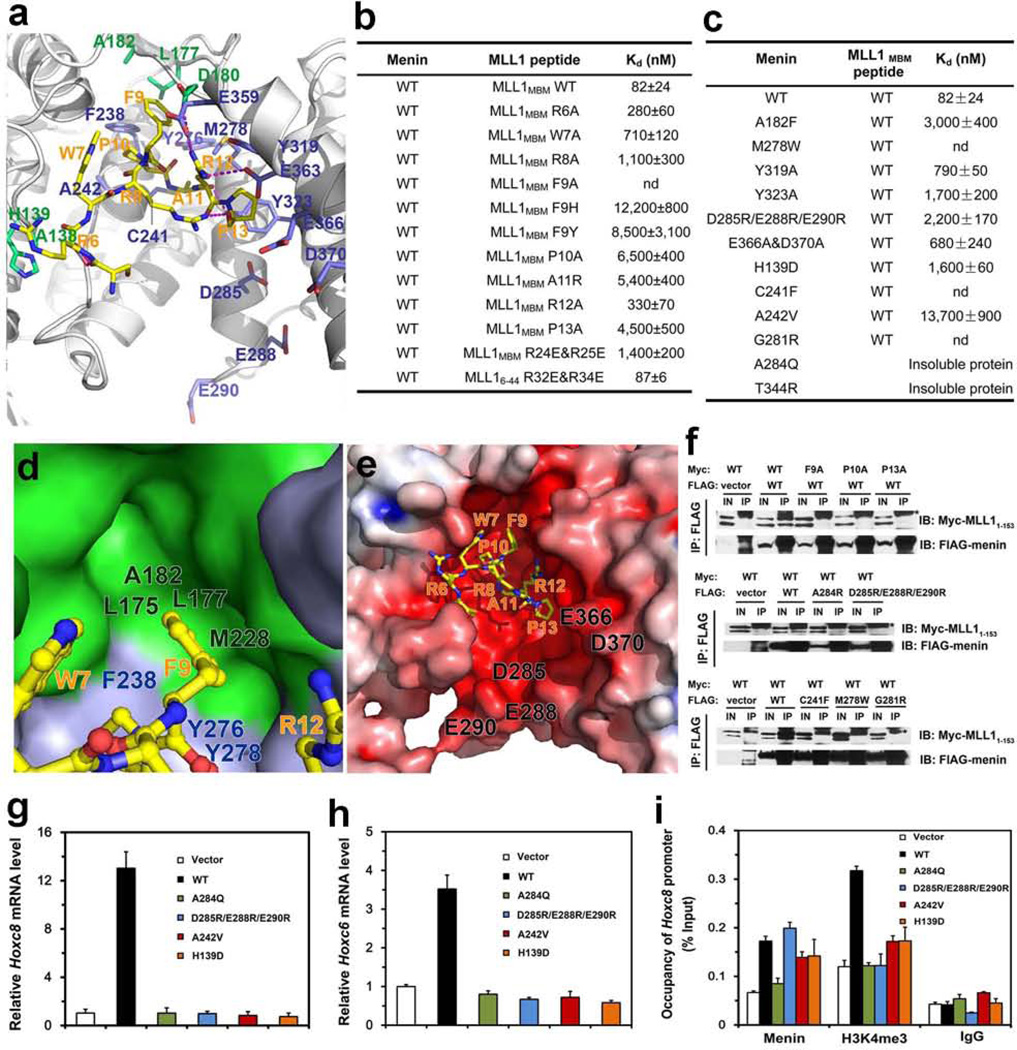
Figure 2. Structural and mutational analyses of the menin-MLL1MBM interface.
a, detailed view of the menin-MLL1MBM interface. MLL1MBM (yellow) and the interacting residues in menin (green: residues in the thumb domain; blue: residues in the palm domain) are presented as stick models. The Menin-MLL1MBM intermolecular hydrogen bonds are shown as magenta dashed lines. b, In vitro ITC binding data of wild-type menin with mutant MLL1MBM (refer to Supplementary Fig. 5). c, In vitro ITC binding data of menin mutants with wild-type MLL1MBM (refer to Supplementary Fig. 6). d, The side chain of MLL1MBM Phe9, in ball-and-stick model (yellow), is nested in a deep hydrophobic pocket of menin formed by the thumb domain (green surface) and the palm domain (blue surface). e, Electrostatic surface potential of the MLL1MBM-binding pocket of menin (positive potential, blue; negative potential, red). f, Co-IP of the mutant menin-MLL11–153 interactions. Lanes marked “IN” represent 2.5% of input cell lysate used for the immunoprecipitation. Asterisks indicate the signals from IgG. Secondary structure prediction analysis showed that the N-terminal region of MLL1 (residues 1–153) is an intrinsically unstructured region (data not shown). Therefore, due to the difficulty of manipulation of full-length MLL1 (3969 residues), the menin-MLL11–153 interaction was used to represent the interaction between menin and full-length MLL1 in the Co-IP assay. g, Expression of Hoxc8 in menin-null MEFs complemented with control vector, wild-type or mutant menin was measured using quantitative real-time RT-PCR. h, Expression of Hoxc6 in menin-null MEFs complemented with control vector, wild type or mutant menin was measured using qRT-PCR. i, Menin binding and H3K4 trimethylation at the Hoxc8 promoter were detected by ChIP.