris-HCl, pH 6.8, 2% SDS, 10% 2-mercaptoethanol, 10% glycerol, and 0.002% bromophenol blue), subjected to protein gel electrophoresis using 4–20% SDS-PAGE, and then transferred to a PVDF membrane. After blocking with TBS containing 5% milk, the membrane was incubated for immunoblotting analysis with a mouse anti-FLAG (Sigma) or anti-c-Myc (Santa Cruz Biotech) monoclonal antibodies, followed by incubating with goat anti-mouse IgG (H+L) secondary antibody (Millipore). The same procedure was also used for the Co-IP experiments for menin and JunD.
Plasmid Construction
To generate recombinant retroviruses, pMX-2x-Flag-menin was constructed by inserting PCR-amplified menin cDNA into the BamHI/NotI site of the retroviral vector pMX-2x-Flag. To generate menin mutants, pMX-2x-Flag-menin was used as a template for site-directed mutagenesis using the QuikChange kit from Qiagen.
Cell lines and cell culture
Menin-null MEFs were cultured in Dulbecco’s modified Eagle’s medium complemented with 10% fetal calf serum and 1% PenStrep. Menin-null MEFs were infected with either vector, wild-type or menin-expressing retroviruses as previously described . The resulting cells were subjected to puromycin selection (2 µg/ml) 72 h post-infection for two days.
Western Blotting
MEFs were lysed in RIPA buffer. The whole cell lysates (20 µg) were separated on a SDS-PAGE gel and the proteins were subsequently transferred to a PVDF membrane. Anti-menin antibody from Bethyl labs (BL342) was used to detect menin.
Real Time qRT-PCR Quantification
Exponentially growing MEFs were seeded at 2×105 cells/100 mm dish and harvested two days later. Total RNA was isolated with a RNeasy minikit from Qiagen. qRT-PCR was performed in an ABI 7500 Real Time PCR system (Applied Biosystems).
ChIP assay
MEFs were cross-linked with 1% formaldehyde for 10 min at 37 °C. Cross-linking was stopped by addition of 125 mM glycine. ChIP assay was performed using the QuikCHIP kit from Imgenex as per manufacturer’s instructions. Antibodies used for ChIP were anti-menin (Bethyl labs), anti-histone H3K4 tri-methyl, anti-histone H3 and IgG (Abcam). Antibody precipitated DNA-protein complex was reverse crosslinked, and the DNA was extracted using phenol/chloroform, and the precipitated DNA was used as the template for PCR. The primers used have been described elsewhere 10.
GST-pull down assay
GST, GST-fused JunD (residues 1–150) and FLAG-tagged JNK3 were expressed in E. coli BL21 (DE3) and were purified to homogeneity. GST-pull down assay was performed by incubating 10 µg of GST or GST-JunD, 40 µg of FLAG-JNK3 and 40 µg of full-length menin with 10 µl of glutathione sepharose 4B beads in binding buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl) at 4°C overnight. Then the beads were extensively washed with binding buffer for four times and the bound proteins were eluted with 10 mM reduced glutathione. After separation on 15% SDS-PAGE, FLAG-tagged JNK3 protein was detected with western blot.
In vitro kinase assay
Wild-type and mutant JunD proteins (residues 1–150) were expressed in E. coli BL21 (DE3) and purified as described above. For the in vitro kinase assay, 1 µg substrate was mixed with 1 µg kinase, 50 µM ATP and with or without 5 µg of full-length menin protein in the kinase buffer (50 mM Tris-HCl, pH 7.5, 20 mM MgCl2, 20 mM β-glycerophosphate, 2 mM DTT and 0.1 mM sodium orthovanadate), and was incubated at 30 °C for 1 h. The reaction mixtures were then separated on 15% SDS-PAGE, and phosphorylated JunD was detected with anti-JunD phosphor-Ser100 antibody (Cell Signaling).
In vivo kinase assay
293T cells were transfected with expression vectors encoding FLAG-tagged menin and c-Myc tagged JunD. After 48 h of transfection, cells were incubated for 30 min in the absence or presence of 10 µg/ml anisomycin (sigma), a potent JNK activator, and then cell lysates were subjected to western blot with anti-JunD phosphor-Ser100 (Cell Signaling), anti-FLAG (Sigma), and anti-c-Myc (Upstate) antibodies.
Cell fractionation
1 × 108 293T cells were collected and washed in cold PBS and hypotonic buffer (10 mM Tris pH 7.3, 10 mM KCl, 1.5 mM MgCl2, 0.2 mM PMSF, and 10 mM β-mercaptoethanol). The cells were then allowed to swell for 15 min in hypotonic buffer. The swelled cells then were homogenized with glass Dounce homogenizer (Wheaton) using the loose pestle until cell membrane lysis was 80%–90%. The nuclei were collected by centrifuging 15 min at 3300 g, resuspended in high salt buffer (600 mM KCl, 20 mM Tris pH 7.4, 25% glycerol, 1.5 mM MgCl2, and 0.2 mM EDTA) and homogenized to break the nuclear membrane. The extracted nuclei were collected by centrifuging 30 min at 25,000 g. The supernatant cell nuclear extracts were fractionated on AKTA Superose 6 gel-filtration column (GE Healthcare). The resulting fractions were resolved by SDS-PAGE and probed with anti-menin antibody (Bethyl).
Supplementary Material
Acknowledgements
We thank Drs. Yong Chen and Wei Deng for help at various stages of the project. M.L. is a Howard Hughes Medical Institute Early Career Scientist. Work was supported by NIH grants (GM 083015-01 to M.L. and R01DK-085121 to X.H.), an American Cancer Society Research Scholar grant (to M.L.) and a Caring for Carcinoid Foundation – AACR Grant to X.H.). The General Medicine and Cancer Institutes Collaborative Access Team has been funded in whole or in part with federal funds from the National Cancer Institute (grant Y1-CO-1020) and the National Institute of General Medical Science (grant Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357.
Footnotes
Accession number
The coordinates and structure factors of menin, the menin-MLL1MBM complex, and the menin-MLLMBM-LBM-LEDGFIBD, and the menin-JunDMBM complex have been deposited in the RCSB Protein Data Bank under accession codes XXXX, XXXX, XXX, and XXXX, respectively.
References
- 1.Chandrasekharappa SC, Teh BT. Functional studies of the MEN1 gene. J Intern Med. 2003;253:606–615. doi: 10.1046/j.1365-2796.2003.01165.x. [DOI] [PubMed] [Google Scholar]
- 2.Busygina V, Kottemann MC, Scott KL, Plon SE, Bale AE. Multiple endocrine neoplasia type 1 interacts with forkhead transcription factor CHES1 in DNA damage response. Cancer Res. 2006;66:8397–8403. doi: 10.1158/0008-5472.CAN-06-0061. [DOI] [PubMed] [Google Scholar]
- 3.Chen G, et al. Menin promotes the Wnt signaling pathway in pancreatic endocrine cells. Mol Cancer Res. 2008;6:1894–1907. doi: 10.1158/1541-7786.MCR-07-2206. [DOI] [PubMed] [Google Scholar]
- 4.Jin S, et al. Menin associates with FANCD2, a protein involved in repair of DNA damage. Cancer Res. 2003;63:4204–4210. [PubMed] [Google Scholar]
- 5.Yang Y, Hua X. In search of tumor suppressing functions of menin. Mol Cell Endocrinol. 2007;265–266:34–41. doi: 10.1016/j.mce.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokoyama A, et al. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 7.Knapp JI, et al. Identification and characterization of JunD missense mutants that lack menin binding. Oncogene. 2000;19:4706–4712. doi: 10.1038/sj.onc.1203832. [DOI] [PubMed] [Google Scholar]
- 8.Agarwal SK, et al. Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell. 1999;96:143–152. doi: 10.1016/s0092-8674(00)80967-8. [DOI] [PubMed] [Google Scholar]
- 9.Groussin L, Bertherat J. Mechanisms of multiple endocrine neoplasia type 1: evidence for regulation of the AP-1 family of transcription factors by menin. Eur J Endocrinol. 1999;141:15–16. doi: 10.1530/eje.0.1410015. [DOI] [PubMed] [Google Scholar]
- 10.Hughes CM, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 11.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama A, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grembecka J, Belcher AM, Hartley T, Cierpicki T. Molecular basis of the mixed lineage leukemia-menin interaction: implications for targeting mixed lineage leukemias. J Biol Chem. 2010;285:40690–40698. doi: 10.1074/jbc.M110.172783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caslini C, et al. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67:7275–7283. doi: 10.1158/0008-5472.CAN-06-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 17.Llano M, Morrison J, Poeschla EM. Virological and cellular roles of the transcriptional coactivator LEDGF/p75. Curr Top Microbiol Immunol. 2009;339:125–1246. doi: 10.1007/978-3-642-02175-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallo A, et al. Menin uncouples Elk-1, JunD and c-Jun phosphorylation from MAP kinase activation. Oncogene. 2002;21:6434–6445. doi: 10.1038/sj.onc.1205822. [DOI] [PubMed] [Google Scholar]
- 19.Yang SH, Whitmarsh AJ, Davis RJ, Sharrocks AD. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. Embo J. 1998;17:1740–1749. doi: 10.1093/emboj/17.6.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yazgan O, Pfarr CM. Regulation of two JunD isoforms by Jun N-terminal kinases. J Biol Chem. 2002;277:29710–29718. doi: 10.1074/jbc.M204552200. [DOI] [PubMed] [Google Scholar]
- 21.Dunn C, Wiltshire C, MacLaren A, Gillespie DA. Molecular mechanism and biological functions of c-Jun N-terminal kinase signalling via the c-Jun transcription factor. Cell Signal. 2002;14:585–593. doi: 10.1016/s0898-6568(01)00275-3. [DOI] [PubMed] [Google Scholar]
- 22.Kaminska B, Pyrzynska B, Ciechomska I, Wisniewska M. Modulation of the composition of AP-1 complex and its impact on transcriptional activity. Acta Neurobiol Exp (Wars) 2000;60:395–402. doi: 10.55782/ane-2000-1358. [DOI] [PubMed] [Google Scholar]
- 23.Agarwal SK, et al. Menin molecular interactions: insights into normal functions and tumorigenesis. Horm Metab Res. 2005;37:369–374. doi: 10.1055/s-2005-870139. [DOI] [PubMed] [Google Scholar]
- 24.Maillard I, et al. Menin regulates the function of hematopoietic stem cells and lymphoid progenitors. Blood. 2009;113:1661–1669. doi: 10.1182/blood-2009-01-135012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method in enzymology. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.De La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods in Enzymology. 1997:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 27.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.